Abstract
Earlier studies have established that infection with HIV-1 subtypes (clades) might differentially influence the neuropathogenesis of HIV-1-associated neurocognitive dysfunction (HAND). HIV-1 Trans activator of transcription protein (Tat) is of considerable significance and plays a major role in the central nervous system (CNS) dysfunction. However, these HIV-1 clades exert diverse cellular effects that leads to neuropathogenic dysfunction has not been well established. We hypothesized that the HIV-1 clade B and clade C Tat proteins effect synaptic plasticity expression in neuroblastoma cells (SK-N-MC) by diverse methods, and accordingly modulates the development of HAND. In the present study, we have analyzed important and highly expressed 84 key human synaptic plasticity genes expression which differentially impact in clade B and clade C Tat treated SK-N-MC cells using RT2 Profile PCR Array human Synaptic Plasticity kit. Observed results demonstrate that out of 84 key synaptic plasticity genes, 36 and 25 synaptic genes were substantially (≥3 fold) up-regulated and 5 and 5 genes considerably (≥3 fold) down-regulated in clade B and clade C Tat treated cells, respectively, compared to the control SK-N-MC. We have also estimated the levels of glutamine and glutamate in HIV-1 clade B and C Tat exposed SK-N-MC cells compared to untreated cells. Our results indicate that levels of glutamate, glutamine and expression of synaptic plasticity genes were highly dysregulated by HIV-1 clade B Tat compared to clade C Tat in SK-N-MC cells. In summary, this study suggests that clade B Tat substantially potentiates neuronal toxicity and further dysregulated synaptic plasticity genes in SK-N-MC may contribute to the severe neuropathogenesis linked with HAND.
Keywords: HIV-subtypes, SK-N-MC, synaptic plasticity, Tat
INTRODUCTION
HIV-1 infection affects the central nervous system (CNS) both directly and indirectly causing neurological impairments, like AIDS dementia complex (ADC) and HIV-associated neurocognitive disorders (HAND), which are characterized by a substantial death of neurons in all areas of the brain [1]. More than 50% HIV positive patients are affected by HAND and it is characterized by progression of cognitive, behavioral and motor abnormalities [2–4]. Earlier studies have established that HIV effectively infects and replicates in the macrophages, microglia, and monocytes in the brain [3, 5]. The HIV infected cells and the activated immune cells in the brain secrete inflammatory cytokines, chemokines and neurotoxic factors that affects neuronal function and ultimately leads to death. Further, HIV-1 Tat (14–16-kDa) protein is known to be released in HIV infection and modulate viral replication and various host cellular and immune dysfunctions [6]. HIV-Tat is well-known to cause oxidative stress and is linked to disruption of the blood- brain barrier [7–9] and induces brain dementia mediated by altered synaptic plasticity. Studies have revealed that HAND associates with decreased dendrite formation and spine density [3] as well as causing the loss of excitatory synapses resulting in learning and memory dysfunction in Tat transgenic rodent models [10, 11]. HIV-1 Tat-induced in vitro neuronal dysfunction result in synaptic loss [12] and neuronal death [3] which may be induced by metabotropic glutamate receptors (mGluRs) leading to increased levels of glutamate release. Studies have also revealed that HIV-1 Tat potentiates and decreases the level of glutamate facilitated by NMDA-evoked increase in intracellular Ca2+ concentration ([Ca2+]i) involved in neuronal toxicity [13, 14].
Globally there are several groups, subgroups and genetic variation in HIV-1 distribution and it predominates in subtypes B found in United States and western countries and subtype C in Asia and South Africa, respectively [15–19]. The major subtypes of HIV-1 clade B and C infections differentially impact the immune and neuronal dysfunctions [20, 21]. We have reported that HIV subtypes or clades differentially influence immune and neuronal dysfunction in HIV viral infection as well as HIV derived gene products like Tat and gp120 proteins [22, 23]. Furthermore, earlier studies have revealed that HIV clade B and C viral infections differentially influence on neuronal plasticity genes [24], however, the exact mechanism of clade-specific HIV-1 Tat protein influence on neuronal plasticity dysfunction is not elucidated yet. In the current study, we hypothesize that the HIV-1 clade B and C Tat differentially affect the neuronal plasticity gene expression in SK-N-MC neuroblastoma cells, which will provide new insights to understand their role in HAND.
MATERIALS AND METHODS
Cell Culture and Reagents
Human neuroblastoma SK-N-MC cells were obtained from ATCC (ATCC Cat # HTB-10). The HIV-1 clade B and C Tat protein were obtained from the National Institutes of Health AIDS Reference Reagent Program (catalog no. 2222) and Diatheva, Fano, Italy, respectively. For the purified recombinant Tat proteins, the purities of clade B and clade C were 95% and 90%, respectively. The functional properties of clade B and C Tat proteins were confirmed by transactivation assay.
SK-N-MC Human Neuroblastoma Cells
Human SK-N-MC human neuroblastoma cells were maintained as described earlier [25] in Minimum Essential Medium (MEM) containing 10% fetal bovine serum, 50 units/ ml penicillin, and 100 µg/ml streptomycin. Medium was obtained from Life Technology (Grand Island, NY), and the cells were grown to 80–90% confluence.
mRNA Isolation and First Strand cDNA Synthesis
After 48 hour HIV-1 clade B and C Tat protein treatment, SK-N-MC human neuroblastoma cells were collected and the pellets used for the mRNA isolation utilizing illustra triplePrep Kit (GE Healthcare Life Sciences, UK; Cat # 28-9425-44) and the on-column DNase treatment step were also included in the procedure. Purity of the RNA was assessed by microspot RNA reader (Synergy HT Multi-Mode Microplate Reader from BioTek, US) and RNAs with an OD260 nm/OD280 nm absorbance ratio of at least 2.0 were utilized for PCR array. One microgram of RNA (Control, clade B and clade C treated) was used for the first strand cDNA synthesis using SABiosciences's RT2 First Strand Kit (Cat # 330401) as per the manufacture’s protocol. Genomic DNA elimination step was conducted before acquiring for reverse transcription.
Human Synaptic Plasticity RT2 Profile PCR Array
Synaptic plasticity gene profiling was done using 96 well format RT2 Profile PCR Array human Synaptic Plasticity kit (SABiosciences, Cat # PAHS-126A-2) utilizing Stratagene Mx3000p qRT-PCR instrument. The human Synaptic Plasticity RT2 Profiler PCR Array interrogates 84 genes related to the human synaptic plasticity. This kit was chosen because it includes diverse genes important in the human synaptic plasticity, including Immediate-Early Response (n = 30), Late Response (n = 2), Long Term Potentiation (n = 28), Long Term Depression (n = 21), Cell Adhesion (n = 9), Extracellular Matrix & Proteolytic Processing (n = 5), CREB Cofactors (n = 10), Neuronal Receptors (n = 19), Postsynaptic Density (n = 15), as well as other genes involved in the synaptic plasticity (n = 2). Few genes have role in several cellular functions listed above. The experiment was repeated three times. Relative abundance of each mRNA species was measured using RT2 SYBR Green/ROX PCR Master mix (SABiosciences, Cat # 330520) and distributed in equal volumes (25 µl) to each well of the real-time PCR arrays. The real-time PCR cycling program (as indicated by the manufacturer) was run on a Stratagene Mx3000p qRT-PCR thermal cycler. The threshold cycle (Ct) of each gene was determined by using the Stratagene MaxPro software. The threshold and baseline were set manually according to the manufacturer's instructions. Ct data were uploaded into the data analysis template on the manufacturer's website (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). The relative expression of each gene in HIV-1 Tat treated SK-N-MC cells was calculated using ΔΔCT method with five housekeeping genes and compared with the expression in control cells. Controls were also included on each array for genomic DNA contamination, RNA quality, and general PCR performance.
Glutamine and Glutamate Assay
The levels of intracellular glutamine were analyzed using a glutamine assay kit (Sigma, MO). Briefly, HIV-1 clade B and C Tat protein were added at 100ng to 2× 106 cells and after the end of incubation period, cells and medium were collected, and an equal concentration of glutamine standards, were incubated with the reaction buffer, the diluent buffer, and the specific enzyme for 1 hour at 37°C. After adding the color reagent, the samples were allowed to develop color for 5 min at room temperature, and then measured O.D at 550 nm using a spectrophotometer according the manufacture protocol. To determine the quantity of glutamine and glutamate, a linear regression analysis of the standard curve was performed.
Measurement of Spine Density and Dendrite Morphology
1,-Dioctadecyl-3,3,3′,3′-Tetramethylindocarbocyanine Perchlorate (DIL) Staining
Established protocols to stain the neuronal cells and measurement of the spine density were utilized with few modifications [24]. In brief, SK-N-MC neuroblastoma cells were grown in Eagle's minimal essential medium containing 10% fetal bovine serum, 5 mM sodium pyruvate, 100 units/ml penicillin, 100 mg/ml streptomycin and retinoic acid at 37°C with 5% CO2. SK-N-MC cells were grown onto 22 mm×50 mm glass coverslips placed in a petri-dish. Cells were treated with HIV-1 clade B and C Tat. After 48 hours, cells were fixed with 4% Formaldehyde in PBS for 30 min at room temperature. The fluorescent membrane tracer 1, 1′-Dioctadecyl-3, 3,3′,3′-tetramethylindocarbocyanine perchlorate (DIL) at 7.5 µg/ml (in PBS) concentration was directly added onto the fixed cultures and allowed to incubate for 90 min at RT. These stained coverslips were placed overnight at 4°C in petri dishes containing PBS before proceeding for confocal microscopy.
Confocal Microscopy
Confocal images were obtained using TCS SP2 Confocal Laser Scanning Microscope (Leica Microsystems, Germany) at 488 nm (100%) illusion of an argon-ion laser using 60× oil immersion objectives with high numeric aperture and 2.5× confocal electronic zoom settings to visualize individual cells and dendrites. Twenty Optical serial sections of 0.14 µm / section (~2.8 µm total) through the cells were captured and reconstructed to yield complete “two dimensional” images of individual cells in focus.
Data Analysis
In the expression studies, a gene was considered differentially regulated if the difference was ≥2–3 fold in comparison with the control [26]. Experiments were conducted at least three times and the values obtained were averaged. All the results were expressed as mean ± s.e.m. Statistical analysis of two groups was calculated by Student's t- test. In case more than two groups were analyzed, one way ANOVA was used followed by Bonferroni's multiple comparison test. Differences were considered significant at p≤0.05. Data analysis was performed with the Statistical Program, GraphPad Prism software (La Jolla, CA).
RESULTS
Human Synaptic Plasticity Genes Expression in HIV-1 Clade B and C Tat Treated SK-N-MC Neuroblastoma Cells
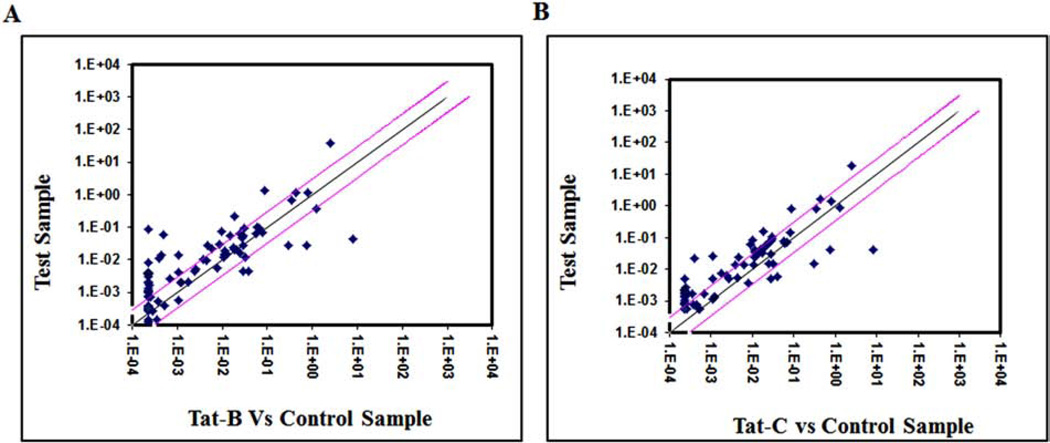
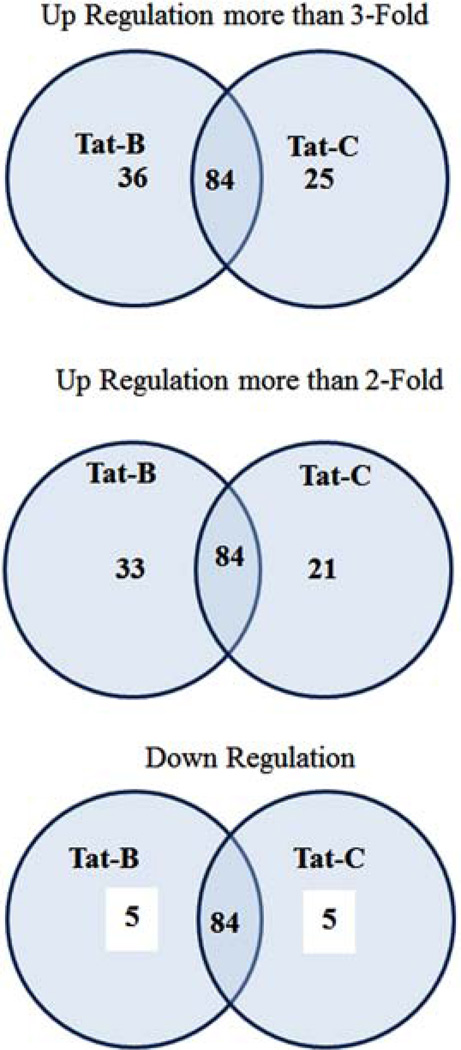
Using RT2 Profile human Synaptic Plasticity PCR Array, we have analyzed the fold change of 84 human synaptic plasticity genes expression (Table 1), in clade B and clade C Tat treated SK-N-MC neuronal cells. The scatterplot graph was plotted to analyze the gene expression results (Fig. 1A, B). Out of these 84 synaptic plasticity genes investigated, remarkably, 69 genes were up regulated and 36 genes were more than ≥3 fold up-regulated in B Tat treated, whereas, in clade C Tat, out of 46 up-regulated genes, 25 were more than ≥3 fold up regulated compared to control cells (Fig. 2). Further, clade B and C Tat treated cells significantly downregulated 5 and 5 genes, respectively (Table 3). These synaptic plasticity genes were divided into 10 groups based on their associated functions representing in immediate-early (IE) response genes, late response (LR) genes, long term depression (LTD), long term potentiation (LTP), CREB cofators, cell adhesion, neuronal receptors, postsynaptic density, extracellular matrix and protelytic processing, and other. Table 3 shows that CREM, NFkB1, RHEB, NCAM and TIMP1 genes were significantly down-regulated than control SK-N-MC. With respect to the control cells, the fold up-regulations of synaptic plasticity genes in clade B treated cells (range 3–54 fold) were significantly higher than clade C treated cells (range 3–28 fold) (Table 2).
Table 1.
Functional grouping of human synaptic plasticity genes.
| 1. lmmediate-Early Response Genes (lEGs) | ARC, BDNF, CEBPB, CEBPD, CREEl, CREM, EGRl, EGR2, EGR3, EGR4, FOS, HOMERl, JUN, JUNE, KLFlO, MMP9 (Gelatinase B), NFKBl, NFKBffi (TR1P9), NGF, NPTX2, NR4Al, NTF3, PCDH8, P!Ml, PLAT (tPA). RELA, RGS2, RHEB, SRF, TNF |
| 2. Late Response Genes | INHBA, SYNPO |
| 3. Long Tenn Potentiation | ADCYl, ADCY8, BDNF, CAMK2A, CAMK2G, CDH2 (N-cadherin), CNRl, GABRAS, GNA11, GRlAl, GR1A2, NMDARl (GRI!Nl/NRl), GRIN2A, GRIN2B, GRIN2C, GRIN2D, MAPKI, MMP9 (Gelatinase B), NTF4, NTRK2, PLCGl, PPPlCA, PPP lCC, PPP3CA, PRKCA, PRKCG, RAB3A, YWHAQ (14-3-3) |
| 4. Long Tenn Depression | GNAIl, GRlAl, GR1A2, GRIA3, GR1A4, GRlPl, GRM1, GRM2, IGFl, MAPKl, NOSl, NGFR. PICKl, PLAT (tPA), PPPlCA, PPPlCC, PPP1R14A (CPI-17), PPP2CA, PPP3CA, PRKCA, PRKGl |
| 5. Cell Adhesion | ADAMlO, CDH2 (N-cadherin), GRIN2A, GRIN2B, NCAMl, PCDH8, PPP2CA, RELN, TNF. |
| 6. Extracellular Matrix & Proteolytic Processing | ADAMlO, MMP9 (Gelatinase B), PLAT (tPA), RELN, TIMPl |
| 7. CREE Cofactors | AKTl, CAMK2G, NMDAR1, GRIN2A, GRIN2B, GRIN2C, GRIN2D, MAPKl (ERK2), PPPlCA, PPPlCC. |
| 8. Neuronal Receptors | EPHB2, GABRAS, GRlAl, GR1A2, GR1A3, GR1A4, NMDARl, GRIN2A, GRIN2B, GRIN2C, GRIN2D, GRMl, GRM2, GRM3, GRM4, GRM5, GRM7, GRM8, NTRK2. |
| 9. Postsynaptic Density | ADAM10, ARC, DLG4 (PSD95), GRlAl, GR1A3, GR1A4, NMDARl, GRIN2A, GRIN2B, GRIN2C, GRM1, GRM3, HOMER, PICK, SYNPO. |
| 10. Others | KIF17, SIRTL |
Fig. (1).
Scatter plot analysis of the changes in synaptic plasticity gene expression in clade B and clade C Tat treated SK-N-MCs. Pair wise comparison of control SK-N-MC (No HIV-Tat proteins treated) and clade B Tat treated SK-N-MCs by scatter plot analysis. Spots associated with individual human synaptic plasticity gene were collected and converted into log10 scale. The central line indicates unchanged gene expression. The synaptic plasticity genes with expression levels higher or lower in HIV-1 clade B Tat (A) and clade C Tat (B) treated SK-NMCs than the control cells are expected to produce dots that deviate from the centerline. The dots are allocated to positions that are above or below than the +3 fold or −3 fold line when the differences are greater than three folds.
Fig. (2).
Venn diagram comparing the number of down-regulated and upregulated synaptic plasticity genes in SK-N-MC neuronal cells treated with HIV-1 clade B and C Tat.
Table 3.
Human synaptic plasticity genes down-regulated in HIV-1 clade B and C Tat treated SK-N-MC (fold down regulation).
| Synaptic Plasticity Genes |
HlV-1 Clade B Tat Fold Change |
HIV-1 Clade C Tat Fold Change |
p < Value |
|---|---|---|---|
| NCAM1 | −3.09 | −3.64 | NS |
| CREM | −6.16 | −5.4 | NS |
| NFKB1 | −8.83 | −6.84 | <0.02 |
| RHEB | −10.64 | −19.21 | <0.001 |
| TIMP1 | −26.76 | −17.8 | <0.001 |
Table 2.
Human synaptic plasticity genes up-regulated in HIV-1 clade B and C Tat treated SK-N-MC (fold up regulation): Out of 84 genes analyzed, only the genes significantly (≥3 fold) up-regulate were shown in this table.
| Synaptic Plasticity Genes | HIV-1 Clade B Tat Fold Change | HIV-1 Clade C Tat Fold Change | p< Value |
|---|---|---|---|
| ADCY1 | 5.47 | 3.2 | <0.01 |
| AKT1 | 7.29 | 3.34 | <0.001 |
| BDNF | 3.47 | 3.08 | NS |
| CAMK2G | 6.22 | 2.93 | <0.01 |
| CEBPB | 6.17 | −1.02 | <0.001 |
| EGR1 | 2.37 | 3.81 | <0.01 |
| EGR2 | 3.47 | 3.29 | NS |
| EGR1 | 9.82 | 7.01 | <0.01 |
| EGR2 | 4.88 | 2.6 | <0.001 |
| EGR3 | 7.65 | 8.93 | <0.05 |
| EGR4 | 5.88 | 4.16 | <0.06 |
| GABRA5 | 11.09 | −1.71 | <0.001 |
| GRIA1 | 4.04 | 1.2 | <0.002 |
| GRIA3 | 23.2 | 13.97 | <0.001 |
| GRIN1 | 8.85 | 7.56 | <0.01 |
| GRIN2B | 4.25 | 1.16 | <0.001 |
| GRIN2C | 4.81 | 3.75 | <0.05 |
| GRIN2D | 3.22 | 1.26 | <0.01 |
| GRM3 | 24.86 | 13.07 | <0.001 |
| GRM4 | 8.32 | 13.91 | <0.001 |
| GRM5 | 3.65 | 1.67 | <0.001 |
| GRM7 | 3.91 | 3.38 | NS |
| JUNB | 4.52 | 5.2 | NS |
| KIF17 | 4.71 | −2.35 | <0.03 |
| KLF10 | 4.81 | 3.75 | <0.05 |
| NFKBIB | 24.86 | 13.07 | <0.001 |
| NGF | 8.32 | 13.91 | <0.001 |
| NGFR | 3.65 | 1.67 | <0.01 |
| NOSI | 17.91 | 3.38 | <0.001 |
| NR4AI | 4.52 | 5.2 | NS |
| NTF4 | 4.52 | 5.2 | NS |
| PMII | 11.2 | 6.53 | <0.001 |
| PPP2CA | 8.55 | 11.78 | <0.01 |
| PRKCA | 3.75 | 2.69 | <0.01 |
| RAB3A | 54.8 | 28.6 | <0.001 |
| RELA | 3.07 | 1.88 | <0.01 |
| RGS2 | 2.4 | 3.63 | <0.05 |
| SIRT1 | 2.78 | 3.55 | NS |
| SYNPO | 5.12 | 5.81 | NS |
| TNF | 6.09 | −1.91 | <0.001 |
Glutamine and Glutamate Levels
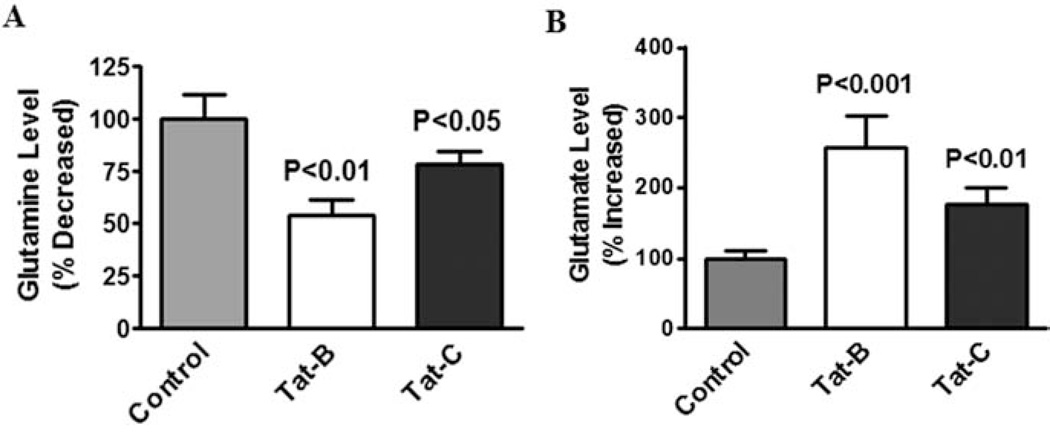
Glutamine synthetase (GS) present in several cellular organs including brain and it play a significant role in the metabolic regulation in glutamate and termination of neurotransmitter function [27]. Studies have shown that neuronal GS inhibition subsequently impact CNS exitotoxicity. In the present study we have investigated the effect of HIV-1 clade B and C Tat induced glutamate formation and glutamine levels in SK-N-MC. Data presented in Fig. 3A shows that both clade B and C Tat (100 ng) treated to SK-N-MC at 48h, significantly decreased the level of glutamine (p<0. 01) compared to clade C Tat (p<0.05) or the untreated control group. Whereas, significantly increased glutamate (p<0.001) when compared to clade C Tat (p<0.05) in SK-N-MC (Fig. 3B).
Fig. (3).
HIV-1 clade B and C Tat protein influence glutamine and glutamate levels. SK-N-MC (2×106 cells/ml) were separately treated with HIV-1 clade B and C Tat at 100 ng/ml for 48 h, and the cell lysates were examined for glutamine (A) and glutamate level (B). Data was expressed as mean±SE of three independent experiments.
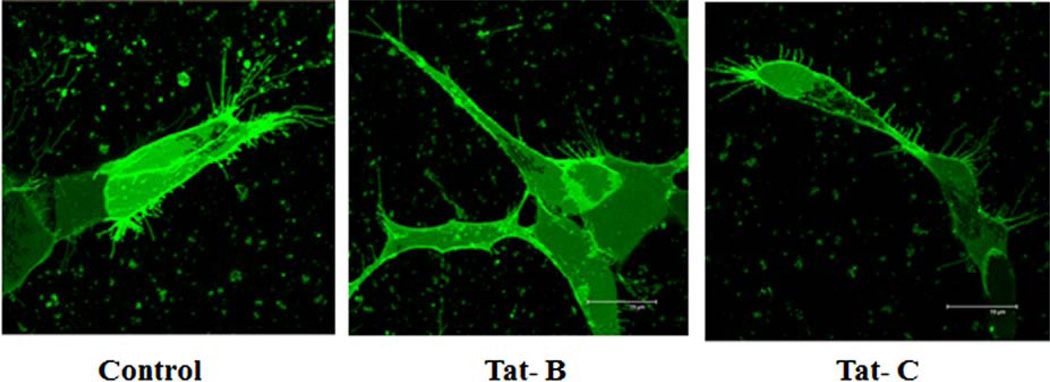
Decreased Spine Density in Clade B and C Tat Treated Neuronal Cells
Confocal microscopy was used to capture the dendrite morphology of control, HIV-1 clade B and C Tat treated cells. Significant spine density (number of spines per dendrite length) and dendrite morphology changes were observed in neuronal cells treated with clade B and C Tat (Fig. 4) compared with untreated control cells. In clade B Tat treated cells, significant decrease in spine density was observed than with clade C Tat treated cells.
Fig. (4).
HIV-1 clade B and C Tat effect on morphology and spine density by confocal images of DiI stained SK-N-MC cells. Control SK-NMC cells: High number of long spines on the dendritic length, high dendrite diameter, total dendrite area and spine area. Clade B Tat treated cells: Loss of spines on the dendrite length was observed than the control and clade C Tat treated SK-N-MC cells. Clade C Tat treated cells: Loss of number of spines was observed than control cells.
DISCUSSION
Our recent studies substantiate other reports in which HIV-1 clade B gp120 and Tat were shown to be more potent in contributing neuropathogesis compared to clade C gp120 and Tat [22, 23, 28–30]. However, there are no reports on HIV-Tat protein role in synaptic genes influencing neuronal dysfunction. In this study, in clade B Tat treated SK-N-MC, 36 synaptic plasticity genes were up-regulated (AKT1, CAMK2G, CEBPB, GABRA5, GRM5, GRIA3, NFKBIB, NOS1, RAB3A and TNF-α etc.) and many other synaptic plasticity genes were dysregulated compared to clade C Tat treated and untreated control cells. In clade C Tat treated SKN- MC, a total of 25 synaptic plasticity genes were upregulated compared to the control cells. It signifies the clade B induced neurotoxicity over clade C by either up-regulating or down regulating the synaptic plasticity genes (Figs. 1, 2 and Table 2). Recently, from our lab, we have reported significant down-regulation of 28 synaptic plasticity genes and up-regulation of 8 genes in HIV-1 clade B infected SKN- MC cells [31]. In the current study we have shown, for the first time, differences in HIV-1 clades by studying modulation of the mGluRs receptor pathway and glutamate activation in SK-N-MC cells. This study provides new insights into how HIV-1 clade B and C Tat proteins differentially modulate neurotoxicity and impact synaptic plasticity expression in SK-N-MC cells. These findings are of substantial importance because it indicates that the mGluRs receptor, NOS1, CaMKs, CREBP and NFkB transcriptional factors and proinflammatory cytokine TNF- α differentially potentiates synaptic plasticity. In addition, our results suggest that the levels of glutamine decreased whereas glutamate levels were increased by HIV-1 clade B and C (Fig. 3A, B), and it may play a role in synaptic plasticity expression.
mGluRs are found in pre- and post-synaptic neurons (in synapses) of the hippocampus, cerebellum, and the cerebral cortex, as well as other parts of the brain and in peripheral tissues. It has been shown that the mGluRs perform multiple functions in the central and peripheral nervous systems and plays major role in learning, memory and anxiety [32, 33]. Previousr studies have demonstrated the role of mGluRs in NMDA Receptor mediated neurotoxicity and excitotoxicity in CNS [34, 35]. Krogh et al., has shown the upregulation of NOS/sGC/PKG pathway by HIV-1 Tat in the hippocampal neuron [36]. Our recent reports also indicated that HIV-1 clade B and C gp120 protein potentiates considerably glutamate as well as NMDAR in astrocytes [23]. However no studies have been focused on HIV subtypes and associated mechanism of mGluRs in human CNS cells. Our results for the first time indicate that HIV-Tat clade differentially potentiate mGluRs mediated synaptic plasticity expression in SK-N-MC.
It is known that HIV-1 infection produced nitric oxide synthase (NOS) affects neuronal function [37]. Increased level of glutamate subsequently reduce glutamine which results in increased NOS which subsequently impact several cellular dysfunctions in CNS [38, 39]. The NOS can be neuroprotective or neurotoxic based on intracellular expression levels [40]. However, NMDAR mediated Ca2+ influx via s-nitrosylation results in improved neuronal survival [41, 42] whereas continuous generation of NOS may be inundated by nitrosative stress following prolonged, persistent, and excessive production of NO, leading to neurotoxicity and cell death. Previous studies have shown that HIV-1 Tat causes an elevation in the production of the NOS in vivo as well as in vitro [43]. Our results indicates that HIV-1 clade B Tat induced NOS1 production potentiate more neuro-pathogenesis than HIV-1 clade C Tat treatment. It is also known that mGluRs can regulate the cellular neurotransmitters mechanisms and neuronal synaptic plasticity depends on NOS [44]. Furthermore, an increased NOS in HIV-associated dementia (HAD) patients has also been reported [45], suggesting an association of mGluRs receptor with neuronal dysfunction. Interestingly, HAD appears to be the most common with HIV-1 clade B infections, whereas much lower level of HAD occurs with HIV-1 clade C infection. This indicates that the prevalence of HAD may be correlated with the difference in subtypes of HIV-1.
Furthermore, our results also show that the induction of mGluRs receptor expression decreased the level of glutamine (Fig. 3A), and significantly increased glutamate formation (Fig. 3B) with HIV-1 clade B Tat protein. In addition, altered spine density has been noticed in clade B Tat treated SK-N-MC cells than control and clade C Tat treated cells. Dendritic spines are part of the major postsynaptic sites for excitatory synaptic transmission (Fig. 4) and can undergo remodeling even in the adult nervous system. These results suggest that HIV-1 clade C Tat is less effective and lack functional activity in mGluRs receptor-mediated induction. Earlier studies have shown that HIV-1 genetic variations play a critical role in affecting HIV-1 infection and differently modify disease progression in the clade B and C [46]. There are differences in sequence and structure of HIV-1 clade B Tat compared to HIV-1 clade C Tat, especially in HIV-1 clade C Tat gene sequence, dicysteine position C30C31 changes the motif and cysteine replaced by serine [20]. These differences might account for the observed differences in the activities of the clade B and C proteins (Fig. 5).
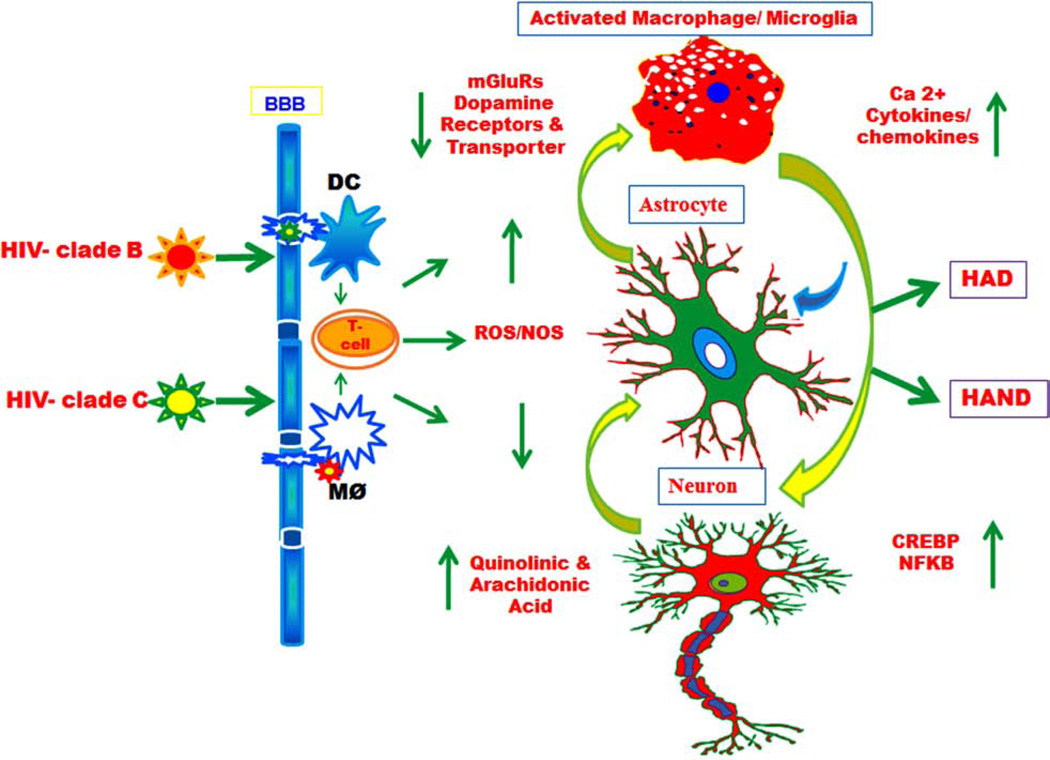
Fig. (5).
Schematic pathway of HIV-1 B and C differentially influence central nervous system. A comprehensive model showing how HIV-1 clade B and C differentially impact signaling through dendritic cells (DC), T cell and macrophages (and the closely related microglial cells) in the brain and their contribution to the pathogenesis of AIDS Dementia Complex (ADC) and neurocognitive disorders (HAND). The viral Trans activator protein (Tat) leads to secretion of neurotoxin QUIN and Arachidonic acid metabolites mediators’ which influence dopamine and mGlu receptor can activate astrocytes, and that may damage neurons differentially.
GRM5 is an important presynaptic regulator of neurotransmission in the mammalian CNS [47]. GABRA5, the main inhibitory neurotransmitter in the vertebrate brain, mediates neuronal inhibition by binding to the GABA/benzodiazepine receptor and opening an integral chloride channel. It plays a major role in chloride transport, signal transduction, synaptic transmission, regulation of neuron apoptosis and neuron development. Zhou et al. (2008) reported that GABA receptor inhibitor considerably suppress NMDAR mediated NO production [48]. Recently from our lab, down-regulation of NMDAR in HIV-1 virus infected with astrocytes was reported [24]. However, neurotransmitters changes due to the influence of NMDAR which is mediated by mGluRs, may explain the dysregulated synaptic plasticity genes in HIV infected cells. Moreover, it is known that HIV infection as well as Tat impact transcriptional factors and inflammatory cytokines [49, 50]. However, we have observed that HIV-1 clade B Tat significantly changed NFkB, CREBP transcription and proinflammatory cytokine TNF-α. Among 10 functional groups, each synaptic plasticity gene has one or more functions. In clade B and C Tat trated cells, 5 highly down regulated synaptic plasticity genes were NCAM1 (3 & 3), CREM (6 & 5 fold), NFKB1 (8 & 6 fold), RHEB (20 & 3 fold) and TIMP1 (26 & 17 fold), respectively (Table 3). GRM5 alters glutamate transmission, cognition, and increases the risk for schizophrenia. GRIA (glutamate receptor, ionotropic, AMPA) assembled from 4 related subunits, Gria1-4 also referred to as predominant excitatory neurotransmitter receptors in the mammalian brain are activated in a variety of normal neurophysiologic processes, acts as a RNA editing (CAG->CGG; Q->R) within the second transmembrane domain, which is thought to provide the channel impermeable to Ca (2+) [51–53].
Over all observed results confirmed that HIV-1 Tat B potentiates significantly and it may play an important role in neuronal plasticity. In support to the current study, from our previous report, in spine density analysis of HIV infected neuronal cells; we have observed that HIV-1 clade B is more neuropathogenic than clade C [24]. This could be due to the HIV-1B derived Tat protein induced neurotoxicity. In conclusion, treatment with HIV-1 clade B and C Tat proteins modulated the NOS1 mediated induction of mGluRs which altered expression of plasticity genes in SK-N-MC cells. Interestingly, we also demonstrated that clade B and C Tat treatment differentially potentiate the synaptic expression, leading to neuronal disease progression. To the best of our knowledge, this is the first report that HIV-1 clade B Tat has a greater impact on the expression of plasticity genes than clade C Tat. Taken together, these results suggest that cladespecific mechanisms in intracellular mGluRs receptormediated NOS might play a critical role in the neuropathogenesis of HANDs.
ACKNOWLEDGEMENTS
Funding Statements: The present study was supported by grants from National Institute of Health (NIH); RO3MH096640 and MH085259.
Biography

Thangavel Samikkannu
Footnotes
Part of this article has been previously published in PLoS ONE 8(4): e61399. doi: 10.1371/journal.pone.0061399.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Gonzalez-Scarano F, Martin-Garcia J. The neuropathogenesis of AIDS. Nat Rev Immunol. 2005;5(1):69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- 2.Antinori A, Trotta MP, Lorenzini P, et al. Virological response to salvage therapy in HIV-infected persons carrying the reverse transcriptase K65R mutation. Antivir Ther. 2007;12(8):1175–1183. [PubMed] [Google Scholar]
- 3.Ellis R, Langford D, Masliah E. HIV and antiretroviral therapy in the brain: neuronal injury and repair. Nat Rev Neurosci. 2007;8(1):33–44. doi: 10.1038/nrn2040. [DOI] [PubMed] [Google Scholar]
- 4.McArthur JC, Brew BJ. HIV-associated neurocognitive disorders: is there a hidden epidemic? Aids. 2010;24(9):1367–1370. doi: 10.1097/QAD.0b013e3283391d56. [DOI] [PubMed] [Google Scholar]
- 5.Nath A, Psooy K, Martin C, et al. Identification of a human immunodeficiency virus type 1 Tat epitope that is neuroexcitatory and neurotoxic. J Virol. 1996;70(3):1475–1480. doi: 10.1128/jvi.70.3.1475-1480.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amarapal P, Tantivanich S, Balachandra K, et al. The role of the Tat gene in the pathogenesis of HIV infection. Southeast Asian J Trop Med Public Health. 2005;36(2):352–361. [PubMed] [Google Scholar]
- 7.Jiang ZG, Piggee C, Heyes MP, et al. Glutamate is a mediator of neurotoxicity in secretions of activated HIV-1-infected macrophages. Journal of Neuroimmunology. 2001;117(12):97–107. doi: 10.1016/s0165-5728(01)00315-0. [DOI] [PubMed] [Google Scholar]
- 8.Nath A. Human immunodeficiency virus (HIV) proteins in neuropathogenesis of HIV dementia. J Infect Dis. 2002;186(Suppl 2):S193–S198. doi: 10.1086/344528. [DOI] [PubMed] [Google Scholar]
- 9.Eugenin EA, King JE, Nath A, et al. HIV-tat induces formation of an LRP-PSD-95- NMDAR-nNOS complex that promotes apoptosis in neurons and astrocytes. Proc Natl Acad Sci USA. 2007;104(9):3438–3443. doi: 10.1073/pnas.0611699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey AN, Sypek EI, Singh HD, Kaufman MJ, McLaughlin JP. Expression of HIV-Tat protein is associated with learning and memory deficits in the mouse. Behavioural Brain Research. 2012;229(1):48–56. doi: 10.1016/j.bbr.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitting S, Ignatowska-Jankowska BM, Bull C, et al. Synaptic Dysfunction in the Hippocampus Accompanies Learning and Memory Deficits in Human Immunodeficiency Virus Type-1 Tat Transgenic Mice. Biological Psychiatry. 2013;73(5):443–453. doi: 10.1016/j.biopsych.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HJ, Martemyanov KA, Thayer SA. Human immunodeficiency virus protein Tat induces synapse loss.via a reversible process that is distinct from cell death. J Neurosci. 2008;28(48):12604–12613. doi: 10.1523/JNEUROSCI.2958-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandra T, Maier W, Konig HG, et al. Molecular interactions of the type 1 human immunodeficiency virus transregulatory protein Tat with N-methyl-d-aspartate receptor subunits. Neuroscience. 2005;134(1):145–153. doi: 10.1016/j.neuroscience.2005.02.049. [DOI] [PubMed] [Google Scholar]
- 14.Haughey NJ, Mattson MP. Calcium dysregulation and neuronal apoptosis by the HIV-1 proteins Tat and gp120. J Acquir Immune Defic Syndr. 2002;31(Suppl 2):S55–S61. doi: 10.1097/00126334-200210012-00005. [DOI] [PubMed] [Google Scholar]
- 15.Robertson DL, Anderson JP, Bradac JA, et al. HIV-1 nomenclature proposal. Science. 2000;288(5463):55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 16.Osmanov S, Pattou C, Walker N, Schwardlander B, Esparza J. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J Acquir Immune Defic Syndr. 2002;29(2):184–190. doi: 10.1097/00042560-200202010-00013. [DOI] [PubMed] [Google Scholar]
- 17.Korber BKC, Foley B, et al. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, NM: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1998. Human retroviruses and AIDS. [Google Scholar]
- 18.Rambaut A, Robertson DL, Pybus OG, Peeters M, Holmes EC. Human immunodeficiency virus: Phylogeny and the origin of HIV-1. Nature. 2001;410(6832):1047–1048. doi: 10.1038/35074179. [DOI] [PubMed] [Google Scholar]
- 19.Myers G, MacInnes K, Korber B. The emergence of simian/human immunodeficiency viruses. AIDS Res Hum Retroviruses. 1992;8(3):373–386. doi: 10.1089/aid.1992.8.373. [DOI] [PubMed] [Google Scholar]
- 20.Mishra M, Vetrivel S, Siddappa NB, Ranga U, Seth P. Clade-specific differences in neurotoxicity of human immunodeficiency virus-1 B and C Tat of human neurons: significance of dicysteine C30C31 motif. Ann Neurol. 2008;63(3):366–376. doi: 10.1002/ana.21292. [DOI] [PubMed] [Google Scholar]
- 21.Campbell GR, Watkins JD, Singh KK, Loret EP, Spector SA. Human immunodeficiency virus type 1 subtype C Tat fails to induce intracellular calcium flux and induces reduced tumor necrosis factor production from monocytes. J Virol. 2007;81(11):5919–5928. doi: 10.1128/JVI.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samikkannu T, Rao KV, Pilakka Kanthikeel S, Subba Rao Atluri V, Agudelo M, Roy U, et al. Immunoneuropathogenesis of HIV-1 clades B and C: Role of redox expression and thiol modification. Free Radic Biol Med. 2014;69:136–144. doi: 10.1016/j.freeradbiomed.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samikkannu T, Agudelo M, Gandhi N, et al. Human immunodeficiency virus type 1 clade B and C gp120 differentially induce neurotoxin arachidonic acid in human astrocytes: implications for neuroAIDS. Journal of NeuroVirology. 2011;17(3):230–238. doi: 10.1007/s13365-011-0026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atluri VS, Kanthikeel SP, Reddy PV, Yndart A, Nair MP. Human synaptic plasticity gene expression profile and dendritic spine density changes in HIV-infected human CNS cells: role in HIV-associated neurocognitive disorders (HAND) PLoS One. 2013;8(4):e61399. doi: 10.1371/journal.pone.0061399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurapati KR, Atluri VS, Samikkannu T, Nair MP. Ashwagandha (Withania somnifera) reverses beta-amyloid1-42 induced toxicity in human neuronal cells: implications in HIV-associated neurocognitive disorders (HAND) PLoS One. 2013;8(10):e77624. doi: 10.1371/journal.pone.0077624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Draghici S. Statistical intelligence: effective analysis of high density microarray data. Drug Discov Today. 2002;7(S55–S63) doi: 10.1016/s1359-6446(02)02292-4. [DOI] [PubMed] [Google Scholar]
- 27.Souba WW. In: Glutamine: Physiology, Biochemistry and Nutrition in Critical Illness (Medical Intelligence Unit) Co RGL, editor. Austin, TX: 1992. [Google Scholar]
- 28.Samikkannu MPNNaT. Differential Regulation of Neurotoxin in HIV Clades: Role of Cocaine and Methamphetamine. Current HIV Research. 2012;10(5):429–434. doi: 10.2174/157016212802138742. [DOI] [PubMed] [Google Scholar]
- 29.Samikkannu T, Rao KK, Gandhi N, Saxena S, Nair MN. Human immunodeficiency virus type 1 clade B and C Tat differentially induce indoleamine 2,3-dioxygenase and serotonin in immature dendritic cells: Implications for neuroAIDS. Journal of NeuroVirology. 2010;16(4):255–263. doi: 10.3109/13550284.2010.497809. [DOI] [PubMed] [Google Scholar]
- 30.Samikkannu T, Saiyed ZM, Rao KVK, et al. Differential Regulation of Indoleamine-2,3-Dioxygenase (IDO) by HIV Type 1 Clade B and C Tat Protein. AIDS Research and Human Retroviruses. 2009;25(3):329–335. doi: 10.1089/aid.2008.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atluri VS, Pilakka-Kanthikeel S, Samikkannu T, et al. Vorinostat positively regulates synaptic plasticity genes expression and spine density in HIV infected neurons: role of nicotine in progression of HIV-associated neurocognitive disorder. Mol Brain. 2014;7:37. doi: 10.1186/1756-6606-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoepp DD. Unveiling the Functions of Presynaptic Metabotropic Glutamate Receptors in the Central Nervous System. Journal of Pharmacology and Experimental Therapeutics. 2001;299(1):12–20. [PubMed] [Google Scholar]
- 33.Bhardwaj A, Northington FJ, Martin LJ, Hanley DF, Traystman RJ, Koehler RC. Characterization of Metabotropic Glutamate Receptor-Mediated Nitric Oxide Production In Vivo. J Cereb Blood Flow Metab. 1997;17(2):153–160. doi: 10.1097/00004647-199702000-00004. [DOI] [PubMed] [Google Scholar]
- 34.Qiang M, Rani CSS, Ticku MK. Neuron-Restrictive Silencer Factor Regulates the N-Methyl-d-aspartate Receptor 2B Subunit Gene in Basal and Ethanol-Induced Gene Expression in Fetal Cortical Neurons. Molecular Pharmacology. 2005;67(6):2115–2125. doi: 10.1124/mol.104.010751. [DOI] [PubMed] [Google Scholar]
- 35.Shih AY, Johnson DA, Wong G, et al. Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. J Neurosci. 2003;23(8):3394–3406. doi: 10.1523/JNEUROSCI.23-08-03394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krogh KA, Wydeven N, Wickman K, Thayer SA. HIV-1 protein Tat produces biphasic changes in NMDA-evoked increases in intracellular Ca2+ concentration via activation of Src kinase and nitric oxide signaling pathways. J Neurochem. 2014;130(5):642–656. doi: 10.1111/jnc.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Jana M, Dasgupta S, et al. Human immunodeficiency virus type 1 (HIV-1) tat induces nitric-oxide synthase in human astroglia. J Biol Chem. 2002;277(42):39312–39319. doi: 10.1074/jbc.M205107200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holscher C. Nitric oxide, the enigmatic neuronal messenger: its role in synaptic plasticity. Trends Neurosci. 1997;20(7):298–303. doi: 10.1016/s0166-2236(97)01065-5. [DOI] [PubMed] [Google Scholar]
- 39.Prast H, Philippu A. Nitric oxide as modulator of neuronal function. Progress in Neurobiology. 2001;64(1):51–68. doi: 10.1016/s0301-0082(00)00044-7. [DOI] [PubMed] [Google Scholar]
- 40.Lipton SA, Choi Y-B, Pan Z-H, et al. A redox-based mechanism for the neuroprotective and neurodestructive effects of nitric oxide and related nitroso-compounds. Nature. 1993;364(6438):626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 41.Choi S, Klingauf J, Tsien RW. Postfusional regulation of cleft glutamate concentration during LTP at /`silent synapses/'. Nat Neurosci. 2000;3(4):330–336. doi: 10.1038/73895. [DOI] [PubMed] [Google Scholar]
- 42.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3(2):193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 43.Pocernich CB, Sultana R, Mohmmad-Abdul H, Nath A, Butterfield DA. HIV-dementia, Tat-induced oxidative stress, and antioxidant therapeutic considerations. Brain Research Reviews. 2005;50(1):14–26. doi: 10.1016/j.brainresrev.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Pinard A, Robitaille R. Nitric oxide dependence of glutamate-mediated modulation at a vertebrate neuromuscular junction. European Journal of Neuroscience. 2008;28(3):577–587. doi: 10.1111/j.1460-9568.2008.06355.x. [DOI] [PubMed] [Google Scholar]
- 45.Vincent VrAM, De Groot CJA, Lucassen PJ, et al. Nitric oxide synthase expression and apoptotic cell death in brains of AIDS and AIDS dementia patients. Aids. 1999;13(3):317–326. doi: 10.1097/00002030-199902250-00003. [DOI] [PubMed] [Google Scholar]
- 46.Satishchandra P, Nalini A, Gourie-Devi M, et al. Profile of neurologic disorders associated with HIV/AIDS from Bangalore, south India (1989–96) Indian J Med Res. 2000;111:14–23. [PubMed] [Google Scholar]
- 47.Gee CE, Peterlik D, Neuhauser C, et al. Blocking Metabotropic Glutamate Receptor Subtype 7 (mGlu7) via the Venus Flytrap Domain (VFTD) Inhibits Amygdala Plasticity, Stress, and Anxiety-related Behavior. Journal of Biological Chemistry. 2014;289(16):10975–10987. doi: 10.1074/jbc.M113.542654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J, Du W, Zhou K, et al. Critical role of TRPC6 channels in the formation of excitatory synapses. Nat Neurosci. 2008;11(7):741–743. doi: 10.1038/nn.2127. [DOI] [PubMed] [Google Scholar]
- 49.Pingle SC, Jajoo S, Mukherjea D, et al. Activation of the adenosine A1 receptor inhibits HIV-1 tat-induced apoptosis by reducing nuclear factor-kappaB activation and inducible nitric-oxide synthase. Mol Pharmacol. 2007;72(4):856–867. doi: 10.1124/mol.106.031427. [DOI] [PubMed] [Google Scholar]
- 50.Sawaya BE, Thatikunta P, Denisova L, et al. Regulation of TNFalpha and TGFbeta-1 gene transcription by HIV-1 Tat in CNS cells. J Neuroimmunol. 1998;87(1–2):33–42. doi: 10.1016/s0165-5728(98)00044-7. [DOI] [PubMed] [Google Scholar]
- 51.Patel S, Hamill TG, Connolly B, et al. Species differences in mGluR5 binding sites in mammalian central nervous system determined using in vitro binding with [18F]F-PEB. Nuclear Medicine and Biology. 2007;34(8):1009–1017. doi: 10.1016/j.nucmedbio.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 52.Nutt S, Kamboj R. Differential RNA editing efficiency of AMPA receptor subunit GluR-2 in human brain. Neuroreport. 1994;5:1679–1683. doi: 10.1097/00001756-199408150-00034. [DOI] [PubMed] [Google Scholar]
- 53.Liu SJ, Zukin RS. Ca2+-permeable AMPA receptors in synaptic plasticity and neuronal death. Trends in Neurosciences. 2007;30(3):126–134. doi: 10.1016/j.tins.2007.01.006. [DOI] [PubMed] [Google Scholar]