Abstract
Purpose
The pathogenesis of age-related macular degeneration (AMD) is associated with systemic and local inflammation. Various studies suggested that viral or bacterial infection may aggravate retinal inflammation in the aged retina. We compared the effects of synthetic viral RNA (poly(I:C)) and viral/bacterial DNA (CpG-ODN) on the expression of genes known to be involved in the development of AMD in retinal pigment epithelial (RPE) cells.
Methods
Cultured human RPE cells were stimulated with poly(I:C; 500 µg/ml) or CpG-ODN (500 nM). Alterations in gene expression and protein secretion were determined with real-time RT–PCR and ELISA, respectively. Phosphorylation of signal transduction molecules was revealed by western blotting.
Results
Poly(I:C) induced gene expression of the pattern recognition receptor TLR3, transcription factors (HIF-1α, p65/NF-κB), the angiogenic factor bFGF, inflammatory factors (IL-1β, IL-6, TNFα, MCP-1, MIP-2), and complement factors (C5, C9, CFB). Poly(I:C) also induced phosphorylation of ERK1/2 and p38 MAPK proteins, and the secretion of bFGF and TNFα from the cells. CpG-ODN induced moderate gene expression of transcription factors (p65/NF-κB, NFAT5) and complement factors (C5, C9), while it had no effect on the expression of various TLR, angiogenic factor, and inflammatory factor genes. The activities of various signal transduction pathways and transcription factors were differentially involved in mediating the poly(I:C)-induced transcriptional activation of distinct genes.
Conclusions
The widespread effects of viral RNA, and the restricted effects of viral/bacterial DNA, on the gene expression pattern of RPE cells may suggest that viral RNA rather than viral/bacterial DNA induces physiologic alterations of RPE cells, which may aggravate inflammation in the aged retina. The data also suggest that selective inhibition of distinct signal transduction pathways or individual transcription factors may not be effective to inhibit viral retinal inflammation.
Introduction
Age-related macular degeneration (AMD) is associated with chronic systemic and local inflammation [1], and with immunological abnormalities such as local activation of the alternative complement cascade [2,3]. Several components of complement and other proteins involved in immune-mediated processes and inflammation are present in drusen [4,5]. Several studies provided evidence for the suggestion that activated macrophages and other inflammatory cells may play a pathogenic role in drusen formation and choroidal neovascularization, hallmarks of dry and wet AMD, respectively [6-8].
Systemic inflammation and macrophage activation associated with retinal inflammation may be induced by multiple factors, including genetic and systemic health factors [9,10]. In addition, various studies using human material and animal models suggested that inflammatory processes in the aged retina might be aggravated by chronic systemic infections. Infectious pathogens, which have been suggested to promote retinal inflammation, include Chlamydia pneumoniae [11–13]; but see [14–17], the cytomegalovirus [18], and the Herpes simplex virus [19]. In addition, it has been suggested that Alu RNA, coded by retrotransposons, may play a role in the degeneration of the retinal pigment epithelium (RPE) in the aged retina [20,21]. It has been shown that injection of viral double-stranded RNA (dsRNA), a component of drusen [20], into the subretinal spaces of mice induces necrosis of the RPE and macrophage infiltration into the outer retina [22].
RPE cells are key players in the first-line retinal defense against invading pathogens, such as viruses and bacteria. Important mediators of microbial recognition are Toll-like receptors (TLRs). TLRs recognize conserved pathogen-associated molecular patterns (PAMPs) produced by viral, bacterial, and fungal pathogens [23]. It has been shown that RPE cells express various TLR subtypes, including TLR2, TLR3, and TLR9 [24-28]. TLR3 is the receptor for dsRNA, an intermediate of virus replication, while TLR9 recognizes unmethylated CpG dinucleotides, which are present at high density in viral and bacterial DNA. TLR2 recognizes certain endogenous antigens and various bacterial, fungal, and viral PAMPs, and mediates the host defense to Gram-positive bacteria and yeast. It has been shown that Chlamydia pneumoniae promotes experimental choroidal neovascularization via the activation of TLR2 in the RPE [26]. The activation of TLR2, TLR3, or TLR9 stimulates the production of proinflammatory cytokines and angiogenic factors, like vascular endothelial growth factor (VEGF), in RPE cells [25-27].
Although viral and/or bacterial infections are suggested to promote retinal inflammation [11,12,18], there is limited knowledge regarding the effects of viral and bacterial PAMPs on RPE cells. Therefore, the objective of the present study was to compare the effects of viral RNA and viral/bacterial DNA on the gene expression of angiogenic, inflammatory, and complement factors in RPE cells. We also compared the effects of viral RNA and viral/bacterial DNA on RPE cell activation by investigation of the gene expression of various transcription factors and of the phophorylation level of intracellular signal transduction molecules. To this end, we stimulated cultured human RPE cells with polyinosinic/polycytidylic acid (poly(I:C)), a synthetic dsRNA analog, and ODN 2006, a synthetic class B CpG oligonucleotide (CpG-ODN), respectively.
Methods
Materials
Tissue culture components and solutions were purchased from Gibco BRL (Paisley, UK). Platelet-derived growth factor (PDGF)-BB was purchased from R&D Systems (Abingdon, UK). The inhibitor of hypoxia-inducible transcription factor 1 (HIF-1) 3-[2-(4-adamantan-1-yl-phenoxy)-acetylamino]-4-hydroxybenzoic acid methyl ester, LY294002, PD98059, and SP600125 were obtained from Calbiochem (Bad Soden, Germany). Stattic was from Enzo Life Science (Lörrach, Germany). Caffeic acid phenethyl ester and SB203580 were from Tocris (Ellisville, MO). ODN 2006 (ODN 7909) was obtained from InvivoGen (San Diego, CA). Poly(I:C) and all other agents used were from Sigma-Aldrich (Taufkirchen, Germany), unless stated otherwise.
The following antibodies were used: a rabbit anti-human zonula occludens (ZO)-1 (1:50; Zymed Laboratory, San Francisco, CA); a rabbit anti-human occludin (1:50; Zymed Laboratory); a monoclonal anti-cytokeratin pan-FITC antibody that recognizes cytokeratins 4, 5, 6, 8, 10, 13, and 18 (1:50, Sigma-Aldrich); a Cy3-conjugated goat anti-rabbit IgG (1:200; Jackson Immuno Research, Suffolk, UK); a rabbit anti-human β-actin (1:2000, Cell Signaling, Frankfurt/M., Germany); rabbit anti-human extracellular signal-regulated kinases 1 and 2 (ERK1/2, p44/p42; 1:1000; Cell Signaling, Frankfurt/M., Germany); a rabbit anti-phosphorylated ERK1/2 (1:1000; Cell Signaling); a rabbit anti-human p38 mitogen-activated protein kinase (p38 MAPK; 1:1000; Cell Signaling); a rabbit anti-human phosphorylated p38 MAPK (1:750; Cell Signaling); a rabbit anti-human protein kinase B (Akt; Cell Signaling; 1:1000); a rabbit anti-human phosphorylated Akt (Cell Signaling; 1:1000); a rabbit anti-human signal transducer and activator of transcription 3 (STAT3; Cell Signaling; 1:1000); a rabbit anti-human phosphorylated STAT3 (Cell Signaling; 1:1000); and anti-rabbit IgG conjugated with alkaline phosphatase (1:2000; Chemicon, Hofheim, Germany).
Cell culture
The study followed the tenets of the Declaration of Helsinki for the use of human subjects. The use of human material was approved by the Ethics Committee of the University of Leipzig (approval #745, 07/25/2011). Human donor eyes were obtained from 12 females and 29 males within 48 h of death with written informed consent from relatives of the donors. The ages of the donors varied between 19 and 85 years (means ± SD, 62.7±19.7 years for females, and 59.3±19.0 years for males). There were no significant differences between data obtained in cells from younger and aged donor eyes, and in cells from both sexes (not shown).
RPE cells were prepared and cultured as following. After removing the vitreous and the retina, RPE cells were mechanically harvested, separated by digestion with 0.05% trypsin and 0.02% EDTA, and washed two times with phosphate-buffered saline. Cells were suspended in complete Ham F-10 medium containing 10% fetal bovine serum, glutamax II, and penicillin/streptomycin, and were cultured in tissue culture flasks (Greiner, Nürtingen, Germany) in 95% air/5% CO2 at 37 °C. Cells of passages 3 to 5 were used. Near-confluent cultures (confluency 80%–90%, reached after 4 days of cultivation) or confluent cultures (5 days of cultivation) were growth-arrested in medium without serum for 16 h, and subsequently, serum-free media with and without test substances were added. The cells were preincubated with the pharmacological inhibitors for 30 min. The confluency of the cultures was evaluated microscopically.
RNA extraction and cDNA synthesis
Total RNA was extracted with the InviTrap Spin Universal RNA Mini Kit (Stratec Molecular, Berlin, Germany). The quality of the RNA was analyzed by agarose gel electrophoresis. The A260/A280 ratio of the optical density was measured using the NanoDrop 1000 device (Peqlab, Erlangen, Germany), and was between 1.8 and 2.1 for all RNA samples, indicating sufficient quality. After treatment with DNase I (Roche, Mannheim, Germany), cDNA was synthesized from 1 µg of total RNA using the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas, St. Leon-Roth, Germany).
RT–PCR
PCR was performed using the Taq PCR Master Mix kit (Qiagen), and the primer pairs are described in Table 1. One µl of the first-strand mixture and 0.25 µM of each gene-specific sense and anti-sense primer were used for amplification in a final volume of 20 µl. Amplification was performed for 40 cycles with the PTC-200 Thermal Cycler (MJ Research, Watertown, MA). Each cycle consisted of 30 s at 94 °C, 60 s at 58 °C, and 1 min at 72 °C.
Table 1. Primer pairs used in PCR experiments. s, sense. as, anti-sense.
| Gene and accession | Primer sequence (5′→3′) | Amplicon (bp) |
|---|---|---|
| ACTB
NM_001101 |
s ATGGCCACGGCTGCTTCCAGC
as CATGGTGGTGCCGCCAGACAG |
237 |
| BFGF
NM_002006 |
s AGAGCGACCCTCACATCAAG
as ACTGCCCAGTTCGTTTCAGT |
234 |
| C3
NM_000064 |
s CTGCAGACCTCAGTGACCAA
as GCTCCGTTTCATCCAGGTAA |
207 |
| C5
NM_001735.2 |
s AGTGTGTGGAAGGGTGGAAG
as GTTCTCTCGGGCTTCAACAG |
222 |
| C9
NM_001737 |
s CAACTGGGCCTCTTCCATAA
as ACCTCCATTTTGGCATGTGT |
184 |
| CFB
NM_001710.5 |
s CTGGAGCACCCTGAAGACTC
as CCGGAGAGTGTAACCGTCAT |
172 |
| CFH
NM_000186.3 |
s CAGCAGTACCATGCCTCAGA
as GGATGCATCTGGGAGTAGGA |
189 |
| HBEGF NM_001945 |
s TGCCTGTAGCTTTCCTGGTCCC
as CCCCACCTCCAACCTTCTCGG |
258 |
| HIF1Α
NM_001530.3 |
s CACAGAAATGGCCTTGTGAA
as CCAAGCAGGTCATAGGTGGT |
214 |
| IL1B
NM_000576 |
s GGGCCTCAAGGAAAAGAATC
as TTCTGCTTGAGAGGTGCTGA |
205 |
| IL6
NM_000600 |
s TACCCCCAGGAGAAGATTCC
as TTTTCTGCCAGTGCCTCTTT |
175 |
| MCP1 NM_002982.3 |
s CCCCAGTCACCTGCTGTTAT
as TGGAATCCTGAACCCACTTC |
171 |
| MIP2
NM_002089.3 |
s CGCCCAAACCGAAGTCAT
as GATTTGCCATTTTTCAGCATCTTT |
111 |
| NFAT5
XM_005255777.1 |
s TCACCATCATCTTCCCACCT
as CTGCAATAGTGCATCGCTGT |
174 |
| RELA NM_001145138.1 |
s ATGGCTTCTATGAGGCTGAG
as GTTGTTGTTGGTCTGGATGC |
128 |
| STAT3 NM_003150.3 |
s ACCTGCAGCAATACCATTGAC
as AAGGTGAGGGACTCAAACTGC |
122 |
| TLR2
NM_003264.3 |
s ATGTCACAGGACAGCACTGG
as TTCTCCACCCAGTAGGCATC |
219 |
| TLR3
NM_003265.2 |
s GCTGGAAAATCTCCAAGAGC
as CTTCCAATTGCGTGAAAAC |
159 |
| TLR9
NM_017442.3 |
s CAGCAGCTCTGCAGTACGTC
as AAGGCCAGGTAATTGTCACG |
224 |
| TNFA
NM_000594 |
s AACCTCCTCTCTGCCATCAA
as CCAAAGTAGACCTGCCCAGA |
185 |
| VEGFA188, 164, 120 NM_003376.5 NM_001287044.1 NM_001025370.2 | s CCTGGTGGACATCTTCCAGGAGTA as CTCACCGCCTCGGCTTGTCACA | 479, 407, 275 |
Real-time RT–PCR
Real-time RT–PCR was performed with the Single-Color Real-Time PCR Detection System (BioRad, Munich, Germany) using the primer pairs described in Table 1. The PCR solution contained 1 µl of cDNA, a specific primer set (0.2 µM each), and 7.5 µl of a 2×mastermix (iQ SYBR Green Supermix, BioRad) in a final volume of 15 µl. The following conditions were used: initial denaturation and enzyme activation (one cycle at 95 °C for 3 min); denaturation, amplification, and quantification for 45 cycles at 95 °C for 30 s, 58 °C for 20 s, and 72 °C for 45 s; and a melting curve, 55 °C with the temperature gradually increased (0.5 °C) up to 95 °C. The amplified samples were analyzed by standard agarose gel electrophoresis. The mRNA expression was normalized to the level of β-actin mRNA. The changes in mRNA expression were calculated according to the 2-ΔΔCT method (CT, cycle threshold), with ΔCT=CTtarget gene - CTactb and ΔΔCT=ΔCTtreatment - ΔCTcontrol.
Western blot analysis
Cells were seeded at 5×105 cells per well in six-well plates in 1.5 ml of complete medium, and were allowed to growth up to a confluency of ~80%. After growth arrest for 16 h, the cells were treated with test substances for 20 min and 1 h, respectively. Then, the medium was removed, the cells were washed twice with prechilled phosphate-buffered saline (pH 7.4; Invitrogen, Paisley, UK), and the monolayer was scraped into 150 µl of lysis buffer (Mammalian Cell Lysis-1 Kit; Sigma). Total cell lysates were centrifuged at 10,000 × g for 10 min, and the supernatants were analyzed by immunoblots. Equal amounts of protein (30 µg) were separated by 10% SDS–PAGE. Immunoblots were probed with primary and secondary antibodies, and immunoreactive bands were visualized using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium.
ELISA
Cells were stimulated with poly(I:C; 500 µg/ml) in a serum-free medium. The supernatants were collected after 6 and 24 h, and the levels of VEGF-A165, basic fibroblast growth factor (bFGF), heparin-binding epidermal growth factor-like growth factor (HB-EGF), and tumor necrosis factor α (TNFα), respectively, in the cultured media (200 µl) were determined with ELISA (R&D Systems).
Immunocytochemistry
For immunolabeling, the cultures were fixed in 1% paraformaldehyde for 20 min on ice. After several washing steps in buffered saline, the cultures were incubated for 15 min in buffered saline containing 1% DMSO and 0.3% Triton X-100. Blocking was performed in buffered saline containing 5% normal goat serum, 1% DMSO, and 0.3% Triton X-100 for 2 h at room temperature, and subsequently, in the primary antibody overnight at 4 °C. After washing in saline plus 1% DMSO and 0.3% Triton X-100, the secondary antibody and Hoechst 33258 were applied for 1 h at room temperature. Images were taken with a confocal laser scanning microscope (LSM 510 Meta; Zeiss, Oberkochen, Germany).
Statistics
For each test, at least three independent experiments using cells from different donors were performed. Data are expressed as means ± SEM. Statistical analysis was made using Prism (Graphpad Software, San Diego, CA). Significance was determined by one-way ANOVA followed by Bonferroni’s multiple comparison test and the Mann–Whitney U test, respectively, and was accepted at p<0.05.
Results
Cultures of human RPE cells
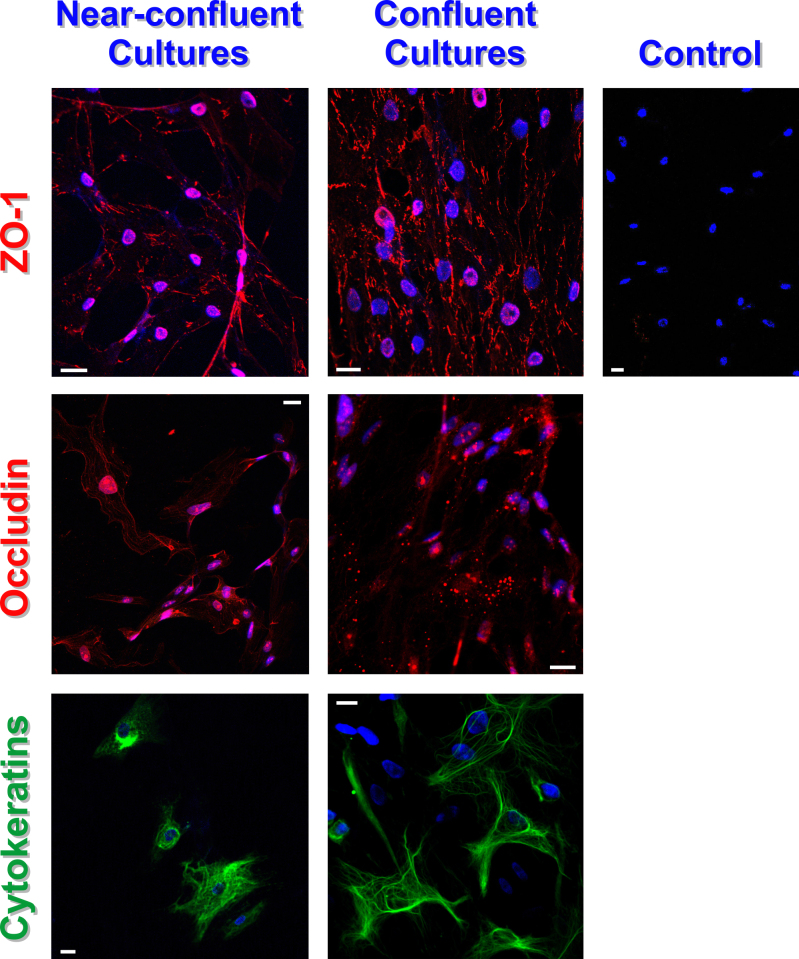
The experiments were performed with near-confluent and confluent cultures of human RPE cells. The epithelial nature of the cells was identified by immunolabeling for cytokeratins (Figure 1). In different near-confluent and confluent cultures, 70%–85% of all cells investigated displayed immunoreactivity for cytokeratins. The expression of the tight junction proteins ZO-1 and occludin (Figure 1) suggests that the cells form tight junctions. The tight junction proteins were distributed in a clustered fashion in the plasma membranes and the nuclei of the cells (Figure 1).
Figure 1.
Immunolabeling of ZO-1 (red), occludin (red), and cytokeratins (green) in near-confluent and confluent cultures of human RPE cells. Cell nuclei were labeled with Hoechst 33258 (blue). The negative control (right) was stained without primary antibodies. Bars, 20 µm.
Expression of TLR genes
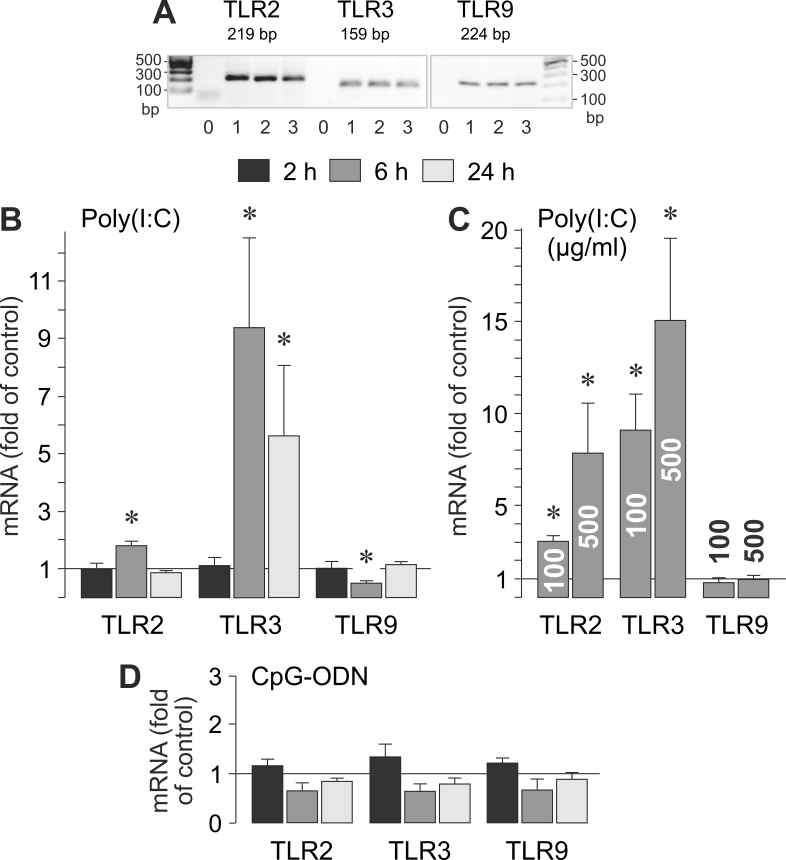
To determine whether viral RNA and viral/bacterial DNA induce alterations in the gene expression patterns of RPE cells, we stimulated the cells with the synthetic viral dsRNA analog poly(I:C) and the viral/bacterial DNA analog CpG-ODN. To examine the effects on the gene expression of pattern recognition receptors, we determined the cellular levels of TLR2, TLR3, and TLR9 mRNAs (Figure 2A). As shown in Figure 2B,C, poly(I:C) induced a significant (p<0.05) time-dependent increase in the expression of the TLR3 gene. In near-confluent cultures, the levels of TLR2 and TLR9 mRNAs were moderately altered in the presence of poly(I:C). In confluent cultures, poly(I:C) induced a significant (p<0.05) increase in the expression of the TLR2 gene and no alteration in the expression of the TLR9 gene (Figure 2C). On the other hand, CpG-ODN did not significantly (p>0.05) alter the cellular levels of TLR2, TLR3, and TLR9 mRNAs (Figure 2D). The data suggest that viral RNA induces upregulation of TLR3 and, to a lower degree, TLR2 in RPE cells.
Figure 2.
Effects of viral RNA and viral/bacterial DNA on the expression of TLR2, TLR3, and TLR9 genes in RPE cells. A: Expression of TLR genes in cells cultured for 2 (1), 6 (2), and 24 h (3), as determined by RT–PCR. Negative controls (0) were done by adding double-distilled water instead of cDNA as a template. B-D: The mRNA levels were determined with real-time RT–PCR analysis after stimulation of the cells for 2, 6, and 24 h (as indicated by the panels of the bars), and are expressed as folds of unstimulated controls. B: Near-confluent cultures were stimulated with poly(I:C; 500 µg/ml). C: Confluent cultures were stimulated with poly(I:C; 100 and 500 µg/ml, respectively). D: Near-confluent cultures were stimulated with CpG-ODN (500 nM). Each bar represents data obtained in 3 to 5 independent RPE cell lines, each from a different human eye donor; experiments with each cell line were carried out in triplicate. Significant difference versus unstimulated controls: *p<0.05.
Expression of transcription factor genes
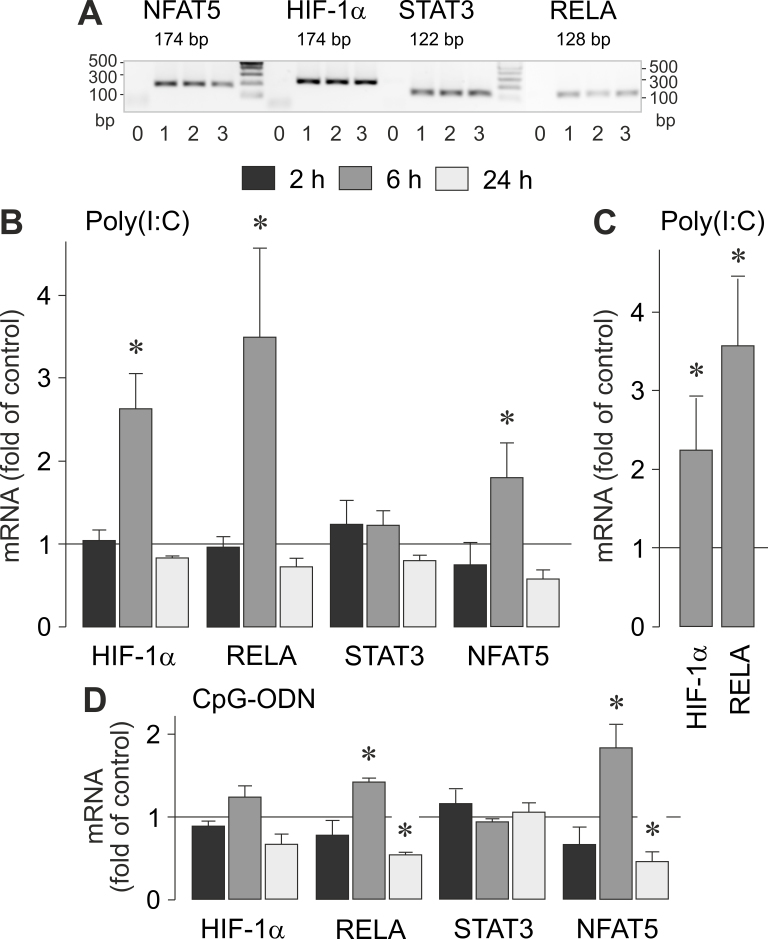
To determine whether viral RNA and viral/bacterial DNA induce activation of RPE cells, we examined the cellular levels of various transcription factor genes (Figure 3A). As shown in Figure 3B,C, stimulation of the cells with the synthetic viral RNA analog poly(I:C) induced time-dependent increases in the gene expression levels of HIF-1α, v-rel avian reticuloendotheliosis viral oncogene homolog A (RELA, the p65 subunit of nuclear factor [NF]-κB), and the nuclear factor of activated T cell 5 (NFAT5). Stimulation with poly(I:C) had no significant (p>0.05) effect on the expression of the STAT3 gene (Figure 3B). Synthetic viral/bacterial DNA (CpG-ODN) induced moderate alterations in the expression of RELA and NFAT5 genes, with up- and downregulation after 6 and 24 h of stimulation, respectively (Figure 3D). The data suggest that both viral RNA and viral/bacterial DNA induce alterations in the transcriptional activation of various transcription factor genes in RPE cells, including HIF-1α (viral RNA), p65/NF-κB (RNA and DNA), and NFAT5 (RNA and DNA).
Figure 3.
Effects of viral RNA and viral/bacterial DNA on the gene expression of the transcription factor proteins HIF-1α, RELA (p65/NF-κB), STAT3, and NFAT5. A: Gene expression of transcription factor proteins in cells cultured for 2 (1), 6 (2), and 24 h (3), as determined by RT–PCR. Negative controls (0) were done by adding double-distilled water instead of cDNA as a template. B-D: The mRNA levels were determined with real-time RT–PCR analysis after stimulation of near-confluent (B, D) and confluent (C) cultures for 2, 6, and 24 h (as indicated by the panels of the bars), and are expressed as fold of unstimulated controls. The cells were stimulated with poly(I:C; 500 µg/ml; B, C) and CpG-ODN (500 nM; D), respectively. Each bar represents data obtained in 3 to 7 independent RPE cell lines, each from a different human eye donor; experiments with each cell line were carried out in triplicate. Significant difference versus unstimulated controls: *p<0.05.
Expression of angiogenic growth factor genes
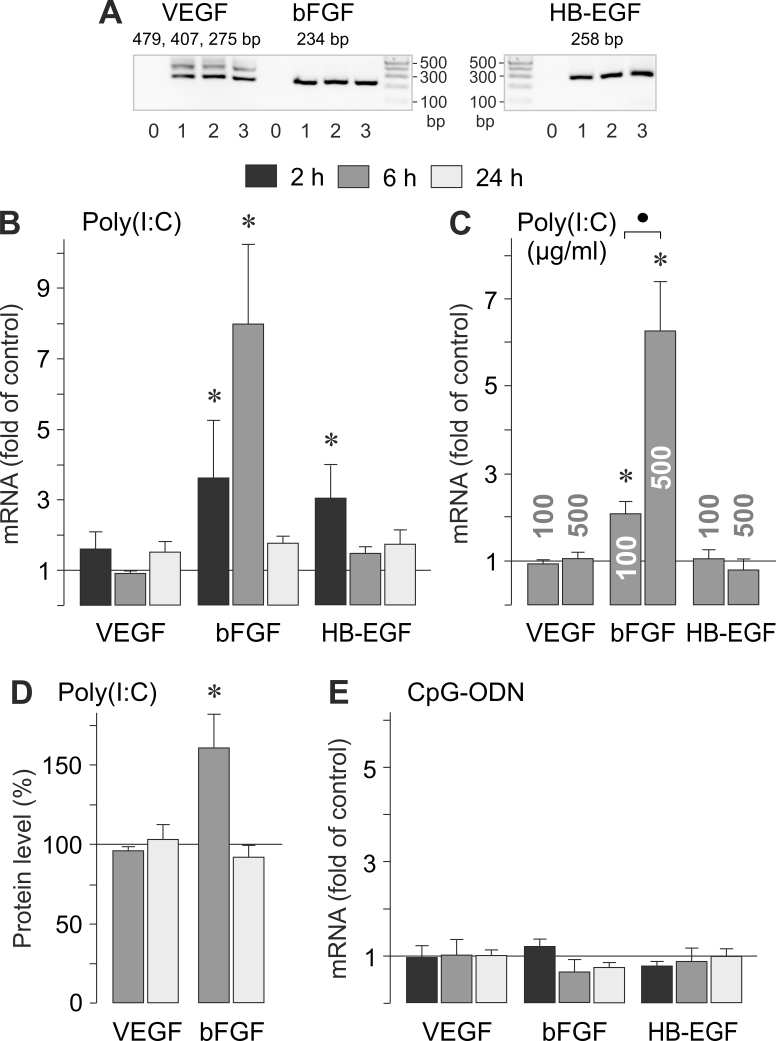
Progression of AMD to the neovascular stage is driven by angiogenic growth factors such as VEGF, bFGF, and HB-EGF, which are produced, for example, by RPE cells [29-31]. We compared the effects of viral RNA and viral/bacterial DNA on the gene expression of these factors in RPE cells (Figure 4A). As shown in Figure 4B,C, the synthetic analog of viral RNA poly(I:C) induced a significant (p<0.05) increase in the expression of the bFGF gene in RPE cells and had no effect on the expression of the VEGF gene. Poly(I:C) also induced a moderate increase in the expression of the HB-EGF gene in non-confluent cultures (Figure 4B). The administration of poly(I:C) also induced a significant (p<0.05) increase in the secretion of the bFGF protein from RPE cells, while it had no effect on the secretion of VEGF protein (Figure 4D). We did not find detectable amounts of soluble HB-EGF protein with ELISA in the media of cells cultured for 6 or 24 h in the absence and presence of poly(I:C; 500 µM; not shown). Stimulation of RPE cells with the synthetic analog of viral/bacterial DNA, CpG-ODN, did not alter the cellular levels of VEGF, bFGF, and HB-EGF mRNAs (Figure 4E). The data suggest that viral RNA induces expression and secretion of bFGF.
Figure 4.
Effects of viral RNA and viral/bacterial DNA on gene expression and the secretion of angiogenic growth factors. The mRNA levels (B, C, E) were determined with real-time RT–PCR analysis after stimulation of the cells for 2, 6, and 24 h (as indicated by the panels of the bars), and are expressed as fold of unstimulated controls. The levels of VEGF-A165 and bFGF proteins (D) were determined with ELISA in the cultured media of cells stimulated for 6 and 24 h, and are expressed in percent of unstimulated controls (100%). A: Expression of growth factor genes in cells cultured for 2 (1), 6 (2), and 24 h (3), as determined by RT–PCR. Negative controls (0) were done by adding double-distilled water instead of cDNA as a template. B, D: Near-confluent cultures were stimulated with poly(I:C; 500 µg/ml). C. Dose-dependent effect of poly(I:C). Confluent cultures were stimulated with poly(I:C; 100 and 500 µg/ml, respectively). E: Near-confluent cultures were stimulated with CpG-ODN (500 nM). Each bar represents data obtained in 3 to 5 independent RPE cell lines, each from a different human eye donor; experiments with each cell line were carried out in triplicate. Significant difference versus unstimulated controls: *p<0.05. ●, p<0.05.
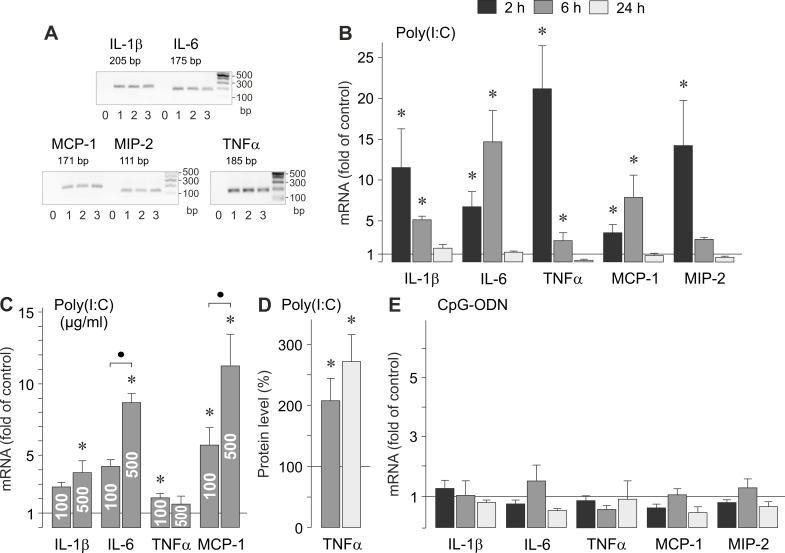
Expression of proinflammatory cytokine genes
AMD is associated with systemic and local inflammation [1]. We compared the effects of viral RNA and viral/bacterial DNA on the gene expression of proinflammatory cytokines in RPE cells (Figure 5A). As shown in Figure 5B,C, stimulation of the cells with the synthetic analog of viral RNA poly(I:C) induced significant (p<0.05) increases in the gene expression of interleukin (IL)-1β, IL-6, TNFα, monocyte chemoattractant protein (MCP)-1, and macrophage inflammatory protein (MIP)-2. The time-dependency of the poly(I:C)-induced gene expression differed among various inflammatory cytokines. While the increases in the IL-1β, TNFα, and MIP-2 mRNA levels were largest after 2 h of stimulation with poly(I:C), the increases in the expression of IL-6 and MCP-1 genes were more pronounced after 6 h of stimulation (Figure 5B). Poly(I:C) also induced a strong increase in the secretion of TNFα protein from RPE cells (Figure 5D). The analog of viral/bacterial DNA, CpG-ODN did not induce significant (p>0.05) alterations in the gene expression of inflammatory cytokines (Figure 5E). The data suggest that viral RNA induces gene expression of various inflammatory cytokines in RPE cells, and the secretion of TNFα.
Figure 5.
Effects of viral RNA and viral/bacterial DNA on the gene expression of inflammatory cytokines. The mRNA levels (B, C, E) were determined with real-time RT–PCR analysis after stimulation of the cells for 2, 6, and 24 h (as indicated by the panels of the bars), and are expressed as fold of unstimulated controls. The level of TNFα protein (D) was determined with ELISA in the cultured media of cells stimulated for 6 and 24 h, and are expressed in percent of unstimulated controls (100%). A: Expression of inflammatory protein genes in cells cultured for 2 (1), 6 (2), and 24 h (3), as determined by RT–PCR. Negative controls (0) were done by adding double-distilled water instead of cDNA as a template. B, D: Near-confluent cultures were stimulated with poly(I:C; 500 µg/ml). C: Dose-dependent effect of poly(I:C). Confluent cultures were stimulated with poly(I:C; 100 and 500 µg/ml, respectively). E: Near-confluent cultures were stimulated with CpG-ODN (500 nM). Each bar represents data obtained in 3 to 5 independent RPE cell lines, each from a different human eye donor; experiments with each cell line were carried out in triplicate. Significant difference versus unstimulated controls: *p<0.05. ●, p<0.05.
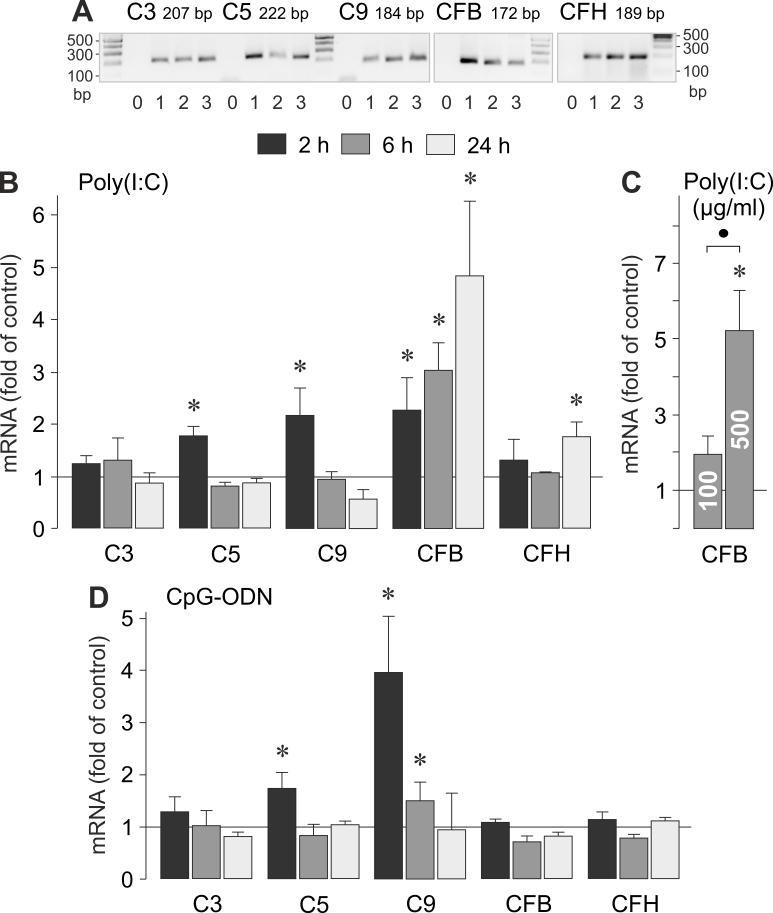
Expression of complement factor genes
Age-related retinal inflammation is associated with local activation of the alternative complement cascade [2,3] and subretinal deposition of activated complement proteins such as C3a and C5a [32]. We compared the effects of viral RNA and viral/bacterial DNA on the gene expression of various complement proteins in RPE cells (Figure 6A). As shown in Figure 6B, the synthetic viral RNA analog poly(I:C) induced significant (p<0.05) time-dependent increases in the expression of C5, C9, CFB, and CFH genes, and had no effect on the expression of the C3 gene. The effect of poly(I:C) was more pronounced in respect to the CFB gene than to the other complement factor genes investigated (Figure 6B,C). Stimulation of RPE cells with the viral/bacterial DNA analog CpG-ODN induced increased expression of C5 and C9 genes, while it had no effect on the expression of C3, CFB, and CFH genes (Figure 6D). The data suggest that both viral RNA and viral/bacterial DNA induce the expression of distinct complement factor genes in RPE cells.
Figure 6.
Effects of viral RNA and viral/bacterial DNA on the expression of complement factor genes. The mRNA levels were determined with real-time RT–PCR analysis after stimulation of the cells for 2, 6, and 24 h (as indicated by the panels of the bars), and are expressed as fold of unstimulated controls. A: Expression of complement factor genes in cells cultured for 2 (1), 6 (2), and 24 h (3), as determined by RT–PCR. Negative controls (0) were done by adding double-distilled water instead of cDNA as a template. B: Near-confluent cultures were stimulated with poly(I:C; 500 µg/ml). C: Dose-dependent effect of poly(I:C) on the expression of the CFB gene. Confluent cultures were stimulated with poly(I:C; 100 and 500 µg/ml, respectively). D: Near-confluent cultures were stimulated with CpG-ODN (500 nM). Each bar represents data obtained in three to five independent experiments using cells from different donors. Each bar represents data obtained in 3 to 5 independent RPE cell lines, each from a different human eye donor; experiments with each cell line were carried out in triplicate: *p<0.05. ●, p<0.05.
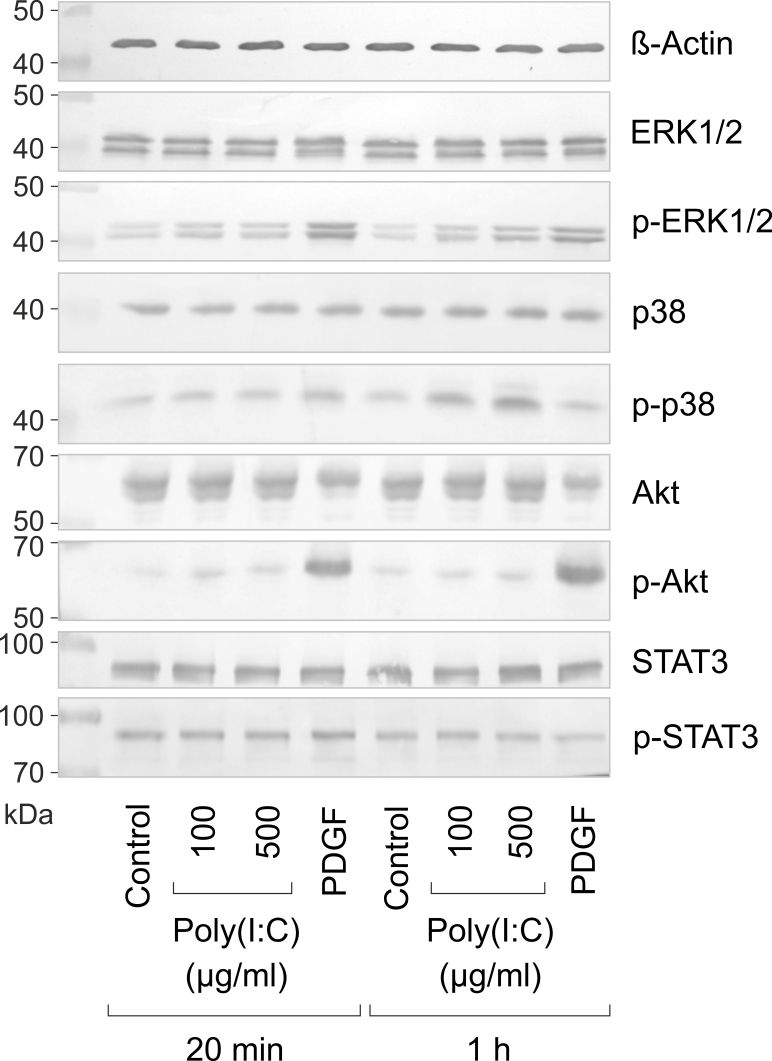
Activation of intracellular signal transduction molecules
We found that the synthetic analog of viral RNA, poly(I:C), induces the expression of various different genes in RPE cells. To determine whether poly(I:C) also induces activation of intracellular signal transduction pathways, we examined with western blot analysis the phosphorylation levels of ERK1/2, p38 MAPK, and Akt proteins. As shown in Figure 7, poly(I:C) induced dose-dependent increases in the phosphorylation levels of ERK1/2 and p38 MAPK proteins. The effects of poly(I:C) were more pronounced after 1 h than after 20 min of stimulation (Figure 7). Poly(I:C) also induced a moderate increase in the phosphorylation level of the Akt protein, which was apparent after 20 min, but not after 1 h of stimulation (Figure 7). In contrast, poly(I:C) did not induce an alteration in the phosphorylation level of STAT3 protein (Figure 7). In agreement with previous studies [33,34], the positive control, PDGF, induced alterations in the phosphorylation level of all proteins investigated (Figure 7). The data suggest that viral RNA induces activation of various signal transduction pathways in RPE cells.
Figure 7.
Effects of viral RNA on the phosphorylation levels of ERK1/2, p38 MAPK, Akt, and STAT3 proteins in RPE cells. Near-confluent cultures were stimulated for 20 min and 1 h, respectively, with 100 and 500 µg/ml of poly(I:C). PDGF (10 ng/ml) was used as a positive control, and β-actin as a control for equal protein loading. Amounts of total proteins are shown above, while amounts of phosphorylated proteins are shown below. Similar results were obtained in three independent experiments using RPE cell lines from different human eye donors.
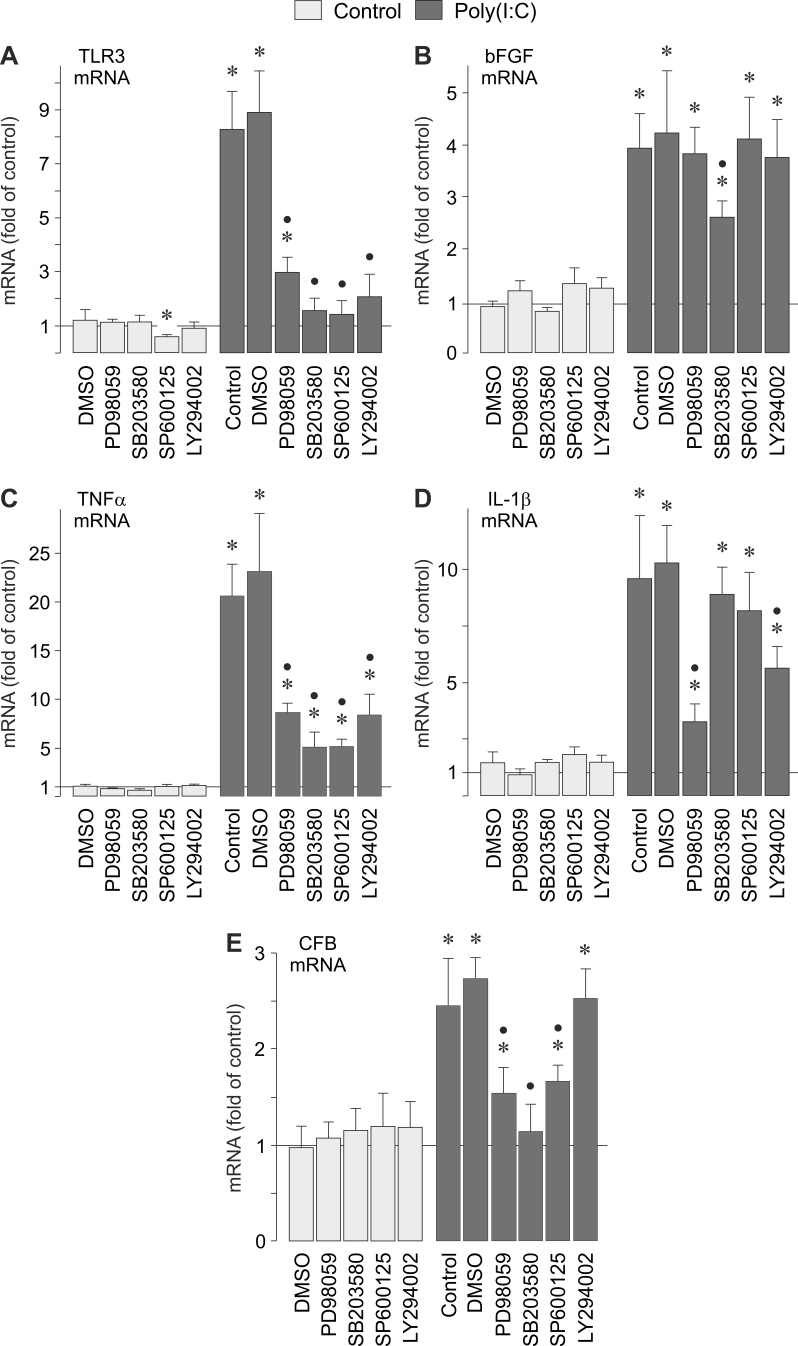
Intracellular signaling involved in poly(I:C)-induced gene expression
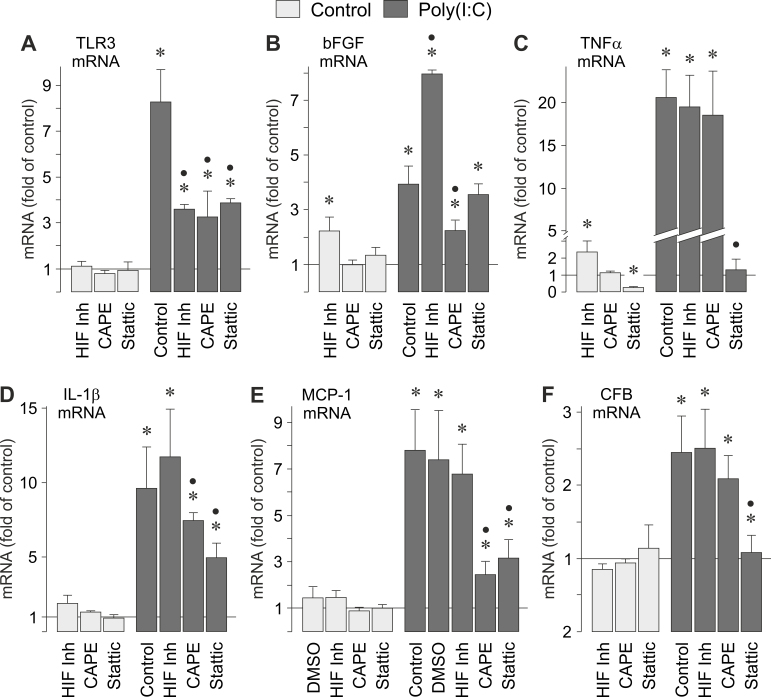
We found that the synthetic analog of viral RNA, poly(I:C), induced activation of different signal transduction molecules in RPE cells (Figure 7). To determine the intracellular signaling that regulates the poly(I:C)-induced gene expression, we tested pharmacological blockers of key intracellular signal transduction molecules. Inhibition of ERK1/2 activation by PD98059 significantly (p<0.05) reduced the poly(I:C)-induced expression of TLR3 (Figure 8A), TNFα (Figure 8C), IL-1β (Figure 8D), and CFB (Figure 8E) genes, while it had no effect on the poly(I:C)-induced expression of the bFGF gene (Figure 8B). The inhibition of p38 MAPK activity by SB203580 resulted in a significant (p<0.05) decrease of the poly(I:C)-induced expression of the following genes: TLR3 (Figure 8A), bFGF (Figure 8B), TNFα (Figure 8C), and CFB (Figure 8E). The inhibition of p38 MAPK activity had no effect on the poly(I:C)-induced expression of the IL-1β gene (Figure 8D). Inhibition of the c-Jun NH2-terminal kinase (JNK) activity by SP600125 resulted in significant (p<0.05) reduction of the poly(I:C)-induced expression of TLR3 (Figure 8A), TNFα (Figure 8C), and CFB (Figure 8E) genes, while it had no effect on the poly(I:C)-induced expression of bFGF (Figure 8B) and IL-1β (Figure 8D) genes. The inhibitor of the phosphatidylinositol-3 kinase (PI3K)-Akt pathway, LY294002, reduced significantly (p<0.05) the poly(I:C)-induced expression of TLR3 (Figure 8A), TNFα (Figure 8C), and IL-1β (Figure 8D) genes, and had no effect on the poly(I:C)-induced expression of bFGF (Figure 8B) and CFB (Figure 8E) genes. The data suggest that the poly(I:C)-induced expression of various genes in RPE cells is differentially regulated by distinct signal transduction pathways.
Figure 8.
Intracellular signaling involved in poly(I:C; 500 µg/ml)-induced gene expression in RPE cells. The cellular levels of the following gene transcripts were determined: TLR3 (A), bFGF (B), TNFα (C), IL-1β (D), and CFB (E) mRNAs. mRNA levels were determined with real-time RT–PCR analysis after stimulation of near-confluent cultures for 2 h (TNFα and IL-1β mRNAs) and 6 h (TLR3, bFGF, and CFB mRNAs), respectively, and are expressed as folds of unstimulated controls. The following blocking agents were tested: the inhibitor of ERK1/2 activation, PD98059 (20 µM); the inhibitor of p38 MAPK activation, SB203580 (10 µM); the JNK inhibitor SP600125 (10 µM); and the inhibitor of PI3K-related kinases, LY294002 (5 µM). Vehicle control was made with DMSO (DMSO; 1:1000). Each bar represents data obtained in 3 to 9 independent RPE cell lines, each from a different human eye donor; experiments with each cell line were carried out in triplicate. Significant difference versus unstimulated controls: *p<0.05. Significant difference versus poly(I:C) controls: ●p<0.05.
Transcription factor activities involved in poly(I:C)-induced gene expression
Because poly(I:C) also induced the expression of various transcription factor genes (Figure 3B), we examined the involvement of transcription factor activities in the poly(I:C)-induced gene expression. To determine the role of HIF-1 activity, we tested an HIF inhibitor [35]. The HIF inhibitor decreased significantly (p<0.05) the poly(I:C)-induced transcriptional activation of the TLR3 gene (Figure 9A), and had no effect on the poly(I:C)-induced expression of TNFα (Figure 9C), IL-1β (Figure 9D), MCP-1 (Figure 9E), and CFB (Figure 9F) genes. The HIF inhibitor increased significantly (p<0.05) the expression of the bFGF gene under control and poly(I:C)-stimulated conditions (Figure 9B). The NF-κB inhibitor, caffeic acid phenethyl ester [36], decreased significantly (p<0.05) the poly(I:C)-induced expression of TLR3 (Figure 9A), bFGF (Figure 9B), and IL-1β (Figure 9D) genes, and had no effect on the poly(I:C)-induced expression of TNFα (Figure 9C), MCP-1 (Figure 9E), and CFB (Figure 9F) genes. The STAT3 inhibitor Stattic [37] reduced significantly (p<0.05) the poly(I:C)-induced expression of TLR3 (Figure 9A), TNFα (Figure 9C), IL-1β (Figure 9D), MCP-1 (Figure 9E), and CFB (Figure 9F) genes. Stattic had no effect on the poly(I:C)-induced expression of the bFGF gene (Figure 9B). The data suggest that the poly(I:C)-induced gene expression in RPE cells is differentially regulated by distinct transcription factor activities.
Figure 9.
Involvement of transcription factor activities in the poly(I:C; 500 µg/ml)-induced gene expression in RPE cells. The cellular levels of the following gene transcripts were determined: TLR3 (A), bFGF (B), TNFα (C), IL-1β (D), MCP-1 (E), and CFB (F) mRNAs. mRNA levels were determined with real-time RT–PCR analysis after stimulation of near-confluent cultures for 2 h (TNFα and IL-1β mRNAs) and 6 h (TLR3, bFGF, MCP-1, and CFB mRNAs), respectively, and are expressed as folds of unstimulated controls. The following blocking agents were tested: an HIF inhibitor (HIF-Inh; 5 µM), the NF-κB inhibitor caffeic acid phenethyl ester (CAPE; 5 µM), and the STAT3 inhibitor Stattic (1 µM). Vehicle control was made with DMSO (DMSO; 1:1000). Each bar represents data obtained in 3 to 8 independent RPE cell lines, each from a different human eye donor; experiments with each cell line were carried out in triplicate. Significant difference versus unstimulated controls: *p<0.05. Significant difference versus poly(I:C) controls: ●p<0.05.
Discussion
AMD is associated with systemic and local inflammation, macrophage activation, and other immunological abnormalities, such as subretinal deposition of activated complement proteins [1,4-7]. It was suggested that microbial pathogens, including bacterial (Chlamydia pneumoniae) and viral pathogens of exogenous (cytomegalovirus, Herpes simplex) and endogenous origin (Alu RNA), may aggravate inflammation in the aged retina [11-13,18-21]. It has been also shown in mice that short-interfering RNAs induce retinal degeneration by activating TLR3 in RPE cells [38]. Conflicting data were published regarding a potential association between AMD and Chlamydia infection [14-17]. The objective of the present study was to compare the effects of viral RNA and viral/bacterial DNA on the gene expression of proteins, which may aggravate inflammatory processes in the aged retina, in cultured human RPE cells. We found that the synthetic analog of viral RNA, poly(I:C), induced upregulation of TLR3 and, to a lower degree, TLR2 in RPE cells (Figure 2B,C). TLR3 is a pattern recognition receptor known to be activated by viral dsRNA [23], and TLR2 recognizes viral antigens, e.g., cytomegalovirus antigens, in addition to bacterial, fungal, and certain endogenous antigens. We also found that viral RNA induced the gene expression of various different proteins in RPE cells, which may aggravate inflammatory processes in situ, including the angiogenic factor bFGF (Figure 4B,C), inflammatory cytokines (Figure 5B,C), and complement proteins (Figure 6B,C). In comparison to the effects of poly(I:C), the effects of the synthetic analog of viral/bacterial DNA, CpG-ODN, on the gene expression of RPE cells were much more restricted. It cannot be fully ruled out that some differences between the effects of viral RNA and viral/bacterial DNA found in the present study result from different concentrations of the agents used. However, the stimulatory effects of CpG-ODN on the expression of distinct genes, e.g., C5 and C9 genes (Figure 6D), may suggest that different concentrations of the agents used are not a major cause of functional differences. The present results are in agreement with a previous study, which showed that activation of TLR3 (the receptor for viral dsRNA) has widespread effects on the production of inflammatory factors in RPE cells, while the activation of TLR9 (which recognizes unmethylated CpG dinucleotides in viral and bacterial DNA) has limited effects [25].
Activated macrophages are proposed to play a pathogenic role in age-related retinal inflammation [6-8]. Subretinal injection of viral RNA in mice induces macrophage infiltration into the outer retina [22]. The present data are in agreement with previous studies, which showed that viral RNA induces the production of inflammatory factors like IL-6 and TNFα, and chemoattractants like MCP-1, in RPE cells [22,24,25,27]. We show that viral RNA also induces the expression of further inflammatory factors like IL-1β and MIP-2 (Figure 5B), as well as secretion of TNFα from RPE cells (Figure 5D). The widespread effects of viral RNA on the expression of inflammatory factors in RPE cells may suggest that inhibition of TLR3 signaling may protect from age-related inflammatory processes in the retina. A protective effect of TLR3 knockdown was recently shown in experimental autoimmune uveitis [27].
Viral RNA (Figure 4B-D), but not viral/bacterial DNA (Figure 4E), induced expression and secretion of the angiogenic factor bFGF in RPE cells. On the other hand, the expression and secretion of VEGF was not stimulated by viral RNA (Figure 4B-D). The lack of an effect of viral/bacterial DNA on the gene expression of VEGF (Figure 4E) is in contradiction to a previous study that showed that Chlamydia pneumoniae antigens induce the expression of the VEGF gene in cultured murine RPE cells [26]. The reason for the different results is unknown and may be related to different stimulation conditions and the use of cells from different species. The stimulatory effects of viral RNA on the expression and secretion of bFGF (Figure 4B-D) suggest that early stages of pathological neovascularization in the aged retina may be aggravated by viral induction of bFGF. This assumption is in agreement with a previous study, which showed that bFGF is an initial stimulator of experimental choroidal neovascularization, while VEGF is important during later phases of angiogenesis [39].
Age-related retinal inflammation is associated with local activation of the alternative complement cascade [2,3] and subretinal deposition of activated complement proteins [32]. The alternative complement pathway is known to be mainly activated by microbial pathogens. In the present study, we show that both viral RNA (Figure 6B,C) and viral/bacterial DNA (Figure 6D) induce the expression of distinct complement factor genes. Among the various complement factor genes activated by viral RNA, the effect on the expression of the CFB gene was more pronounced (Figure 6B). CFB is the main activator of the alternative complement pathway; overproduction of CFB in RPE cells may result in local overactivation of the complement system.
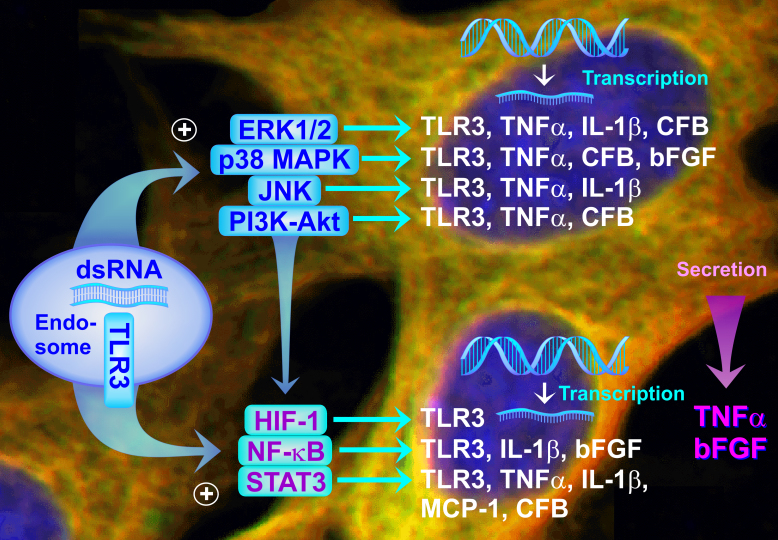
Viral RNA induces the activation of RPE cells, as indicated by the increased phosphorylation levels of intracellular signal transduction molecules and elevated expression of distinct transcription factor genes (Figure 10). The activities of ERK1/2, p38 MAPK, JNK, and PI3K, and of various transcription factors, are differentially involved in mediating the poly(I:C)-induced transcriptional activation of distinct genes (Figure 10). The data suggest that selective inhibition of distinct intracellular signal transduction pathways or of individual transcription factors may not be effective to inhibit viral inflammation of the retina.
Figure 10.
Schematic summary of the effects of viral dsRNA on the gene expression of cultured human RPE cells. Activation of the pattern recognition receptor TLR3 by dsRNA results in activation of various signal transduction pathways (e.g., ERK1/2, p38 MAPK, JNK, PI3K-Akt) which are differentially involved in inducing the gene expression of TLR3, inflammatory factors such as TNFα and IL-1β, complement factors such as CFB, and the angiogenic factor bFGF. The expression of various genes is differentially induced by the transcription factors HIF-1, NF-κB, and STAT3. In addition to gene expression induction, dsRNA induces the secretion of inflammatory (TNFα) and angiogenic factors (bFGF) from the cells. It remains to be determined whether dsRNA also induces a secretion of further proteins, such as CFB, IL-1β, and MCP-1.
We found different time dependencies of the poly(I:C)-induced gene expression of various angiogenic (Figure 4B) and inflammatory proteins (Figure 5B). The different time-dependencies well reflect the classification of TLR-induced genes into primary response genes, like the TNFα gene, and secondary response genes, like the IL-6 gene (Figure 5B) [40]. Differences in the time dependency of transcriptional gene activation were shown to result from different modes of association of transcription factors with target genes and the requirements of de novo protein synthesis and nucleosome remodeling.
In the present study, we used near-confluent and confluent RPE cell cultures. Both types of cultures expressed the RPE cell marker cytokeratins and the tight junctional proteins ZO-1 and occludin (Figure 1). The expression of these proteins suggests that the cells used display distinct features of differentiated RPE cells. With the exception of TLR9 gene expression (Figure 2C), we found similar dsRNA-induced transcriptional alterations in near-confluent and confluent RPE cell cultures, including the altered expression of TLR2 and TLR3 genes (Figure 2C), HIF-1α and p65/NF-κB genes (Figure 3C), the bFGF gene (Figure 4C), the CFB gene (Figure 6C), and different inflammatory factor genes (Figure 5C). The similar results may suggest that near-confluent cultures can be used to investigate some aspects of RPE cell physiology, which may have relevance for the understanding of pathogenic mechanisms in the aged retina. However, whether the present results have relevance for the understanding of the RPE cell physiology in vivo remains to be determined in future investigations. Nevertheless, the present results may serve as basis for further research regarding a possible contribution of viral infections to the development of age-related retinal inflammation.
The present results suggest that viral RNA, rather than viral/bacterial DNA, induces activation and alterations in gene expression of RPE cells, which may aggravate retinal inflammation (Figure 10). The results support previous suggestions that viral RNA, either of exogenous (cytomegalovirus, Herpes simplex) or endogenous origin (Alu RNA), may play a role in the aggravation of inflammatory processes in the retina [18-21]. However, further research is required to prove whether the retinal/subretinal presence of viral or Alu RNA contributes to retinal inflammation in the aged retina.
Acknowledgments
The authors thank Ute Weinbrecht for excellent technical assistance. This study was supported by grants from the Deutsche Forschungsgemeinschaft (KO 1547/7–1 to L.K.) and the Geschwister Freter Stiftung (Hannover, Germany).
References
- 1.Cheung CM, Wong TY. Is age-related macular degeneration a manifestation of systemic disease? New prospects for early intervention and treatment. J Intern Med. 2014;276:140–53. doi: 10.1111/joim.12227. [DOI] [PubMed] [Google Scholar]
- 2.Johnson LV, Leitner WP, Staples MK, Anderson DH. Complement activation and inflammatory processes in Drusen formation and age related macular degeneration. Exp Eye Res. 2001;73:887–96. doi: 10.1006/exer.2001.1094. [DOI] [PubMed] [Google Scholar]
- 3.Anderson DH, Mullins RF, Hageman GS, Johnson LV. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002;134:411–31. doi: 10.1016/s0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- 4.Anderson DH, Radeke MJ, Gallo NB, Chapin EA, Johnson PT, Curletti CR, Hancox LS, Hu J, Ebright JN, Malek G, Hauser MA, Rickman CB, Bok D, Hageman GS, Johnson LV. The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog Retin Eye Res. 2010;29:95–112. doi: 10.1016/j.preteyeres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stanton CM, Wright AF. Inflammatory biomarkers for AMD. Adv Exp Med Biol. 2014;801:251–7. doi: 10.1007/978-1-4614-3209-8_32. [DOI] [PubMed] [Google Scholar]
- 6.Penfold PL, Madigan MC, Gillies MC, Provis JM. Immunological and aetiological aspects of macular degeneration. Prog Retin Eye Res. 2001;20:385–414. doi: 10.1016/s1350-9462(00)00025-2. [DOI] [PubMed] [Google Scholar]
- 7.Cousins SW, Espinosa-Heidmann DG, Csaky KG. Monocyte activation in patients with age-related macular degeneration: a biomarker of risk for choroidal neovascularization? Arch Ophthalmol. 2004;122:1013–8. doi: 10.1001/archopht.122.7.1013. [DOI] [PubMed] [Google Scholar]
- 8.Cousins SW, Espinosa-Heidmann DG, Miller DM, Pereira-Simon S, Hernandez EP, Chien H, Meier-Jewett C, Dix RD. Macrophage activation associated with chronic murine cytomegalovirus infection results in more severe experimental choroidal neovascularization. PLoS Pathog. 2012;8:e1002671. doi: 10.1371/journal.ppat.1002671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammond CJ, Webster AR, Snieder H, Bird AC, Gilbert CE, Spector TD. Genetic influence on early age-related maculopathy: a twin study. Ophthalmology. 2002;109:730–6. doi: 10.1016/s0161-6420(01)01049-1. [DOI] [PubMed] [Google Scholar]
- 10.Ambati J, Ambati BK, Yoo SH, Ianchulev S, Adamis AP. Age-related macular degeneration: etiology, pathogenesis, and therapeutic strategies. Surv Ophthalmol. 2003;48:257–93. doi: 10.1016/s0039-6257(03)00030-4. [DOI] [PubMed] [Google Scholar]
- 11.Ishida O, Oku H, Ikeda T, Nishimura M, Kawagoe K, Nakamura K. Is Chlamydia pneumoniae infection a risk factor for age related macular degeneration? Br J Ophthalmol. 2003;87:523–4. doi: 10.1136/bjo.87.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalayoglu MV, Bula D, Arroyo J, Gragoudas ES, D'Amico D, Miller JW. Identification of Chlamydia pneumoniae within human choroidal neovascular membranes secondary to age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2005;243:1080–90. doi: 10.1007/s00417-005-1169-y. [DOI] [PubMed] [Google Scholar]
- 13.Robman L, Mahdi O, McCarty C, Dimitrov P, Tikellis G, McNeil J, Byrne G, Taylor H, Guymer R. Exposure to Chlamydia pneumoniae infection and progression of age-related macular degeneration. Am J Epidemiol. 2005;161:1013–9. doi: 10.1093/aje/kwi130. [DOI] [PubMed] [Google Scholar]
- 14.Kessler W, Jantos CA, Dreier J, Pavlovic S. Chlamydia pneumoniae is not detectable in subretinal neovascular membranes in the exudative stage of age-related macular degeneration. Acta Ophthalmol Scand. 2006;84:333–7. doi: 10.1111/j.1600-0420.2005.00591.x. [DOI] [PubMed] [Google Scholar]
- 15.Guymer R, Robman L. Chlamydia pneumoniae and age-related macular degeneration: a role in pathogenesis or merely a chance association? Clin Experiment Ophthalmol. 2007;35:89–93. doi: 10.1111/j.1442-9071.2006.01392.x. [DOI] [PubMed] [Google Scholar]
- 16.Haas P, Steindl K, Schmid-Kubista KE, Aggermann T, Krugluger W, Hageman GS, Binder S. Complement factor H gene polymorphisms and Chlamydia pneumoniae infection in age-related macular degeneration. Eye (Lond) 2009;23:2228–32. doi: 10.1038/eye.2008.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khandhadia S, Foster S, Cree A, Griffiths H, Osmond C, Goverdhan S, Lotery A. Chlamydia infection status, genotype, and age-related macular degeneration. Mol Vis. 2012;18:29–37. [PMC free article] [PubMed] [Google Scholar]
- 18.Miller DM, Espinosa-Heidmann DG, Legra J, Dubovy SR, Sũner IJ, Sedmak DD, Dix RD, Cousins SW. The association of prior cytomegalovirus infection with neovascular age-related macular degeneration. Am J Ophthalmol. 2004;138:323–8. doi: 10.1016/j.ajo.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Mukhamed'ianova ASh, Aznabaev RA, Aznabaeva AF. Clinical and immunological factors of the onset and development of age-related macular degeneration. Vestn Oftalmol. 2014;130:9–13. Article in Russian. [PubMed] [Google Scholar]
- 20.Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, Karikó K, Yoo JW, Lee DK, Hadziahmetovic M, Song Y, Misra S, Chaudhuri G, Buaas FW, Braun RE, Hinton DR, Zhang Q, Grossniklaus HE, Provis JM, Madigan MC, Milam AH, Justice NL, Albuquerque RJ, Blandford AD, Bogdanovich S, Hirano Y, Witta J, Fuchs E, Littman DR, Ambati BK, Rudin CM, Chong MM, Provost P, Kugel JF, Goodrich JA, Dunaief JL, Baffi JZ, Ambati J. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–30. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, Albuquerque RJ, Hauswirth WW, Chiodo VA, Kugel JF, Goodrich JA, Ponicsan SL, Chaudhuri G, Murphy MP, Dunaief JL, Ambati BK, Ogura Y, Yoo JW, Lee DK, Provost P, Hinton DR, Núñez G, Baffi JZ, Kleinman ME, Ambati J. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–59. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami Y, Matsumoto H, Roh M, Giani A, Kataoka K, Morizane Y, Kayama M, Thanos A, Nakatake S, Notomi S, Hisatomi T, Ikeda Y, Ishibashi T, Connor KM, Miller JW, Vavvas DG. Programmed necrosis, not apoptosis, is a key mediator of cell loss and DAMP-mediated inflammation in dsRNA-induced retinal degeneration. Cell Death Differ. 2014;21:270–7. doi: 10.1038/cdd.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trinchieri G, Sher A. Cooperation of Toll-like receptor signals in innate immune defence. Nat Rev Immunol. 2007;7:179–90. doi: 10.1038/nri2038. [DOI] [PubMed] [Google Scholar]
- 24.Kumar MV, Nagineni CN, Chin MS, Hooks JJ, Detrick B. Innate immunity in the retina: Toll-like receptor (TLR) signaling in human retinal pigment epithelial cells. J Neuroimmunol. 2004;153:7–15. doi: 10.1016/j.jneuroim.2004.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebihara N, Chen L, Tokura T, Ushio H, Iwatsu M, Murakami A. Distinct functions between Toll-like receptors 3 and 9 in retinal pigment epithelial cells. Ophthalmic Res. 2007;39:155–63. doi: 10.1159/000103235. [DOI] [PubMed] [Google Scholar]
- 26.Fujimoto T, Sonoda KH, Hijioka K, Sato K, Takeda A, Hasegawa E, Oshima Y, Ishibashi T. Choroidal neovascularization enhanced by Chlamydia pneumoniae via Toll-like receptor 2 in the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2010;51:4694–702. doi: 10.1167/iovs.09-4464. [DOI] [PubMed] [Google Scholar]
- 27.Chen S, Yan H, Sun B, Zuo A, Liang D. Subretinal transfection of chitosan-loaded TLR3-siRNA for the treatment of experimental autoimmune uveitis. Eur J Pharm Biopharm. 2013;85:726–35. doi: 10.1016/j.ejpb.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Klettner A, Hamann T, Schlüter K, Lucius R, Roider J. Retinal pigment epithelium cells alter the pro-inflammatory response of retinal microglia to TLR-3 stimulation. Acta Ophthalmol (Copenh) 2014;92:e621–9. doi: 10.1111/aos.12472. [DOI] [PubMed] [Google Scholar]
- 29.Hollborn M, Iandiev I, Seifert M, Schnurrbusch UEK, Wolf S, Wiedemann P, Bringmann A, Kohen L. Expression of HB-EGF by retinal pigment epithelial cells in vitreoretinal proliferative disease. Curr Eye Res. 2006;31:863–74. doi: 10.1080/02713680600888807. [DOI] [PubMed] [Google Scholar]
- 30.Hollborn M, Stathopoulos C, Wiedemann P, Kohen L, Bringmann A. Positive feedback regulation between MMP-9 and VEGF in human RPE cells. Invest Ophthalmol Vis Sci. 2007;48:4360–7. doi: 10.1167/iovs.06-1234. [DOI] [PubMed] [Google Scholar]
- 31.Hollborn M, Petto C, Steffen A, Trettner S, Bendig A, Wiedemann P, Bringmann A, Kohen L. Effects of thrombin on RPE cells are mediated by transactivation of growth factor receptors. Invest Ophthalmol Vis Sci. 2009;50:4452–9. doi: 10.1167/iovs.08-3194. [DOI] [PubMed] [Google Scholar]
- 32.Nozaki M, Raisler BJ, Sakurai E, Sarma JV, Barnum SR, Lambris JD, Chen Y, Zhang K, Ambati BK, Baffi JZ, Ambati J. Drusen complement components C3a and C5a promote choroidal neovascularization. Proc Natl Acad Sci USA. 2006;103:2328–33. doi: 10.1073/pnas.0408835103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollborn M, Bringmann A, Faude F, Wiedemann P, Kohen L. Signaling pathways involved in PDGF-evoked cellular responses in human RPE cells. Biochem Biophys Res Commun. 2006;344:912–9. doi: 10.1016/j.bbrc.2006.03.185. [DOI] [PubMed] [Google Scholar]
- 34.Hollborn M, Tenckhoff S, Seifert M, Köhler S, Wiedemann P, Bringmann A, Kohen L. Human retinal epithelium produces and responds to placenta growth factor. Graefes Arch Clin Exp Ophthalmol. 2006;244:732–41. doi: 10.1007/s00417-005-0154-9. [DOI] [PubMed] [Google Scholar]
- 35.Lee K, Lee JH, Boovanahalli SK, Jin Y, Lee M, Jin X, Kim JH, Hong YS, Lee JJ. (Aryloxyacetylamino)benzoic acid analogues: a new class of hypoxia-inducible factor-1 inhibitors. J Med Chem. 2007;50:1675–84. doi: 10.1021/jm0610292. [DOI] [PubMed] [Google Scholar]
- 36.Natarajan K, Singh S, Burke TR, Jr, Grunberger D, Aggarwal BB. Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-κB. Proc Natl Acad Sci USA. 1996;93:9090–5. doi: 10.1073/pnas.93.17.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13:1235–42. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 38.Kleinman ME, Kaneko H, Cho WG, Dridi S, Fowler BJ, Blandford AD, Albuquerque RJ, Hirano Y, Terasaki H, Kondo M, Fujita T, Ambati BK, Tarallo V, Gelfand BD, Bogdanovich S, Baffi JZ, Ambati J. Short-interfering RNAs induce retinal degeneration via TLR3 and IRF3. Mol Ther. 2012;20:101–8. doi: 10.1038/mt.2011.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu W, Criswell MH, Fong SL, Temm CJ, Rajashekhar G, Cornell TL, Clauss MA. Differences in the temporal expression of regulatory growth factors during choroidal neovascular development. Exp Eye Res. 2009;88:79–91. doi: 10.1016/j.exer.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 40.Saccani S, Pantano S, Natoli G. Two waves of nuclear factor κB recruitment to target promoters. J Exp Med. 2001;193:1351–9. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]