Abstract
We investigated whether the gut microbiota differed in 48 postmenopausal breast cancer case patients, pretreatment, vs 48 control patients. Microbiota profiles in fecal DNA were determined by Illumina sequencing and taxonomy of 16S rRNA genes. Estrogens were quantified in urine. Case-control comparisons employed linear and unconditional logistic regression of microbiota α-diversity (PD_whole tree) and UniFrac analysis of β-diversity, with two-sided statistical tests. Total estrogens correlated with α-diversity in control patients (Spearman Rho = 0.37, P = .009) but not case patients (Spearman Rho = 0.04, P = .77). Compared with control patients, case patients had statistically significantly altered microbiota composition (β-diversity, P = .006) and lower α-diversity (P = .004). Adjusted for estrogens and other covariates, odds ratio of cancer was 0.50 (95% confidence interval = 0.30 to 0.85) per α-diversity tertile. Differences in specific taxa were not statistically significant when adjusted for multiple comparisons. This pilot study shows that postmenopausal women with breast cancer have altered composition and estrogen-independent low diversity of their gut microbiota. Whether these affect breast cancer risk and prognosis is unknown.
In addition to traditional factors (1–3), breast cancer risk for postmenopausal women is directly related to level of endogenous estrogens and differences in estrogen metabolism (4–11). The gut microbiota modulates estrogen homeostasis through enterohepatic circulation, with large differences among individuals (12–16). The microbiota also modulates many other metabolic and immunologic pathways (17–19). Independent of estrogen levels, cancer risk is increased with metabolic syndrome through growth factors like insulin (20,21). Inflammation probably also contributes (22), as use of nonsteroidal anti-inflammatory drugs was associated with a 20% to 30% reduced risk of postmenopausal breast cancer in some studies, albeit with inconsistency by estrogen receptor (ER) tumor expression (23–29). Noting that gut microbial differences could affect breast cancer risk through several pathways, herein we tested the hypothesis that the gut microbiota of postmenopausal women with incident breast cancer, pretreatment, differs from control women.
Following review and approval by the respective institutional review boards, with signed informed consent 48 female members of Kaiser Permanente Colorado ages 50 to 74 years who were scheduled for treatment of biopsy-proven breast cancer and 48 normal-mammography women provided data, urine (without preservative), and feces (in RNAlater), frozen at home and thereafter below -80°C until use (30–33). Microbiota profiles in fecal DNA were determined by amplification, Illumina sequencing, and taxonomy of 16S rRNA genes (34–38). Urinary estrogens and estrogen metabolites (EMs) were quantified by liquid chromatography/tandem mass spectrometry (39). Microbiota alpha diversity was estimated as follows. Richness: number of unique species-level taxa, unadjusted for their relative abundances. Chao1: richness, but bias-corrected for rare (singleton, doubleton) taxa. Phylogenetic diversity (PD)_whole tree: sum of the branch lengths of a phylogenetic tree constructed from all taxa in the sample. Shannon index: a conservative estimate that adjusts for relative abundance of each taxon and that is defined as (negative) the sum over taxa of the product of the relative abundance of each taxon times the natural logarithm of its relative abundance. Estrogen associations with microbiota alpha diversity were tested by Spearman rank-order correlation. Alpha diversity associations with case-control status were tested by linear and unconditional logistic regression, with adjustment for age, body mass index (BMI), and total estrogens. Composition of the microbiota (beta diversity) was compared by unweighted and weighted UniFrac analysis of the distance matrix with 10 000 permutations (40). All statistical tests were two-sided, and a P value of less than .05 was considered statistically significant. Detailed methods can be found in the Supplementary Materials (available online).
Case patients and control patients were 86% non-Hispanic white with mean age 62 years (SD = 6.86), mean BMI of 28 (SD 1.07), and equivalent reproductive and menstrual histories (Supplementary Table 1, available online). Two case patients (excluded from some analyses) and no control patient reported receiving an antibiotic within the previous two weeks. Two cancers were stage 3, ten at stage 2, 25 at stage 1, and 11 in situ (American Joint Committee on Cancer, Collaborative Staging Version 2.04) (41). Forty-two tumors were ER-positive, 37 were progesterone receptor–positive, and five were HER2-positive.
All mean urinary estrogens were two-fold higher in the case patients, although these were not statistically significant (P ≥ .10) (Table 1). In control patients, fecal microbiota alpha diversity (phylogenetic diversity [PD]_whole tree) correlated directly with total estrogens (Spearman Rho = 0.37, P = .009). In contrast, PD_whole tree was not correlated with total estrogens in case patients (Spearman Rho = 0.04, P = .77). PD_whole tree was weakly correlated with EM:parent estrogen ratio in control patients (Spearman Rho = 0.26, P = .08). There was no such correlation between PD_whole tree and EM:parent estrogen ratio in case patients (Rho = -0.11, P = .45).
Table 1.
Urinary estrogens and estrogen metabolites (EM) and fecal microbiome alpha diversity in postmenopausal breast cancer cases and controls
| Variable/outcome | Case patients, N=48 | Control patients, N=48 | P* | ||
|---|---|---|---|---|---|
| Estrogen, EM levels, mean (SD)† | |||||
| Total estrogens and EM | 45.40 (106.94) | 22.36 (17.79) | .12 | ||
| Parent estrogens | 16.89 (44.38) | 7.30 (5.93) | .12 | ||
| Estrone | 12.97 (31.76) | 5.83 (4.81) | .11 | ||
| Estradiol | 3.92 (12.86) | 1.47 (1.31) | .17 | ||
| Total EM | 28.51 (63.33) | 15.07 (12.42) | .15 | ||
| 2-Hydroxylation pathway | 12.94 (28.83) | 6.51 (5.59) | .10 | ||
| 16-Hydroxylation pathway | 14.43 (32.12) | 7.99 (6.81) | .21 | ||
| 4-Hydroxylation pathway | 1.13 (2.41) | .56 (.47) | .11 | ||
| Estrogen, EM ratios | |||||
| EM/parent | 2.15 (.87) | 2.35 (2.01) | .56 | ||
| 2-pathway/parent | .97 (.40) | .96 (.44) | .90 | ||
| 16-pathway/parent | 1.10 (.45) | 1.30 (1.59) | .41 | ||
| 4-pathway/parent | .09 (.04) | .09 (.04) | .95 | ||
| 2-pathway/16-pathway | .89 (.16) | .85 (.15) | .14 | ||
| Fecal microbiome richness, alpha diversity, mean (SE) | |||||
| No. observed species | 78.6 (23.1) | 91.2 (16.6) | .004 | ||
| Chao1 | 909.5 (24.4) | 1053.8 (174.9) | .001 | ||
| PD_whole tree | 33.1 (7.9) | 37.5 (6.1) | .004 | ||
| Shannon index | 6.0 (.7) | 6.2 (.6) | .09 | ||
| Breast cancer risk, by tertile, odds ratio (95% confidence intervals)‡ | Tertile 1 | Tertile 2 | Tertile 3 | ||
| No. observed species | .50 (.30 to .84) | Referent | .24 | .29 | |
| Chao1 | .53 (.31 to .89) | Referent | .28 | .30 | |
| PD_whole tree | .50 (.30 to .85) | Referent | .20 | .29 | |
| Shannon index | .83 (.50 to 1.37) | Referent | .89 | .68 | |
* Linear regression with (log-transformed estrogen and EM values) and adjustment for age.
† Picomoles/mg creatinine.
‡ Logistic regression, tertiles among controls, adjusted for age, body mass index, and total urine estrogen level.
The fecal microbiota of case patients, compared with control patients, had statistically significantly lower alpha diversity (P ≤ .004), except Shannon index (P = .09) (Table 1). Adjusted for age, BMI, and total urine estrogens, the breast cancer odds ratio (ORadj) per tertile category increase in PD_whole tree was 0.50 (95% confidence interval [CI] = 0.30 to 0.85), and nearly identical for richness and Chao1 (Table 1). The association was not linear. Rather, compared with lowest-tertile levels, breast cancer ORadj was reduced 70% to 80% with both middle-tertile and highest-tertile levels of these measures (Table 1 and Figure 1A).
Figure 1.
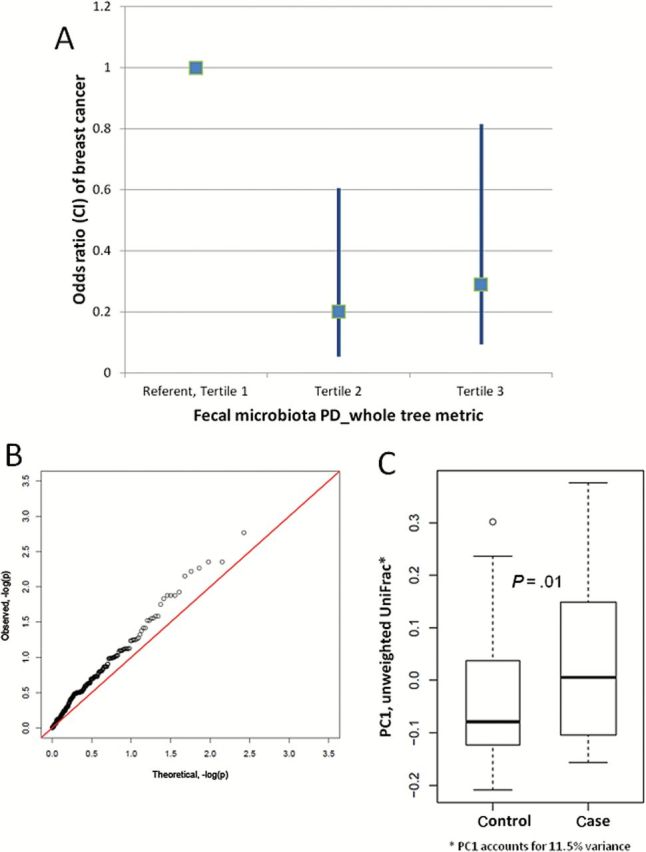
Fecal microbiota differences between postmenopausal breast cancer case patients and control patients. A) Odds ratio (square) and 95% confidence interval (bar) of breast cancer by tertile of alpha diversity (phylogenetic diversity [PD]_whole tree). Odds ratios by tertile are presented. B) Beta diversity, quantile-quantile plot of two-sided Wilcoxon rank-sum P values for all genus-level taxa. The distribution (x-axis expected, y-axis observed) diverges from the null (diagonal line) for many taxa. C) Beta diversity, distribution of the first principal coordinate values (PC1, 11.5% of the variance*) of the unweighted UniFrac distance matrix. Boxes are the interquartile range; median values are bands within the boxes; whiskers are 1.5-times the IQR; open circle is an outlier value. CI = confidence interval; PD = phylogenetic diversity.
Fecal microbiota composition (beta diversity) also differed between case patients and control patients overall (unweighted UniFrac P = .009), across all genus-level taxa (Figure 1B), and on the first principal coordinate (PC1) of the beta diversity distance matrix (P = .01) (Figure 1C). The difference was larger with exclusion of the two antibiotic-exposed case patients (unweighted UniFrac P = .006). Without adjustment for multiple comparisons, relative abundance of several taxa differed between case patients and control patients by 1% or more (Supplementary Table 2, available online). Particularly in the order Clostridiales, case patients had higher levels of Clostridiaceae, Faecalibacterium, and Ruminococcaceae; and they had lower levels of Dorea and Lachnospiraceae (Supplementary Figure 1, available online).
This population-based case-control study found that the fecal microbiota was less diverse and compositionally different in postmenopausal women who were awaiting treatment for biopsy-proven breast cancer compared with similar women without breast cancer. As expected (5–11), the cancer case patients also had higher levels of systemic estrogens, although these were not statistically significant in this small study. Importantly, the difference in estrogens, and statistical adjustment for this difference, did not alter the cancer-microbiota association.
Our findings are consistent with the 40-year literature on the gut microbiota’s effects on systemic estrogens (12–16) and with our previously observed correlations of alpha diversity with systemic estrogen level and EM:parent estrogen ratio (16,33). The novel implication of our current study is that breast cancer was statistically significantly associated with other functions of the gut microbiota, unrelated to systemic estrogen levels. Low gut microbial diversity occurs with adiposity, insulin resistance, dyslipidemia, leukocytosis, and elevated C-reactive protein (17), some of which are associated with breast cancer (20,42–47).
The infant’s gut microbial composition may influence breast cancer risk in adulthood. In both mice and people, microbiota composition is acquired directly from the mother during birth (48–53). The distinct microbiota of adults who were born by cesarean vs vaginal delivery (54), as well as the similarity of microbial composition within adult dizygotic twin pairs (55–57), implies that composition is stable for decades. Breast cancer risk is affected by obscure early-life effects that also are transmitted through the maternal line (3). Such maternal effects could reflect differences in systemic estrogens, but genetic determinants of estrogen levels have been inconsistent (58–60). We postulate an effect for maternal transmission of the microbiota.
The strengths of our study include representative control patients, careful clinical staging and histopathology, optimal specimens collected prior to treatment among case patients, state-of-the-art assays, and rigorous statistical analysis methods. Weaknesses include the small sample size, which precluded assessment of minor taxa and of interactions between microbiota metrics and known risk factors, particularly estrogens. Estrogen parameters were correlated with microbiota alpha diversity, although only in control patients (16,33). Our case-control design is another important weakness, precluding exclusion of reverse causality—that cancer caused the microbiota distinction. To minimize this possibility, our case patients received only an outpatient breast biopsy, no surgical or systemic therapy prior to specimen collection. Excluding case patients who reported antibiotic exposure, perhaps biopsy-related, had no major impact on the microbiota associations.
In summary, postmenopausal women with newly diagnosed breast cancer had a fecal microbiota that was less diverse and compositionally different compared with similar women without breast cancer. The cancer case patients also had higher levels of urinary estrogens, but these were independent of microbiota differences. The findings imply that the gut microbiota may affect breast cancer risk and may do so through estrogen-independent pathways.
Funding
This work was supported by the Intramural Research Program of the National Cancer Institute at the National Institutes of Health (Z01CP010214).
Supplementary Material
The study funder had no role in the design of the study, the collection, analysis, or interpretation of the data, the writing of the manuscript, nor the decision to submit the manuscript for publication.
The authors are grateful to Jennifer McCance for coordinating the field work, to Elaine Flores and Mollie Miedzinski for assisting with the field work, to Mike Humphrys for coordinating the microbiota testing, to Kathleen Spaid for urine creatinine testing, and to especially the study participants.
References
- 1. Colditz GA, Baer HJ, Tamimi RM. Breast Cancer. In: Schottenfeld D, Fraumeni JF, Jr, eds. Cancer Epidemiology and Prevention. Third ed New York, NY: Oxford Univerity Press, Inc; 2006: 995–1012. [Google Scholar]
- 2. Falkenberry SS, Legare RD. Risk factors for breast cancer. Obstet Gynecol Clin North Am. 2002;29(1):159–172. [DOI] [PubMed] [Google Scholar]
- 3. Weinberg CR, Shi M, DeRoo LA, Taylor JA, Sandler DP, Umbach DM. Asymmetry in family history implicates nonstandard genetic mechanisms: application to the genetics of breast cancer. PLoS Genet. 2014;10(3):e1004174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Samavat H, Kurzer MS. Estrogen metabolism and breast cancer. Cancer Lett. 2015;356(2):231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Key TJ, Appleby PN, Reeves GK, et al. Circulating sex hormones and breast cancer risk factors in postmenopausal women: reanalysis of 13 studies. Br J Cancer. 2011;105(5):709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fuhrman BJ, Schairer C, Gail MH, et al. Estrogen metabolism and risk of breast cancer in postmenopausal women. J Natl Cancer Inst. 2012;104(4):326–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eliassen AH, Spiegelman D, Xu X, et al. Urinary estrogens and estrogen metabolites and subsequent risk of breast cancer among premenopausal women. Cancer Res. 2012;72(3):696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dallal CM, Tice JA, Buist DS, et al. Estrogen metabolism and breast cancer risk among postmenopausal women: a case-cohort study within B~FIT. Carcinogenesis. 2014;35(2):346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rinaldi S, Key TJ, Peeters PH, et al. Anthropometric measures, endogenous sex steroids and breast cancer risk in postmenopausal women: a study within the EPIC cohort. Internat J Cancer. 2006;118(11):2832–2839. [DOI] [PubMed] [Google Scholar]
- 10. Arslan AA, Koenig KL, Lenner P, et al. Circulating estrogen metabolites and risk of breast cancer in postmenopausal women. Cancer Epidemiol Biomarkers Prevention. 2014;23(7):1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Falk RT, Brinton LA, Dorgan JF, et al. Relationship of serum estrogens and estrogen metabolites to postmenopausal breast cancer risk: a nested case-control study. Breast Cancer Res. 2013;15(2):R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adlercreutz H, Pulkkinen MO, Hamalainen EK, Korpela JT. Studies on the role of intestinal bacteria in metabolism of synthetic and natural steroid hormones. J Steroid Biochem. 1984;20(1):217–229. [DOI] [PubMed] [Google Scholar]
- 13. Heimer GM, Englund DE. Enterohepatic recirculation of oestriol studied in cholecystectomized and non-cholecystectomized menopausal women. Ups J Med Sci. 1984;89(2):107–115. [DOI] [PubMed] [Google Scholar]
- 14. Goldin BR, Adlercreutz H, Gorbach SL, et al. Estrogen excretion patterns and plasma levels in vegetarian and omnivorous women. N Engl J Med. 1982;307(25):1542–1547. [DOI] [PubMed] [Google Scholar]
- 15. Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe. 2011;10(4):324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flores R, Shi J, Fuhrman B, et al. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med. 2012;10:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. [DOI] [PubMed] [Google Scholar]
- 18. Delzenne NM, Neyrinck AM, Backhed F, Cani PD. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat Rev Endocrinol. 2011;7(11):639–646. [DOI] [PubMed] [Google Scholar]
- 19. Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14(7):676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gunter MJ, Xie X, Xue X, et al. Breast cancer risk in metabolically healthy but overweight postmenopausal women. Cancer Res. 2015;75(2):270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Esposito K, Chiodini P, Capuano A, et al. Metabolic syndrome and postmenopausal breast cancer: systematic review and meta-analysis. Menopause. 2013;20(12):1301–1309. [DOI] [PubMed] [Google Scholar]
- 22. Howe LR, Subbaramaiah K, Hudis CA, Dannenberg AJ. Molecular pathways: adipose inflammation as a mediator of obesity-associated cancer. Clin Cancer Res. 2013;19(22):6074–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang X, Smith-Warner SA, Collins LC, Rosner B, Willett WC, Hankinson SE. Use of aspirin, other nonsteroidal anti-inflammatory drugs, and acetaminophen and postmenopausal breast cancer incidence. J Clin Oncol. 2012;30(28):3468–3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bardia A, Olson JE, Vachon CM, et al. Effect of aspirin and other NSAIDs on postmenopausal breast cancer incidence by hormone receptor status: results from a prospective cohort study. Breast Cancer Res Treat. 2011;126(1):149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marshall SF, Bernstein L, Anton-Culver H, et al. Nonsteroidal anti-inflammatory drug use and breast cancer risk by stage and hormone receptor status. J Natl Cancer Inst. 2005;97(11):805–812. [DOI] [PubMed] [Google Scholar]
- 26. Gierach GL, Lacey JV, Jr, Schatzkin A, et al. Nonsteroidal anti-inflammatory drugs and breast cancer risk in the National Institutes of Health-AARP Diet and Health Study. Breast Cancer Res. 2008;10(2):R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harris RE, Chlebowski RT, Jackson RD, et al. Breast cancer and nonsteroidal anti-inflammatory drugs: prospective results from the Women’s Health Initiative. Cancer Res. 2003;63(18):6096–6101. [PubMed] [Google Scholar]
- 28. Terry MB, Gammon MD, Zhang FF, et al. Association of frequency and duration of aspirin use and hormone receptor status with breast cancer risk. JAMA. 2004;291(20):2433–2440. [DOI] [PubMed] [Google Scholar]
- 29. Rahme E, Ghosn J, Dasgupta K, Rajan R, Hudson M. Association between frequent use of nonsteroidal anti-inflammatory drugs and breast cancer. BMC Cancer. 2005;5:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feigelson HS, Bischoff K, Ardini MA, et al. Feasibility of self-collection of fecal specimens by randomly sampled women for health-related studies of the gut microbiome. BMC Res Notes. 2014;7:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Flores R, Shi J, Gail MH, Ravel J, Goedert JJ. Assessment of the human faecal microbiota: I. Measurement and reproducibility of selected enzymatic activities. Eur J Clin lnvest. 2012;42(8):848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Flores R, Shi J, Gail MH, Gajer P, Ravel J, Goedert JJ. Assessment of the human faecal microbiota: II. Reproducibility and associations of 16S rRNA pyrosequences. Eur J Clin Invest. 2012;42(8):855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fuhrman BJ, Feigelson HS, Flores R, et al. Associations of the Fecal Microbiome With urinary estrogens and estrogen metabolites in postmenopausal women. J Clin Endocrinol Metab. 2014;99(12):4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. [DOI] [PubMed] [Google Scholar]
- 35. Consortium HM. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fadrosh DW, Ma B, Gajer P, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environmen Microbiol. 2007;73(16):5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu X, Keefer LK, Ziegler RG, Veenstra TD. A liquid chromatography-mass spectrometry method for the quantitative analysis of urinary endogenous estrogen metabolites. Nat Protocols. 2007;2(6):1350–1355. [DOI] [PubMed] [Google Scholar]
- 40. Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5(2):169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Howlader N, Chen VW, Ries LA, et al. Overview of breast cancer collaborative stage data items--their definitions, quality, usage, and clinical implications: a review of SEER data for 2004–2010. Cancer. 2014;120 Suppl 23:3771–3780. [DOI] [PubMed] [Google Scholar]
- 42. Margolis KL, Rodabough RJ, Thomson CA, Lopez AM, McTiernan A. Prospective study of leukocyte count as a predictor of incident breast, colorectal, endometrial, and lung cancer and mortality in postmenopausal women. Arch Intern Med. 2007;167(17):1837–1844. [DOI] [PubMed] [Google Scholar]
- 43. Zhang SM, Lin J, Cook NR, et al. C-reactive protein and risk of breast cancer. J Natl Cancer Inst. 2007;99(11):890–894. [DOI] [PubMed] [Google Scholar]
- 44. Ollberding NJ, Kim Y, Shvetsov YB, et al. Prediagnostic leptin, adiponectin, C-reactive protein, and the risk of postmenopausal breast cancer. Cancer Prev Res (Phila). 2013;6(3):188–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hong T, Liu A, Cai D, et al. Preoperative serum C-reactive protein levels and early breast cancer by BMI and menopausal status. Cancer Invest. 2013;31(4):279–285. [DOI] [PubMed] [Google Scholar]
- 46. Dossus L, Jimenez-Corona A, Romieu I, et al. C-reactive protein and postmenopausal breast cancer risk: results from the E3N cohort study. Cancer Causes Control. 2014;25(4):533–539. [DOI] [PubMed] [Google Scholar]
- 47. Wang G, Li N, Chang S, et al. A prospective follow-up study of the relationship between C-reactive protein and human cancer risk in the Chinese Kailuan Female Cohort. Cancer Epidemiol Biomarkers Prev. 2015;24(2):459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol. 2014;5:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Adlerberth I, Lindberg E, Aberg N, et al. Reduced enterobacterial and increased staphylococcal colonization of the infantile bowel: an effect of hygienic lifestyle? Pediatr Res. 2006;59(1):96–101. [DOI] [PubMed] [Google Scholar]
- 51. Biasucci G, Benenati B, Morelli L, Bessi E, Boehm G. Cesarean delivery may affect the early biodiversity of intestinal bacteria. J Nutr. 2008;138(9):1796S–1800S. [DOI] [PubMed] [Google Scholar]
- 52. Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–521. [DOI] [PubMed] [Google Scholar]
- 53. Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goedert JJ, Hua X, Yu G, Shi J. Diversity and composition of the adult fecal microbiome associated with history of cesarean birth or appendectomy: Analysis of the American Gut Project. EBioMedicine. 2014;1(2–3): 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cozen W, Yu G, Gail MH, et al. Fecal microbiota diversity in survivors of adolescent/young adult Hodgkin lymphoma: A study of twins. Br J Cancer. 2013;108(5):1163–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dunning AM, Dowsett M, Healey CS, et al. Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J Natl Cancer Inst. 2004;96(12):936–945. [DOI] [PubMed] [Google Scholar]
- 59. Pineda B, Garcia-Perez MA, Cano A, Lluch A, Eroles P. Associations between aromatase CYP19 rs10046 polymorphism and breast cancer risk: from a case-control to a meta-analysis of 20,098 subjects. PloS One. 2013;8(1):e53902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Prescott J, Thompson DJ, Kraft P, et al. Genome-wide association study of circulating estradiol, testosterone, and sex hormone-binding globulin in postmenopausal women. PloS One. 2012;7(6):e37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


