Abstract
The gastrointestinal (GI) microbiota forms a mutualistic relationship with the host through complex and dynamic interactions. Because of the complexity and interindividual variation of the GI microbiota, investigating how members of the microbiota interact with each other, as well as with the host, is daunting. The altered Schaedler flora (ASF) is a model community of eight microorganisms that was developed by R.P. Orcutt and has been in use since the late 1970s. The eight microorganisms composing the ASF were all derived from mice, can be cultured in vitro, and are stably passed through multiple generations (at least 15 years or more by the authors) in gnotobiotic mice continually bred in isolator facilities. With the limitations associated with conventional, mono- or biassociated, and germfree mice, use of mice colonized with a consortium of known bacteria that naturally inhabit the murine gut offers a powerful system to investigate mechanisms governing host–microbiota relationships, and how members of the GI microbiota interact with one another. The ASF community offers significant advantages to study homeostatic as well as disease-related interactions by taking advantage of a well-defined, limited community of microorganisms. For example, quantification and spatial distribution of individual members, microbial genetic manipulation, genomic-scale analysis, and identification of microorganism-specific host immune responses are all achievable using the ASF model. This review compiles highlights associated with the 37-year history of the ASF, including descriptions of its continued use in biomedical research to elucidate the complexities of host-microbiome interactions in health and disease.
Keywords: ASF, gnotobiotic, microbiota
The Gut Microbiota
It is well established that the number of bacteria living in our gastrointestinal (GI) tract outnumber our own cells 10 to 1, with values that reach 1011 cells/gram contents in the colon (Eckburg et al. 2005; Luckey 1972; Savage 1977; Whitman et al. 1998). The mutualistic relationship with the host has co-evolved to form a dynamic interaction between the gut microbiota and the host. In healthy individuals, the microbiota thrives in a largely stable environment with continuous nutrient delivery, and in which the host benefits from microbial contributions to metabolism, vitamin production, immune system development, and competitive exclusion of pathogens (Ley et al. 2006; O'Hara and Shanahan 2006).
Determining how specific autochthonous microorganisms contribute to the health of the mammalian host remains a significant challenge due to the vast diversity of microbial species in the gut, which is colonized with several hundred species and subspecies (Eckburg et al. 2005; Savage 1977). Members of the microbiota are also not evenly distributed throughout the GI tract, but are concentrated in many niches throughout the alimentary tract from the lumen to the mucosa (Dubos et al. 1965; Eckburg et al. 2005; Savage et al. 1968; Zoetendal et al. 2002). Understanding how microorganisms have adapted to colonize specific niches is also challenging due to the varying environmental conditions that can be encountered throughout the GI tract, including pH, nutrient availability, host immunity, and bacterial cell density.
Mice as Models for the Gut Microbiota
Mice are the most common mammalian model with which to examine the mutualistic relationship between the host and the microbial world living within us. Similar to humans, mice have a complex gut microbiota dominated by Firmicutes and Bacteroidetes, along with members of the Proteobacteria, as well as other phyla that respond to perturbations such as diet and antibiotics (Dethlefsen et al. 2007; Ley et al. 2006). The murine microbiota can be manipulated in ways not possible with humans, including generation of gnotobiotic mice encompassing both germfree and defined microbiota mice. In addition to their overall complexity, conventionally reared mice have been demonstrated to have a variable microbiota between individuals, as well as significant cage-to-cage variation (McCafferty et al. 2013). Differences in the GI microbiota within the same strain or stock of mouse obtained from different vendors have been shown to influence research results; for example, the presence or absence of segmented filamentous bacteria have been shown to influence the development of Th17 cells (Ivanov et al. 2008). Additional examples demonstrating the influence of the microbiota on host responses have been reported. As reviewed recently by Schoeb and Bullard (2012), for example, phenotypic variation in murine inflammatory bowel disease models is a consequence of the differences in the microbiota.
Although this diversity in the GI microbiota makes commercially available mice similar to the humans who also have a variable GI microbiota between individuals, microbial diversity makes it difficult to draw conclusions between studies regarding how changes in the microbiota affect the host when the study subjects do not have a uniform microbiota as a point of reference. Thus, defined microbiota models offer the advantage of a known and consistent standard upon which to determine how manipulation of GI microorganisms affect physiological processes in the host.
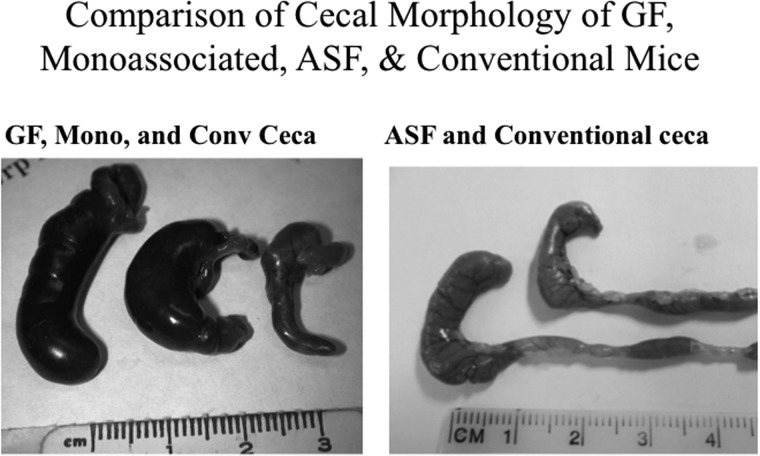
Gnotobiotics and, more specifically, the generation of germ-free mice, have provided the basis to allow investigators the ability to study host–microbe as well as microbe–microbe interactions following the purposeful introduction of selected microorganisms or microbial communities. Germfree animals, however, are not representative of a normal host because they have an underdeveloped mucosal and systemic immune systems characterized by reduced serum leukocytes and antibody (Bauer et al. 1963; Horowitz et al. 1964; Thompson and Trexler 1971). Germfree mice also display anatomical alterations (Figure 1) (i.e., an enlarged cecum), along with histological, physiological, and metabolic differences (Lesher et al. 1964; Thompson and Trexler 1971). Again, it should be realized that mono- or biassociated models lack the community dynamics of a complex microbiota, and the resultant host–microbial interactions would not be representative of those interactions that naturally occur in the conventional mammalian gut.
Figure 1.
Morphological features of the ceca from gnotobiotic and conventional mice. Left panel: Representative images of the cecum excised from a germfree (left), a monoassociated (center), or a conventional (right) C3H/HeN mouse. Right panel: Representative images of the cecum excised from an ASF (left) or conventional (right) C3H/HeN mouse.
Germfree mice have also been used to develop defined microbiota models by colonizing them with a limited number of bacterial species isolated from humans. These “humanized” mice, however, have been utilized mainly for short-term biomedical research studies. In this regard, it is unclear how bacteria from a human source would adapt during long-term colonization and vertical transmission in murine hosts in order to maintain genetic and numerical stability, or organizational structure (Faith et al. 2013; Turnbaugh et al. 2009).
Given the limitations of conventional, germfree, or highly contrived defined-community mouse models, use of mice with a consortium of known bacteria that normally inhabit the murine GI tract offers a powerful system to understand how expression of specific genes within the microbiome affect the host, as well as to determine how members of the GI microbial community interact with one another. The altered Schaedler flora (ASF) represents a gnotobiotic mouse model that has been in use for decades (Table 1) and that offers significant advantages to study microbiome–host interactions.
Table 1.
Metadata characterizing the eight members of the altered Schaedler flora (ASF)
| ASF 356a | ASF 360a | ASF 361a | ASF 457 | ASF 492 | ASF 500 | ASF 502 | ASF 519a | |
|---|---|---|---|---|---|---|---|---|
| Mouse strainb | Nelson Collins Swiss (NCS) | NCS | NCS | NCS | CD-1 | CD-1 | CD-1 | NCS |
| Date of isolation | Early 1960s | Early 1960s | Early 1960s | Early 1960s | Early 1970s | Early 1970s | Early 1970s | Early 1960s |
| Geographic location | Rockefeller University | Rockefeller University | Rockefeller University | Rockefeller University | Charles River Laboratories | Charles River Laboratories | Charles River Laboratories | Rockefeller University |
| Scientist | Russell Schaedler | Russell Schaedler | Russell Schaedler | Russell Schaedler | Roger Orcutt | Roger Orcutt | Roger Orcutt | Russell Schaedler |
| Source | Intestine/Cecum | Stomach | Stomach | Intestine/Cecum | Intestine/cecum | Intestine/cecum | Intestine/Cecum | Intestine/Cecum |
| Taxonomy | Clostridium species | Lactobacillus intestinalis | Lactobacillus murinus | Mucispirillum schaedleri | Eubacterium plexicaudatum | Pseudoflavonifactor species | Clostridium species | Parabacteroides goldsteinii |
| Genome size (Mb) | 2.91 | 2.01 | 2.17 | 2.33 | 6.51 | 3.70 | 6.48 | 6.87 |
| GenBank Accession No. | AQFQ00000000.1 | AQFR00000000.1 | AQFs00000000.1 | AYGZ00000000.1 | AQFT00000000.1 | AYJP00000000.1 | AQFU00000000.1 | AQFV00000000.1 |
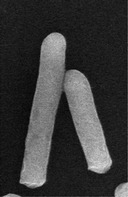
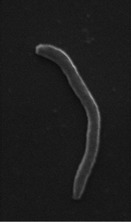
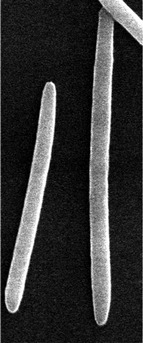
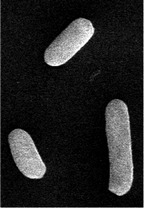
| Cellular morphology |  |
 |
 |
 |
 |
 |
 |
 |
aMembers of the original Schaedler flora.
bMouse strain from which the organism was isolated.
Development of the ASF
In 1965, Russell W. Schaedler developed a defined microbiota mouse colony in which the mice were colonized with two Lactobacillus spp., an anaerobic Streptococcus sp. (Group N), a strain of bacteroides, an Enterococcus sp., and a coliform strain, all of which were isolated from Nelson Collins Swiss mice (Schaedler et al. 1965). The microbial population was maintained from dam to offspring, and the GI tissue of the mice resembled that of a mouse with conventional microbiota, but not that of a germfree mouse (Figure 1) (Schaedler et al. 1965). The bacteria comprising this defined microbial community (Schaedler flora) were supplied to animal vendors for use as a base microbiota for newly derived germfree rodents as a means to provide a competitive niche to limit or prevent colonization of GI opportunistic bacterial pathogens (Trexler and Orcutt 1999). A limitation of the Schaedler flora, however, was the presence of facultative anaerobes (specifically, Escherichia coli and Streptococcus fecalis) that typically outgrew isolator contaminants when cultured aerobically (Baker 1966).
In 1978, at the behest of the National Cancer Institute, Roger P. Orcutt, whose PhD mentor was Russell W. Schaedler, developed a refined microbiota based on the Schaedler flora to be used by contract suppliers (Orcutt et al. 1987; Trexler and Orcutt 1999). He replaced four of the original bacterial species from the Schaedler flora with microorganisms isolated from CD-1 mice (Trexler and Orcutt 1999). The new organisms represented major constituents of the autotchotonous microbiota of the mouse, as opposed to the original Schaedler facultative anaerobes which only reach significant levels in infant mice during their second week of life before the extremely oxygen-sensitive (EOS) anaerobes arrive as the mice start to ingest solid food on day 11 after birth. Three of the newly introduced bacterial species were EOS anaerobes, selected for morphological differences and ease of identification with direct phase microscopy of cecal contents or a fecal smear (Trexler and Orcutt 1999). The final addition was a spiral-shaped microorganism (i.e., ASF 457) that was originally isolated by Dr. Schaedler, using differential filtration through a 0.45 μm millipore filter. This species was later named Mucispirillum schaedleri in his honor (Robertson et al. 2005).
The new microbial community derived by Dr. Orcutt (Table 1) consisted of eight anaerobic bacteria of which only the two lactobacilli could grow aerobically, allowing for detection of contamination under aerobic growth conditions. This was not possible in the original Schaedler flora formulation due to the presence of bacterial species that would replicate aerobically. The ASF members do not have cocci or spore-forming blunt-ended rods, which are the predominant contaminants in gnotobiotic isolators (Baker 1966). The ASF establish stable GI colonization in the mouse, although ASF 360 (Lactobacillus intestinalis) may disappear depending on the mouse's diet. The impact of the diet on the microbiota was noted in the 1960s when Dubos and Schaedler observed that ASF 360 was unable to be cultured when NCS mice bearing the Schaedler flora were fed a 15% casein diet (Dubos and Schaedler 1962; Dubos et al. 1965).
This new ASF microbiota was utilized by major vendors of commercial mice and rats (e.g., Charles River Laboratories, Harlan Laboratories, Simonsen Laboratories, and Taconic Biosciences) as all had contracts with the National Cancer Institute. The new set of eight microorganisms (i.e., the ASF) eliminated the problems that were voiced by these NCI supplies that were associated with the original Schaedler “cocktails” (Trexler and Orcutt 1999). Many vendors including Taconic Biosciences and Charles River Laboratories continue to use the ASF in their isolator foundation breeding colonies to provide a baseline microbiota prior to introduction into barrier production.
The ASF community is representative of a normal murine GI microbiota that can be stably maintained for generations under gnotobiotic conditions (Alexander et al. 2006; Sarma-Rupavtarm et al. 2004; Stehr et al. 2009). The ASF longitudinally distribute through the GI tract and differentially occupy niches similar to a complex GI microbiota; and ASF-colonized mice are healthy, with a normal immune system and GI function (Orcutt et al. 1987; Sarma-Rupavtarm et al. 2004). Each microorganism can be individually evaluated, as organism-specific polymerase chain reaction (PCR) primers have now been developed and used in research studies (Alexander et al. 2006; Sarma-Rupavtarm et al. 2004). Each of the ASF can be individually cultivated in vitro, allowing the production of bacterial antigen that can be used to evaluate organism-specific immune responses, and offer the opportunity for genetic manipulation of the individual ASF species (Jergens et al. 2007). Thus, this model is relevant to investigate specific members of the bacterial community, as well as the impact of the ASF on the host immune system.
Phylogeny of the ASF
In 1999, the 16S rRNA genes of the ASF were sequenced, providing new phylogenetic information about each of the ASF members (Dewhirst et al. 1999). From this, ASF 361 was found to be identical to the type strains of both L. murinus and L. animalis, while ASF 360 was determined to be a novel Lactobacillus similar to L. acidophilus and L. lactis (Dewhirst et al. 1999). ASF 519 was most closely related to Porphyromonas and aligned with an unnamed genus, while ASF 457 was determined to be in a distinct phylum (Dewhirst et al. 1999). The remaining four ASF members comprised a group of gram-positive bacteria possessing a low G/C content (Dewhirst et al. 1999). ASF 492 was identical to Eubacterium plexicaudatum, and ASF 500 was not closely related to any other microorganism known at the time (Dewhirst et al. 1999). With the availability of additional DNA sequence-based taxonomic information, ASF 519 was recently identified as Parabacteroides goldsteinii, ASF 360 as Lactobacillus intestinalis, and ASF 500 has been more specifically identified as a species of Pseudoflavonifractor within the order Clostridiales (Duca et al. 2014; Momose et al. 2011; Park and Itoh 2005). Also, ASF 457 is now classified as a novel bacterium, Mucispirillum schaedleri, in the phylum Deferribacteres (Robertson et al. 2005), while whole-genome sequencing enabled further classification of ASF 502 as a Clostridium sp. (Wannemuehler et al. 2014).
Near complete genome sequences of each of the ASF members have recently been reported (Table 1), revealing the genetic composition of an entire gut microbiome (Wannemuehler et al. 2014). This resource will provide unprecedented opportunities to understand how gene expression and metabolic activity respond to changes to the mammalian host. For example, transcriptome analysis of the ASF bacteria can be used to interrogate how the GI community responds to changes in diet, genetic background, or pathogenic bacterial infection. In addition, ongoing work in the authors’ laboratories includes resequencing the ASF genomes in an effort to identify heritable genetic alterations within the ASF in response to changes (e.g., diet, inflammation, additional microbes) to the host's GI microenvironment.
In Vivo Evaluation of the ASF
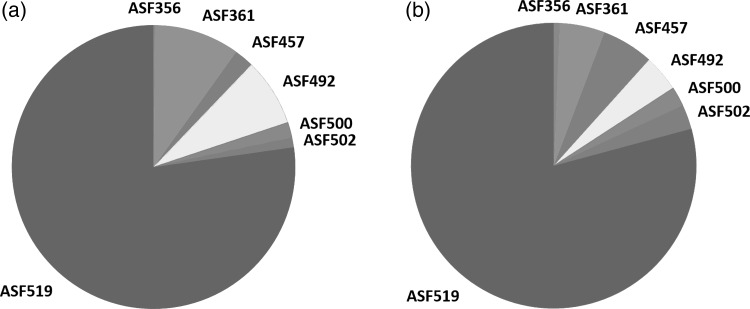
Quantitative, real-time PCR (qPCR) has been developed to monitor the absolute abundance of each ASF member (Sarma-Rupavtarm et al. 2004). For example, Sarma-Rupavtarm and colleagues evaluated the distribution of the ASF in the GI tract of 6-week-old C.B-17 SCID mice and noted that P. goldsteinii (ASF 519) was the dominant microorganism, with Clostridium sp. (ASF 502), Mucispirillum sp. (ASF 457), and Lactobacillus sp. (ASF 361) as the next most prevalent organisms (Sarma-Rupavtarm et al. 2004). An independent study also revealed ASF 519 as the dominant organism, and ASF 519 and 502 have previously been reported to be the dominant organisms from fecal samples (Geuking et al. 2011; Sarma-Rupavtarm et al. 2004). More recently, use of 16S rRNA gene sequencing on the Illumina platform confirms the taxonomic distributions identified in previous studies (Figure 2). Also, consistent with previous studies, ASF 360 is too low in abundance to routinely be detected by 16S sequencing but is detectable with organism-specific PCR from stomach contents. This high throughput approach facilitates analysis of a larger number of samples than is feasible using qPCR and, thus, will be useful to study changes in the microbiota during longitudinal studies in great detail.
Figure 2.
Stability and relative abundance of the altered Schaedler flora (ASF) in gnotobiotic C3H/HeN mice bred at Iowa State University between 2005 and 2013. (a) Pie chart representing the relative abundance of the members of the ASF in feces collected and archived in 2005. (b) Pie chart representing the relative abundance of the members of the ASF in feces collected and archived in 2013. The relative distribution of taxonomic classes were determined by 16s ribosomal gene sequence analysis (V4 variable region) on the Illumina MiSeq platform. While the relative abundance of ASF 360 was below limits of detection by species-specific 16s amplicon sequencing, this species was detectable by end-point PCR analysis of contents from the upper GI tract.
Fluorescence in situ hybridization (FISH) can be used to monitor changes in bacterial mucosal association. Using a colony of gnotobiotic ASF-colonized C3H/HeN mice continuously maintained for 14 years, the Wannemuehler laboratory at Iowa State University has utilized FISH to evaluate spatial distribution of the ASF following the colonization of E. coli (Figure 3). Mucispirillum schaedleri (ASF 457) has been demonstrated to be a mucous colonizer in the mouse GI tract and also translocates to the liver (Robertson et al. 2005); the fusiform ASF organisms are likely to be differentially associated with the mucous layer of the GI tract as previously suggested (Savage et al. 1971).
Figure 3.
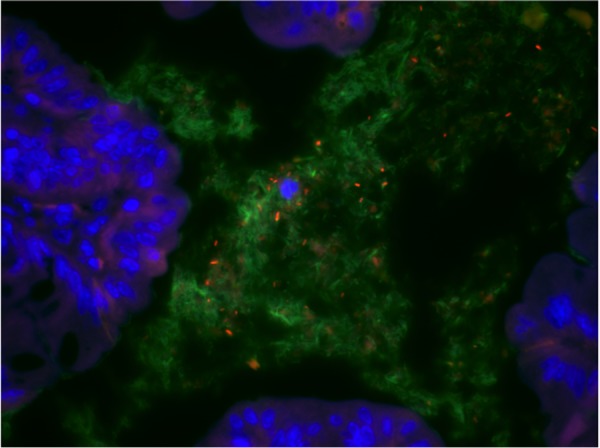
Photomicrograph depicting the use of fluorescent in situ hybridization (FISH) to evaluate the spatial distribution of the altered Schaedler flora (ASF) relative to the mucosal epithelium (DAPI stain - blue) of the proximal colon in a C3H/HeN mouse colonized with the ASF and Escherichia coli. The EUB338-FITC probe (green) was used as a nonspecific probe to detect the ASF with a species-specific Cy3-labeled probe for E. coli (orange).
Several studies have evaluated the ASF in multiple genetic backgrounds of mice, revealing that GI colonization dynamics are influenced by host genetic background, sex, and age of the mice, though the community structure largely remains stable over time (Alexander et al. 2006; Ge et al. 2006; Sarma-Rupavtarm et al. 2004; Stehr et al. 2009). Longitudinal evaluation demonstrates that oxygen availability affects colonization dynamics, with aerotolerant Lactobacillus (ASF 361) dominating in the upper GI tract, stomach, and esophagus, and obligate anaerobes more heavily colonizing the cecum and colon (Alexander et al. 2006; Sarma-Rupavtarm et al. 2004). ASF 360 (Lactobacillus) was detected in very low numbers (Alexander et al. 2006; Sarma-Rupavtarm et al. 2004) in the proximal alimentary tract (i.e., esophagus). Also, fecal samples were not necessarily equivalent to samples isolated from within different compartments of the gastrointestinal tract (Alexander et al. 2006; Sarma-Rupavtarm et al. 2004).
When gnotobiotic ASF mice are conventionalized, the ASF remain stable components of the complex microbial community (Alexander et al. 2006; Stehr et al. 2009). Conversely, colonization of gnotobiotic ASF mice with H. hepaticus has revealed quantitative changes in the community structure with the introduction of the new microorganism (Ge et al. 2006).
ASF as a Microbial Model Community
The ASF provides a well-defined, limited community of microorganisms available for manipulation, quantification, and organism-specific immune responses. This model has been used for microbiological, immunological, and metabolic studies. The ASF have mostly been used in mice, but also in rats and pigs (Laycock et al. 2012; Rath et al. 1996). However, colonization of gnotobiotic pigs was unreliable, as one might expect, because pigs have their own autochthonous microbiota (Laycock et al. 2012).
Mice colonized with the ASF can be useful to study the impact of enterobacteria colonization on a model microbial community because the ASF is devoid of proteobacteria . The ASF was used to colonize germfree mice, resulting in mice harboring a low-complexity microbiota (LCM) (i.e., raised under barrier conditions) in order to study colonization resistance of Salmonella enterica (Stecher et al. 2010). These LCM mice were easily colonized with S. enterica and developed pathological changes intermediate between conventional and germfree mice. When the authors cohoused the LCM mice with a mouse harboring a conventional microbiota, the LCM mice developed colonization resistance toward S. enterica as evidenced by lower colony-forming units (CFU) of S. enterica in fecal and cecal samples (Stecher et al. 2010). The most abundant ASF members identified in these studies were ASF 519 and ASF 500 (Stecher et al. 2010).
The authors have demonstrated the ability to stably colonized Helicobacter bilis, Campylobacter jejuni, and different E. coli strains in gnotobiotic C3H/HeN ASF-colonized mice. Microorganisms that typically do not stably colonize mice with a conventional microbiota have been shown to chronically colonize mice with an established ASF. Four strains of C. jejuni stably colonized for at least 7 weeks and LF82, an adherent invasive E. coli strain associated with Crohn's disease, have been stably maintained across six successive generations of mice (manuscripts in preparation). Additionally, a study using Helicobacter felis demonstrated stable colonization of ASF mice, whereas this microorganism was lost from conventional microbiota mice (Schmitz et al. 2011).
The ASF-colonized mice have been used to evaluate the dynamics of the mucosal IgA response to the resident microbiota (Hapfelmeier et al. 2010). Recently, ASF has also been used to determine that the colonization of the GI tract with bacteria in the postnatal period is necessary for development of the enteric nervous system (ENS). In fact, the ENS of ASF-colonized mice was similar to that found in conventionally reared, SPF animals (Collins et al. 2014). When colonizing germfree MyD88-deficient non-obese diabetic (NOD) mice with the ASF, a reduced type 1 diabetes (T1D) phenotype, similar to conventional SPF mice, was observed compared with the germfree, MyD88-deficient NOD mice (Wen et al. 2008). This demonstrates that the ASF community is sufficient to reduce the tendency to develop T1D mellitus. The ASF has also been used in the HLA-B27 transgenic rat model of colitis to colonize germfree rats, demonstrating that the microbiota, especially Bacteroides species, contributed to colitis in this model (Rath et al. 1996).
The ASF has been used as a representative subset of the GI microbiota in order to evaluate the changes in the distribution of the microbial community following GI perturbation. For example, colonization of conventional mice with H. pylori was shown to increase the numbers of several members of the ASF (356, 361, 457, and 500) detected in the stomach by PCR analysis (Whary et al. 2014). In contrast, co-infection of the mice with Heligmosomoides polygyrus prevented the H. pylori-induced detection of these same four ASF members in the stomach (Whary et al. 2014). It should be stated that the detection of three colonic members of the ASF (356, 457, and 500) in the stomach by PCR may be due to their transient presence as a result of coprophagy. Another study with H. pylori used a subset of the ASF to augment gastric cancer progression mouse model (Lertpiriyapong et al. 2014). Collectively, the use of the ASF as representatives of the lower bowel microbiota demonstrated that spatial changes in microbial colonization along the GI tract likely contributed to ongoing inflammation and development of precancerous dysplasia.
ASF Microbiological Functions In Vivo
The importance of the microbiota with respect to contributing to the nutritional needs of the host is well documented. For example, the GI microbiota produce short chain fatty acids (SCFAs) such as butyrate by digestion of dietary fiber, and butyrate has been described as a major energy source for colonic epithelial cells. In addition, Garrett and colleagues have demonstrated that SCFAs are capable of attenuating the severity of colitis (Smith et al. 2013). With respect to using the ASF to study production of SCFAs, ASF 356 and ASF 492 have been shown to produce large amounts of SCFAs in vivo and in vitro (Smith et al. 2013). Because the ASF genomes have now been sequenced, it is now possible to evaluate the metabolic pathways responsible for production of SCFAs and to predict metabolic changes (i.e., metabolites) that would be associated with dietary changes or those associated with disease. The ASF is a stable community, suggesting that each member of the ASF contributes metabolically to the homeostasis of this model microbiota and/or to that of the host (Figure 2). In this regard, ASF-colonized animals have been used to evaluate the role of intestinal bacteria in the metabolic activation of nitrotoluene into a genotoxin. It was demonstrated that ASF-colonized animals were less efficient at metabolizing the compound (Doolittle et al. 1983). Members of the ASF have been used to evaluate foraging of host metabolites by the microbiota, demonstrating that a simplified microbiota was not as efficient as a complex microbiota at utilizing these host-derived compounds (e.g., mucins) (Berry et al. 2013).
ASF and Gut Immunology Research
ASF colonization has been used in several studies to evaluate components of mucosal immunity including CD4+ T cells, IgA, and RegIII-γ. ASF-colonized mice have also been used to demonstrate that microbial antigens are necessary for homeostatic development of mucosal T cells in the colonic lamina propria; specifically, the ASF was shown to be sufficient to induce regulatory T cells (iTREG cells) and to induce both spontaneous and homeostatic T cell proliferation that was essential to establish GI immune homeostasis (Feng et al. 2010). Furthermore, germfree mice reconstituted with the ASF lacked Th17 cells and had increased proportions of FoxP3+ TREG cells in the small intestinal lamina propria (Ivanov et al. 2008). Because of the complexity of a conventional microbiota, it is difficult to evaluate the role of the GI microbiota in the development of mutualistic immune adaptations. To address these limitations, Geuking and colleagues used ASF-colonized mice to study the maintenance of mutualistic CD4+ T cells response (Geuking et al. 2011). Following treatment of these ASF-colonized mice with low-dose dextran sulfate sodium (DSS), the colonic lamina proprial T cells responses were characterized by the presence of iTREG cells and the absence of demonstrable Th1 or Th17 cells, which resulted in the maintenance of mucosal homeostasis (Geuking et al. 2011). While ASF 519 was the dominant organism in the GI microbiota of these mice, colonization with ASF 519 alone, unlike B. fragilis, was unable to induce an expansion of the iTREG cell population, demonstrating the necessity for a community of organisms to establish mucosal homeostasis (Geuking et al. 2011; Round and Mazmanian 2010). Additionally, a later study demonstrated that ASF colonization was sufficient to induce thymic stromal lymphopoietin (TSLP) expression that was critical for the prevention of the exaggerated production of IL-17A in the colonic lamina propria (Geuking et al. 2011; Mosconi et al. 2013).
The colonization of the GI tract with the ASF alone was sufficient to stimulate the development of mucosal immunity as demonstrated by Ivanov and colleagues (2008). These authors were able to show that reconstitution of germfree mice with the ASF induces the production of natural secretory IgA in the gut at levels equivalent to that detected in SPF mice; however, the number of IgA+ plasmablasts in the small intestinal lamina propria were reduced compared with SPF mice (Cong et al. 2009; Hapfelmeier et al. 2010; Ivanov et al. 2008). Another study investigated the dynamics of microbiota-specific secretory IgA and used ASF colonized mice to demonstrate that IgA specificity is determined by the immunodominant antigens of the resident microbiota. Moreover, the loss of transient microorganisms correlates with the attrition of the respective antigen-specific IgA-secreting plasma cells in the lamina propria (Hapfelmeier et al. 2010). Cong and colleagues utilized ASF mice to demonstrate that IgA-mediated immune exclusion of bacterial flagellin was antigen-specific and was not polyspecific (Cong et al. 2009). While several of the ASF express flagella, ASF-colonized mice were useful in these studies because they possess a limited bacterial antigen load due to the small size of this bacterial community and, thus, lack preexisting antibody to antigen(s) not present (i.e., anti-CBir1 flagellin antibody) (Cong et al. 2009). Because the ASF promotes the development of mucosal immunity, the limited complexity of the ASF community allows colonization with new microorganisms (e.g., Proteobacteria) unrelated to the existing community and can thus be used to evaluate antigen-specific mucosal IgA to the newly introduced organism within the context of a resident microbial community (Cong et al. 2009).
There are >170 genes now identified as susceptibility markers for inflammatory bowel diseases (IBDs), and polymorphisms or deficiencies in Nod1 and Nod2 have been directly linked to IBD susceptibility (Jostins et al. 2012). In this regard, SPF and ASF-colonized Nod1−/−; Nod2−/− mice were used to evaluate the impact of the microbial complexity on intestinal barrier function and susceptibility to DSS-induced colitis (Natividad et al. 2012). In comparison to an SPF microbiota, Natividad and colleagues demonstrated that colonization of Nod1−/−; Nod2−/− mice with the ASF attenuated the severity of DSS-induced colitis. While the ASF-colonized Nod1−/−; Nod2−/− mice presented with increased epithelial permeability, there was no evidence of spontaneous colitis. In addition, ASF colonization was sufficient to normalize RegIII-γ expression when compared with Nod1;Nod2 sufficient mice (Natividad et al. 2012). The same authors subsequently demonstrated that ASF-colonized mice expressed RegIII-γ at lower levels when compared with mice with a complex SPF microbiota (Natividad et al. 2013). Collectively, these studies indicate that ASF-colonized mice can be used to interrogate the role of the microbiota in the onset of IBD and that the ASF is a microbial community lacking “colitogenic” bacteria that initiate spontaneous IBD as seen in other genetic models of IBD. Due to the reduced microbial complexity, ASF-colonized mice can also be used to evaluate the benefits of probiotic bacterial species to attenuate disease in genetically susceptible colitic mice (Natividad et al. 2013).
ASF and Colitis Research
IL-10−/− mice maintained under traditional housing conditions develop severe enterocolitis, while SPF IL-10−/− mice (i.e., likely colonized by Helicobacter species) develop localized colitis (Kuhn et al. 1993). Because the ASF was designed to be free of any members of the Proteobacteria (e.g., Helicobacter, Campylobacter, Escherichia), ASF-colonized mice can be used to evaluate the impact of specific members of the Proteobacteria on the development of colitis in genetically susceptible mice. Unlike conventionally reared IL-10−/− mice, ASF-colonized IL-10−/− mice fail to develop colitis or exhibit very mild disease, emphasizing the impact of the microbiota in this colitis model (Stehr et al. 2009). The ASF community has proven useful as a microbial GI community to evaluate the underlying mechanisms by which introduction of microbial provocateurs directly contributes to the onset of colitis. For example, it has been demonstrated that SPF IL-10−/− mice colonized with Helicobacter trogontum developed typhlocolitis shortly after infection; in addition, the relative abundance of some of the ASF members decreased during the H. trogontum-induced inflammatory response, consistent with the observed dysbiosis in IBD patients (Whary et al. 2006). Using ASF-colonized SCID mice, Cahill and colleagues demonstrated that Helicobacter hepaticus alone was insufficient to trigger IBD-like colitis in the absence of CD45RBHigh CD4+ T cells (Cahill et al. 1997). Because of their defined nature, ASF-colonized mice have been used to demonstrate that there are decided interactions between the host immune system, inflammatory triggers provided by individual bacterial provocateurs, and onset of colitis in genetically susceptible mice.
Induction of typhlocolitis in gnotobiotic ASF mice colonized with either H. bilis or Brachyspira hyodysenteriae was accompanied by the induction of ASF-specific serum antibody. In contrast, the severity of typhlocolitis in the B. hyodysenteriae-infected ASF mice was less severe and more variable than that observed in H. bilis-colonized ASF mice (Jergens et al. 2006). As the previous studies focused on the host response to the provocateur (Cahill et al. 1997; Whary et al. 2006), more recent studies have focused on the host response to the ASF ( Jergens et al. 2007). Because each of the ASF is cultivable in complex media including brain heart infusion medium or Schaedler's medium supplemented with bovine serum, host immune responses to the “entire” microbiota can be evaluated. In another study, gnotobiotic ASF mice colonized with H. bilis demonstrated increased ASF-specific serum antibody and CD4+ T cell responses (Jergens et al. 2007). A subsequent study using a similar study design used microarray analysis to demonstrate differential mucosal gene expression associated with fatty acid metabolism and detoxification in a time course following H. bilis colonization (Liu et al. 2009). The addition of low-dose DSS treatment to induce colitis demonstrated that H. bilis increases the host's sensitivity to a mild colitic insult by initiating inflammation and disrupting mucosal homeostasis (Liu et al. 2011).
Limitations of the ASF Model
With a limited, defined microbiota, ASF mice lack the functional redundancy and metabolic capacity of a conventional microbiota (Norin and Midtvedt 2010). It could be argued that one limitation of the ASF community is that it may be considered dysbiotic (i.e., limited diversity). Regardless, it is evident that ASF-colonized mice are immunologically, reproductively, and metabolically similar to conventional mice. The limited nature of the ASF should allow investigators to evaluate the in vivo effect of other bacterial species (e.g., segmented filamentous bacteria) on the community dynamics as well as mucosal homeostasis (i.e., development of Th17 cells). In contrast, additional iterations of the ASF community to further limit the diversity and metabolic capabilities of this consortium of microorganisms would also be informative. Recently, this has been successfully addressed by colonizing mice with three ASF members, ASF356 Clostridium species, ASF361 Lactobacillus murinus, and ASF519 Parabacteroides goldsteinii, to study colonization dynamics in a mouse model of H. pylori-induced gastric cancer (Lertpiriyapong et al. 2014). While it can be informative to further reduce the GI microbiota, it would be extremely laborious to construct “altered” ASF communities that lack one or more of the ASF microorganisms to evaluate the factorial interactions of this model community. Lastly, even though all members of the ASF community can be cultivated in vitro (e.g., BHI plus serum or Schaedler's medium plus serum), there have been no published works describing the replication of the EOS members (356, 492, 500, and 502) on a defined medium. Therefore, there is a need to develop better in vitro culture techniques to fully realize the potential that this model community can provide with respect to bacterial–bacterial interactions and bacterial–host interactions.
Conclusions
The usefulness of the ASF model is multidimensional. The ASF bacteria form a stable community that is effectively transmitted from dam to each offspring over the long term (MJW, personal observation – stable colonization for 14 years at Iowa State University). In addition, as all microorganisms can be cultured in vitro, bacterial antigen can be prepared to test immune responses to individual organisms by evaluating for antigen-specific antibody or ex vivo cell stimulation (Jergens et al. 2006, 2007). With the availability of ASF genome sequences, every microorganism within the community can be detected and quantified by qPCR and interrogated by amplicon sequencing, and expression of individual genes of interest can be monitored. Because the ASF community possesses eight bacterial species, this can be viewed as both an advantage and a limitation of this model community. The ASF offers the advantage of studying a stable consortium of bacterial species that can be used to understand the “operating system” of the gut microbiome that describes and potentially predicts community dynamics following a variety of perturbations including diet; introduction of other bacteria, fungi, or viruses; and induction of inflammation, as well as the influence of the host genetic background on the gut community. The limitations of the ASF must also be realized in as much that this consortium is less complex than the microbiota of conventional animals and may be limited in its ability to fully predict the impact of a microbial community on the health of the host. Although the original Schaedler flora is celebrating its 50th anniversary, the development of new techniques and technologies along with the availability of defined microbiota rodent models offers unique opportunities to study host–microbiota interactions well beyond what may have been envisioned by Schaedler in 1965.
Acknowledgements
This work was supported by the National Institutes of Health (R01GM099537 to GJP, AEJ, and MJW; and R01OD011141, R01CA093405, P30ES002109, and P01CA028842 to JGF) and the Kenneth Rainin Foundation (to AEJ, MJW and GJP). The authors would also like to acknowledge Dr. Tim Brenza for his assistance with preparing scanning electron photomicrographs.
References
- Alexander AD, Orcutt RP, Henry JC, Baker J, Jr., Bissahoyo AC, Threadgill DW. 2006. Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm Genome 17:1093–1104. [DOI] [PubMed] [Google Scholar]
- Baker DE. 1966. The commercial production of mice with a specified flora. Natl Cancer Inst Monogr 20:161–166. [PubMed] [Google Scholar]
- Bauer H, Horowitz RE, Levenson SM, Popper H. 1963. The response of the lymphatic tissue to the microbial flora. Studies on germfree mice. Am J Pathol 42:471–483. [PMC free article] [PubMed] [Google Scholar]
- Berry D, Stecher B, Schintlmeister A, Reichert J, Brugiroux S, Wild B, Wanek W, Richter A, Rauch I, Decker T, Loy A, Wagner M. 2013. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci USA 110:4720–4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill RJ, Foltz CJ, Fox JG, Dangler CA, Powrie F, Schauer DB. 1997. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun 65:3126–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J, Borojevic R, Verdu EF, Huizinga JD, Ratcliffe EM. 2014. Intestinal microbiota influence the early postnatal development of the enteric nervous system. Neurogastroenterol Motil 26:98–107. [DOI] [PubMed] [Google Scholar]
- Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. 2009. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci USA 106:19256–19261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. 2007. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449:811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chien CC, Paster BJ, Ericson RL, Orcutt RP, Schauer DB, Fox JG. 1999. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol 65:3287–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle DJ, Sherrill JM, Butterworth BE. 1983. Influence of intestinal bacteria, sex of the animal, and position of the nitro group on the hepatic genotoxicity of nitrotoluene isomers in vivo. Cancer Res 43:2836–2842. [PubMed] [Google Scholar]
- Dubos R, Schaedler RW, Costello R, Hoet P. 1965. Indigenous, normal, and autochthonous flora of the gastrointestinal tract. J Exp Med 122:67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos RJ, Schaedler RW. 1962. The effect of diet on the fecal bacterial flora of mice and on their resistance to infection. J Exp Med 115:1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca FA, Sakar Y, Lepage P, Devime F, Langelier B, Dore J, Covasa M. 2014. Replication of obesity and associated signaling pathways through transfer of microbiota from obese-prone rats. Diabetes 63:1624–1636. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng T, Wang L, Schoeb TR, Elson CO, Cong Y. 2010. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J Exp Med 207:1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Feng Y, Taylor NS, Ohtani M, Polz MF, Schauer DB, Fox JG. 2006. Colonization dynamics of altered Schaedler flora is influenced by gender, aging, and Helicobacter hepaticus infection in the intestines of Swiss Webster mice. Appl Environ Microbiol 72:5100–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ. 2011. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34:794–806. [DOI] [PubMed] [Google Scholar]
- Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, Geuking MB, Curtiss R, 3rd, McCoy KD, Macpherson AJ. 2010. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328:1705–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz RE, Bauer H, Paronetto F, Abrams GD, Watkins KC, Popper H. 1964. The response of the lymphatic tissue to bacterial antigen. Studies in germfree mice. Am J Pathol 44:747–761. [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. 2008. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jergens AE, Dorn A, Wilson J, Dingbaum K, Henderson A, Liu Z, Hostetter J, Evans RB, Wannemuehler MJ. 2006. Induction of differential immune reactivity to members of the flora of gnotobiotic mice following colonization with Helicobacter bilis or Brachyspira hyodysenteriae. Microbes Infect 8:1602–1610. [DOI] [PubMed] [Google Scholar]
- Jergens AE, Wilson-Welder JH, Dorn A, Henderson A, Liu Z, Evans RB, Hostetter J, Wannemuehler MJ. 2007. Helicobacter bilis triggers persistent immune reactivity to antigens derived from the commensal bacteria in gnotobiotic C3H/HeN mice. Gut 56:934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D'Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. 2012. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491:119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. 1993. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75:263–274. [DOI] [PubMed] [Google Scholar]
- Laycock G, Sait L, Inman C, Lewis M, Smidt H, van Diemen P, Jorgensen F, Stevens M, Bailey M. 2012. A defined intestinal colonization microbiota for gnotobiotic pigs. Vet Immunol Immunopathol 149(3–4):216–224. [DOI] [PubMed] [Google Scholar]
- Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, Ge Z, Wang TC, Fox JG. 2014. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut 63:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesher S, Walburg HE, Jr., Sacher GA., Jr. 1964. Generation cycle in the duodenal crypt cells of germ-free and conventional mice. Nature 202:884–886. [DOI] [PubMed] [Google Scholar]
- Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848. [DOI] [PubMed] [Google Scholar]
- Liu Z, Henderson AL, Nettleton D, Wilson-Welder JH, Hostetter JM, Ramer-Tait A, Jergens AE, Wannemuehler MJ. 2009. Mucosal gene expression profiles following the colonization of immunocompetent defined-flora C3H mice with Helicobacter bilis: a prelude to typhlocolitis. Microbes Infect 11:374–383. [DOI] [PubMed] [Google Scholar]
- Liu Z, Ramer-Tait AE, Henderson AL, Demirkale CY, Nettleton D, Wang C, Hostetter JM, Jergens AE, Wannemuehler MJ. 2011. Helicobacter bilis colonization enhances susceptibility to typhlocolitis following an inflammatory trigger. Dig Dis Sci 56:2838–2848. [DOI] [PubMed] [Google Scholar]
- Luckey TD. 1972. Introduction to intestinal microecology. Am J Clin Nutr 25:1292–1294. [DOI] [PubMed] [Google Scholar]
- McCafferty J, Muhlbauer M, Gharaibeh RZ, Arthur JC, Perez-Chanona E, Sha W, Jobin C, Fodor AA. 2013. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. Isme J 7:2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose Y, Park SH, Miyamoto Y, Itoh K. 2011. Design of species-specific oligonucleotide probes for the detection of Bacteroides and Parabacteroides by fluorescence in situ hybridization and their application to the analysis of mouse caecal Bacteroides-Parabacteroides microbiota. J Appl Microbiol 111:176–184. [DOI] [PubMed] [Google Scholar]
- Mosconi I, Geuking MB, Zaiss MM, Massacand JC, Aschwanden C, Kwong Chung CK, McCoy KD, Harris NL. 2013. Intestinal bacteria induce TSLP to promote mutualistic T-cell responses. Mucosal Immunol 6:1157–1167. [DOI] [PubMed] [Google Scholar]
- Natividad JM, Hayes CL, Motta JP, Jury J, Galipeau HJ, Philip V, Garcia-Rodenas CL, Kiyama H, Bercik P, Verdu EF. 2013. Differential induction of antimicrobial REGIII by the intestinal microbiota and Bifidobacterium breve NCC2950. Appl Environ Microbiol 79:7745–7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natividad JM, Petit V, Huang X, de Palma G, Jury J, Sanz Y, Philpott D, Garcia Rodenas CL, McCoy KD, Verdu EF. 2012. Commensal and probiotic bacteria influence intestinal barrier function and susceptibility to colitis in Nod1−/−; Nod2−/− mice. Inflamm Bowel Dis 18:1434–1446. [DOI] [PubMed] [Google Scholar]
- Norin E, Midtvedt T. 2010. Intestinal microflora functions in laboratory mice claimed to harbor a “normal” intestinal microflora. Is the SPF concept running out of date? Anaerobe 16:311–313. [DOI] [PubMed] [Google Scholar]
- O'Hara AM, Shanahan F. 2006. The gut flora as a forgotten organ. EMBO Rep 7:688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orcutt RP, Gianni FJ, Judge RJ. 1987. Development of an “altered Schaedler flora” for NCI gnotobiotic rodents. Microecology and Therapy 17:59. [Google Scholar]
- Park SH, Itoh K. 2005. Species-specific oligonucleotide probes for the detection and identification of Lactobacillus isolated from mouse faeces. J Appl Microbiol 99:51–57. [DOI] [PubMed] [Google Scholar]
- Rath HC, Herfarth HH, Ikeda JS, Grenther WB, Hamm TE, Jr., Balish E, Taurog JD, Hammer RE, Wilson KH, Sartor RB. 1996. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human ß2 microglobulin transgenic rats. J Clin Invest 98:945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson BR, O'Rourke JL, Neilan BA, Vandamme P, On SL, Fox JG, Lee A. 2005. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol 55:1199–1204. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. 2010. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA 107:12204–12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma-Rupavtarm RB, Ge Z, Schauer DB, Fox JG, Polz MF. 2004. Spatial distribution and stability of the eight microbial species of the altered schaedler flora in the mouse gastrointestinal tract. Appl Environ Microbiol 70:2791–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC. 1977. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31:107–133. [DOI] [PubMed] [Google Scholar]
- Savage DC, Dubos R, Schaedler RW. 1968. The gastrointestinal epithelium and its autochthonous bacterial flora. J Exp Med 127:67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC, McAllister JS, Davis CP. 1971. Anaerobic bacteria on the mucosal epithelium of the murine large bowel. Infect Immun 1971:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedler RW, Dubs R, Costello R. 1965. Association of germfree mice with bacteria isolated from normal mice. J Exp Med 122:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz JM, Durham CG, Schoeb TR, Soltau TD, Wolf KJ, Tanner SM, McCracken VJ, Lorenz RG. 2011. Helicobacter felis-associated gastric disease in microbiota-restricted mice. J Histochem Cytochem 59:826–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeb TR, Bullard DC. 2012. Microbial and histopathologic considerations in the use of mouse models of inflammatory bowel diseases. Inflamm Bowel Dis 18:1558–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Chaffron S, Kappeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, McCoy KD, von Mering C, Macpherson AJ, Hardt WD. 2010. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog 6:e1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehr M, Greweling MC, Tischer S, Singh M, Blocker H, Monner DA, Muller W. 2009. Charles River altered Schaedler flora (CRASF) remained stable for four years in a mouse colony housed in individually ventilated cages. Lab Anim 43:362–370. [DOI] [PubMed] [Google Scholar]
- Thompson GR, Trexler PC. 1971. Gastrointestinal structure and function in germ-free or gnotobiotic animals. Gut 12:230–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trexler PC, Orcutt RP. 1999. Development of gnotobiotics and contamination control in laboratory animal science. In: Macpherson CW, Mattingly SF, eds. 50 Years of Laboratory Animal Science. Memphis: American Association for Laboratory Animal Science; p. 121–128. [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannemuehler MJ, Overstreet AM, Ward DV, Phillips GJ. 2014. Draft genome sequences of the altered schaedler flora, a defined bacterial community from gnotobiotic mice. Genome Announc 2:e00287–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. 2008. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455:1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whary MT, Danon SJ, Feng Y, Ge Z, Sundina N, Ng V, Taylor NS, Rogers AB, Fox JG. 2006. Rapid onset of ulcerative typhlocolitis in B6.129P2-IL10tm1Cgn (IL-10−/−) mice infected with Helicobacter trogontum is associated with decreased colonization by altered Schaedler's flora. Infect Immun 74:6615–6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whary MT, Muthupalani S, Ge Z, Feng Y, Lofgren J, Shi HN, Taylor NS, Correa P, Versalovic J, Wang TC, Fox JG. 2014. Helminth co-infection in Helicobacter pylori infected INS-GAS mice attenuates gastric premalignant lesions of epithelial dysplasia and glandular atrophy and preserves colonization resistance of the stomach to lower bowel microbiota. Microbes Infect 16:345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95:6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 68:3401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]