Abstract
Eukaryotic organisms are colonized by rich and dynamic communities of microbes, both internally (e.g., in the gastrointestinal and respiratory tracts) and externally (e.g., on skin and external mucosal surfaces). The vast majority of bacterial microbes reside in the lower gastrointestinal (GI) tract, and it is estimated that the gut of a healthy human is home to some 100 trillion bacteria, roughly an order of magnitude greater than the number of host somatic cells. The development of culture-independent methods to characterize the gut microbiota (GM) has spurred a renewed interest in its role in host health and disease. Indeed, associations have been identified between various changes in the composition of the GM and an extensive list of diseases, both enteric and systemic. Animal models provide a means whereby causal relationships between characteristic differences in the GM and diseases or conditions can be formally tested using genetically identical animals in highly controlled environments. Clearly, the GM and its interactions with the host and myriad environmental factors are exceedingly complex, and it is rare that a single microbial taxon associates with, much less causes, a phenotype with perfect sensitivity and specificity. Moreover, while the exact numbers are the subject of debate, it is well recognized that only a minority of gut bacteria can be successfully cultured ex vivo. Thus, to perform studies investigating causal roles of the GM in animal model phenotypes, researchers need clever techniques to experimentally manipulate the GM of animals, and several ingenious methods of doing so have been developed, each providing its own type of information and with its own set of advantages and drawbacks. The current review will focus on the various means of experimentally manipulating the GM of research animals, drawing attention to the factors that would aid a researcher in selecting an experimental approach, and with an emphasis on mice and rats, the primary model species used to evaluate the contribution of the GM to a disease phenotype.
Keywords: gut microbiota, metagenomics, microbiome, model phenotype
Introduction
Eukaryotic organisms are colonized by rich and dynamic communities of microbes, both internally (e.g., in the gastrointestinal and respiratory tracts) and externally (e.g., on skin and external mucosal surfaces). These communities, comprising not just prokaryotic cells but also eukaryotes, viruses, and archaea, are dynamic tissues with a rapid collective rate of turnover. The vast majority of bacterial microbes reside in the lower gastrointestinal (GI) tract, and it is estimated that the gut of a healthy human is home to some 100 trillion bacteria (Whitman et al. 1998), roughly an order of magnitude greater than the number of host somatic cells. Strict obligate anaerobes dominate the gut microbiota (GM) (Backhed et al. 2004; Gordon and Dubos 1970; Harris et al. 1976) and, due to their extreme oxygen sensitivity, only a small minority of gut microbes can be cultured (Bleich and Hansen 2012; Hugenholtz 2002). The development of culture-independent methods to characterize the GM has spurred a renewed interest in its role in host health and disease. Indeed, associations have been identified between various changes in the composition of the GM and an extensive list of diseases, both enteric and systemic (Clemente et al. 2012; de Vos and de Vos 2012). That being said, all but a handful of the studies demonstrating those associations provide purely correlative data. Due to the ethical issues of experimental testing on humans and the relative novelty of the molecular methods used to characterize the GM, very little causation has been shown. Additionally, data generated in humans are obfuscated by the influence of environmental factors (e.g., diet [Claesson et al. 2012; Wu et al. 2011], activity level, physical and psychological stress [Konturek et al. 2011], tobacco and alcohol consumption [Biedermann et al. 2013; Queipo-Ortuno et al. 2012] as well as several genetic loci [Benson et al. 2010; Khachatryan et al. 2008], capable of modulating the GM.)
Animal models provide a means whereby causal relationships between characteristic differences in the GM and diseases or conditions can be formally tested using genetically identical animals in highly controlled environments. A clear distinction should be made here regarding the difference between the classical “one pathogen” model of infectious disease and investigations of the resident microbiota. Whereas evidence of causality in the former is historically founded on Koch's postulates, this rubric quickly falls apart when applied to studies of the GM. The first postulate is that the causative agent is found in all affected, and no unaffected, hosts. Clearly, the GM and its interactions with the host and myriad environmental factors are exceedingly complex, and it is rare that a single microbial taxon associates with, much less causes, a phenotype with perfect sensitivity and specificity. Moreover, the second and third of Koch's postulates assume the ability to culture the microbe in question. While the exact numbers are the subject of debate, it is well recognized that only a minority of gut bacteria can be successfully cultured ex vivo. Thus, to perform studies investigating causal roles of the GM in animal model phenotypes, researchers need clever techniques to experimentally manipulate the GM of animals, and several ingenious methods of doing so have been developed, each providing its own type of information and with its own set of advantages and drawbacks. The current review will focus on the various means of experimentally manipulating the GM of research animals, with a focus on mice and rats and drawing attention to the factors that would aid a researcher in selecting an experimental approach.
Factors Affecting the Composition of the Gut Microbiota
Prior to discussion of methods of intentionally altering the GM, it is worthwhile to consider husbandry-related factors capable of effecting change in the GM of research animals. Practically speaking, such factors need to be considered as sources of inadvertent manipulation of the GM. Certain factors, such as antibiotics and rederivation, may also however provide a framework to be exploited for experimental manipulation.
Arguably, the greatest single factor in determining the composition of the GM in most instances is the birth dam, which is the source of microbes colonizing the pups beginning immediately at parturition. In a closed breeding colony, the gut microbiota remains fairly consistent over generations (Chung et al. 2012). However, in the event that new breeders, particularly dams, are introduced into a colony, one must recognize the potential for them to possess a GM that is intrinsically different from that of the existing population. While the dam is the primary nidus of microbes seeding the pups, the presence of a sire harboring a different GM may present a confounding experimental factor due to horizontal transfer (see Cohousing below).
An often-overlooked situation in which a qualitatively different GM may be introduced into a colony is rederivation, typically performed via surgical transfer of embryos into a surrogate dam selected for favorable fecundity and maternal care. Rederivation has become commonplace in three settings: the population of new research facilities, the resuscitation of cryopreserved genetic backgrounds, and selective rederivation of a strain to eliminate certain unwanted pathogens. Regardless of the setting, the GM of the surrogate dam becomes the indigenous GM of the newly rederived animals. Using an unweighted hierarchical clustering analysis of denaturing gradient gel electrophoresis profiles, Friswell and colleagues demonstrated that offspring resulting from allogeneic embryos cotransplanted into a surrogate dam showed a 93% concordance between each other and the birth dam (Friswell et al. 2010). Less intensive methods of eliminating unwanted bacteria (e.g., Helicobacter spp.), such as cross-fostering, will intuitively have similar results although the amount of time between birth and placement on a surrogate will likely affect the degree to which the endpoint GM of the pup mirrors that of the biological dam or surrogate dam.
As has been reported in humans (Biasucci et al. 2010; Dominguez-Bello et al. 2010), mode of delivery likely has an influence on the composition of the murine GM. Vaginal delivery typically seeds offspring with microbes normally found in the maternal vaginal or intestinal microbiota (e.g., Lactobacillus spp., Bifidobacterium spp.) while Cesarian delivery tends to result in greater proportions of microbes normally associated with the external body surfaces (e.g., Staphylococcus spp., Corynebacterium spp., and Propionibacterium spp.). These differences are more than superficial as exposure to exogenous lipopolysaccharides during vaginal delivery induces epithelial tolerance in the offspring, resulting in differential innate immune responses between mice delivered via the two modes (Lotz et al. 2006).
The composition of the GM can shift rapidly and globally in response to abrupt changes in the macromolecular content of diet. Due to insolubility or a lack of the necessary hydrolases, complex plant polysaccharides are often refractory to digestion in the small intestines. As such, undigested polysaccharides reach the colon and serve as a major energy source for the GM. Not surprisingly, the enzymatic capacity of any singular bacterial taxon is a major determinant of fitness within its environment (Sonnenburg et al. 2010). While this has been elegantly demonstrated on the level of individual species, diet clearly places selective pressures on the GM at higher phylogenetic levels as well. Of the greater than 50 bacterial phyla identified to date (Schloss and Handelsman 2004), the GM of mammals is dominated by two, the Firmicutes and Bacteroidetes. In general, increases in the Firmicutes:Bacteroidetes ratio are associated with obesity and increased food intake (Turnbaugh et al. 2006). While it appears that obesity per se, even in the absence of increased consumption of diet, can result in such shifts (Ley et al. 2005), there is also evidence that such shifts enhance the ability of the GM to harvest energy from ingesta (Turnbaugh et al. 2006), resulting in a positive feedback loop. Thus, any factor capable of modulating the ratio of these two phyla (e.g., unwarranted consumption of breeder chow) could ostensibly initiate a gradual shift, perhaps occurring over multiple generations, toward a phenotype of increased energy harvest and adiposity. Assuming that most researchers already control for the macromolecular content of feed in their studies by simply feeding the same diet to all subjects, one must also remember that, while the protein, fat, and fiber content of most commercially formulated rodent chows are within a specified range, the grains and protein sources used to mill the food are subject to the constant fluctuations of the commodities market. Studies performed months or years apart from each other may include data generated from animals consuming diets milled from different components. It is entirely plausible that the interactions may exist between such differences in dietary ingredients and the microbiota.
In addition to the composition of the diet, autoclaving and irradiation are also variables that influence the microbial exposure to research animals. Indeed, the label of one commercially available irradiated rodent chow lists the amount of thermophilic and mesophilic, aerobic and anaerobic, spores recovered from “control” chow, suggesting that nonirradiated feed is a likely source of multiple bacterial and fungal species. Anecdotally, segmented filamentous bacteria, unclassified bacteria in the family Clostridiaceae with a significant impact on several mouse models of disease (Denning et al. 2011; Lee et al. 2011; Wu et al. 2010), can also be acquired from food contaminated with spores.
Several groups have demonstrated substantial differences between the GM of mice purchased from different commercial sources (Denning et al. 2011; Hufeldt et al. 2010; Ivanov et al. 2008), and our own studies (Ericsson, Davis et al. 2014) have attempted to characterize the nature and extent of those differences. One of the most recognized differences is the presence or absence of segmented filamentous bacteria (SFB). While there are undoubtedly exceptions, mice from The Jackson Laboratory are largely free of SFB, while most mice from other commercial sources are endemically colonized. SFB has a well-recognized influence on the ontogeny of the mucosal immune system and has been shown to significantly affect several mouse models of both enteric and systemic disease (Denning et al. 2011; Ivanov et al. 2009; Lee et al. 2011; Wu et al. 2010). For a more detailed description of the physiological effects of SFB, readers are referred to a recent review on the topic (Ericsson, Hagan et al. 2014).
There are obviously many other factors capable or suspected of modulating the microbiota of research animals including psychological stress (Cryan and Dinan 2012; O'Mahony et al. 2009), type of caging, type of bedding, frequency of bedding change, and water source. That said, it is clearly beyond the scope of the current review to discuss each in turn, and the reader is referred to other manuscripts in the current edition for additional details.
Impact of the Gut Microbiota on Variability and Reproducibility of Disease Models
There is now little debate among physicians and biomedical researchers regarding the impact of the GM on human health. Intuitively, the GM of research animals can similarly affect the phenotype of models of human disease. While this has been recognized anecdotally for many years, only recently have reports appeared in the literature with substantive data to support this hypothesis (Denning et al. 2011; Ivanov et al. 2008; Robosky et al. 2005; Rohde et al. 2007; Yang et al. 2013). Frequently, an investigator will experience a loss or change in phenotype when an animal colony is relocated to a different institution or undergoes a change in husbandry. Scenarios such as these raise the specter of a GM-mediated effect, although the authors readily acknowledge the potential influence of myriad other factors as well. To convincingly document changes in the GM, researchers must have the foresight to have collected fecal samples prior to the change in phenotype. While few investigators routinely bank fecal samples “just in case” their model phenotype is lost, the authors would recommend doing so prior to any foreseen changes in husbandry (e.g., diet change) or relocation to a different institution. In addition to colony-wide changes in phenotype, it stands to reason that the inherent variability of the GM within a colony of mice may be reflected in the phenotypes of those models with GM-dependent mechanisms. For example, APCmin/+ mice, a model of colorectal cancer purportedly susceptible to GM-mediated effects on tumor load (Li et al. 2012; Mai et al. 2007), develop a highly divergent number of intestinal tumors. Could the difference between individual mice that eventually develop 75 or 25 intestinal tumors be related to differences in the composition of, or metabolites produced by, the GM? This is an important consideration due to the pathogenic mechanisms that may be revealed and also to the implications for animal welfare. If it is possible to reduce the variance of the GM within research animals via strict attention to and control of those variables capable of modulating the GM, it may be feasible to reduce the variance seen in the phenotype, potentially reducing the sample size needed to achieve adequate statistical power.
There is also the larger picture of biomedical research in general. The National Institutes of Health (NIH) has recently acknowledged issues of poor reproducibility in biomedical research, particularly in preclinical studies using animal models (Collins and Tabak 2014; Perrin 2014). Several editorials (Begley and Ellis 2012; Hutchinson and Kirk 2011; Prinz et al. 2011) have lamented the costs associated with the inability to reproduce preclinical studies, including high attrition rates of candidate drugs, and a wide range of contributing factors has been identified. While several have recommended inclusion of both sexes in animal studies (Perrin 2014), division of litters between treatment groups, and thorough documentation of all husbandry variables (Kilkenny et al. 2010) as means of enhancing reproducibility of biomedical research, only a handful of forward-thinking individuals in the laboratory animal community have advocated for reporting, much less standardizing, the GM of research animals (Bleich and Hansen 2012). While it may not be practical to require documentation of the GM composition for all published animal-based research, factors known or suspected to modulate the GM (e.g., animal source, sex, dietary formulation, housing density, time of sample collection) should be reported in as much detail as is practical, whenever possible. Additionally, as the associated cost and availability of next-generation sequencing decreases and increases, respectively, it may become more common to document the composition of model animals’ GM in research reports.
Methods to Manipulate the Gut Microbiota
Gnotobiotic, Defined Microbiota
Gnotobiotics or gnotobiology is the study of animals that are free of all microorganisms or colonized only by known species. The term arises from the Greek gnotos, meaning “known,” and bios, meaning “life” (Rahija 2007). Terms pertinent to any discussion of gnotobiology include:
Axenic or germfree (GF): an animal free of all microorganisms including bacteria, viruses (with the exception of endogenous retroviruses), fungi, protozoa, and other parasites.
Monoxenic or mono-associated animals: animals colonized by only one microbial species. These animals are generated by reconstituting GF mice with a single agent.
Defined microbiota (flora) animals: animals with a defined set of microorganisms. Again, these are generated by reconstituting GF mice with “cocktails” or consortia of bacteria or other agents.
GF mice may also be reconstituted with more complex microbiota that are not completely defined. Sources for reconstitution may include other mice or xenobiotic sources including humans and other animal species.
Of note, the terms GF and axenic are not synonymous with gnotobiotic, as the latter includes both GF animals and animals associated with defined microbiota.
While Reyniers had developed GF rats in 1946 (Reyniers et al. 1946), the use of gnotobiotic animals began in earnest in 1959 with the first successful rearing of GF mice (Pleasants 1959). This was followed by an explosion of technology development and optimization of procedures (reviewed in Rahija 2007) that included development of isolator and sterilization procedures for housing these mice, adaptations of hysterectomy and hysterotomy techniques to allow for transfer of pups into a GF environment, assessment of axenic animal nutritional needs, development of diets fortified with vitamins normally produced by bacteria (e.g., vitamin K) at sufficient levels to withstand autoclaving, and development of procedures to maintain isolator integrity and monitor GF status. Early research demonstrated that animals could survive in a germfree environment and that microbiota were key components of several models. Moreover, germfree mice facilitated the elimination of pathogens in commercial colonies. To this end, in the early 1960s, the LOBUND (Laboratories of Bacteriology, University of Notre Dame) Institute made GF mice and rats available to the research community and partnered with several animal vendors to develop specific pathogen-free colonies that are now commonplace in biomedical research (Rahija 2007). The field maintained a small core of scientists until recently when the explosion in the interest in microbiota resulted in a resurgence of gnotobiology. Using key words such as GF, axenic, and Schaedler, a search of the literature reveals a steady increase with a collective doubling in the number of manuscripts over the past 10 years. Many institutions across the country are now establishing gnotobiotic core facilities to address the needs of research.
An extensive discussion of gnotobiotic technology is beyond the scope of this manuscript, but the basic steps are as follows (Rahija 2007). One must first have some means to maintain mice in a sterile environment; most often this is accomplished using flexible film isolators. Surrogate GF mice can then be purchased and placed into the isolator using established procedures that maintain integrity of the isolator and GF status of the mouse. If these mice are to be used as study mice, the project may progress. If it is desired to render mice with specific mutations axenic, the purchased mice can be used as surrogate dams. Donor and surrogate mice are time-mated, with the surrogate timed to have pups 1 to 2 days before the donor. When parturition of the donor nears, pups are removed via hysterotomy or hysterectomy and sterilely transferred into the isolator and fostered onto the surrogate dam. Throughout production and maintenance, mice must be monitored to ensure that they have maintained their GF status. Likewise, the isolator must be monitored to ensure that no breaks have occurred.
Germfree Mice
In studies of microbiota, GF mice are ideal for asking whether or not microbiota, in general, plays any role in a model phenotype. If GF mice fail to develop the phenotype seen in their conventionally raised counterparts, microbiota can be implicated in the development of this phenotype. The use of these mice has been critical to our understanding of how intestinal microbes can influence health and disease, especially when coupled with mono-association, defined microbiota, or humanized microbiota strategies (Donohoe et al. 2014; Goodman et al. 2011; Nguyen et al. 2015; Ridaura et al. 2013) (see below). Data from GF mouse studies must also be interpreted in context as several normal host physiologic parameters are altered in these mice. For example, GF mice have underdeveloped immune systems (Atarashi et al. 2011; Gaboriau-Routhiau et al. 2009; Helgeland et al. 1996; Ivanov et al. 2009; Macpherson and Harris 2004; Umesaki et al. 1993), slower intestinal epithelial turnover (Savage et al. 1981), differences in epithelial gene expression (Chowdhury et al. 2007; Hooper et al. 2001), differing nutritional requirements, less body fat despite increased consumption (Backhed et al. 2004), and markedly enlarged ceca. The latter may lead to death from volvulus or may indirectly lower reproductive performance, presumably due to competition for space with the gravid uterus. GF mice are also markedly susceptible to infection by opportunistic agents, should a break in containment occur. Of note, the National Gnotobiotic Rodent Resource Center is an NIH-funded center that can aid investigators in studies using GF or gnotobiotic mice (http://www.med.unc.edu/ngrrc). Alternatively, many institutions have or are creating in-house gnotobiotic facilities. These are relatively easy to set up, but it is prudent to do a thorough cost analysis before doing so because there are added expenses of creating, maintaining, and monitoring GF mice as compared with conventional mice.
Mono-Associated Mice
GF mice are also important to microbiota research because they can be reconstituted with agents ranging from a single bacterium (mono-associated) to defined microbiota (e.g., modified Schaedler's flora) to complex microbiota to xenografted microbiota (e.g., human microbiota). To create mono-associated mice, GF mice generated and maintained as described above are inoculated with a pure bacterial culture, usually by gastric gavage. Mono-associated mice allow for the study of responses to a single agent (most commonly bacterial) or identification of bacterial species responsible for specific bacterial products (Wikoff et al. 2009). A notable example of the use of mono-associated mice in understanding host:microbe interactions comes from studies that identified SFB as a key component of the intestinal microbiota that promotes the development of Th17 cells (Gaboriau-Routhiau et al. 2009; Ivanov et al. 2009). Similarly, Bacteroides fragilis, through the production of polysaccharide A, was shown to be important in the induction of T regulatory cells (Mazmanian et al. 2005; Round et al. 2011). In another interesting model that exploits mono-association, GF mice are colonized by a triple mutant Escherichia coli that colonizes the intestinal tract for up to 48 hours but cannot divide and persist in vivo (Hapfelmeier et al. 2010). This model allows for the study of events initiated by microbes that do not require continuous presence of microbes. Recent studies by Kernbauer and colleagues have taken this strategy beyond the study of resident bacteria. To this end, these investigators showed that an enteric virus, murine norovirus, provides some beneficial functions of resident mutualistic bacteria (Kernbauer et al. 2014). While important data have been generated from studies using mono-associated mice, results should also be interpreted with caution because, like GF mice, physiologic and immunologic processes in these mice may be very different when compared with mice raised with a complex microbiota (Chung et al. 2012). Moreover, as has been shown in several models of inflammatory bowel disease (reviewed in Nell et al. 2010), the presence of agents in isolation may modulate disease phenotypes very differently than when present in the context of other microbiota. As an example, IL-10-deficient mice raised in a conventional setting develop mild large intestinal inflammation that does not develop when these mice are raised GF (Sellon et al. 1998). When mice raised in a conventional setting are colonized by Helicobacter hepaticus, the resulting inflammation is exacerbated, resulting in a model of human inflammatory bowel disease that has greatly advanced our understanding of these devastating diseases (Cahill et al. 1997; Kullberg et al. 1998). However, mono-association of GF IL-10-deficient mice with H. hepaticus results in no intestinal inflammation (Dieleman et al. 2000). This, as well as studies using defined microbiota mice, has led to the belief that some bacteria such as the rodent helicobacters serve as provocateurs for inflammation initiated against other microbial species (Fox 2007; Jergens et al. 2007; Jiang et al. 2002; Liu et al. 2011). Collectively, these findings indicate that any beneficial or detrimental process ascribed to a microbe in mono-associated mice should be interpreted only after performing similar studies where the agent in question is in the presence of complex microbiota (Fritz et al. 2013). The combination of such approaches has great potential to unravel the complex interactions of intestinal microbiota.
Defined Microbiota (flora) Mice
To circumvent some of the physiologic disadvantages of GF and mono-associated mice while still maintaining a controlled microbiota, mice reconstituted with defined microbiota were established. Schaedler and colleagues (1965a, b) initiated these studies by defining key cultivatable components of the intestinal microbiota and then experimentally inoculating GF mice with various “cocktails” of aerobes and anaerobes. The standardized cocktail that emerged was colloquially referred to as “Schaedler defined flora.” This cocktail was refined and standardized by Orcutt and colleagues (Orcutt et al. 1987), resulting in “altered Schaedler's flora” (ASF) that is now most commonly used in gnotobiotic research. The current makeup of ASF is (Dewhirst et al. 1999; Robertson et al. 2005; Sarma-Rupavtarm et al. 2004; Wannemuehler et al. 2014)
ASF356, Clostridium sp.
ASF360, Lactobacillus sp.
ASF361, Lactobacillus murinus
ASF457, Mucispirillum schaedleri
ASF492, Eubacterium plexicaudatum
ASF500, Firmicutes bacterium
ASF502, Clostridium sp.
ASF519, Parabacteroides sp.
Mice reconstituted with ASF or similar defined microbiota offer several advantages to microbiota-based research. Like GF mice, they are very well defined but, unlike GF mice, they develop a mucosal immune system, have cecal volumes approaching those of conventionally raised mice, and have normal reproductive performance. Disadvantages of using ASF-reconstituted mice center on the simplicity of this microbiota, which does not recapitulate the interactions that may occur in complex microbiota that typifies human populations. Like GF mice, these mice must also be generated in isolators and monitored routinely for the presence of appropriate microbiota and the lack of contaminants. To reduce costs, defined microbiota mice can be generated in isolators but then removed for study. This approach still requires strict biosecurity for study mice, including housing in ventilated racks and appropriated barrier husbandry practices for cage changing, sanitation, animal handling, etc. These mice also must be monitored regularly for shifts in their status. If a shift occurs, their status can be changed to restricted microbiota (flora), which may still maintain the original intent.
Humanized and Other Xenografted Microbiota
GF mice may also serve as recipients of xenografted microbiota, most commonly from human fecal samples. While mice and humans share common bacterial phyla in their intestinal microbiota, there are marked differences at the genus and species level (Kibe et al. 2005; Ley et al. 2005; Turnbaugh et al. 2009). Thus, the use of humanized microbiota may offer a more directly translatable model of human microbiota and dysbiosis. Humanizing the microbiota of mice usually involves gavage of either a fresh or frozen fecal sample so that the entire complex microbiota is transferred (Turnbaugh et al. 2009). When transferring complex microbiota, much of the bacterial diversity is reproduced, and this humanized microbiota can be stably transferred to subsequent generations. However, it should be noted that not all agents present ultimately colonize recipient mice (Kibe et al. 2005; Turnbaugh et al. 2009). This is likely due, at least in part, to loss of certain species during transit through the gastrointestinal tract and the fact that the genetics of the recipient mouse contribute to shaping their microbiota (Chung et al. 2012; Gootenberg and Turnbaugh 2011; Kibe et al. 2005). Host responses of mice to humanized microbiota should also be interpreted in context as the metabolome of these mice differs from those of conventionally raised mice (Marcobal et al. 2013). In addition, Chung and colleagues (Chung et al. 2012) showed that colonization of mice with human or rat microbiota did not restore immune cell numbers to the levels seen in the normal mouse intestines. Moreover, the predominant T helper cell in mice reconstituted with human microbiota was of the Th2 type, whereas intestines of mice colonized by autochthonous microbiota are predominated by Th17 cells. Regardless, the use of mice with humanized microbiota is expanding, a very provocative example being studies from the Gordon laboratory examining obesity (Turnbaugh et al. 2006, 2008, 2009). In these studies, transfer of microbiota from a lean twin conferred a lean phenotype in recipient mice, whereas transfer of microbiota from an obese twin resulted in obesity in the recipient mouse. These studies have been greatly expanded to look at diet–obesity interactions and are beginning to unearth putative culprits in these processes. The concept of humanizing the microbiota of mice can also be coupled with the use of defined microbiota. For example, studies of obesity have been further refined to define microbes correlating with lean phenotypes by transferring a defined consortium of bacteria (Goodman et al. 2009; McNulty et al. 2011; Ridaura et al. 2013).
Collectively, an overall advantage of using gnotobiotics to manipulate microbiomes is optimal control through use of strictly defined microbiota. In addition, the ability to inoculate GF mice with complex microbiota offers a unique tool to assess host:microbiota evolution and interactions, and may yield results more translatable to the microbiota donor species. Of note, GF mice are often reconstituted with defined or complex microbiota at weaning or adulthood and then used as study subjects. Data from these studies must be interpreted with the caveat that they do not account for early life events that may occur if the mouse is immediately exposed at birth. Therefore, consideration should be given to use of progeny if early life events are of importance. An overall disadvantage is the costs associated with producing, maintaining, and monitoring these mice. Moreover, results from gnotobiotic mice must be interpreted with caution because the lack of microbiota in GF mice or lowered richness and diversity of microbiota in defined microbiota mice do not fully recapitulate the complex interactions that occur within the complex microbiota that exists in conventionally raised mice or the humans that they are intended to model. Studies using gnotobiotic animals should perhaps best be performed in multiple microbiota statuses. For example, GF mice may implicate microbiota, mono-association studies may aid in identification of putative causative agents of disease, and studies using defined microbiota and ultimately highly complex microbiota may confirm the role of these causative agents.
Complex, Undefined Microbiota
The systems described above allow for reductionist approaches to examine the influence of individual or relatively simple consortia of microbes. Studies utilizing these approaches are highly informative when applied to specific questions regarding the interaction between a taxon of interest and the host. As mentioned above, these approaches are often most useful when considered in tandem with studies utilizing mice harboring a complex microbiota. Animals harboring a defined microbiota, such as ASF-colonized mice, are undeniably artificial, and the complex interplay that exists between the hundreds of different species present in a normal GM is absent. Metabolic cross-feeding (Pfeiffer and Bonhoeffer 2004) is an essential homeostatic mechanism in the GM of humans (Belenguer et al. 2006), and model systems that fail to account for this may produce data that are not translatable to the human condition. The two most common methods of generating mice that possess a desired complex GM are cross-fostering and rederivation.
Cross-Fostering
Cross-fostering, an affordable and simple means of generating mice or rats with a desired GM, requires foster dams harboring that GM. Following timed matings of mice possessing the desired genotype and the prospective foster dam, pups must then be transferred from the former to the latter as soon as possible after parturition. Pups then acquire the GM of the foster dam gradually and in a physiologically natural manner. As with cross-fostering to eliminate bacterial pathogens, one of the most frustrating possible outcomes is cannibalism of the pups by the foster dam. Anecdotally, this is more common with primiparous dams but can be encountered with experienced dams as well. Effective means of reducing cannibalism include minimized handling and activity in the animal room, and pharmacological intervention (Carter et al. 2002). In our experience, the use of static microisolator caging as opposed to cages on individually ventilated racks may also decrease cannibalism. Additionally, it may be beneficial to leave a few of the biological pups with the foster dam when placing foster pups in the cage. This has the added benefit of providing control animals harboring the same GM as the newly cross-fostered pups, but with a wild type (WT) or other genotype. Using cross-fostering, amid a variety of other methods to manipulate the GM, Garrett and colleagues showed that mice lacking the transcription factor T-bet develop a colitogenic GM capable of inducing disease in T-bet-sufficient hosts (Garrett et al. 2007). The colitogenic (Couturier-Maillard et al. 2013; Elinav et al. 2011) or protective (Fuhrer et al. 2010) capacity of the GM in several other models has been similarly demonstrated via cross-fostering.
Rederivation
Rederivation via surgical embryo transfer (ET) can be used to render mice axenic (via Cesarian delivery of pups and transfer to a GF foster dam) or to eliminate pathogens incapable of transmission in utero (via embryo transfer into, and natural parturition by, a surrogate dam determined to be pathogen free). Rederivation via surgical implantation of embryos or morulae possessing the desired genotype into surrogate dams harboring the desired GM produces pups similar to those generated via cross-fostering with a few differences worth noting. As mentioned above, cross-fostering requires transfer of pups to a timed-mating foster dam as soon as possible following parturition, and there is typically a brief but unavoidable period during which pups are exposed to the GM of the biological dam. The effects of this transient exposure on the ontogeny of the pups′ GM is unknown. Pups generated via ET avoid that short period and are exposed to the maternal GM beginning immediately at birth. Rederivation requires considerable expertise and specialized equipment and facilities. When performed as a means to resuscitate cryopreserved germplasm or eliminate pathogens, it is common practice to use an outbred stock (or hybrid of inbred strains) with favorable reproductive indices as surrogate dams. In the context of transferring GM, however, researchers may need to use inbred surrogate dams (harboring the GM of interest), thus making the ET procedure more technically challenging.
One important consideration in the context of either cross-fostering or rederivation is the possibility of confounding maternal effects. Phenotypic differences detected in isogenic pups born to dams harboring distinct GM may be associated with differences in the GM, or they may be due to differing levels of maternal care or epigenetic factors such as methylation status (Turecki and Meaney 2014). One solution to this potential confounder, used in a series of eloquent studies by Friswell and colleagues (Friswell et al. 2010), is the cotransfer of allogeneic embryos into a single surrogate dam.
Antibiotic Administration
The GM influences the ontogeny of the mucosal immune system (Bauer et al. 2006; Eberl and Lochner 2009) and host metabolic phenotype (Backhed 2011; Cox et al. 2014). Thus, to eliminate the effect of differences present from birth, it may be desirable to experimentally effect changes in the GM of adult animals, and there are several effective methods of doing so. Frequently, initial evidence supporting a role for the GM in a phenotype of interest is provided by comparison of antibiotic-treated and untreated animals. If treatment abrogates (or exacerbates) a model phenotype, this may support some undefined role for the GM. An underlying assumption is that the antibiotic does not possess an effect on the model phenotype, independent of the microbiota. Thus, proper control groups, and often multiple different antibiotic regimens, are necessary to demonstrate that any difference detected between groups is indeed a result of the effects of treatment on the GM. Additionally, it must be remembered that the various members of a healthy GM exist in equilibrium with each other, generating and consuming metabolic substrates at a steady-state level (Belenguer et al. 2006; Samuel and Gordon 2006). The removal of one small portion of that microbial syntrophy via antibiotics, while perhaps not directly involved in the phenotype under study, may lead to subsequent changes in the larger microbial population associated with a change in phenotype. Thus, changes in phenotype associated with antibiotic-mediated changes in the GM must be interpreted cautiously, resisting the temptation to ascribe direct causality to the alterations detected in the microbial profile. Alternatively, transcriptome-based studies highlight the possibility of making a type II error when evaluating the impact of antibiotics on the GM; even in the absence of overt changes in the composition of the GM, antibiotics and other xenobiotics may significantly alter the transcriptional activity of the GM (Maurice et al. 2013).
Long before the development of the molecular techniques used to characterize the GM, researchers were using antibiotic administration to assess the impact of the GM on host susceptibility to pathogens (Bohnhoff et al. 1954; Meynell 1955), a phenomenon referred to as “colonization resistance” (van der Waaij et al. 1971). There are several informative studies related to the extent and duration of effect of various antibiotics on the composition of the murine GM (Antonopoulos et al. 2009; Hill et al. 2010; Hoentjen et al. 2003; Robinson and Young 2010; Sekirov et al. 2008). As the goal of antibiotics in the present context is to test for an influence of the gut microbiota, researchers typically rely on broad spectrum, bactericidal antibiotics that can be administered orally. Frequently, multiple drugs with complementary spectra are administered in combination to enhance overall efficacy, differences in which will affect the remaining bacterial load in the gut, as well as the composition of the GM following discontinuation of antibiotics. Antonopoulos and colleagues showed that IL-10-deficient C57BL/6J mice fed chow containing combined amoxicillin/metronidazole/bismuth (AMB) had greatly reduced levels of the two dominant phyla, Firmicutes and Bacteroidetes, resulting in an increased relative abundance of Proteobacteria (Antonopoulos et al. 2009). Specifically, two sequences annotated to the family Enterobacteriaceae accounted for 93% of sequences recovered from antibiotic-treated mice. Of note, when mice were allowed to recover from treatment for 2 weeks, the GM returned to a conformation very similar to that present prior to treatment, and the proteobacterial sequences that dominated the GM of treated mice constituted less than 1% of the GM following the recovery period. However, treatment with the third-generation cephalosporin cefoperazone resulted in a greater than 4000-fold reduction in the overall number of 16S genes detected via real-time polymerase chain reaction (PCR), obviating 16S rRNA sequencing. In contrast to the AMB cocktail, mice allowed to recover from cefoperazone experienced an incomplete restitution of the GM, primarily due to sustained reduction in the relative abundance of bacteria in the phylum Bacteroidetes. Moreover, an addendum from the same group investigated the effects of vancomycin, an antibiotic with selective efficacy against Gram-positive bacteria. While the richness of the GM was significantly reduced during treatment with vancomycin, the overall bacterial load was unchanged, suggesting that the niche left by susceptible Gram-positive microbes was filled by some compensating, resistant taxa. Collectively, these data highlight differential and long-lasting effects of antibiotic treatment dependent on the target spectrum of the drug. The different activities of antibiotics can also be exploited to gain information regarding the influence of broad classes of bacteria. In the work leading up to the discovery of SFB as a key microbial factor capable of promoting and enhancing mucosal TH17 immune responses (Ivanov et al. 2009), Ivanov and colleagues treated mice with multiple individual antibiotics, or combinations thereof, in an effort to narrow the pool of candidate microbes responsible for the phenotype (Ivanov et al. 2008). Treatment with either vancomycin or ampicillin, but not a combination of metronidazole and neomycin, significantly decreased the abundance of IL-17-producing cells, suggesting that the microbe(s) associated with the phenotype were Gram positive. However, these data also demonstrate the limitations of this approach. Metronidazole has excellent efficacy against anaerobic microbes, and SFB are typically regarded as obligate anaerobes; thus, treatment with metronidazole and neomycin should, ostensibly, have eliminated the phenotype. These studies raise another important consideration, matching regional effects of certain antibiotics and differences in the microbiota throughout the gastrointestinal tract (GIT). Hoentjen and colleagues demonstrated in IL-10-deficient mice that, depending on the antibiotic regimen used, lesion scores were differentially reduced in the cecum or colon and levels of different taxa were affected (Hoentjen et al. 2003). While this may seem academic, it would be a critical consideration in the case of microbes such as SFB that inhabit only a very small segment of the GIT.
Taking things to an extreme, antibiotics can be administered chronically to adult rodents, resulting in a host approximating a GF status (Bongers et al. 2014; Rakoff-Nahoum et al. 2004). Historically, laboratory animal veterinarians have been reluctant to prescribe oral antibiotics to rodents for fear of inducing dysbiosis (loss of lack of microbial diversity resulting in clinical disease). While the authors readily acknowledge this very real potential sequela, the risk of inducing clostridial overgrowth and dysbiosis via antibiotics may paradoxically be increased through the use of selective antibiotics or short treatment durations. One could speculate that the broad spectrum and chronicity of use eliminate not just mutualistic resident microbiota but also opportunistic pathogens because mice treated this way can apparently survive with no untoward effects (Bongers et al. 2014).
Our own studies suggest that one must also consider differences in the richness and diversity of the GM prior to administration of antibiotics. Following 3 days of oral amoxicillin/clavulanic acid and streptomycin administration, 16S rRNA amplicons generated from fecal DNA from BALB/cJ mice annotated almost entirely to Zea luxurians and the order Streptophyta, presumably residual plant-origin DNA introduced via feed (unpublished data, Figure 1). This interpretation is corroborated by the work of Hill and colleagues (2010). Conversely, analysis of identically treated BALB/cAnNHsd, which harbored a much richer and more diverse GM prior to antibiotics, indicated a less complete eradication of microbes. Thus, the degree to which antibiotics exert their effects on the host GM cannot simply be extrapolated from the literature but must be confirmed in the relevant experimental setting and host.
Figure 1.
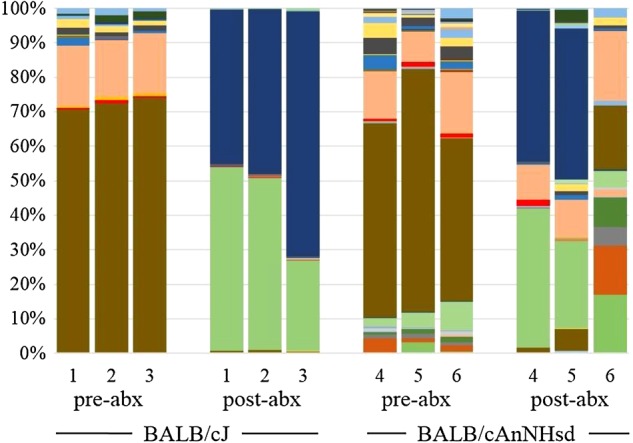
Bar charts depicting the relative abundance of operational taxonomic units detected via 16S rRNA amplicon sequencing in the feces of three BALB/cJ (1-3) and three BALB/cAnNHsd (4-6) mice before (pre-abx) and after (post-abx) 3 consecutive days of oral streptomycin and amoxicillin/clavulanic acid administration. Greater than 97% of DNA recovered from BALB/cJ mice post-abx was specific for either order Streptophyta (light green) or Zea luxurains (dark blue), presumably residual plan-origin DNA from the diet, while substantial amounts of microbial DNA were still present in the feces of identically treated BALB/cAnNHsd mice.
Fecal Microbiota Transfer
Following reduction of the autochthonous bacteria via antibiotics or beginning from an axenic state, it is possible to repopulate the GIT with a desired complex microbial population via fecal microbiota transfer (FMT). In humans, FMT (also called fecal bacteriotherapy or transfaunation depending on the context) has gained considerable attention as a simple but highly effective treatment for overgrowth of Clostridium difficile. Interestingly, this practice has existed for many centuries in human medicine (Zhang et al. 2012) and is also used in veterinary medicine as a treatment for dysbiosis, primarily in horses (Naylor and Dunkel 2009), guinea pigs, and rabbits (DeCubellis and Graham 2013). Experimentally, FMT allows for prospective evaluation of the effects of complex microbial populations on a model system. When performed using axenic mice, there is little difficulty in constitution of recipients with the microbes of interest, and such methods have been applied in seminal studies on the influence of the microbiota on the gut–brain axis (Bercik et al. 2011; Collins et al. 2013), metabolic rate and adiposity (Backhed et al. 2004; Turnbaugh et al. 2006), and determinants of host fitness (Rawls et al. 2006; Seedorf et al. 2014). The use of FMT in GF mice also allows for a period of life in which mice are lacking the microbial stimulation necessary for normal ontogeny of the immune system. When performed at different postnatal time-points, or in conjunction with cohorts colonized with the same GM from birth, one can evaluate the impact of early host:microbe interactions in the programming of various immunological and metabolic rheostats (Hansen et al. 2012; Kelly et al. 2007).
The administration of FMT to recipients already harboring a complex GM requires the use of antibiotics antecedent to FMT to create a niche for newly introduced microbes. In the absence of antibiotics, the existing complex microbiota exerts a strong colonization resistance against novel resident microbes, just as it protects the host against invading pathogens. Unpublished work from our own lab demonstrated that the GM of recipient mice, in the absence of prior antibiotic treatment, was qualitatively unchanged following FMT, even when multiple gavages were administered. As mentioned previously, one must then consider not just the antibiotic spectrum and mode of action but also the endogenous GM in recipient mice prior to antibiotic administration. There is also evidence that the differential susceptibility of various inbred mice to pathogens may be due to genetically determined differences in the GM of those strains. Work from the Finlay lab at the University of British Columbia used FMT to convincingly show that the GM of mouse strains considered resistant or susceptible to Citrobacter rodentium-mediated disease was capable of transferring those respective phenotypes in a reciprocal fashion (Willing et al. 2011). It is likely that similar differences will influence the success of FMT with resident microbe populations.
Cohousing
One of the simplest methods of assessing the influence of a complex GM on a recognized phenotype is cohousing of affected and unaffected animals already harboring complex microbial populations. Transfer of the phenotype of interest to previously unaffected animals following cohabitation with affected animals is built on the assumption that whatever component of the GM confers the phenotype on affected animals will be transferred to cagemates, presumably via stochastic exposure to GM-associated microbes in the environment or outright coprophagy. In the event that a phenotype is reliant on a single bacterial species, cohousing provides a simple means of demonstrating transmissibility via the GM (Ivanov et al. 2008). Disadvantages of cohousing as a means of evaluating the influence of the GM include its reliance on passive transfer of microbes and a relatively incomplete transfer of microbes. Definitive studies performed by Campbell and colleagues demonstrated that Bray-Curtis dissimilarity indices of most isogenic cagemates are within the range observed for mice housed in isolation (Campbell et al. 2012). Similarly, in genetically dissimilar mice cohoused in various ratios beginning post-weaning, interstrain variation outweighs caging effects (Campbell et al. 2012). That said, the above studies evaluated transfer between unmanipulated, albeit genetically distinct, mice with microbial profiles not expected to vary dramatically from the “normal” baseline microbiota found in untreated WT mice. It should be noted that, when cohousing genetically manipulated and WT mice, there is an increased likelihood of greater dissimilarity between the two microbial profiles. Intuitively, such a scenario would favor transfer of microbiota, or at least one's ability to detect transfer, and this is indeed what is seen in many instances (Henao-Mejia et al. 2012; Zenewicz et al. 2013).
Cohousing offers several logistical advantages to other methods of altering an established complex GM, including minimal cost and necessary expertise. Additionally, despite the incomplete transfer of GM, this method has been used extensively and to great effect. While a lack of phenotype transfer between cohoused mice does not necessarily obviate a contribution of the GM to the phenotype, positive transmission of the phenotype between cohoused mice provides strong support for a microbial influence (Bel et al. 2014; Ivanov et al. 2008; Vijay-Kumar et al. 2010).
Cohousing can also be performed using combinations of colonized and GF mice as a means of evaluating the “fitness” of microbial communities to colonize the GF animals (Seedorf et al. 2014), while allowing for acquisition of microbes in a somewhat natural manner, as opposed to experimental administration of microbes. In this setting, the GF mice represent an open niche for bacteria, removing any colonization resistance provided by an endogenous microbiota. As with FMT, GF mice can be cohoused with colonized mice at various ages to determine the effects of early life events in the adult host (Hansen et al. 2012).
Other Species
As in most areas of biomedical research, mice are the most commonly used laboratory animal species in studies of the GM. There are, however, instances in which other species are preferable. Moreover, corroboration of findings in multiple host species suggests a strong effect that may be translatable to humans. Rats provide a biological system similar to mice but large enough to better accommodate certain experimental techniques (e.g., colonoscopy or surgical manipulations) and possessing certain physiological parameters more closely related to those of humans. Rats can be maintained germfree (Gustafsson 1948) but are less common as a gnotobiotic species, likely due to the high costs associated with housing and husbandry. That said, however, with the recent development of nuclease-based methods of manipulating the rat genome (Peng et al. 2014), rats may become more widely used in studies of the GM.
One other species gaining favor in the GM research community is zebrafish (Danio rerio). Zebrafish are much more cost effective with regard to housing and breeding efficiency. Additionally, zebrafish can be rendered axenic for studies requiring gnotobiotic hosts (Rawls et al. 2004, 2006). Limitations of zebrafish in GM-related studies include the dissimilarity of zebrafish GM (Roeselers et al. 2011), anatomy (Wallace and Pack 2003), and physiology to that of mammalian hosts, and difficulty in collecting serial samples from the same fish over time.
Summary
In summary, with the growing interest in microbiota, several techniques to manipulate these complex populations have resurfaced (e.g., germfree mice, defined microbiota mice), been developed (e.g., humanized microbiota in germfree mice), or been refined (e.g., fecal transplants). Technology and concepts surrounding microbiota are rapidly expanding with inclusion of more complex defined “cocktails” and combining both host and microbiota factors into exciting experimental designs. Moving forward, the microbiota research community faces a number of challenges. For example, the vast majority of intestinal microbes remain uncultivable. Can novel culture methods or creative strategies to selectively eliminate targeted agents be developed? Moreover, is there need for a standardized complex microbiota and, if so, how will its composition be determined, and how would animals be maintained such that the GM remained consistent? Are microbiota of existing commercial colonies of sufficient richness and diversity to appropriately mimic the human condition? What common husbandry variables such as bedding, diet, and housing affect the microbiota, and how do we avoid institutional microbiota drift to optimize reproducibility among studies? Can microbiota be banked adequately for future studies? Are there strategies to decrease the expense of gnotobiotic housing, sequencing, transcriptomics, metabolomics, and bioinformatics, and are novel statistical methods required for these complex datasets? How do other possible intestinal inhabitants including viruses, protozoa, and fungi affect the shaping of the bacterial microbiota and influence model phenotypes? All of these questions face the biomedical research and laboratory animal community but, if the rapid expansion that has occurred in the past decade is any predictor, these challenges will ultimately be surmounted and exciting revelations about health and disease are on the horizon.
Acknowledgments
Authors (A. C. Ericsson and C. L. Franklin) and preparation of this manuscript were partially supported by NIH grants U42OD010918 and P40OD011062.
References
- Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun 77:2367–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F. 2011. Programming of host metabolism by the gut microbiota. Ann Nutr Metab 58(Suppl 2):44–52. [DOI] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101:15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer E, Williams BA, Smidt H, Verstegen MW, Mosenthin R. 2006. Influence of the gastrointestinal microbiota on development of the immune system in young animals. Curr Issues Intest Microbiol 7:35–51. [PubMed] [Google Scholar]
- Begley CG, Ellis LM. 2012. Drug development: Raise standards for preclinical cancer research. Nature 483:531–533. [DOI] [PubMed] [Google Scholar]
- Bel S, Elkis Y, Elifantz H, Koren O, Ben-Hamo R, Lerer-Goldshtein T, Rahimi R, Ben Horin S, Nyska A, Shpungin S, Nir U. 2014. Reprogrammed and transmissible intestinal microbiota confer diminished susceptibility to induced colitis in TMF-/- mice. Proc Natl Acad Sci USA 111:4964–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ. 2006. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72:3593–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA 107:18933–18938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, Verdu EF, Collins SM. 2011. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology 141:599–609, 609 e591–593. [DOI] [PubMed] [Google Scholar]
- Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. 2010. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev 86(Suppl 1):13–15. [DOI] [PubMed] [Google Scholar]
- Biedermann L, Zeitz J, Mwinyi J, Sutter-Minder E, Rehman A, Ott SJ, Steurer-Stey C, Frei A, Frei P, Scharl M, Loessner MJ, Vavricka SR, Fried M, Schreiber S, Schuppler M, Rogler G. 2013. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One 8:e59260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleich A, Hansen AK. 2012. Time to include the gut microbiota in the hygienic standardisation of laboratory rodents. Comp Immunol Microbiol Infect Dis 35:81–92. [DOI] [PubMed] [Google Scholar]
- Bohnhoff M, Drake BL, Miller CP. 1954. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med 86:132–137. [DOI] [PubMed] [Google Scholar]
- Bongers G, Pacer ME, Geraldino TH, Chen L, He Z, Hashimoto D, Furtado GC, Ochando J, Kelley KA, Clemente JC, Merad M, van Bakel H, Lira SA. 2014. Interplay of host microbiota, genetic perturbations, and inflammation promotes local development of intestinal neoplasms in mice. J Exp Med 211:457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill RJ, Foltz CJ, Fox JG, Dangler CA, Powrie F, Schauer DB. 1997. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun 65:3126–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JH, Foster CM, Vishnivetskaya T, Campbell AG, Yang ZK, Wymore A, Palumbo AV, Chesler EJ, Podar M. 2012. Host genetic and environmental effects on mouse intestinal microbiota. ISME J 6:2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter DB, Kennett MJ, Franklin CL. 2002. Use of perphenazine to control cannibalism in DBA/1 mice. Comp Med 52:452–455. [PubMed] [Google Scholar]
- Chowdhury SR, King DE, Willing BP, Band MR, Beever JE, Lane AB, Loor JJ, Marini JC, Rund LA, Schook LB, Van Kessel AG, Gaskins HR. 2007. Transcriptome profiling of the small intestinal epithelium in germfree versus conventional piglets. BMC Genomics 8:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149:1578–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O'Sullivan O, Fitzgerald GF, Deane J, O'Connor M, Harnedy N, O'Connor K, O'Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O'Toole PW. 2012. Gut microbiota composition correlates with diet and health in the elderly. Nature 488:178–184. [DOI] [PubMed] [Google Scholar]
- Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Tabak LA. 2014. Policy: NIH plans to enhance reproducibility. Nature 505:612–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SM, Kassam Z, Bercik P. 2013. The adoptive transfer of behavioral phenotype via the intestinal microbiota: experimental evidence and clinical implications. Curr Opin Microbiol 16:240–245. [DOI] [PubMed] [Google Scholar]
- Couturier-Maillard A, Secher T, Rehman A, Normand S, De Arcangelis A, Haesler R, Huot L, Grandjean T, Bressenot A, Delanoye-Crespin A, Gaillot O, Schreiber S, Lemoine Y, Ryffel B, Hot D, Nunez G, Chen G, Rosenstiel P, Chamaillard M. 2013. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest 123:700–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, Zarate Rodriguez JG, Rogers AB, Robine N, Loke P, Blaser MJ. 2014. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158:705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan JF, Dinan TG. 2012. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 13:701–712. [DOI] [PubMed] [Google Scholar]
- de Vos WM, de Vos EA. 2012. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev 70(Suppl 1):S45–S56. [DOI] [PubMed] [Google Scholar]
- DeCubellis J, Graham J. 2013. Gastrointestinal disease in guinea pigs and rabbits. Vet Clin North Am Exot Anim Pract 16:421–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning TL, Norris BA, Medina-Contreras O, Manicassamy S, Geem D, Madan R, Karp CL, Pulendran B. 2011. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J Immunol 187:733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chien CC, Paster BJ, Ericson RL, Orcutt RP, Schauer DB, Fox JG. 1999. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol 65:3287–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieleman LA, Arends A, Tonkonogy SL, Goerres MS, Craft DW, Grenther W, Sellon RK, Balish E, Sartor RB. 2000. Helicobacter hepaticus does not induce or potentiate colitis in interleukin-10-deficient mice. Infect Immun 68:5107–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 107:11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe DR, Holley D, Collins LB, Montgomery SA, Whitmore AC, Hillhouse A, Curry KP, Renner SW, Greenwalt A, Ryan EP, Godfrey V, Heise MT, Threadgill DS, Han A, Swenberg JA, Threadgill DW, Bultman SJ. 2014. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov 4:1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G, Lochner M. 2009. The development of intestinal lymphoid tissues at the interface of self and microbiota. Mucosal Immunol 2:478–485. [DOI] [PubMed] [Google Scholar]
- Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 145:745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, Davis JW, Spollen W, Bivens N, Givan S, Hagan CE, McIntosh M, Franklin CL. 2014. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One 10:e116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, Hagan CE, Davis DJ, Franklin CL. 2014. Segmented filamentous bacteria: commensal microbes with potential effects on research. Comp Med 64:90–98. [PMC free article] [PubMed] [Google Scholar]
- Fox JG. 2007. Helicobacter bilis: bacterial provocateur orchestrates host immune responses to commensal flora in a model of inflammatory bowel disease. Gut 56:898–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friswell MK, Gika H, Stratford IJ, Theodoridis G, Telfer B, Wilson ID, McBain AJ. 2010. Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS One 5:e8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JV, Desai MS, Shah P, Schneider JG, Wilmes P. 2013. From meta-omics to causality: experimental models for human microbiome research. Microbiome 1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer A, Sprenger N, Kurakevich E, Borsig L, Chassard C, Hennet T. 2010. Milk sialyllactose influences colitis in mice through selective intestinal bacterial colonization. J Exp Med 207:2843–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31:677–689. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. 2007. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131:33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. 2011. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci USA 108:6252–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. 2009. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6:279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg DB, Turnbaugh PJ. 2011. Companion animals symposium: humanized animal models of the microbiome. J Anim Sci 89:1531–1537. [DOI] [PubMed] [Google Scholar]
- Gordon JH, Dubos R. 1970. The anaerobic bacterial flora of the mouse cecum. J Exp Med 132:251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson BE. 1948. Germfree rearing of rats. Acta Path et Microbiol Scand suppl. 73.
- Hansen CH, Nielsen DS, Kverka M, Zakostelska Z, Klimesova K, Hudcovic T, Tlaskalova-Hogenova H, Hansen AK. 2012. Patterns of early gut colonization shape future immune responses of the host. PLoS One 7:e34043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, Geuking MB, Curtiss R 3rd, McCoy KD, Macpherson AJ. 2010. Reversible microbial colonization of germfree mice reveals the dynamics of IgA immune responses. Science 328:1705–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Reddy CA, Carter GR. 1976. Anaerobic bacteria from the large intestine of mice. Appl Environ Microbiol 31:907–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgeland L, Vaage JT, Rolstad B, Midtvedt T, Brandtzaeg P. 1996. Microbial colonization influences composition and T-cell receptor V beta repertoire of intraepithelial lymphocytes in rat intestine. Immunology 89:494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, Camporez JP, Shulman GI, Gordon JI, Hoffman HM, Flavell RA. 2012. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482:179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. 2010. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol 3:148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoentjen F, Harmsen HJ, Braat H, Torrice CD, Mann BA, Sartor RB, Dieleman LA. 2003. Antibiotics with a selective aerobic or anaerobic spectrum have different therapeutic activities in various regions of the colon in interleukin 10 gene deficient mice. Gut 52:1721–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881–884. [DOI] [PubMed] [Google Scholar]
- Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, Hansen AK. 2010. Variation in the gut microbiota of laboratory mice is related to both genetic and environmental factors. Comp Med 60:336–347. [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P. 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol 3:REVIEWS0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L, Kirk R. 2011. High drug attrition rates--where are we going wrong? Nat Rev Clin Oncol 8:189–190. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. 2008. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 4:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jergens AE, Wilson-Welder JH, Dorn A, Henderson A, Liu Z, Evans RB, Hostetter J, Wannemuehler MJ. 2007. Helicobacter bilis triggers persistent immune reactivity to antigens derived from the commensal bacteria in gnotobiotic C3H/HeN mice. Gut 56:934–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HQ, Kushnir N, Thurnheer MC, Bos NA, Cebra JJ. 2002. Monoassociation of SCID mice with Helicobacter muridarum, but not four other enterics, provokes IBD upon receipt of T cells. Gastroenterology 122:1346–1354. [DOI] [PubMed] [Google Scholar]
- Kelly D, King T, Aminov R. 2007. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat Res 622:58–69. [DOI] [PubMed] [Google Scholar]
- Kernbauer E, Ding Y, Cadwell K. 2014. An enteric virus can replace the beneficial function of commensal bacteria. Nature 516:94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khachatryan ZA, Ktsoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, Aminov RI. 2008. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS One 3:e3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibe R, Sakamoto M, Yokota H, Ishikawa H, Aiba Y, Koga Y, Benno Y. 2005. Movement and fixation of intestinal microbiota after administration of human feces to germfree mice. Appl Environ Microbiol 71:3171–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. 2010. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol 8:e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konturek PC, Brzozowski T, Konturek SJ. 2011. Stress and the gut: pathophysiology, clinical consequences, diagnostic approach and treatment options. J Physiol Pharmacol 62:591–599.22314561 [Google Scholar]
- Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, Cheever A, Jankovic D, Sher A. 1998. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun 66:5157–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. 2011. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA 108(Suppl 1):4615–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102:11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kundu P, Seow SW, de Matos CT, Aronsson L, Chin KC, Karre K, Pettersson S, Greicius G. 2012. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APCMin/+ mice. Carcinogenesis 33:1231–1238. [DOI] [PubMed] [Google Scholar]
- Liu Z, Ramer-Tait AE, Henderson AL, Demirkale CY, Nettleton D, Wang C, Hostetter JM, Jergens AE, Wannemuehler MJ. 2011. Helicobacter bilis colonization enhances susceptibility to Typhlocolitis following an inflammatory trigger. Dig Dis Sci 56:2838–2848. [DOI] [PubMed] [Google Scholar]
- Lotz M, Gutle D, Walther S, Menard S, Bogdan C, Hornef MW. 2006. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J Exp Med 203:973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Harris NL. 2004. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 4:478–485. [DOI] [PubMed] [Google Scholar]
- Mai V, Colbert LH, Perkins SN, Schatzkin A, Hursting SD. 2007. Intestinal microbiota: a potential diet-responsive prevention target in ApcMin mice. Mol Carcinog 46:42–48. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Kashyap PC, Nelson TA, Aronov PA, Donia MS, Spormann A, Fischbach MA, Sonnenburg JL. 2013. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J 7:1933–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice CF, Haiser HJ, Turnbaugh PJ. 2013. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107–118. [DOI] [PubMed] [Google Scholar]
- McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, Chervaux C, Knights D, Lozupone CA, Knight R, Duncan AE, Bain JR, Muehlbauer MJ, Newgard CB, Heath AC, Gordon JI. 2011. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med 3:106ra106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meynell GG. 1955. Some factors affecting the resistance of mice to oral infection by Salmonella typhimurium. Proc R Soc Med 48:916–918. [Google Scholar]
- Naylor RJ, Dunkel B. 2009. The treatment of diarrhoea in the adult horse. Equine Vet Educ 21:494–504. [Google Scholar]
- Nell S, Suerbaum S, Josenhans C. 2010. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat Rev Microbiol 8:564–577. [DOI] [PubMed] [Google Scholar]
- Nguyen TLA, Vieira-Silva S, Liston A, Raes J. 2015. How informative is the mouse for human gut microbiota research? Disease Models and Mechanisms 8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony SM, Marchesi JR, Scully P, Codling C, Ceolho AM, Quigley EM, Cryan JF, Dinan TG. 2009. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry 65:263–267. [DOI] [PubMed] [Google Scholar]
- Orcutt RP, Gianni FJ, Judge RJ. 1987. Development of an “altered” Schaedler flora for NCI gnotobiotic rodents. Microecol Ther 17:59. [Google Scholar]
- Peng Y, Clark KJ, Campbell JM, Panetta MR, Guo Y, Ekker SC. 2014. Making designer mutants in model organisms. Development 141:4042–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin S. 2014. Preclinical research: Make mouse studies work. Nature 507:423–425. [DOI] [PubMed] [Google Scholar]
- Pfeiffer T, Bonhoeffer S. 2004. Evolution of cross-feeding in microbial populations. Am Nat 163:E126–E135. [DOI] [PubMed] [Google Scholar]
- Pleasants JR. 1959. Rearing germfree cesarean-born rats, mice, and rabbits through weaning. Ann N Y Acad Sci 78:116–126. [DOI] [PubMed] [Google Scholar]
- Prinz F, Schlange T, Asadullah K. 2011. Believe it or not: how much can we rely on published data on potential drug targets? Nat Rev Drug Discov 10:712. [DOI] [PubMed] [Google Scholar]
- Queipo-Ortuno MI, Boto-Ordonez M, Murri M, Gomez-Zumaquero JM, Clemente-Postigo M, Estruch R, Cardona Diaz F, Andres-Lacueva C, Tinahones FJ. 2012. Influence of red wine polyphenols and ethanol on the gut microbiota ecology and biochemical biomarkers. Am J Clin Nutr 95:1323–1334. [DOI] [PubMed] [Google Scholar]
- Rahija RJ. 2007. Gnotobiotics. In: Fox JG, Barthold SW, Davisson MT, Newcomer CE, Quimby FW, Smith AL, eds. The Mouse in Biomedical Research. 2nd ed. Boston: Academic Press; p. 217–233. [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118:229–241. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germfree recipients reveal host habitat selection. Cell 127:423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Samuel BS, Gordon JI. 2004. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA 101:4596–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyniers JA, Trexler PC, Ervin RF. 1946. Rearing germfree albino rats. Lobund Reports 1–84. [PubMed]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Iikayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson BR, O'Rourke JL, Neilan BA, Vandamme P, On SL, Fox JG, Lee A. 2005. Mucispirillum schaedleri gen. nov., sp. nov., a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol 55(Pt 3):1199–1204. [DOI] [PubMed] [Google Scholar]
- Robinson CJ, Young VB. 2010. Antibiotic administration alters the community structure of the gastrointestinal micobiota. Gut Microbes 1:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robosky LC, Wells DF, Egnash LA, Manning ML, Reily MD, Robertson DG. 2005. Metabonomic identification of two distinct phenotypes in Sprague-Dawley (Crl:CD(SD)) rats. Toxicol Sci 87:277–284. [DOI] [PubMed] [Google Scholar]
- Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. 2011. Evidence for a core gut microbiota in the zebrafish. ISME J 5:1595–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde CM, Wells DF, Robosky LC, Manning ML, Clifford CB, Reily MD, Robertson DG. 2007. Metabonomic evaluation of Schaedler altered microflora rats. Chem Res Toxicol 20:1388–1392. [DOI] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332:974–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel BS, Gordon JI. 2006. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci USA 103:10011–10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma-Rupavtarm RB, Ge Z, Schauer DB, Fox JG, Polz MF. 2004. Spatial distribution and stability of the eight microbial species of the altered schaedler flora in the mouse gastrointestinal tract. Appl Environ Microbiol 70:2791–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC, Siegel JE, Snellen JE, Whitt DD. 1981. Transit time of epithelial cells in the small intestines of germfree mice and ex-germfree mice associated with indigenous microorganisms. Appl Environ Microbiol 42:996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedler RW, Dubos R, Costello R. 1965a. The development of the bacterial flora in the gastrointestinal tract of mice. J Exp Med 122:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedler RW, Dubos R, Costello R. 1965b. Association of germfree mice with bacteria isolated from normal mice. J Exp Med 122:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. 2004. Status of the microbial census. Microbiol Mol Biol Rev 68:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf H, Griffin NW, Ridaura VK, Reyes A, Cheng J, Rey FE, Smith MI, Simon GM, Scheffrahn RH, Woebken D, Spormann AM, Van Treuren W, Ursell LK, Pirrung M, Robbins-Pianka A, Cantarel BL, Lombard V, Henrissat B, Knight R, Gordon JI. 2014. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell 159:253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Tam NM, Jogova M, Robertson ML, Li Y, Lupp C, Finlay BB. 2008. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun 76:4726–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. 1998. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun 66:5224–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecki G, Meaney MJ. 2014. Effects of the Social Environment and Stress on Glucocorticoid Receptor Gene Methylation: A Systematic Review. Biol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3:213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. 2009. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 1:6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesaki Y, Setoyama H, Matsumoto S, Okada Y. 1993. Expansion of alpha beta T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germfree mice and its independence from thymus. Immunology 79:32–37. [PMC free article] [PubMed] [Google Scholar]
- van der Waaij D, Berghuis-de Vries JM, Lekkerkerk L-v. 1971. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg (Lond) 69:405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. 2010. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328:228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace KN, Pack M. 2003. Unique and conserved aspects of gut development in zebrafish. Dev Biol 255:12–29. [DOI] [PubMed] [Google Scholar]
- Wannemuehler MJ, Overstreet AM, Ward DV, Phillips GJ. 2014. Draft genome sequences of the altered schaedler flora, a defined bacterial community from gnotobiotic mice. Genome Announc 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95:6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA 106:3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing BP, Vacharaksa A, Croxen M, Thanachayanont T, Finlay BB. 2011. Altering host resistance to infections through microbial transplantation. PLoS One 6:e26988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. 2010. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32:815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I, Eibach D, Kops F, Brenneke B, Woltemate S, Schulze J, Bleich A, Gruber AD, Muthupalani S, Fox JG, Josenhans C, Suerbaum S. 2013. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS One 8:e70783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenewicz LA, Yin X, Wang G, Elinav E, Hao L, Zhao L, Flavell RA. 2013. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol 190:5306–5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Luo W, Shi Y, Fan Z, Ji G. 2012. Should we standardize the 1,700-year-old fecal microbiota transplantation? Am J Gastroenterol 107:1755 author reply p 1755–1756. [DOI] [PubMed] [Google Scholar]


