Abstract
Purpose
To ensure the correct interpretation of the results of quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) from ovarian tissue cryopreserved by vitrification, it is critical to normalize expression levels to a reference gene with stable messenger RNA (mRNA) expression in the vitrified/warmed ovarian tissue. The aim of this work was to identify suitable reference genes for qRT-PCR analysis during ovarian cryopreservation by vitrification.
Methods
GeNorm, NormFinder, comparative Delta-CT, and BestKeeper were used to analyze the expression and stability of the 14 reference genes GAPDH, ABL1, ACTB, CDKN1A, GPER, GUSB, HPRT1, HSP90AB1, IPO8, PPIA, RPL4, RPL30, TBP, and UPAR.
Results
Our results indicated that ACTB and RPL4 were relatively stable reference genes in vitrified/warmed ovaries.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-015-0503-5) contains supplementary material, which is available to authorized users.
Keywords: Ovarian vitrification, Reference gene, Stability, Expression
Introduction
Radiotherapy and chemotherapy are the main methods to treat cancer, and 90 % of children and adolescent patients with ovarian cancer have hope for a cure [1, 2]. However, ovarian function is damaged by radiotherapy and chemotherapy, which can cause subfertility or infertility. In the past decades, ovarian cryopreservation by vitrification was developed and has been used to preserve the ovaries during radiotherapy and chemotherapy. The ovary is then orthotopically transplanted when the cancer is cured. Previous studies suggested that the expression of most genes changed during ovarian vitrification [3, 4]. Additionally, the expression of the reference gene GAPDH decreased in vitrified/warmed ovarian tissue compared with non-vitrified ovaries [5, 6]. Whether expression of other reference genes changed during ovarian vitrification is unknown. Therefore, the stability of reference genes should be evaluated for standard normalization of the expression of target genes. Hence, 14 genes were chosen, and expression was detected in vitrified/warmed ovaries. The stability of these genes was analyzed by GeNorm, NormFinder, and BestKeeper.
Materials and methods
Ovarian tissue and treatments
Four-week-old C57BL/6J mice were purchased from the Ningxia Medical University Animal Center and were maintained at (24 ± 2 °C) in a light-controlled room (12 h light/12 h darkness) with free access to food and water. All procedures were performed according to the Committee for the Ethics on Animal Care and Experiments at Ningxia Medical University. The four stages of the estrus cycle were tracked by vaginal smear according to previous studies [7, 8], and mice in dioestrus were used for this study.
Sixteen 4-week-old mice in dioestrus (DE) were sacrificed by dislocation after anesthesia, and the bilateral ovaries were collected and randomly divided into three groups as follows: (A) control group (CG)—fresh ovaries were collected from the mice and preserved in liquid nitrogen for RNA extraction; (B) vitrified/warmed groups—the ovaries underwent vitrified cryopreservation without any further treatment; and (C) CPA (cryoprotectant)-treated group—fresh ovaries were equilibrated with CPA and warmed but not vitrified (solution-treated ovaries).
Vitrified cryopreservation procedure
The preparation of the basic medium, culture, freezing, and thawing solutions is described in supplemental doc1, and the vitrified cryopreservation procedure was performed. Briefly, the process mainly consists of pre-culturing in culture solution for 1 h at 37 °C with 5 % CO2. The mixture was pre-equilibrated for 8 min with pre-equilibration solution, and osmotic equilibrium was achieved after 3.5 min of incubation in the vitrification solution. The tissue was preserved in liquid nitrogen for at least 1 day, warmed for 10 min with a gradient thawing solution, and post-cultured for 1 h at 37 °C with 5 % CO2.
Selection of candidate reference genes and primer design
Fourteen candidate genes, GAPDH, ABL1, ACTB, CDKN1A, GPER, GUSB, HPRT1, HSP90AB1, IPO8, PPIA, RPL4, RPL30, TBP, and UPAR, were chosen for evaluation in the four groups. The primer sequences for all 14 candidate reference genes are shown in Table 1.
Table 1.
qRT-PCR primers
| Target gene | GenBank accession no. | Primer sequence | Product size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|
| ABL1 | NM_001271730.1 | F: 5′-CAGGTTTATGAGCTGCTGGA-3′ | 146 | 60 |
| R: 5′-GTTTCAAAGGCTTGGTGGAT-3′ | ||||
| ACTB | NM_007393.3 | F: 5′-CCTTCCTTCTTGGGTATGGA-3′ | 81 | 60 |
| R: 5′-ACGGATGTCAACGTCACACT-3′ | ||||
| CDKN1A | NM_001111099.1 | F: 5′-CAAAGTGTGCCGTTGTCTCT-3′ | 125 | 60 |
| R: 5′-TCTCCGTGACGAAGTCAAAG-3′ | ||||
| GPER | NM_029771.3 | F: 5′-GCTTCTGCTTTGCTGATGTC-3′ | 101 | 60 |
| R: 5′-CACGATGAGGGAGTAGCAGA-3′ | ||||
| GUSB | NM_010368.1 | F: 5′-GGGTCAATAAGCACGAGGAT-3′ | 116 | 60 |
| R: 5′-GTGGCTGGTACGAAAGGAAT-3 | ||||
| HPRT1 | NM_013556.2 | F: 5′-AGGACCTCTCGAAGTGTTGG-3′ | 115 | 60 |
| R: 5′-CGTGATTCAAATCCCTGAAG-3 | ||||
| HSP90AB1 | NM_008302.3 | F: 5′-GACATCACGCAGGAGGAGTA-3′ | 109 | 60 |
| R: 5′-CCCTGAATTCCAACTGACCT-3 | ||||
| IPO8 | NM_001082223.1 | F: 5′-AACAGACCCGAACTTTGACC-3′ | 122 | 60 |
| R: 5′-GATTCTGCAGGAACAGCTCA-3 | ||||
| PPIA | NM_008907.1 | F: 5′-CAGGGTGGTGACTTTACACG-3′ | 136 | 60 |
| R: 5′-TTGTGTTTGGTCCAGCATTT-3 | ||||
| RPL4 | NM_024212.4 | F: 5′-ATCTGGACGGAGAGTGCTTT-3′ | 115 | 60 |
| R: 5′-GGTCGGTGTTCATCATCTTG-3 | ||||
| RPL30 | NM_001163485.1 | F: 5′-GAAGTACGTGCTGGGCTACA-3′ | 95 | 60 |
| R: 5′-TCCTCAAAGCTGGACAGTTG-3 | ||||
| TBP | NM_013684.3 | F: 5′-ACTTCGTGCAAGAAATGCTG-3′ | 131 | 60 |
| R: 5′-CTTCACTCTTGGCTCCTGTG-3 | ||||
| UPAR | NM_011113.3 | F: 5′-TCATCAGCCTGACAGAGACC-3′ | 136 | 60 |
| R: 5′-CTCTCACAGCTCTGGTCCAA-3 | ||||
| GAPDH | NM_001289726.1 | F: 5′-ACAACTTTGGCATTGTGGAA-3′ | 133 | 60 |
| R: 5′-GATGCAGGGATGATGTTCTG-3 |
Extraction of total RNA and cDNA synthesis
TRIZOL reagent (Invitrogen Co., Carlsbad, CA, USA) was used for RNA isolation from 32 tissue samples, and the process was performed as previously described [7, 8]. RNA samples were dissolved with RNase-free DNaseI (Promega, Fitchburg, WI, USA) before reverse transcription. The concentration and purity of all RNA samples were checked using a NanoDrop Spectrophotometer ND-1000 (Saveen Werner, Limhamn, Sweden), and the A260/280 was 1.8~2.0. The integrity of the RNA samples was analyzed by 2 % agarose gel electrophoresis.
The complementary DNA (cDNA) was synthesized in a 20-μL reaction system according to the instructions for the TRAN PrimeScript RT Reagent Kit (Perfect for Real Time) (TRAN Biotechnology, Beijing, China). All the cDNA samples were diluted to 50 ng mL−1 for the following quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) reaction and stored at −20 °C until use.
Real-time PCR
An ABI 7500 Fast Real Time PCR machine and the default program (95 °C for 30 s, followed by 40 cycles at 95 °C for 5 s and 60 °C for 30 s) were employed for qRT-PCR with a reaction volume of 20 μL in an optical 96-well plate. Ten microliters of SYBR Green qPCR SuperMix (TRAN), 10 μM of each primer, 2 μL of the final cDNA, 0.4 μL Passive Reference Dye II (50×), and 6.8 μL of RNase-free water were added to the reaction mixture. A control was included in each plate using 2.0 μL of RNase-free water as the template.
GeNorm analysis
The GeNorm algorithm was used to identify the reference genes with the most stable expression in different tissues or culture conditions [9]. The software defines two parameters to quantify stability, expression stability (M value), and pairwise variation (V value). The gene with the lowest M value is considered to have the most stable expression. V values were proposed as a guide to determine the optimal number of candidate reference genes required for normalization [9] using a 0.15 cutoff value. If the V value is below or equal to 0.15, it is not necessary to include additional reference genes for normalization.
BestKeeper analysis
BestKeeper software is a tool to determine the most stable reference genes based on the analysis of the correlation coefficient of all possible pairs of candidate reference genes [10]. The software determines the BestKeeper index (BI), which is the geometric mean of the Ct values of highly correlated candidate reference genes [10]. The reference genes are identified as the most stable genes when they exhibit the lowest standard deviation (SD) and highest correlation coefficient (r). Genes with an SD greater than 1 are considered unacceptable.
NormFinder analysis
The NormFinder program is a Visual Basic application tool for Microsoft Excel used to select reference genes among a set of candidate genes for optimal normalization [11]. As in the GeNorm method, the gene with the lowest stability value (M) is the most stably expressed gene. NormFinder takes intragroup and intergroup variations in stability into account, ranking the best two reference genes for normalization.
Statistical analysis
The expression levels of the 14 reference genes in three groups (three biological duplicates and three different conditions) were determined by the number of cycles (Cq) needed for amplification-related fluorescence to reach a specific threshold level of detection. Following quantitative PCR (qPCR) data collection, Cq values were converted to relative quantities using the following formula: 2(−deltaCq), in which (deltaCq) = the corresponding Cq value − minimum Cq. Relative quantities were used for GeNorm and NormFinder, while BestKeeper analysis was performed based on raw Cq values [9–12]. The expression stability of the reference genes was ranked using the three different Microsoft Excel-based software programs described above (GeNorm, NormFinder, and BestKeeper).
All experiments were replicated at least three times for each group, and the data are presented as the mean ± SEM. Data were analyzed with one-way ANOVA followed by Fisher’s least significant difference (FLSD) test with SPSS software (version 13.0; SPSS, Inc., Chicago, IL, USA). Differences were considered significant at P < 0.05.
Results
Expression profiles of candidate reference genes in mice ovarian tissue
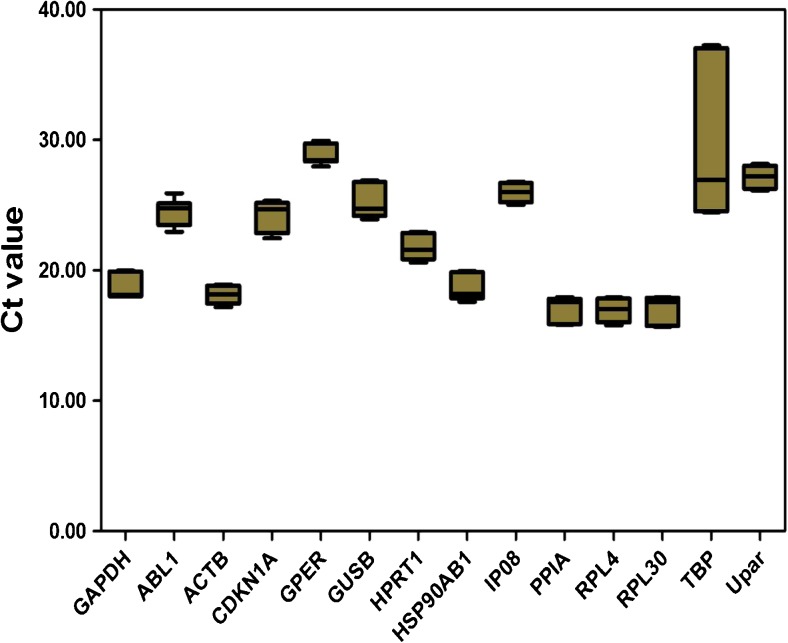
Cycle threshold (Ct) values were used to compare the transcript abundance of the selected genes in different samples, assuming equivalent Ct values for identical transcript amounts because an equal quantity of total RNA was used in all qPCR reactions [8]. The mean Ct values of the 14 reference genes varied from 15.68 to 37.01, with the lowest and highest Ct values obtained from RPL4 (Ct 16.96) and TBP (Ct 29.24) (Fig. 1). RPL4 and RPL30 showed the most abundant expression levels (lowest Ct value), followed by PPIA (mean Ct 17.11), ACTB (mean Ct 18.10), HSP90AB1 (mean Ct 18.62), and GAPDH (mean Ct 18.70). The remaining eight reference genes had a Ct value of 21 or higher.
Fig. 1.
Expression levels of 14 candidate reference genes with frozen ovarian tissue in mice. Values are given as the cycle threshold (Ct) and are inversely proportional to the amount of template. The expression levels of the genes studied are shown as box-and-whisker plots
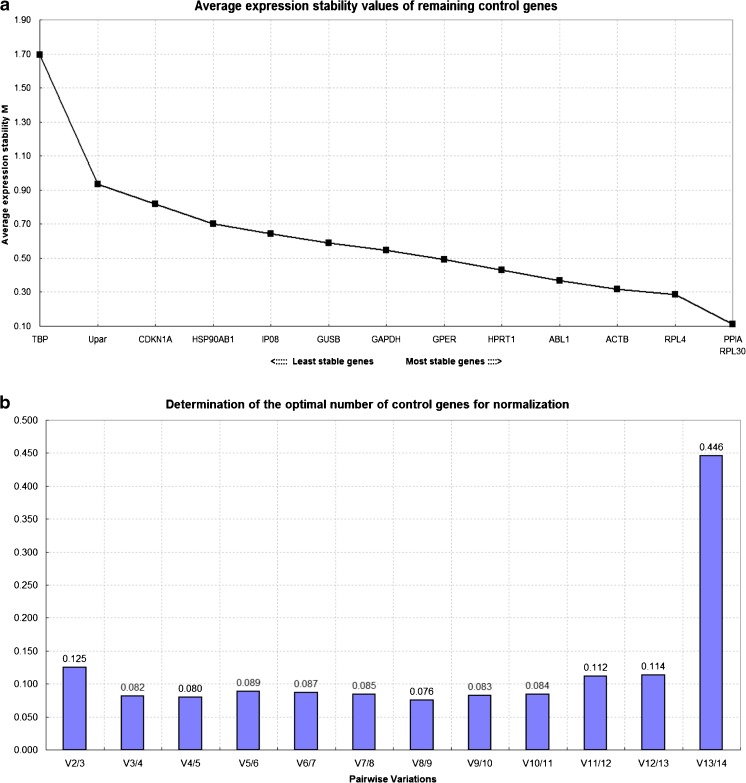
GeNorm analysis
The candidate reference genes had M values (stability values) below 1.5 except for TBP, which had an M value of 1.54 (Fig. 2a). Therefore, the TBP gene was considered unsuitable for normalization by GeNorm. PPIA and RPL30 had the lowest M values (0.10) (Fig. 2a), followed by RPL4, ACTB, ABL1, and HPRT1, which had average expression stability with M values below 0.5. The remaining eight reference genes had M values below 1.0 and so can be used as reference genes. GeNorm also predicts the optimal number of reference genes for accurate representation of gene expression based on the calculation of the pairwise variation (Vn/Vn + 1) between sequential normalization factors (NFn and NFn + 1). A cutoff value of 0.15 is used; below this value, there is no requirement for any other reference gene [9]. The study showed that V8/9 = 0.076 (Fig. 2b), so GeNorm analysis suggested that PPIA, RPL30, RPL4, ACTB, ABL1, HPRT1, GPER, and GAPDH are the most suitable genes to normalize messenger RNA (mRNA) expression for all samples (Fig. 2b). The GeNorm analysis ranked the stability as follows, from most to least stable: PPIA = RPL30 > RPL4 > ACTB > ABL1 > HPRT1 > GPER > GAPDH > GUSB > IPO8 > HSP90AB1 > CDKN1A > UPAR > TBP (p < 0.05) (Fig. 2a).
Fig. 2.
Stability values (a) and pairwise variation values (b) of reference genes obtained by GeNorm. A lower M value indicates more stable expression. Note that PPIA and RPL30 have a low M stability value (0.1) (a). Average pairwise variation is calculated between the normalization factors NFn and NFn + 1 to indicate whether the inclusion of an extra reference gene adds to the stability of the normalization factor (b)
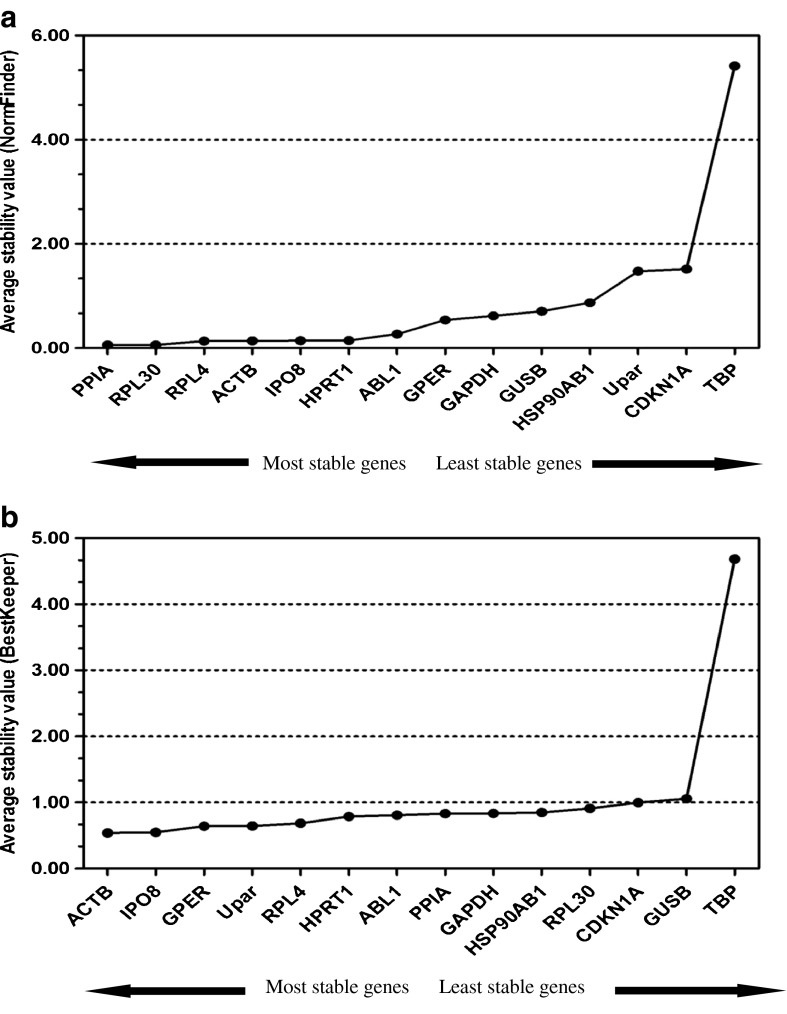
NormFinder analysis
NormFinder analyzes candidate reference genes according to a stable expression value. Consistent results were obtained using NormFinder. In the NormFinder analysis, the most stable reference genes (lowest M value) for the studied samples were PPIA and RPL30 (0.063) followed by RPL4 (0.136), ACTB (0.140), IPO8 (0.142), HPRT1 (0.144), and ABL1 (0.268) (Fig. 3a), making them the most suitable genes to normalize mRNA expression in our samples. The NormFinder analysis ranked gene expression stability from most to least stable as follows: PPIA = RPL30 > RPL4 > ACTB > IPO8 > HPRT1 > ABL1 > GPER > GAPDH > GUSB > HSP90AB1 > UPAR > CDKN1A > TBP (p < 0.05) (Fig. 3a).
Fig. 3.
Average expression stability values of the candidate reference genes for FSH intervention using frozen ovarian tissue in mice by NormFinder (a) and BestKeeper (b). Note that a lower stability value indicates more stable expression for NormFinder. PPIA and RPL30 have the lowest stability values (a). Lower SD and SD [±x-fold] indicate more stable expression for BestKeeper software. BestKeeper showed that ACTB and IPO8 are the most stable reference genes (b)
BestKeeper analysis
BestKeeper analysis determined which genes exhibited the lowest (CV six standard deviations) to judge the stability of gene expression (Table 2). ACTB had the lowest Ct value standard deviation (SD) across the groups (mean Ct ± SD 18.09 ± 0.54), followed by IPO8 (mean Ct ± SD 25.97 ± 0.55), GPER (mean Ct ± SD 28.83 ± 0.64), UPAR (mean Ct ± SD 27.15 ± 0.65), RPL4 (mean Ct ± SD 16.94 ± 0.69), and HPRT1 (mean Ct ± SD 21.77 ± 0.79) (Table 2). ACTB was the most stable reference gene, with the highest coefficient of correlation (Fig. 3b). IPO8, GPER, and UPAR also had a high coefficient of correlation, but the combination of this coefficient and their SD values was inferior to that of ACTB. Thus, BestKeeper ranked the stability from most to least stable as follows: ACTB > IPO8 > GPER > UPAR > RPL4 > HPRT1 > ABL1 > PPIA > GAPDH > HSP90AB1 > RPL30 > CDKN1A > GUSB > TBP (p < 0.05). The results from GeNorm analysis, NormFinder analysis, and BestKeeper analysis were further checked by the online software http://www.leonxie.com/referencegene.php, and the results suggested that the most stable genes are ACTB and RPL4 (Fig. 4).
Table 2.
Descriptive and correlation analysis of the candidate RGs obtained by BestKeeper
| CP data of housekeeping genes by BestKeeper | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GAPDH | ABL1 | ACTB | CDKN1A | GPER | GUSB | HPRT1 | HSP90AB1 | IP08 | PPIA | RPL4 | RPL30 | TBP | UPAR | |
| n | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 | 9 |
| gM (Ct) | 18.68 | 24.51 | 18.09 | 24.21 | 28.83 | 25.22 | 21.77 | 18.60 | 25.97 | 17.08 | 16.94 | 17.06 | 28.83 | 27.15 |
| aM (Ct) | 18.70 | 24.53 | 18.10 | 24.23 | 28.84 | 25.24 | 21.79 | 18.62 | 25.98 | 17.11 | 16.96 | 17.09 | 29.24 | 27.16 |
| Min (Ct) | 18.01 | 22.97 | 17.18 | 22.46 | 27.96 | 23.94 | 20.61 | 17.59 | 25.06 | 15.81 | 15.80 | 15.69 | 24.47 | 26.13 |
| Max (Ct) | 19.98 | 25.93 | 18.91 | 25.33 | 29.93 | 26.91 | 22.97 | 19.96 | 26.82 | 17.94 | 17.93 | 17.93 | 37.01 | 28.14 |
| SD (±Ct) | 0.84 | 0.81 | 0.54 | 1.00 | 0.64 | 1.06 | 0.79 | 0.85 | 0.55 | 0.83 | 0.69 | 0.91 | 4.68 | 0.65 |
| CV (% Ct) | 4.47 | 3.30 | 2.98 | 4.12 | 2.22 | 4.18 | 3.62 | 4.56 | 2.11 | 4.86 | 4.04 | 5.33 | 16.02 | 2.38 |
| Min (x-fold) | −1.59 | −2.91 | −1.88 | −3.36 | −1.83 | −2.43 | −2.24 | −2.02 | −1.89 | −2.41 | −2.21 | −2.59 | −20.43 | −2.02 |
| Max (x-fold) | 2.47 | 2.67 | 1.77 | 2.18 | 2.15 | 3.23 | 2.30 | 2.56 | 1.80 | 1.81 | 1.98 | 1.83 | 291.01 | 1.99 |
| SD (±x-fold) | 1.78 | 1.75 | 1.45 | 2.00 | 1.56 | 2.08 | 1.73 | 1.80 | 1.46 | 1.78 | 1.61 | 1.88 | 25.69 | 1.56 |
Fig. 4.
All data are comprehensively analyzed by the online software. The optimal stable reference genes were furtherly analyzed by online software http://www.leonxie.com/referencegene.php. The ACTB and RPL4 are optimal reference genes
Discussion
Currently, qRT-PCR is often used to study the expression patterns of single genes because of its high sensitivity and specificity [13]. For accurate estimation of the expression of target genes, estimates are normalized relative to internally expressed reference genes. These genes must be equally expressed among the states of interest for normalization to reduce nonspecific variation. Such variation may be the result of different amounts of template RNA or different efficiencies in cDNA synthesis, for example. In addition, suitable reference genes should be expressed at intensities similar to those of the target genes [14]. Traditionally, normalization is done using a single housekeeping gene [13]. However, several studies have revealed that these traditional reference genes are often not as stably expressed as expected, leading to misinterpretation of biological data [15, 16]. Consequently, new and more stably expressed reference genes have been successfully identified in a variety of organisms [17], and the optimal reference genes were chosen to analyze the biological data accurately.
This study evaluated 14 of the traditional reference genes, which are involved in basic biochemical metabolism in all cells, and the stability was closely related with its structure and characteristics. HSP90AB1 belongs to the heat shock protein family, so its expression is easily affected by temperature and pressure. RPL4 and RPL30 belong to the ribosomal protein family, and the structure is very stable. ACTB is a cytoskeleton actin protein, and this kind of protein fiber network structure is very important in maintaining cell morphology; the influence of external factors is low [16, 17]. GUSB, HPRT1, PPIA, and GAPDH are enzymes encoding [16, 17] esters for TBP [16]; IPO8, UPAR, and GPER are receptors [17]; ABL1 is a receptor kinase [17]; and CDKN1A is a protease inhibitor [17]. Therefore, their stability is also different during different conditions, such as ovarian cryopreservation. The GeNorm and NormFinder analysis results are basically identical, with the stability of internal candidate genes ranked as follows from most to least stable: PPIA = RPL30 > RPL4 > ACTB. BestKeeper analysis ranked the stability of internal candidate genes from most to least stable as follows: ACTB > UPAR > PPIA > IPO8. From the above Ct values, RPL4, RPL30, PPIA, and ACTB were the most abundantly expressed, so the stability of reference candidate genes could be related to their expression levels. Specifically, if the Ct value is high, the stability of internal candidate genes may be low. Reference genes involved in different biological processes may also have varied stabilities. The enzymatic activity of PPIA may affect its stability as a reference gene. ACTB (actin, beta) is related to cell motility, structure, and integrity; it is a structural protein [16] and has good stability, likely because outside conditions do not generally affect cell structure. To track small changes in gene expression, use of single internal genes often does not produce accurate quantitative results, so two or more internal genes should be used for reference calibration.
In conclusion, ACTB and RPL4 are the most suitable reference genes. GeNorm, NormFinder, and BestKeeper analyses showed that TBP is the least stable reference gene.
Electronic supplementary material
Preparation of medium (DOC 41 kb)
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81260110). Science and technology research project in Ningxia([2012]17).
Footnotes
Capsule This work firstly suggested that the genes of ACTB and RPL4 were optimal reference genes in vitrified/warmed ovaries.
Shan Yuanyuan and Su Qin equally contributed to this work.
Contributor Information
Yang Yanzhou, Email: alnord820119@163.com.
Pei Xiuying, Email: peixiuying@163.com.
References
- 1.Pieters R. Acute lymphoblastic leukaemia in children and adolescents: chance of cure now higher than 80% Ned Tijdschr Geneeskd. 2010;154:A1577. [PubMed] [Google Scholar]
- 2.Tanzler E, Morris CG, Kirwan JM, Amdur RJ, Mendenhall WM. Outcomes of WHO grade I meningiomas receiving definitive or postoperative radiotherapy. Int J Radiat Oncol Biol Phys. 2011;79:508–513. doi: 10.1016/j.ijrobp.2009.11.032. [DOI] [PubMed] [Google Scholar]
- 3.Fatehi R, Ebrahimi B, Shahhosseini M, Farrokhi A, Fathi R. Effect of ovarian tissue vitrification method on mice preantral follicular development and gene expression. Theriogenology. 2014;81:302–308. doi: 10.1016/j.theriogenology.2013.09.029. [DOI] [PubMed] [Google Scholar]
- 4.Abdollahi M, Salehnia M, Salehpour S, Ghorbanmehr N. Human ovarian tissue vitrification/warming has minor effect on the expression of apoptosis-related genes. Iran Biomed J. 2013;17(4):179–186. doi: 10.6091/ibj.1243.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isachenko V, Lapidus I, Isachenko E, Krivokharchenko A, Kreienberg R, Woriedh M, et al. Human ovarian tissue vitrification versus conventional freezing: morphological, endocrinological, and molecular biological evaluation. Reproduction. 2009;138:319–327. doi: 10.1530/REP-09-0039. [DOI] [PubMed] [Google Scholar]
- 6.Lee RK, Li SH, Lu CH, Ho HY, Chen YJ, Yeh HI. Abnormally low expression of connexin 37 and connexin 43 in subcutaneously transplanted cryopreserved mouse ovarian tissue. J Assist Reprod Genet. 2008;25:489–497. doi: 10.1007/s10815-008-9264-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Jin Y, Lin P, Hu L, Cui C, Li X, et al. The expression and localization of LRF in the female reproductive tract of cycling mice throughout the estrous cycle. J Immunoassay Immunochem. 2013;34:313–322. doi: 10.1080/15321819.2012.732169. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y, Ma H, Ma W, Sun M, Pu J, Yu J, et al. Expression and localization of SerpinB11 in mouse uteri during peri-implantation and the estrous cycle. Cell Tissue Res. 2014;357:373–380. doi: 10.1007/s00441-014-1829-5. [DOI] [PubMed] [Google Scholar]
- 9.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. [DOI] [PMC free article] [PubMed]
- 10.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 11.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, Zhang L, Li W, Han S, Yang W, Qi L. Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PLoS One. 2013;8:e53196. doi: 10.1371/journal.pone.0053196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 14.Zemp N, Minder A, Widmer A. Identification of internal reference genes for gene expression normalization between the two sexes in dioecious white Campion. PLoS One. 2014;9:e92893. doi: 10.1371/journal.pone.0092893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu J, Bian L, Zhao L, Dong Z, Gao X, Luan H, et al. Identification of genes for normalization of quantitative real-time PCR data in ovarian tissues. Acta Biochim Biophys Sin (Shanghai) 2010;42:568–574. doi: 10.1093/abbs/gmq062. [DOI] [PubMed] [Google Scholar]
- 16.Kolkova Z, Arakelyan A, Casslén B, Hansson S, Kriegova E. Normalizing to GADPH jeopardises correct quantification of gene expression in ovarian tumours—IPO8 and RPL4 are reliable reference genes. J Ovarian Res. 2013;6:60. doi: 10.1186/1757-2215-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li YL, Ye F, Hu Y, Lu WG, Xie X. Identification of suitable reference genes for gene expression studies of human serous ovarian cancer by real-time polymerase chain reaction. Anal Biochem. 2009;394:110–116. doi: 10.1016/j.ab.2009.07.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Preparation of medium (DOC 41 kb)