Significance
Fragile X syndrome (FXS) is a frequent mental disorder characterized by intellectual disability and other symptoms including autism. The disease gene-encoded protein FMRP regulates activity-dependent translation of a large number of mRNAs in neurons. We used quantitative mass spectrometry to systematically compare protein expression in neocortical synaptic fractions between Fmr1 (fragile X mental retardation 1) knockout (KO) and wild-type mice during adolescence and adulthood. We discovered an upregulation of a large number of synaptic proteins in young KO mice but not in adult ones. Many of the upregulated proteins are correlated with an increased protein synthesis in KO neurons. This study provides a greatly expanded view of protein-level changes in FXS and identifies a previously unrecognized developmental dynamics in FXS pathogenesis.
Keywords: fragile X, quantitative mass spectrometry, synaptic protein synthesis, autism, stable isotope labeling
Abstract
Fragile X syndrome (FXS) is an X-linked neurodevelopmental disorder characterized by severe intellectual disability and other symptoms including autism. Although caused by the silencing of a single gene, Fmr1 (fragile X mental retardation 1), the complexity of FXS pathogenesis is amplified because the encoded protein, FMRP, regulates the activity-dependent translation of numerous mRNAs. Although the mRNAs that associate with FMRP have been extensively studied, little is known regarding the proteins whose expression levels are altered, directly or indirectly, by loss of FMRP during brain development. Here we systematically measured protein expression in neocortical synaptic fractions from Fmr1 knockout (KO) and wild-type (WT) mice at both adolescent and adult stages. Although hundreds of proteins are up-regulated in the absence of FMRP in young mice, this up-regulation is largely diminished in adulthood. Up-regulated proteins included previously unidentified as well as known targets involved in synapse formation and function and brain development and others linked to intellectual disability and autism. Comparison with putative FMRP target mRNAs and autism susceptibility genes revealed substantial overlap, consistent with the idea that the autism endophenotype of FXS is due to a “multiple hit” effect of FMRP loss, particularly within the PSD95 interactome. Through studies of de novo protein synthesis in primary cortical neurons from KO and WT mice, we found that neurons lacking FMRP produce nascent proteins at higher rates, many of which are synaptic proteins and encoded by FMRP target mRNAs. Our results provide a greatly expanded view of protein changes in FXS and identify age-dependent effects of FMRP in shaping the neuronal proteome.
Fragile X syndrome (FXS) is an X-linked monogenic disorder that leads to highly debilitating changes in neurodevelopment. Affected individuals exhibit mental retardation, attention deficit and hyperactivity, anxiety, autism spectrum behaviors, and other symptoms that compound overall impairment (1). In the vast majority of cases, FXS is caused by an mRNA-dependent epigenetic silencing of the Fmr1 (fragile X mental retardation 1) gene (2), which occurs secondarily to a CGG repeat expansion in the 5′ UTR region of Fmr1 (3) and results in absence of the encoded protein FMRP. FMRP is an RNA-binding protein that regulates several aspects of mRNA translation (1), transport (4), and stability (5) in neurons. Substantial evidence indicates that FMRP is particularly critical as a suppressor of activity-dependent mRNA translation at glutamatergic synapses (6, 7) and that loss of this function results in abnormalities in dendritic spine shape and several forms of long-term synaptic plasticity (8, 9). In addition, significant changes have been described regarding the structure and/or function of other synaptic systems, including GABAergic and endocannabinoid synapses (10, 11). Loss of FMRP expression has been recently linked to abnormalities in adult neurogenesis, migration, neural differentiation, and cortical maturation (12).
Although monogenetic in origin, the molecular etiology of FXS is likely to be highly complex because FMRP binds to a large number of mRNAs in neurons (13, 14). Adding to this complexity are the effects of FMRP on the biogenesis and function of some miRNAs (15) and its regulation of targets in the hub of key signal transduction pathways [such as PI3K (16, 17) and PIKE (17) in the mTOR pathway] that affect the translation of mRNAs that are not direct FMRP targets. Prior efforts to comprehensively understand FMRP function have focused on identifying mRNA targets in a high-throughput manner. Large numbers of mRNAs associated with FMRP were identified initially using microarray and other approaches (13). More recently, high-throughput sequencing of mRNAs cross-linked to FMRP in mouse brain polysome fractions and retrieved by immunoprecipitation (HITS-CLIP) identified over 800 high-confidence FMRP-associated transcripts (18). These data included many mRNAs corresponding to synaptic proteins and autism susceptibility genes and led to the idea that FMRP exerts its translational repression by stalling ribosomes during the elongation step on its target mRNAs (18). Although valuable, such studies are not able to capture secondary effects of FMRP loss on the proteome, and they may render both false-positives and false-negatives resulting, respectively, from potential “bystander” roles of FMRP and effects that occur outside the context of polysomes (e.g., mRNA stability and transport) (4, 5). Moreover, because it is possible that FMRP may promote synaptic expression of some of its targets through effects on mRNA stability or transport (4, 5), it cannot be reliably inferred that a direct target of FMRP is up or down-regulated in FXS. Such complexities can be captured only by direct quantification of changes at the protein level. Prior work by our group and others used proteomic methods to identify proteome changes in synaptic fractions from Fmr1 knockout (KO) primary neurons and in dFmr1 Drosophila (19, 20). However, these studies likely underestimated the extent of proteome shifts caused by loss of FMRP.
In the present study, we used a panel of quantitative proteomic approaches to generate the most comprehensive view to date of synaptic protein expression changes that occur in vivo in the Fmr1 KO mouse model. Moreover, we apply these methods at two developmental time points: postnatal day 17 (P17), within a period of peak synaptogenesis (21), and adulthood (P45). Stable isotope labeling in mammals (SILAM) enables complete labeling of amino acids with 15N in the whole animal, including rodents (22, 23). This method has been successfully applied to analyze synaptic proteome dynamics in developing brain (24), to quantify neuronal proteome phosphorylation events in neurons (25), and to study protein turnover and identify long-lived proteins in neuronal nuclear pore complexes (26). To further differentiate the role of de novo translation in protein changes from alterations in other processes such as mRNA stability, protein degradation, or trafficking, stable isotope pulsed labeling (pSILAC) has been developed to measure genome-scale protein synthesis rates (27). However, applying pSILAC in cultured primary neurons has not been implemented, presumably due to the low protein turnover in this cell type (19). With these combined approaches, we define a set of nearly 1,000 proteins that are differentially expressed in Fmr1 cortical synapses at P17, including a diverse array of proteins involved in synaptic development and function, metabolism, and a large number corresponding to FMRP HITS-CLIP targets and autism susceptibility genes. Moreover, we obtained evidence that the dominant shift in expression is up-regulation in the Fmr1 KO, due to enhanced translation, and that this effect is strongly age-dependent, being almost entirely absent at P45.
Results
Age-Dependent Changes in the Profile of Synaptic Protein Expression in Fmr1 KO Mice.
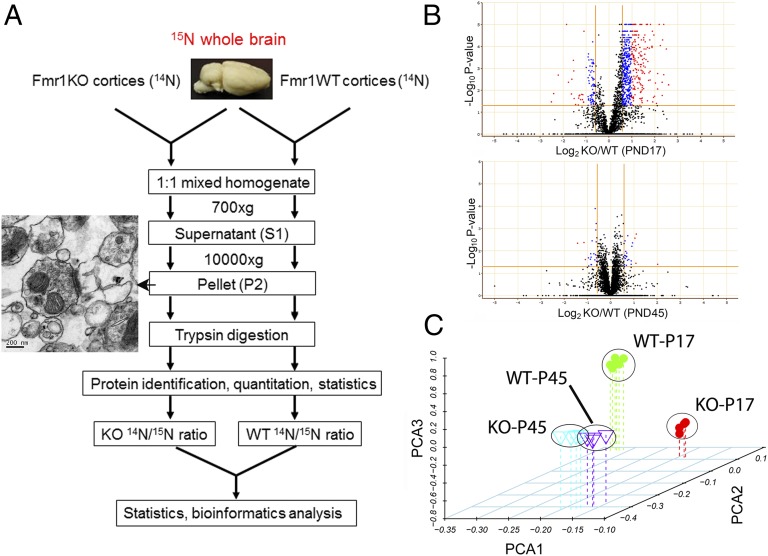
Using a combination of 15N-enriched adult mouse (SILAM) whole brain as a common internal standard and high-resolution mass spectrometry, we quantified proteins from crude synaptosome-enriched samples (Fig. 1A). To ensure the best possible comparison between conditions, neocortical homogenates derived from Fmr1 KO and WT mice at P17 or P45 were independently mixed with the common 15N-enriched brain homogenate for simultaneous preparation of crude synaptic fractionations (designated as P2 as described in Materials and Methods), protein extraction, and tryptic digestion (Fig. 1A; WT-P17, n = 6; KO-P17, n = 4; WT-P45, n = 4; KO-P45, n = 4). The enrichment of synaptic components in the cortical fraction used for proteomic analyses was evident by transmission electron microscopy images, as well as by Western blot analysis of synaptic proteins in sequential fractions (Fig. 1A and Fig. S1). In total, over 5,000 proteins were quantified by high-throughput mass spectrometric analysis of tryptic peptides derived from these fractions, and 2,600 proteins were common among all four experimental conditions. ANOVA analysis followed by a Benjamini–Horchberg post hoc test identified 999 proteins that were differentially expressed with high significance (P < 0.05, Dataset S1) in P17 KO cortical synaptic fractions. A volcano plot of all of the proteins quantified under the four experimental conditions shows a dramatic shift of significantly expressed proteins toward up-regulation in KO cortical fractions at P17, which occupy 90% of all of the changed proteins. Surprisingly, this trend is greatly diminished at P45 (Fig. 1B). Principal component analysis (PCA) clearly shows that, in the first component, KO and WT cortices are poorly separated at P45 but well separated at P17 (Fig. 1C), indicating that genotype differences result in dramatic changes in protein expression profiles during early brain development, but not after brain maturation. These data suggested that the majority of protein expression changes that may be involved in FXS pathogenesis are up-regulated and occur at a young age when active synaptogenesis is ongoing.
Fig. 1.
Quantitative proteomic analysis of synaptosomes from Fmr1 KO mice reveals age-dependent changes in protein expression patterns between WT and KO mice. (A) Schematic representation of the quantitative proteomic method used in this study. 15N-enriched amino acid-labeled mice whole-brain lysates were used as a common internal standard for the relative quantification. Enrichment of synaptic structures was indicated by a representative transmission electron microscopy image. (B) Volcano plots show a dramatic age-dependent difference in differentially expressed proteins in Fmr1 KO mice cortices comparing P17 and P45. (C) Principal component analysis of proteins quantified in all four experimental groups shows that the protein expression profiles of KO mice are well separated in young mice but not in older ones.
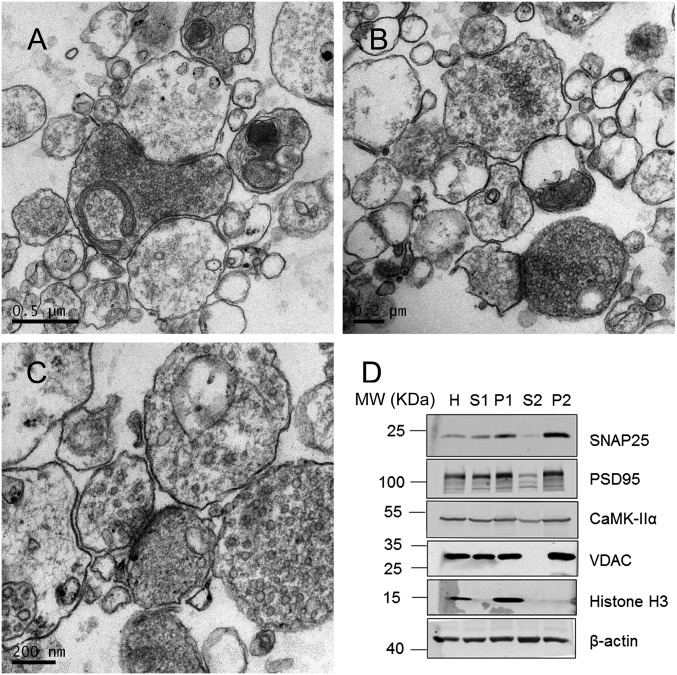
Fig. S1.
Related to Fig. 1. (A–C) Additional transmission electron micrographs in a variety of magnifications showing the enrichment of synaptic structure. (D) Western blot analysis of different fractions of the synaptic preparation from mouse brain used in the SILAM experiment. Enrichment of presynaptic (SNAP25), postsynaptic (PSD25), and mitochondrial proteins (VDAC) in P2 fractions is evident. In contrast, depletion of nuclear components (H3) in P2 fractions is also evident.
Validation of Quantitative Proteomic Results.
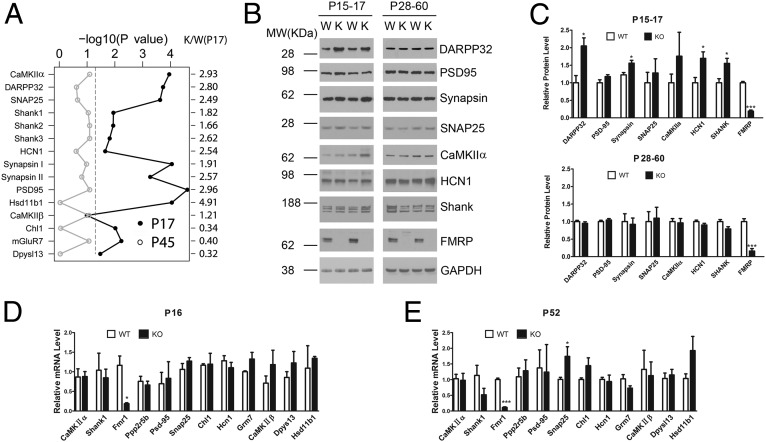
From the SILAM dataset we selected 15 proteins representing core components in presynaptic vesicles (synapsin 1 and 2, SNAP25); postsynaptic channels and receptors (HCN1, mGluR7); signaling molecules (CaMKIIα, DARPP32); synaptic scaffold proteins (Shanks, PSD95, Dpysl3); adhesion molecules (Chl1); and metabolic enzymes (Hsd11b1) and plotted their significance values comparing the two age groups with the relative protein expression ratios between KO and WT at P17 (Fig. 2A, Right). With the exception of Chl1, Dpysl3, and mGluR7, which were significantly down-regulated at P17 in the Fmr1 KO, a majority of these proteins were significantly up-regulated at P17. However, none of the changes evident at P17 were present at P45. To validate the quantitative proteomic results, we used Western blotting to probe a panel of pre- and postsynaptic proteins in cortical synaptic fractions (Fig. 2B). Comparing adolescent (P17) KO and WT littermates, we observed up-regulation in the presynaptic vesicle component synapsin 1 and two postsynaptic proteins in KO cortical synaptic fractions: the cyclic nucleotide-gated potassium channel HCN1 and the Shank family proteins (Fig. 2B and statistical test in Fig. 2C). DARPP32 (Ppp1r1b), a bifunctional regulator important in dopamine signal transduction (28), was also greatly up-regulated in the KO at this stage. These observations are consistent with the quantitative proteomic results. However, we did not observe statistically significant changes in other synaptic proteins including PSD95, CaMKIIα, and SNAP25, although CaMKIIα trended upward in the KO (Fig. 2C). Consistent with age-dependent changes we observed using SILAM, none of the proteins tested by Western blot exhibited significant up-regulation in cortical synaptic fractions from mature mice (age between 1 and 2 mo); HCN1 and SHANK family proteins in particular trended slightly downward. To test whether the increase in some of the synaptic proteins in young mouse cortex is the result of increased transcription activity, we performed quantitative RT-PCR on mouse cortices, covering both ages. The results show that except for Fmr1 itself and SNAP25 in P52, all other transcripts are unchanged in KO mice in both young and adult age groups (Fig. 2 D and E). To add another layer of validation, we measured intrinsic electrophysiological properties of pyramidal neurons in layer II/III of the somatosensory cortex from P17 mouse using whole-cell patch configuration. We find that input resistance is significantly reduced, consistent with elevated expression of HCN1 (Fig. S2). Taken together, these results indicate that, consistent with previous studies, lack of FMRP expression predominantly influences synaptic protein expression at the level of translational control.
Fig. 2.
Validation of a subset of significantly changed proteins. (A) Log-transformed significance values against their expression levels of 15 proteins involved in synaptic structure or transmission are plotted comparing P17 (closed circle) and P45 (open circle). (B) Western blot analysis of eight synaptic proteins from cortical synaptic preparations indicates an overall up-regulation in P17 mice but no change in P45 mice. (C) Bar graph shows statistical test of the Western blot data in B (Student’s t test, *P < 0.05, ***P < 0.0005, n = 4∼5). (D and E) Real-time PCR analysis comparing the mRNA levels of 12 synaptic proteins between WT and KO in P16 (D) and P52 (E) (Student’s t test, *P < 0.05, ***P < 0.0005, n = 4).
Fig. S2.
Summary of intrinsic electrophysiological properties of pyramidal cells recorded in whole-cell patch configuration in layer II/III of somatosensory cortex (S2). (A) Sample of voltage traces evoked in a somatosensory cortical pyramidal neuron recorded in brain slice of a WT mouse in response to 350-ms depolarizing and hyperpolarizing current pulses. Note that the action potentials are followed by fast and slow after-hyperpolarizations that control the firing rate of the neurons. (B–F) Graphical summaries of input resistance (B), spike threshold (C), resting membrane potential (D), spike width (E), and rheobase current (F). Pyramidal cells in Fmr1 KO mice showed a significant reduction in input resistance (P = 0.02, t test), consistent with elevated expression of HCN1.
Differentially Expressed Proteins in Fmr1 KO Mice Show Gene Ontology Enrichment in Nervous System Development and Function.
Gene ontology analysis of proteins differentially expressed in P17 KO synaptic fractions yielded three main observations. In the cellular components category, proteins localized to the cytoplasm, mitochondria, and plasma membrane occupy a large portion, together representing 63% of changed proteins (Fig. 3A, Upper). In the molecular functions category, enzymes, transporters, and kinases are enriched (Fig. 3A, Lower). Further statistical enrichment analysis using Fisher’s exact test identified distinct functional categories that are significantly enriched, including nervous system development and function, cell-to-cell signaling and function, cellular assembly, and organization as the most statistically enriched (Fig. 3B). To gain further insight into the functions and interactions of the differentially expressed proteins in P17 KO synaptic fractions, we mapped the genes representing these proteins to the CORUM database (29), which contains curated data on 453 mouse protein complexes. Within this data set, 111 proteins from 183 complexes corresponded to proteins changed at P17. Notably, four of the six components in the postsynaptic GluRδ-2 receptor complex, including autism-susceptible genes Shank1, Shank2, and homer (30–32), and four of the five components in the NDMA receptor complex were significantly up-regulated. All four components in the presynaptic vesicle SNARE complex were also significantly up-regulated. Our significantly up-regulated proteins also covered the Cask–Dlg1 complex, a scaffolding protein complex that organizes postsynaptic receptors and ion channels as well as the Psd3-Actn1 and Profilin-2 complexes, protein assemblies that reshape the actin cytoskeletons in response to extracellular stimuli (Fig. 3C). Many members of the MAGUK. family of scaffolding proteins were up-regulated, including Dlg1, Mpp2 (Dlg2), Dlgap2, SAPAP2, and others that mediate pre- and postsynaptic organization of channels and signaling complexes. Thus, differentially expressed proteins in Fmr1 KO cortical synapses, mostly up-regulated, participate in shaping the developing nervous system and determining the structure and function of glutamatergic synapses.
Fig. 3.
Bioinformatics analysis of differentially expressed proteins in Fmr1 KO mice. (A) Pie chart shows the gene ontology of changed proteins in the category of cellular localization (Upper) and molecular function (Lower); the components in each category were displayed as a percentage of proteins. (B) Functional enrichment analysis of changed proteins by Fisher’s exact test. The top 12 most significant functional categories were plotted with the x axis representing significance values. (C) Representative protein complexes identified in the CORUM database from the significantly altered proteins in PND17. Red color indicates significantly up-regulated proteins in KO mice. (D) Comparison of the genes from all quantified proteins and significantly changed proteins to gene entries from SFARI autism database, gene-to-cognition database (G2Cdb:PSP), as well as from FMRP target dataset by Darnell et al. (18). Proteins listed in the green oval are autism-associated gene products that are up-regulated in KO mice and intersect with all three data sets. The P value represents the enrichment odds ratio of the overlap as determined by χ2 test.
Comparison of Fmr1 KO Synaptic Proteome Shifts Relative to Known Synaptic Proteins, Putative FMRP Targets, and Autism Susceptibility Genes.
We further compared our quantitative results with existing datasets of synaptic proteins, putative FMRP target mRNAs, and autism-associated genes (Fig. 3D). The gene-to-cognition postsynaptic proteome (G2Cdb:PSP) database listed 1,121 postsynaptic proteins, of which our quantitative data covered 78%. Of our 999 significantly changed proteins, 298 were present in the G2Cdb:PSP, covering one-third of the significantly changed proteins. Darnell et al. used cross-linking immunoprecipitation and RNA-seq to identify 842 direct FMRP mRNA targets associated with polyribosomes (18). We compared these 842 protein-coding genes with our dataset and found 379 proteins in common, among which 120 (32%, P = 0.0003, a significant enrichment as tested by χ2) were significantly changed at P17, almost all of them being up-regulated. The SFARI autism database listed 621 genes associated with autism spectrum disorders, among which 221 (36%) were covered by our quantitative data and 48 (22%) corresponded to proteins that were significantly altered in P17 cortical synaptic fractions (Dataset S2). Thus, it is possible that loss of FMRP leads to the frequently observed autism endophenotype by triggering a “multiple hit” effect on the expression of autism-related genes. Among the autism-associated gene products significantly altered in P17 KO cortical synapses are well-characterized Shank family proteins (Shank -1, -2, and -3), Dlg4 (PSD95), Dlgap2, Grin2b, Syngap1, and poorly understood proteins such as Iqsec2 and Lrrc7 (represented inside the oval in Fig. 3D). Finally, we compiled a list of all three databases and our P17 dataset, representing a core set of synaptic proteins that are encoded by autism-associated genes, bound at the mRNA level by FMRP and expressed abnormally in Fmr1 KO synaptic fractions (Dataset S2). These proteins include Stxbp1 and Syn1 (synapsin 1) that also show altered synthesis rates as assessed by following pSILAC experiments.
Gene Coexpression Modules in Fmr1 KO Mice Show Connectivity Enriched in Synaptic Structure.
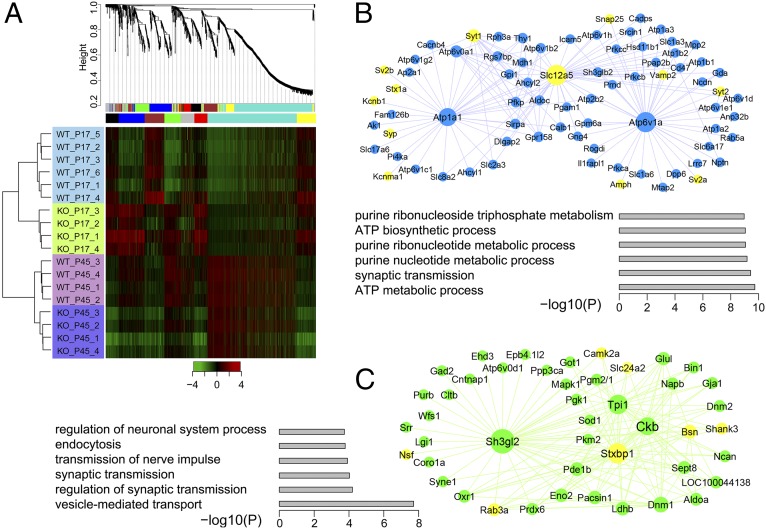
We applied weighted gene coexpression network analysis (WGCNA) (33) to identify groups of proteins in cortical synapses that exhibit high correlation in expression patterns and to understand the differences of these correlations between WT and KO cortices. Unsupervised hierarchical clustering of 1,524 quantified proteins across all experimental conditions in at least three biological replicates revealed that the two genotypes in the same age group are more closely clustered together (Fig. 4A), indicating that the effects of age on the clustering are larger than the effects of the genotypes compared. Nevertheless, a heat map of protein expression ratios calculated in reference to 15N-enriched brain showed significant differences between KO and WT cortical synaptic preparations at the younger age (P17), and this difference is largely eliminated when reaching adulthood (P45; Fig. 4A). The branches in the dendrogram correspond to groups of proteins (modules) the expression profiles of which are highly correlated across samples; in total, seven distinct modules were identified and are represented here by different colors. Although age was the dominant driver of clustering, Fmr1 KO-related changes in protein expression impacted the resolved modules in significant ways at both hub and satellite positions. The blue module is enriched with proteins involved in ATP metabolic process and synaptic transmission with the hub proteins Atp1a1, Atp6v1a, and Slc12a5 (KCC2) all significantly changed at P17. Presynaptic vesicle components altered in the KO, such as syntaxin1a, Snap25, Vamp2, and several ATPases, are marked yellow (Fig. 4B). Proteins from this module are closely related to synaptic vesicle release. Remarkably, 12 proteins from this module that are altered in the KO also overlap with the list of autism-associated genes (SFARI), such as Prkcb, Hsd11b1, Dpp6, Lrrc7 (Densin 180), and others (Dataset S2). The green module is enriched in proteins involved in synaptic transmission, vesicle transport, endocytosis, and regulation of neurological system process (Fig. 4C). Hub proteins in this module that are altered in the KO include Sh3gl2, Ckb, Stxbp1, and others that, together with many of the node proteins, are involved in presynaptic vesicle transport and postsynaptic structure organization, which are in yellow. Proteins in both the blue and the red modules displayed a substantial trend of up-regulation in P17 KO cortical synaptic fractions, whereas proteins in other modules each showed distinct patterns (Fig. S3). For example, the turquoise module is composed of predominantly mitochondrial proteins. The expression of these module proteins is increased in adulthood compared with adolescence regardless of the genotype, possibly reflecting an increased demand for energy metabolism and ATP production in the adult brain.
Fig. 4.
WGCNA of protein expression in Fmr1 KO mice. (A) Hierarchical clustering analysis using WGCNA shows coexpressed protein modules labeled by different colors. These modules are subsequently rearranged, and the heatmap shows that the protein expression in P17 displays distinct patterns compared to that of P45 (B) Network connections of the blue modules contain proteins greatly up-regulated in KO mice in P17. Enlarged nodes represent hub proteins, and the yellow color highlights presynaptic proteins. (C) Network connections of the green module contain proteins greatly up-regulated in KO mice in PND17. Enlarged nodes represent hub proteins, and the yellow color highlights pre- and postsynaptic proteins. Functional enrichment of the module proteins and their statistical significance are represented by bar graphs in B and C, respectively.
Fig. S3.
WGCNA of protein expression in Fmr1 KO mice (related to Fig. 4). WGCNA identified a total of seven protein module networks, in addition to the two networks displayed in Fig. 4, the remaining five networks represent a variety of biological functions and are displayed here.
Based on these modules, we constructed a model of protein expression abnormalities at fragile X cortical synapses (Fig. S4) that may impact the function of key protein assemblies, including many presynaptic vesicle components, scaffold proteins, postsynaptic receptors, synaptic adhesion molecules such as neurexin and neuroligin, as well as signal transduction molecules such as protein kinase A (PKA) and ERK, all of which appear up-regulated. Although not a comprehensive representation of our findings on synaptic protein changes in KO cortical synaptic fractions, this subset may provide a molecular basis for the frequently observed abnormalities in synaptic form and function in fragile X brain and model systems.
Fig. S4.
Construction of major key synaptic components that are up-regulated in Fmr1 KO mice cortices (related to Fig. 4).
FMRP Deficiency Results in Increased Synthesis of Synaptic Proteins.
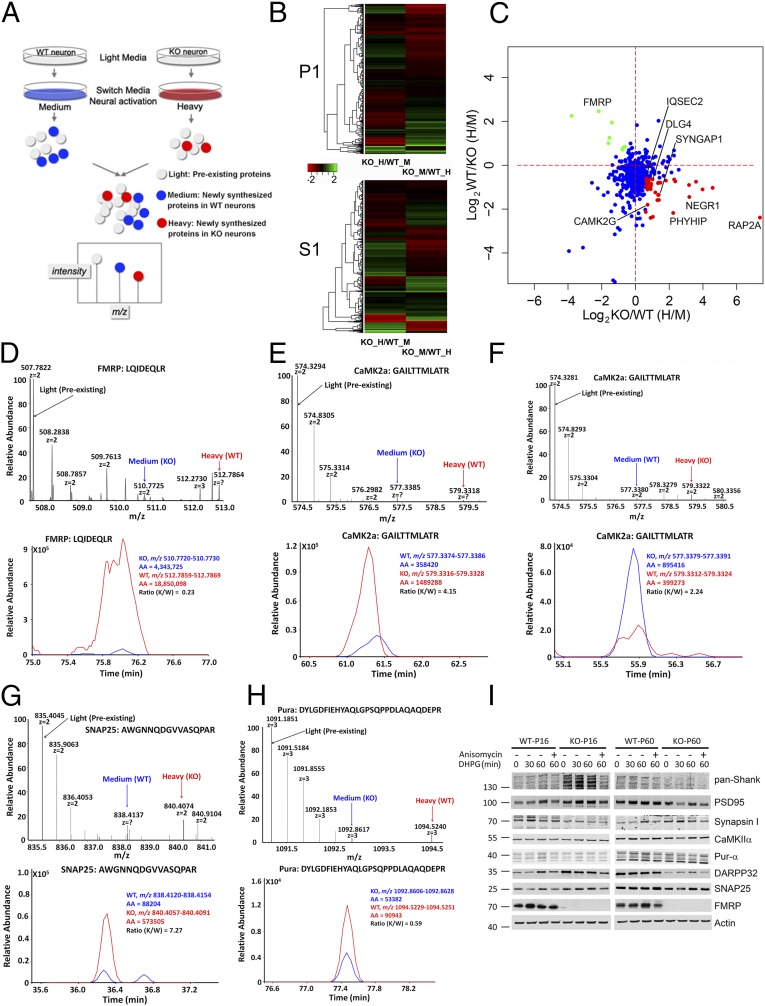
To investigate on a broad scale whether protein changes detected in Fmr1 KO synaptic fractions by SILAM-based proteomics reflect altered rates of de novo protein synthesis, we applied a recently developed pulsed stable isotope labeling approach—pSILAC (27)—to primary cortical neurons. We modified this approach such that proteins synthesized in WT and KO neurons incorporate different heavy isotopes, a strategy that makes quantification of newly synthesized proteins less dependent on the absolute level of isotope incorporation in primary neurons. As depicted in Fig. 5A, we grew WT and KO neurons under regular culture conditions initially. After switching WT and KO neurons to “medium heavy” and “heavy” arginine-/lysine-containing media, respectively, we stimulated translation by treating neurons with (S)-3,5-dihydroxyphenylglycine (DHPG), an agonist of group I metabotropic glutamate receptors (mGluRs) that have been linked to phenotypically relevant exaggerations in global translation rates and FMRP target synthesis in FXS model systems (1, 34). WT and KO neuronal lysates were then combined in synaptosomal preparation buffer, and soluble and pellet fractions were generated for LC-MS/MS analysis. With the aforementioned labeling strategy, newly synthesized proteins from WT neurons should be labeled with medium heavy, and those from KO neurons with heavy isotopes. In the second experiment, the labeling was inversed so that we would observe opposite ratios between heavy and medium heavy labeled peptides if there were any differences.
Fig. 5.
Pulsed SILAC reveals increased protein synthesis upon mGluR5 activation in cortical neurons dissociated from Fmr1 KO mice. (A) Schematic representation of the pulsed-SILAC approach applied in this experiment, whereby newly synthesized proteins in primary neurons dissociated from the two genotypes are encoded by medium heavy or heavy isotope-labeled lysine and arginine. (B) The heatmap shows the visualized nascent protein ratios comparing KO to WT. P1: 800-g pellet; S1: 800-g supernatant. (C) Correlation plot of another round of forward and reverse pSILAC experiments with 5-min DHPG stimulation. Several “core” FMRP target proteins show increased synthesis in KO neurons and are labeled in red. (D) Precursor ion spectrum and reconstructed chromatogram of an FMRP peptide shows nascent protein synthesis from KO (blue) or WT (red) neurons. (E and F) Similar to D but shows the same CaMKIIα peptide with reciprocal nascent protein ratios between KO and WT neurons. (G and H) Similar to D but shows peptides identifying SNAP25 and Pur-α, respectively. (I) Western blot analysis of acute cortical slices from WT or KO mice at two different ages shows distinct responses in protein expression to mGluR5 stimulation and that the increase in protein expression can be blocked by anisomycin, suggesting that the increases in protein levels are dependent on protein synthesis.
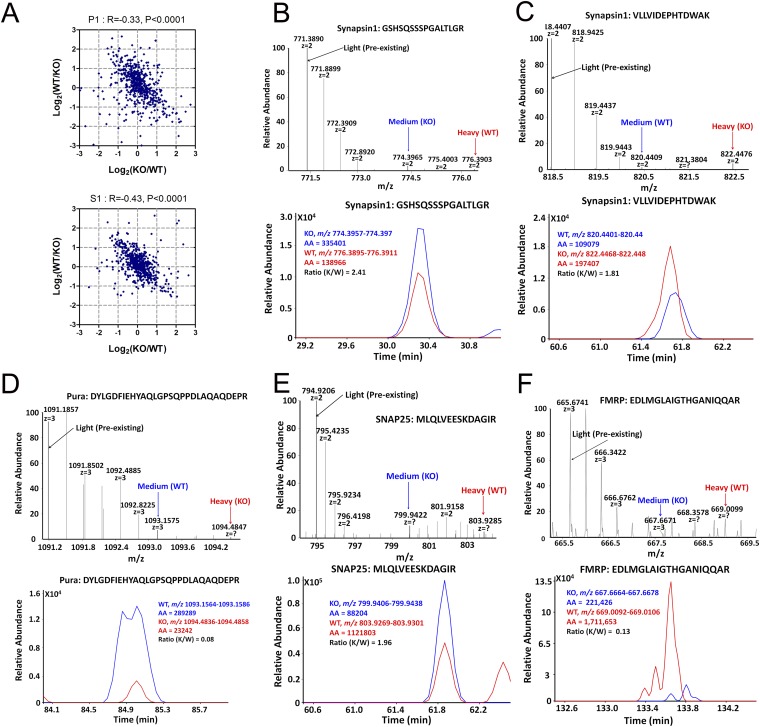
As expected, a heat map rendering of log-transformed labeling ratios revealed opposite changes in a large set of proteins (Fig. 5B). The peptide ratios representing relative levels of new protein synthesis in WT and KO neurons displayed a statistically significant correlation between these reciprocal labeling methods in both soluble and pellet fractions (Fig. S5A). To avoid potential epileptogenic activities caused by long-term DHPG stimulation, and to avoid nascent proteins undergoing possible degradation after longer pulse, we performed another round of forward and reverse pSILAC experiments in which neurons were stimulated with DHPG for 5 min and harvested 55 min after cessation of treatment by washout. A correlation plot of the two experiments showed consistency in many of the synaptic proteins identified as increased synthesis in KO neurons (Fig. 5C). Illustrative of this correspondence, a peptide derived from the postsynaptic kinase CaMKIIα showed inverse heavy-to-medium heavy isotope ratios in the two replicate experiments (Fig. 5 E and F). Confirming the sensitivity of our approach, high-resolution MS1 spectra (Fig. 5D, Upper) and reconstructed chromatograms (Fig. 5D, Lower) revealed that FMRP itself was newly synthesized in WT synaptic neurons in response to mGluR activation (indicated by red arrow), corroborating prior reports (35, 36). In KO neurons, there was a weak signal of nascent peptides (indicated by blue arrow), albeit nearly at the noise levels (also in Fig. S5F). Representative presynaptic proteins SNAP25 (Fig. 5G and Fig. S5E) and synapsin 1 (Fig. S5 B and C) were synthesized at a much higher rate in KO neurons than in WT neurons after treatment with DHPG, consistent with the notion that lack of FMRP exaggerated mGluR-induced translation (1). Unexpectedly, the purine-rich single-stranded DNA element-binding protein A (Pur-α) displayed a sharply reduced de novo synthesis in KO neurons compared with WT neurons (Fig. 5H and Fig. S5D). The pSILAC results for selected synaptic proteins with short DHPG treatment were presented in Fig. S6, in which all trends of increased nascent proteins in KO neurons were consistent with the results from longer DHPG treatment. In comparing the pSILAC dataset with the set of significantly changed proteins detected by SILAM, we found that 30% of the proteins that showed changes in de novo synthesis with mGluR stimulation were in the SILAM P17 dataset. This level of overlap is considerable, given prior findings that many FMRP targets are refractory to further stimulation in the Fmr1 KO (37). Indeed, when focusing on bona fide synaptic proteins, this correspondence increased to 70% (Dataset S2), thus supporting the hypothesis that increased synthesis rates account for synaptic protein expression changes in KO neurons. These data may also offer a view of proteins that are particularly vulnerable to mGluR-induced dysregulation in FXS.
Fig. S5.
Pulsed SILAC reveals increased protein synthesis upon mGluR5 activation in cortical neurons dissociated from Fmr1 KO mouse (related to Fig. 5). (A) Overall correlation of the two replicate experiments that had the labeling strategy swapped, so that a negative correlation in either the S1 or P1 fraction was observed. (B and C) Precursor ion spectrum and reconstructed chromatogram of two different synapsin 1 peptides display reciprocal heavy to medium-heavy ratios in the label swap experiments indicating reproducibility of the approach. (D–F) Other peptides representing Pur-α, SNAP25, and FMRP, respectively.
Fig. S6.
Pulsed SILAC reveals increased protein synthesis upon mGluR5 activation in cortical neurons dissociated from Fmr1 KO mouse, related to Fig. 5. (A) Precursor ion spectrum and reconstructed chromatogram of CaMK2a (A), synapsin 1 (B), SNAP25 (E), Stxbp1 (F), and Iqsec2 (G) show increased synthesis of these proteins in KO neurons. (C and D) A CaMK2b peptide shows reciprocal ratios from the label swap experiments and also shows increased protein synthesis in KO neurons.
To further test our observation of age-dependent changes in FMRP-dependent protein synthesis, we acutely cultured cortical slices from WT and KO mice in P17 and P45 age groups. After treatment with DHPG for 30 or 60 min, we analyzed the expression levels of synaptic proteins by Western blot. Except for synapsin 1, Shank family proteins, PSD95, CaMKIIα, SNAP25, DARPP32, and others all show increased expression after DHPG stimulation in a protein synthesis-dependent manner in P17 slices, and this trend of increase diminished in P45 slices. Also consistent with the SILAM results, the expression level of Pur-α is completely opposite. Together, these results are highly supportive of our proteomic data.
Discussion
In this study, we used state of the art proteomic approaches to generate the most expansive view to date of in vivo changes in cortical synaptic protein expression in the Fmr1 KO mouse. A principle emerging from this work is that the influence of FMRP on the synaptic proteome is age-dependent: i.e., a large number of proteins are up-regulated in the absence of FMRP at a young age, when active synaptogenesis is ongoing, but these differences are largely absent in adulthood. Furthermore, we found that neurons derived from KO mice produce many nascent synaptic proteins at a higher rate than that of WT mice, many of which are reportedly direct FMRP targets. Given that FMRP expression declines during later brain maturation (38, 39), it may be that the age-dependent differences that we observed between WT and KO synaptic proteomes reflect a declining influence of FMRP (in WT) on nascent protein synthesis at the elongation step.
Among our list of significantly changed proteins in young mice, many play key roles in postsynaptic structure and signaling. For example, we observed increases in diverse scaffolding proteins that interact within the postsynaptic elements to localize and modulate receptors, regulate F-actin assemblies, and create platforms of signaling molecules. Included in this set are MAGUK. family members, homer1, and Shanks 1–3. That all shanks were elevated at P17 (by SILAM and Western) is of particular interest, given that their abnormal expression leads to changes in postsynaptic density structure, AMPA receptor currents, and autism-like features in mice (40, 41). Of note, it has been demonstrated that lack of Shank2 expression leads to up-regulation of synaptic Shank3 (40), and there is evidence that accumulation of Shank3 due to deficient ubiquitination causes autism-like phenotypes (42). Other up-regulated proteins with structural functions include actin regulatory proteins, such as synaptopodin and its binding partner α-actinin 1. Overall, the increases in synaptic proteins with structural functions at P17 suggest an improper deposition of these components onto the postsynaptic membrane, which may play a role in the neuropathological findings in FXS patients of increases in spine density (8). However, the overabundance of PSD scaffolding proteins, which can promote synaptic maturation, appears to be at odds with consistent findings of abnormally long, thin, and tortuous spine profiles in FXS (43). In addition, our results revealed up-regulation of many presynaptic proteins, adding weight to the idea that FMRP has phenotypically relevant presyntaptic functions (44). We observed increases in presynaptic vesicle components, including synapsin 1, VAMP2, SNAP25, syntaxin1, and the syntaxin regulators Stxbp 1 and 5, and Snph, which binds free syntaxin. Recent studies have provided genetic evidence that genes encoding presynaptic release factors, especially SNARE complexes genes, may play important roles in susceptibility of attention-deficit/hyperactivity disorder (45, 46) and autism, two frequent endophenotypes of FXS (47, 48). Together, these data raise the prospect of altered active zone structure and release dynamics at cortical synapses in FXS and may help explain abnormalities in presynaptic function seen at hippocampal synapses in the Fmr1 KO (44).
An analysis of autism-related gene products in our P17 dataset revealed a signaling module that may be particularly important in the autism endophenotype of FXS. Autism occurs with high frequency in FXS patients [30–50% (49)], and there is evidence of mechanistic convergence between pathways dysregulated by loss of FMRP and those disrupted by autism-associated gene mutations and copy number variations. We sought to define a core set of autism susceptibility genes that are dysregulated at the translational level by loss of FMRP. To do this, we compiled a list of genes at the intersection of four datasets: our set of proteins that significantly changed at P17, the G2C synaptic protein database, the list of FMRP target mRNAs defined by HITS-CLIP, and the SFARI database of autism susceptibility genes. We identified 19 proteins that are synaptic, encoded by autism-associated genes, bound at the mRNA level by FMRP, and expressed abnormally in Fmr1 KO synaptic fractions (Dataset S2); all were up-regulated. Intriguingly, all nine postsynaptic proteins on this list are in the PSD95 interactome (50) (www.string-db.org), including Dlg4 (PSD95), Lrrc7 (Densin180), Shanks 1–3, Iqsec2 (BRAG1), Dlgap2 (SAPA2), GRIN2B (NMDA receptor 2b), and Syngap1 (Fig. 3D). Gene duplications in Dlgap2 have been linked to autism (51), and this effect may be recapitulated in FXS due to translational disinhibition. Moreover, Lrrc7 (52) and Iqsec2 (53) regulate mGluR signaling, long-term depression, and AMPA receptor internalization, raising the possibility that their up-regulation contributes to the lower efficacy state seen in cortical synapses in FXS. More generally, these data may indicate that FXS involves a skewed PSD95-based signaling module that shares mechanistic features with some forms of sporadic autism.
The extent of intersection between our SILAM set of up-regulated proteins and the HITS-CLIP set of putative FMRP target mRNAs—120 up-regulated proteins were HITS-CLIP targets—highlights a critical issue in FXS: How many phenotypically relevant gene expression changes can be accounted for directly by loss of FMRP translational inhibition? There is general agreement that both the mTOR and the ERK pathways of translational control show heightened activation in Fmr1 KO mouse neurons and human FXS lymphocytes. This could lead to broad protein expression changes that are secondary to FMRP loss, but of no less relevance phenotypically. The relative distribution of protein changes between these two broad types of dysregulation may determine the efficacy of emerging therapeutic strategies aimed at normalizing translation signaling (54).
Although most of the focus in FXS research has historically been on the molecular and functional pathology of glutamatergic synapses, recent work has established that several other synaptic systems are perturbed. Our SILAM data may inform on the mechanistic underpinnings of these changes. Reductions in GABAergic synapses and function have been observed in FXS and result in cortical network hyperexcitability (55). We detected significant increases in SLC12A2 (NKCC1, 1.9-fold) and SLC12A5 (KCC2, 2.3-fold), transporters that move Cl− into and out of neurons, respectively. A developmental shift in the expression of these proteins toward SLC12A5 regulates the transition of GABAergic transmission from depolarizing to hyperpolarizing. The diuretic NKCC1 inhibitor bumetanide has been proposed as a potential therapeutic in FXS and autism (56), and our data corroborate recent work suggesting that FXS may indeed involve an improper balance of SLC12A2 and SLC12A5 (10). Interestingly, at P45, both proteins were comparably expressed in WT and KO synapses, suggesting that the ratio imbalance evident in young cortex may continue into adulthood even as absolute levels of each decrease. We also found evidence for alterations in endocannabinoid transmission. Monoglyceride lipase 1 (Mgll), a serine hydrolase that degrades the endocannabinoid 2-arachidonoylglycerol (2-AG), was up-regulated 3.7-fold in KO cortical synaptic fractions, which may account for some forms of altered cortical prefrontal cortical plasticity in the KO (11). It has also been reported that FMRP is a positive modulator of dopamine signaling in prefrontal cortical neurons by sequestration and therefore enhances dopamine-mediated PKA activation; in Fmr1 KO mice this modulation is attenuated (57). We found a nearly threefold up-regulation of DARPP32 (SILAM and Western), a potent protein phosphatase 1 inhibitor upon phosphorylation at Thr34 by PKA. This observation may provide an alternative explanation for the attenuated dopamine signaling in the prefrontal cortex. It will be interesting for further studies to test the phosphorylation state of DARPP32.
Dendritic channelopathies have emerged as a contributing factor to cortical hyperexcitability in Fmr1 KO mice, an electrophysiological change that could account for the sensory motor hyperactivity seen in FXS. A recent study found a reduction and dysfunction of h- and BKca channels in dendrites (58). They found a statistically significant reduction of HCN1 in the primary sensory barrel cortex in 4-wk-old KO mice, which is seemingly in contrast to our results that HCN1 was up-regulated in the cortical synaptic preparation from Fmr1 KO mice in P17 and accompanied by a reduced input resistance. We speculate that age differences (P17 mice cortices used in our study), brain region differences (we used the whole cortex), and differences in sample preparation (we enriched synaptosomes) could account for this discrepancy. In fact, both our SILAM and Western blot results suggest a downtrend in P45 cortices. It is also notable that HCN1 is up-regulated in hippocampus of Fmr1 KO mice (59). In addition to HCN1, we found a panel of dysregulated ion channels, including Kcnma1, Kcnj10, and Grin2b. The abnormal expression of these channels in brain is strongly associated with many neuropsychiatric disorders, including autism (ref. 60 and SFARI). It is reasonable to propose that abnormal expression of these and other K+ channels alters the properties of cortical and subcortical networks underlying behavioral phenotypes and that they may represent novel opportunities for pharmacotherapy (61).
Several aspects of our data bear on the mGluR theory of FXS. On a gross level, our finding that the vast majority of protein expression changes in Fmr1 cortical synapses involve up-regulation supports the view that FMRP acts mainly as a translational suppressor at synapses. Recent studies have shown that FMRP gates translation downstream of multiple synaptic receptors, including Gq-coupled receptors for glutamate (i.e., mGluRs), acetylcholine, and other transmitters, as well as the brain-derived neurotrophic factor receptor TrkB (18). Our additional finding that 70% of up-regulated synaptic proteins in the “core” components exhibit higher rates of de novo synthesis during mGluR stimulation (Dataset S2) suggests that endogenous mGluR activity may induce a significant portion of the FMRP-regulated cortical synaptic proteome. These findings also challenge the general view that translation of FMRP-regulated mRNAs is refractory to further stimulation by activation of mGluRs in the Fmr1 KO. Interestingly, mGluR stimulation of P17 slices up-regulated many presynaptic proteins, such as Stxbp1 and Synapsin 1. It will be important to determine how unique these proteome responses are to mGluR stimulation. As noted above, we also detected increases in several proteins that promote mGluR signaling and/or synaptic localization, including Densin 180 (LRRC7), Homer 1, and Iqsec2 (BRAG1). Together, our findings are compatible with the idea that mGluR signaling is elevated in FXS and that this alters the synaptic proteome in a phenotypically relevant manner.
Analogous to traditional pulse-chase labeling of radioactive isotope-coded methionine, we adopted a pulsed SILAC approach (27) to allow stable isotope-coded lysine and arginine to incorporate into nascent proteins. Due to its postmitotic nature, protein synthesis in primary neurons is in general at a lower rate than mitotic cells. Indeed, our previous study showed that stable isotope labeling of cultured neurons is far from complete even after 3 wk in culture. Activation of mGluRs by DHPG has been demonstrated to rapidly increase the levels of a number of synaptic proteins (62) in the timescale of 1 h. However, we failed to observe any obvious nascent polypeptides at 30 min or 1 h poststimulation. We therefore used 6 h after DHPG stimulation and found subtle but robust nascent polypeptides. Although we could not rule out the possibility that newly synthesized proteins are rapidly degraded, our focus on identifying differences in nascent polypeptides indeed revealed a panel of synaptic proteins that are differentially synthesized in KO neurons, and these findings are largely consistent with the steady-state synaptic proteome levels in KO cortices. Recent technical development has enabled selective enrichment of nascent proteins using unnatural amino acid azidohomoalanine in place of methionine to incorporate into nascent proteins (63). It would be interesting for further studies to apply this approach to label nascent proteins in Fmr1 KO mice in vivo, especially in the context of a variety of challenging behavioral tasks.
Our data provide an in vivo landscape of abnormally expressed proteins in the absence of FMRP that are critical in cortical synapse formation and neural transmission; these proteins influence a wide range of central nervous system functions. The age dependence of these alterations suggests that the efficacy of some types of targeted therapeutics may be limited to the developmental period.
Materials and Methods
Animals.
All experiments involving animal treatment and care were performed following the Institutional Animal Care and Use Committee protocols from the East China Normal University (ECNU). FVB.129P-Fmr1tm1Cgr (Fmr1 KO) mice were kindly provided by Zhiqi Xiong and were originally obtained from the Jackson Laboratory ECNU animal facility. The Fmr1 KO mice were crossed with wild-type FVB mice for multiple generations to obtain Fmr1 KO and wild-type littermates. All animals used for comparison were age- and sex-matched.
Quantitative Proteomics.
We used mouse whole brain fully labeled with 15N-enriched amino acids (SILAM) as a common internal standard for quantitative proteomic analysis, as described previously for rat (24). Although mice were used in experiments described here, the basic labeling procedure remains the same. Briefly, female mice were fed a 15N-labeled protein diet starting after weaning and remained on the 15N diet through pregnancy and weaning of pups. On P45, the pups were anesthetized with halothane, and the brains were quickly removed and frozen with liquid nitrogen. The brains were homogenized in a motor-driven Teflon-glass homogenizer with 20 strokes in a buffer containing 0.32 M sucrose, 4 mM Hepes, and protease inhibitors, and the homogenates were stored at –80 °C. The 15N enrichment was determined to be 94% using a previously described protocol (64). The 15N-enriched mouse brain homogenates were then mixed at a 1:1 protein ratio with cortical homogenates from either Fmr1 KO or wild-type mice. Crude synaptosomes were prepared according to previously published protocol (19). Briefly, the homogenates were centrifuged at 700 × g for 15 min, followed by centrifuging the resulting supernatant fractions (S1) at 10,000 × g for 15 min. The resuspended pellets (P2) were used as crude synaptosomal fractions.
Statistics and Bioinformatics Analysis.
Significant differences in protein expression were determined using analysis of variances (ANOVA), and P values were then adjusted with the Benjamini–Hochberg method.
The coexpression network was constructed based on protein expression levels of mouse-brain samples using the WGCNA package in R software described by Zhang and Horvath (65). Briefly, a Pearson correlation matrix was first created by pairwise comparisons between all pairs of proteins across all mass spectrometric samples and was then transformed to an adjacency matrix with a soft thresholding power of 14. With the adjacency matrix, the topological overlap of a pair of proteins reflecting the relative interconnectivity between proteins was calculated, and then a topological overlap dissimilarity measure (1-topological overlap) in conjunction with average linkage hierarchical clustering was used to identify biologically meaningful modules of proteins with highly similar coexpression relationship. Using the Dynamic Hybrid Tree Cut algorithm and a minimum module size of 75 proteins, we identified a total of seven modules. The protein expression profile of each module is summarized in Dataset S1. The network of each module was visualized with Cytoscape v3.0.1.
The expression levels of all proteins from the 18 samples were analyzed with PCA. The first three principle components (PC1–3) contained the most useful information for these data, accounting for 47.8, 41.3, and 3.6% of the variance, respectively. The PC1 identified strongly reflects differential expression of proteins between WT and KO mice, whereas PC2 explains the differential expression between ages.
SI Materials and Methods
Pulsed Stable-Isotope Labeling of Primary Neurons (pSILAC).
Neocortices from newborn Fmr1 KO or WT littermate mice were dissected and dissociated, and neurons were cultured in Neurobasal media based on a previously described protocol (19). The neurons were plated at a density of 60,000 cells/cm2 in 10-cm dishes and maintained in Neurobasal media supplemented with B27 (Invitrogen), penicillin (50 μg/mL), streptomycin (50 U/mL), and glutamine (2 mM). After 10 d in culture, KO neurons were switched into Neurobasal media containing D(4)-lysine and 13C(6)-arginine (medium-heavy), and WT neurons were switched into Neurobasal media containing 13C(6)15N(2)-lysine and 13C(6) 15N(4)-arginine (heavy) for 5 h and then stimulated with DHPG in the presence of DL-2-Amino-5-phosphonovaleric acid (AP5) for 5 min. DHPG and AP5 were washed out by switching to the same medium-heavy or heavy medium without the drugs and incubated for 55 min before harvesting. In a “reverse” pSILAC experiment, the media switch was swapped such that the KO neurons received heavy media and the WT neurons received medium-heavy media. In total we performed two “forward” and two “reverse” biological experiments. After stimulation, the neurons were collected with Hepes-buffered sucrose [10 mM Hepes, pH 7.4, 0.32 M sucrose, protease inhibitor mixtures (Roche), 2 mM NaF, 1 mM Na3VO4] and then homogenized and centrifuged at 700 × g for 10 min to separate the nuclei and cell debri (P1) from the cytosolic and light membrane fractions (S1). Both the P1 and the S1 fractions were used for further analysis. Stable isotope-labeled arginine and lysine were purchased from Cambridge Isotope Laboratory.
Analysis by Multidimensional Protein Identification Technology.
For each condition, 50 μg of synaptosomal proteins were solublized in 8 M urea/RapiGest (Waters), reduced with 10 mM DTT, alkylated with 10 mM iodoacetomide, diluted 4× with 100 mM Tris⋅HCl, and then digested with trypsin overnight at an enzyme-to-substrate ratio of 1:100. After digestion, the pH was adjusted to about 2.5 using 90% formic acid.
For quantitative proteomic analysis, two 12-step Multidimensional Protein Identification Technology (66) experiments were performed to maximize the proteome coverage. Peptides were pressure-loaded onto a 100-µm i.d. fused silica capillary column packed with strong cation exchanger (SCX, Whatman) and a C18 material (Phenomenex) with the SCX end fritted with immobilized Kasil 1624 (PQ). After desalting, an analytical column of 100-µm i.d. capillary packed with another C18 material was attached to the SCX end with a ZDV union, and the column was placed in line with an HPLC pump as a nanospray ionization source and interfaced with an LTQ-Orbitrap Velos mass spectrometer. The buffers used were the following: 5% (all bases for buffer concentrations are vol/vol) acetonitrile/0.1% formic acid (buffer A); 80% acetonitrile/0.1% formic acid (buffer B); and 500 mM ammonium acetate/5% acetonitrile/0.1% formic acid (buffer C). The 12-step elution consisted of a 100-min gradient from 0 to 100% buffer B, and each of the remaining steps are composed of 5 min of step-increased salt buffer (buffer C), a 10-min gradient from 0 to 15% buffer B, and a 90-min gradient from 15 to 45% buffer B, followed by a 15-min gradient increase to 100% buffer B, and a 5-min reversal of gradient to 100% buffer A. As peptides were eluted from the microcapillary column, they were electrosprayed directly into the LTQ-Orbitrap Velos mass spectrometer (Thermo) with the application of a distal 2.4-kV spray voltage. A cycle of one full scan with 60,000 resolution at 400 m/z by Orbitrap (400–1,400 m/z) followed by 10 data-dependent MS/MS scans was repeated continuously throughout the entire process.
MS/MS spectra were analyzed using the following software analysis protocol. MS/MS spectra were searched with the SEQUEST algorithm (67) against the European Bioinformatics Institute mouse International Protein Index database (ftp://ftp.ebi.ac.uk/pub/databases/IPI/, version 3.85, releasing date June 2011) concatenated to a decoy database in which the sequence for each entry in the original database was reversed. SEQUEST results were assembled and filtered using the DTASelect program with a peptide false-positive score of 5%. Only proteins with two peptide hits or single hits with over 10 spectra counts were accepted. All peptides identified are at least half tryptic. Under such filtering conditions, the estimated false-positive rate at the protein level is about 0.2% based on decoy database hits.
Quantitative Real-Time PCR.
Total RNA was isolated from mouse brain using RNeasy Mini Kit (Qiagen). Isolated RNA (3 μg) was reverse-transcribed using Oligo(dt) primer (Invitrogen). Primers were designed from the National Center for Biotechnology website and then synthesized, followed by a regular PCR to evaluate its specificity. Quantitative real-time PCR was performed using the SYBR Green Master Mix (Promega) with an Agilent Mx3000P system. The reaction system contains 1×PCR mix, 0.2 μM primer and cDNA equivalent to 12.5 ng RNA in a total volume of 10 μL. The amplification conditions were as follows: 2 min at 95 °C, followed by 40 cycles of PCR for 15 s at 95 °C, 60 s at 60 °C. During the extension the PCR machine monitored the amplification by quantitative analysis of the fluorescence emission. The housekeeping gene GAPDH was used as the internal control, and all of the reactions were run in duplicates with four biological replications. The cycle threshold (CT) values were calculated by the software, and the mean CT values were taken into account for quantitative analysis. CT values for each gene was normalized by subtracting the mean CT value of GAPDH to get a ΔCT value, and then the 2−ΔΔCT method was used to obtain a final value for statistical analysis using the GraphPad Prism software.
Acute Cortical Slice Culture.
Cortical slices from Fmr1 KO or wild-type mice were prepared according to the procedure described by Hou et al. (36). Coronal slices of 300 μm were cut and submerged in saline solution (110 mM sucrose, 60 mM NaCl, 3 mM KCl, 1.25 mM NaH2PO4, 28 mM NaHCO3, 0.5 mM CaCl2, 7 mM MgCl2, 5 mM d-glucose, pH 7.4, saturated with 95% O2/5% CO2) for 30 min at room temperature and placed into artificial cerebralspinal fluid (ACSF, 125 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 25 mM NaHCO3, 2 mM CaCl2, 1 mM MgCl2, 25 mM d-glucose, pH 7.4, and saturated with 95% O2/5% CO2) for 1 h. Slices were then stimulated with 50 μM DHPG for 30 or 60 min. Some slices were preincubated with 10 μM of anisomycin before applying DHPG to inhibit de novo protein synthesis. After stimulation, the slices were snap-frozen into liquid nitrogen for immunoblot analysis.
Electrophysiological Recording.
Coronal slices 350 μm thick containing cortex were made with a vibratome and preincubated in oxygenated ACSF at 30 °C for at least 1 h before transfer to a recording chamber in which they were superfused (2∼3 mL/min) with oxygenated ACSF while maintained at 30 °C. The whole-cell patch-clamp recordings were performed with Multiclamp 700B amplifier (Axon Instruments). Pyramidal neurons were recorded from layer II/III of somatosensory cortex (68). Patch pipettes with resistances of 5–8 MΩ were filled with the following solution (in mM): 120 K-gluconate, 5 NaCl, 1 MgCl 2, 0.2 EGTA, 10 Hepes, 2 Mg ATP, 0.1 Na 3-GTP, and 10 sodium phosphocreatine (adjusted to pH 7.2 with KOH).
Access resistance <30 MΩ was considered acceptable. Only neurons that had a resting membrane potential more negative than −60 mV were considered for the analysis, and neurons were discarded if access resistance changed more than 15% during an experiment. Data were filtered at 1 kHz and digitized at 10 kHz. Input resistance was determined from the linear slope (between −60 and −80 mV) of the current-voltage (I-V) relationship. Action potentials (APs) were detected in response to suprathreshold current injections from the resting membrane potential of the cell. Depolarizing currents (350-ms duration) were delivered starting from 120 pA with increments of 5 pA until an AP was evoked. The rheobase was defined as the minimum current required to evoke an action potential. The AP voltage threshold (V threshold in mV) was defined as the first point on the rising phase of the spike at which the change in voltage exceeded 60 mV/ms. The spike amplitude was quantified as the difference between the V threshold and the peak voltage. The duration of the AP was measured at the threshold voltage. The spike width was measured at 1/2 of the maximum spike amplitude (from the V threshold level). Recorded neurons responded with a single action potential at the onset of a depolarizing current pulse even to a strong depolarizing current.
Immunoblotting.
For immunoblot analysis, 30 µg of protein was boiled in lithium dodecyl sulfate buffer and analyzed by Western blot using the following antibodies: rabbit against DARPP32 (1:1,000), Synapsin 1 (1:500), SNAP25 (1:1,000), and Pur-α (1:1,000) and mouse antibodies against PSD95 (1:2,000), CaMKIIα (1:1,000), HCN1 (1:1,000), pan-Shank (1:1,000), FMRP (1:1,000), and GAPDH (1:1,000). Western blot analyses were performed on samples from three separate experiments. The blots were scanned and band intensities were measured using Adobe Photoshop CS5, followed by Student’s t test to assess the statistical significance.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants BIMR P30 NS057096, 5R01 MH067880, and P41 GM103533 (to J.R.Y.), and by a Shanghai Institute of Biochemistry and Cell Biology new investigator grant, Shanghai Pujiang Talent Project, and from the East China Normal University National “985” project grant (to L.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Data have been provided in the MS RAW Data Repository, sealion.scripps.edu/pint/?project=261ff98f356429fa784dba44d184742c.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502258112/-/DCSupplemental.
References
- 1.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27(7):370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 2.Colak D, et al. Promoter-bound trinucleotide repeat mRNA drives epigenetic silencing in fragile X syndrome. Science. 2014;343(6174):1002–1005. doi: 10.1126/science.1245831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoogeveen AT, Oostra BA. The fragile X syndrome. J Inherit Metab Dis. 1997;20(2):139–151. doi: 10.1023/a:1005392319533. [DOI] [PubMed] [Google Scholar]
- 4.Kao DI, Aldridge GM, Weiler IJ, Greenough WT. Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proc Natl Acad Sci USA. 2010;107(35):15601–15606. doi: 10.1073/pnas.1010564107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zalfa F, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci. 2007;10(5):578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Napoli I, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134(6):1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Pfeiffer BE, Huber KM. Fragile X mental retardation protein induces synapse loss through acute postsynaptic translational regulation. J Neurosci. 2007;27(12):3120–3130. doi: 10.1523/JNEUROSCI.0054-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comery TA, et al. Abnormal dendritic spines in fragile X knockout mice: Maturation and pruning deficits. Proc Natl Acad Sci USA. 1997;94(10):5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci USA. 2002;99(11):7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Q, Nomura T, Xu J, Contractor A. The developmental switch in GABA polarity is delayed in fragile X mice. J Neurosci. 2014;34(2):446–450. doi: 10.1523/JNEUROSCI.4447-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung KM, et al. Uncoupling of the endocannabinoid signalling complex in a mouse model of fragile X syndrome. Nat Commun. 2012;3:1080. doi: 10.1038/ncomms2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Till SM. The developmental roles of FMRP. Biochem Soc Trans. 2010;38(2):507–510. doi: 10.1042/BST0380507. [DOI] [PubMed] [Google Scholar]
- 13.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107(4):477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 14.Ascano M, Jr, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492(7429):382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65(3):373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross C, et al. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J Neurosci. 2010;30(32):10624–10638. doi: 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma A, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30(2):694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao L, Park SK, Xu T, Vanderklish P, Yates JR., III Quantitative proteomic analysis of primary neurons reveals diverse changes in synaptic protein content in fmr1 knockout mice. Proc Natl Acad Sci USA. 2008;105(40):15281–15286. doi: 10.1073/pnas.0804678105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monzo K, Dowd SR, Minden JS, Sisson JC. Proteomic analysis reveals CCT is a target of Fragile X mental retardation protein regulation in Drosophila. Dev Biol. 2010;340(2):408–418. doi: 10.1016/j.ydbio.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman JS, et al. Netrin-1 promotes excitatory synaptogenesis between cortical neurons by initiating synapse assembly. J Neurosci. 2013;33(44):17278–17289. doi: 10.1523/JNEUROSCI.1085-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClatchy DB, Dong MQ, Wu CC, Venable JD, Yates JR., III 15N metabolic labeling of mammalian tissue with slow protein turnover. J Proteome Res. 2007;6(5):2005–2010. doi: 10.1021/pr060599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR., III Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem. 2004;76(17):4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 24.McClatchy DB, Liao L, Park SK, Venable JD, Yates JR. Quantification of the synaptosomal proteome of the rat cerebellum during post-natal development. Genome Res. 2007;17(9):1378–1388. doi: 10.1101/gr.6375007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao L, et al. 15N-labeled brain enables quantification of proteome and phosphoproteome in cultured primary neurons. J Proteome Res. 2012;11(2):1341–1353. doi: 10.1021/pr200987h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toyama BH, et al. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell. 2013;154(5):971–982. doi: 10.1016/j.cell.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 28.Napolitano F, et al. Role of aberrant striatal dopamine D1 receptor/cAMP/protein kinase A/DARPP32 signaling in the paradoxical calming effect of amphetamine. J Neurosci. 2010;30(33):11043–11056. doi: 10.1523/JNEUROSCI.1682-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruepp A, et al. CORUM: The comprehensive resource of mammalian protein complexes—2009. Nucleic Acids Res. 2010;38(Database issue):D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sungur AO, Vörckel KJ, Schwarting RK, Wöhr M. Repetitive behaviors in the Shank1 knockout mouse model for autism spectrum disorder: Developmental aspects and effects of social context. J Neurosci Methods. 2014;234:92–100. doi: 10.1016/j.jneumeth.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Berkel S, et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010;42(6):489–491. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- 32.Fatemi SH, et al. Impairment of fragile X mental retardation protein-metabotropic glutamate receptor 5 signaling and its downstream cognates ras-related C3 botulinum toxin substrate 1, amyloid beta A4 precursor protein, striatal-enriched protein tyrosine phosphatase, and homer 1, in autism: A postmortem study in cerebellar vermis and superior frontal cortex. Mol Autism. 2013;4(1):21. doi: 10.1186/2040-2392-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aschrafi A, Cunningham BA, Edelman GM, Vanderklish PW. The fragile X mental retardation protein and group I metabotropic glutamate receptors regulate levels of mRNA granules in brain. Proc Natl Acad Sci USA. 2005;102(6):2180–2185. doi: 10.1073/pnas.0409803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiler IJ, et al. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proc Natl Acad Sci USA. 1997;94(10):5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou L, et al. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51(4):441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 37.Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci. 2010;30(46):15616–15627. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu R, et al. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc Natl Acad Sci USA. 2004;101(42):15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Till SM, et al. Altered maturation of the primary somatosensory cortex in a mouse model of fragile X syndrome. Hum Mol Genet. 2012;21(10):2143–2156. doi: 10.1093/hmg/dds030. [DOI] [PubMed] [Google Scholar]
- 40.Hayashi MK, et al. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell. 2009;137(1):159–171. doi: 10.1016/j.cell.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bangash MA, et al. Enhanced polyubiquitination of Shank3 and NMDA receptor in a mouse model of autism. Cell. 2011;145(5):758–772. doi: 10.1016/j.cell.2011.03.052. [Retracted in Cell 2013 152(1-2):367.] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Lee JY, Kwak M, Lee PC. Impairment of social behavior and communication in mice lacking the Uba6-dependent ubiquitin activation system. Behav Brain Res. 2015;281:78–85. doi: 10.1016/j.bbr.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 43.He CX, Portera-Cailliau C. The trouble with spines in fragile X syndrome: Density, maturity and plasticity. Neuroscience. 2013;251:120–128. doi: 10.1016/j.neuroscience.2012.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng PY, Sojka D, Klyachko VA. Abnormal presynaptic short-term plasticity and information processing in a mouse model of fragile X syndrome. J Neurosci. 2011;31(30):10971–10982. doi: 10.1523/JNEUROSCI.2021-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gálvez JM, et al. Evidence of association between SNAP25 gene and attention deficit hyperactivity disorder in a Latin American sample. Atten Defic Hyperact Disord. 2014;6(1):19–23. doi: 10.1007/s12402-013-0123-9. [DOI] [PubMed] [Google Scholar]
- 46.Gao Q, et al. Synaptosome-related (SNARE) genes and their interactions contribute to the susceptibility and working memory of attention-deficit/hyperactivity disorder in males. Prog Neuropsychopharmacol Biol Psychiatry. 2015;57:132–139. doi: 10.1016/j.pnpbp.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 47.Farzin F, et al. Autism spectrum disorders and attention-deficit/hyperactivity disorder in boys with the fragile X premutation. J Dev Behav Pediatr. 2006;27(2) Suppl:S137–S144. doi: 10.1097/00004703-200604002-00012. [DOI] [PubMed] [Google Scholar]
- 48.Hagerman RJ. Lessons from fragile X regarding neurobiology, autism, and neurodegeneration. J Dev Behav Pediatr. 2006;27(1):63–74. doi: 10.1097/00004703-200602000-00012. [DOI] [PubMed] [Google Scholar]
- 49.Belmonte MK, Bourgeron T. Fragile X syndrome and autism at the intersection of genetic and neural networks. Nat Neurosci. 2006;9(10):1221–1225. doi: 10.1038/nn1765. [DOI] [PubMed] [Google Scholar]
- 50.Dosemeci A, et al. Composition of the synaptic PSD-95 complex. Mol Cell Proteomics. 2007;6(10):1749–1760. doi: 10.1074/mcp.M700040-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7304):368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlisle HJ, et al. Deletion of densin-180 results in abnormal behaviors associated with mental illness and reduces mGluR5 and DISC1 in the postsynaptic density fraction. J Neurosci. 2011;31(45):16194–16207. doi: 10.1523/JNEUROSCI.5877-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myers KR, et al. Arf6-GEF BRAG1 regulates JNK-mediated synaptic removal of GluA1-containing AMPA receptors: A new mechanism for nonsyndromic X-linked mental disorder. J Neurosci. 2012;32(34):11716–11726. doi: 10.1523/JNEUROSCI.1942-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michalon A, et al. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74(1):49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cea-Del Rio CA, Huntsman MM. The contribution of inhibitory interneurons to circuit dysfunction in Fragile X Syndrome. Front Cell Neurosci. 2014;8:245. doi: 10.3389/fncel.2014.00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lemonnier E, et al. Treating Fragile X syndrome with the diuretic bumetanide: A case report. Acta Paediatr. 2013;102(6):e288–e290. doi: 10.1111/apa.12235. [DOI] [PubMed] [Google Scholar]
- 57.Wang H, et al. FMRP acts as a key messenger for dopamine modulation in the forebrain. Neuron. 2008;59(4):634–647. doi: 10.1016/j.neuron.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, et al. Dendritic channelopathies contribute to neocortical and sensory hyperexcitability in Fmr1(-/y) mice. Nat Neurosci. 2014;17(12):1701–1709. doi: 10.1038/nn.3864. [DOI] [PubMed] [Google Scholar]
- 59.Brager DH, Akhavan AR, Johnston D. Impaired dendritic expression and plasticity of h-channels in the fmr1(-/y) mouse model of fragile X syndrome. Cell Reports. 2012;1(3):225–233. doi: 10.1016/j.celrep.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levine JB, Morrow EM, Berdichevsky Y, Martin GE. BKca channel in autism and mental retardation. Am J Psychiatry. 2007;164(6):977–978; author reply 978–979. doi: 10.1176/ajp.2007.164.6.977a. [DOI] [PubMed] [Google Scholar]
- 61.Myrick LK, et al. Independent role for presynaptic FMRP revealed by an FMR1 missense mutation associated with intellectual disability and seizures. Proc Natl Acad Sci USA. 2015;112(4):949–956. doi: 10.1073/pnas.1423094112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59(1):84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dieterich DC, et al. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2(3):532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- 64.MacCoss MJ, Wu CC, Matthews DE, Yates JR., III Measurement of the isotope enrichment of stable isotope-labeled proteins using high-resolution mass spectra of peptides. Anal Chem. 2005;77(23):7646–7653. doi: 10.1021/ac0508393. [DOI] [PubMed] [Google Scholar]
- 65.Zhang B, Horvath S. 2005. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4:17.
- 66.Wolters DA, Washburn MP, Yates JR., III An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73(23):5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 67.Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 68.Blanton MG, Lo Turco JJ, Kriegstein AR. Whole cell recording from neurons in slices of reptilian and mammalian cerebral cortex. J Neurosci Methods. 1989;30(3):203–210. doi: 10.1016/0165-0270(89)90131-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.