Abstract
Recently, we determined that BioR, the GntR family of transcription factor, acts as a repressor for biotin metabolism exclusively distributed in certain species of α-proteobacteria, including the zoonotic agent Brucella melitensis and the plant pathogen Agrobacterium tumefaciens. However, the scenario is unusual in Paracoccus denitrificans, another closely related member of the same phylum α-proteobacteria featuring with denitrification. Not only does it encode two BioR homologs Pden_1431 and Pden_2922 (designated as BioR1 and BioR2, respectively), but also has six predictive BioR-recognizable sites (the two bioR homolog each has one site, whereas the two bio operons (bioBFDAGC and bioYB) each contains two tandem BioR boxes). It raised the possibility that unexpected complexity is present in BioR-mediated biotin regulation. Here we report that this is the case. The identity of the purified BioR proteins (BioR1 and BioR2) was confirmed with LC-QToF-MS. Phylogenetic analyses combined with GC percentage raised a possibility that the bioR2 gene might be acquired by horizontal gene transfer. Gel shift assays revealed that the predicted BioR-binding sites are functional for the two BioR homologs, in much similarity to the scenario seen with the BioR site of A. tumefaciens bioBFDAZ. Using the A. tumefaciens reporter system carrying a plasmid-borne LacZ fusion, we revealed that the two homologs of P. denitrificans BioR are functional repressors for biotin metabolism. As anticipated, not only does the addition of exogenous biotin stimulate efficiently the expression of bioYB operon encoding biotin transport/uptake system BioY, but also inhibits the transcription of the bioBFDAGC operon resembling the de novo biotin synthetic pathway. EMSA-based screening failed to demonstrate that the biotin-related metabolite is involved in BioR-DNA interplay, which is consistent with our former observation with Brucella BioR. Our finding defined a complex regulatory network for biotin metabolism in P. denitrificans by two BioR proteins.
Keywords: BioR, biotin, Paracoccus denitrificans
Introduction
Biotin (vitamin H), a sulfur-containing fatty acid derivative, functions as the covalently bound enzyme cofactor that is required by three domains of life (Beckett 2007). The representative biotin-requiring enzyme refers to the AccB subunit (i.e., biotin carboxyl carrier protein, BCCP) of acetyl-CoA carboxylase (ACC), catalyzing the first committed step of fatty acid biosynthesis (Chakravartty and Cronan 2012). To account for such kinds of metabolic requirement for the biotin cofactor, bacteria seemed to have developed two different strategies, one of which is BioY transporter-based scavenging route (Rodionov et al. 2002; Guillen-Navarro et al. 2005; Hebbeln et al. 2007), and the other is de novo synthesis pathway dependent of a full enzyme kit (BioF, BioA, BioD, and BioB) (Fig.1A) (Beckett 2007, 2009). Given the fact that biotin is an energetically expensive molecule in that its de novo biosynthesis requires 20 ATP equivalents, it is reasonable that different organisms have evolved diversified mechanisms to tightly negotiate its production and/or utilization (Streit and Entcheva 2003; Guillen-Navarro et al. 2005; Beckett 2007).
Figure 1.
A working model proposed for biotin metabolism and BioR-mediated regulation in Paracoccus denitrificans. (A) Schematic diagram for the two bacterial biotin-acquiring strategies (biotin biosynthetic pathway and biotin transport/uptake route). (B) Two half-reactions of BirA-proceeded AccB biotinylation. (C) BioR represses biotin biosynthesis pathway in Agrobacterium tumefaciens. (D) Negative autoregulation of BioR and its repression of both biotin biosynthesis pathway and biotin transport system in Brucella. (E) Complex regulation network of biotin metabolism by two BioR proteins in P. denitrificans. KAPA, 7-keto-8-aminopelargonic acid; DAPA, 7, 8-diaminopelargonic acid; DTB, dethiobiotin; AMTOB, S-adenosyl-2-oxo-4-methylthiobutyric acid; SAM, S-adenosyl methionine. BioF, 7-keto-8-amino pelargonic acid synthase; BioA, 7,8-diaminopelargonic acid aminotransferase; BioD, dethiobiotin synthase; BioB, biotin synthase; BirA, biotin protein ligase.
To the best of our knowledge, no less than three types of regulatory factors have been attributed to biotin metabolism (Beckett 2007, 2009; Brune et al. 2012; Feng et al. 2013a,b; Tang et al. 2014). First, the prototypical regulatory mechanism for bacterial biotin synthesis is derived from extensive studies with Escherichia coli (Beckett 2007; Chakravartty and Cronan 2012), in which the central player is the bi-functional BirA protein. The E. coli birA protein product is unusual, in that it not only functions as a repressor for biotin synthesis route (Barker and Campbell 1981b; Brown et al. 2004; Beckett 2007, 2009), but also acts as the enzymatic activity of biotin protein ligase (BPL) (Fig.1B) (Barker and Campbell 1981a; Cronan 1989; Brown et al. 2004). Given the fact whether the BPL enzyme has the N-terminal DNA-binding domain or not, two groups have been categorized (Rodionov et al. 2002). Unlike Group II BPL retaining DNA-binding activity (generally referred to BirA), Group I BPL acts solely as biotin attachment enzymes due to the lacking of the N-terminal winged helix-turn-helix DNA-binding motif (Chapman-Smith and Cronan 1999; Henke and Cronan 2014). As the paradigm group II BPL, the E. coli BirA thus has the ability to physiologically sense the intracellular levels of both biotin and unbiotinylated biotin accepting protein BCCP (Cronan 1989; Beckett 2005, 2007). Moreover, the regulatory role of E. coli BirA depends on the presence of ligand biotinoyl-5′-AMP (biotinyl-adenylate), the product of the first ligase half reaction that is the intermediate of the BirA-catalyzed ligation (Fig.1B) (Ke et al. 2012). Unlike the scenarios seen in E. coli carrying the bi-functional BirA regulatory protein, some organisms (e.g., α-proteobacteria) only encode Group I BPL with sole ligase activity, suggesting that an alternative transcription factor might exist to compensate the loss of regulatory function for the mono-functional BPL enzyme (Rodionov et al. 2002). This hypothesis was furthered by Rodionov and Gelfand (2006), using the approach of computational prediction. In 2013, we provided integrative experimental evidence that BioR, the GntR family of transcription factor, represses expression of bio operon relevant to biotin metabolism in both the plant pathogen Agrobacterium tumefaciens (Feng et al. 2013b) (Fig.1C) and the zoonotic agent Brucella melitensis (Feng et al. 2013a) (Fig.1D). Relative to the paradigm BirA mechanism that is a single protein model, our findings suggested a new biotin sensing machinery: the two-protein paradigm of BirA and BioR. Very recently, we and others established the second two-protein paradigm for bacterial biotin sensing, in which a new TetR-type transcription factor, referred to BioQ, is recruited in Mycobacterium smegmatis (Tang et al. 2014) and Corynebacterium glutamicum (Brune et al. 2012). Surprisingly, no direct evidence was found in supporting that DNA binding of BioR (and/or BioQ) requires the participation of biotin metabolites (Feng et al. 2013a,b; Tang et al. 2014), which is far different from scenarios seen with BirA proteins of E. coli (Brown et al. 2004; Chakravartty and Cronan 2012) and Bacillus (Henke and Cronan 2014).
Paracoccus is taxonomically referred to a genus of the Rhodobacteraceae, and comprises a diversified set of species, one of which is Paracoccus denitrificans (http://en.wikipedia.org/wiki/Paracoccus) (Ludwig et al. 1993; Rainey et al. 1999). As a nonmotile coccoid soil organism from α-subdivision of the phylum proteobacteria, P. denitrificans is well known in its unusual ability of denitrification (reducing nitrate to dinitrogen), and growth under the condition of hyper gravity (Baker et al. 1998). The announcement of genomic sequences for P. denitrificans such as strain PD1222 (http://genome.jgi-psf.org/parde/parde.home.html) (Siddavattam et al. 2011) greatly facilitated the development of being a model organism for extensive investigations of molecular mechanism (endosymbiotic theory) implicated into denitrifications and possible ancestors for the eukaryotic mitochondrion (http://en.wikipedia.org/wiki/Paracoccus_denitrificans) (John and Whatley 1975). In views of genomic contents, we noted that P. denitrificans is unusual in that the gene duplication and/or redundancy (especially two BioR orthologs) is prevalent in the context of biotin metabolism, unlike the scenarios observed with its close relatives A. tumefaciens and B. melitensis (Fig.2). Also, totally six putative BioR-recognizable palindromes were predicted (http://regprecise.lbl.gov/RegPrecise/regulon.jsp?regulon_id=53141) (Rodionov and Gelfand 2006; Feng et al. 2013a; Novichkov et al. 2013), implying unexpected complexity in BioR-mediated regulation of biotin metabolism in P. denitrificans. The question we raised is why P. denitrificans evolves such kind of complicated network for biotin metabolism and regulation. Is there any physiological/ecological requirement for this regulatory system in adaptation to its growing/inhabiting niches?
Figure 2.
Genetic loci of bio operons and BioR signals. (A) Genomic organization of bio operon in plant pathogen Agrobacterium tumefaciens. (B) Genomic organization of bio operon in zoonotic pathogen Brucella melitensis. (C) Genomic organization of bio operon in Paracoccus denitrificans. (D) Sequence logo for the BioR-binidng sites. The sequence logo is generated using WebLogo (http://weblogo.berkeley.edu/logo.cgi).
In this paper, we are attempting to address the above questions. We report that (1) extraordinary copies of biotin metabolism-related genes in P. denitrificans are acquired through possible events of horizontal gene transfer (HGT); (2) two BioR homologs are functional in biotin regulation/sensing; (3) unprecedent complexity is present in the BioR-mediated regulatory network for biotin metabolism (Fig.1E).
Experimental Procedures
Bacterial strains and growth conditions
In addition to PD1222, the wild type of P. denitrificans, all of the bacterial strains used here were E. coli K-12 derivatives (Table1). The media are separately LB medium (10 g of tryptone, 5 g of yeast extract and 10 g of NaCl per liter), and rich broth (RBO medium; 10 g of tryptone, 1 g of yeast extract, and 5 g of NaCl per liter). Antibiotics were supplemented as follows (in mg/L): sodium ampicillin, 100; kanamycin sulfate, 50; tetracycline HCl, 15; and chloramphenicol, 20.
Table 1.
Strains and plasmids in this study
| Strains or plasmids | Relevant characteristics | References or origins |
|---|---|---|
| Strains | ||
| Topo10 | A cloning Escherichia coli host (F−, ΔlacX74) | Invitrogen |
| BL21(DE3) | An expression E. coli host | Lab stock |
| FYJ179 | Agrobacterium tumefaciens NTL4 | Feng et al. (2013b) |
| FYJ284 | NTL4, ΔbioR::Km, ΔbioBFDA | Feng et al. (2013a,b) |
| FYJ291 | FYJ284 (NTL4, ΔbioR::Km, ΔbioBFDA) carrying pRG-PbioBat | Feng et al. (2013a,b) |
| PD1222 | The wild-type strain of Paracoccus denitrificans | ATCC |
| FYJ347 | Topo carrying pET28-bioRpd1 | This work |
| FYJ350 | Topo carrying pET28-bioRpd2 | This work |
| FYJ351 | BL21 (DE3) carrying pET28-bioRpd1 | This work |
| FYJ354 | BL21 (DE3) carrying pET28-bioRpd2 | This work |
| FYJ376 | FYJ291 carrying pSRKGm-bioRpd1 | This work |
| FYJ377 | FYJ291 carrying pSRKGm-bioRpd2 | This work |
| Plasmids | ||
| pET28(a) | Commercial T7-driven expression vector, KmR | Novagen |
| pET28- bioRpd1 | pET28(a) carrying P. denitrificans bioRpd1 gene, KmR | This work |
| pET28- bioRpd2 | pET28(a) carrying P. denitrificans bioRpd2 gene, KmR | This work |
| pSRKGm | Broad host range expression vector with the tightly regulated promoter | Feng et al. (2013b) |
| pSRK-bioRpd1 | pSRKGm encoding P. denitrificans bioRpd1 gene, GmR | This work |
| pSRK-bioRpd2 | pSRKGm encoding P. denitrificans bioRpd2 gene, GmR | This work |
| pRG970 | Low copy transcriptional promoter-less lacZ/Gus bi-directional fusion vector, SpcR | Van den Eede et al. (1992); van Dillewijn et al. (2001) |
| pRG-PbioBat | pRG970 encoding the A. tumefaciens bioBFDAZ promoter region | Feng et al. (2013a,b) |
ATCC, American Type Culture Collection.
Paracoccus denitrificans (Table1) was grown in minimal medium containing (per liter) 6.0 g of K2HPO4, 4.0 g of KH2PO4, 0.15 g of sodium molybdate, 0.2 g of MgSO4·7H2O, 0.04 g of CaCl2, 0.001 g of MnSO4·2H2O, and 1.1 g of FeSO4·7H2O with 1.6 g of NH4Cl as the nitrogen source (Zhao et al. 2013; Kumar et al. 2014). Cultures were grown aerobically at 30°C with or without Biotin (100 mmol/L) in mineral medium supplemented with glucose (20 mmol/L) as the carbon source.
Plasmids and genetic manipulations
The two bioR genes (pden_1431 and pden_2922) of P. denitrificans were amplified with PCR and cloned into the expression vector pET28(a), giving the recombinant plasmids pET28-bioRpd1 and pET28-bioRpd2, respectively (Table1). To prepare the appropriate BioR proteins, the corresponding expression plasmids (pET28-bioRpd1 and pET28-bioRpd2) were transformed into the strain BL21(DE3) (Feng and Cronan 2009b). To examine the role of bioR in vivo, the two genes were inserted into pSRKGm, the broad host range expression vector, generating the chimeric plasmids pSRKG-bioR1 and pSRKG-bioR2, respectively (Table1). The recipient strain is a reporter strain FYJ291 we recently developed (Feng et al. 2013b), which is the ΔbioR::Km mutant of A. tumefaciens carrying pRG-PbioBat, a plasmid-borne LacZ transcriptional fusion (Table1). All the acquired plasmids were confirmed by both PCR detection and direct DNA sequencing.
Expression and purification of BioR protein
Both BioR1 and BioR2 of P. denitriifcans were overexpressed using prokaryotic expression system with induction of 0.3 mmol/L isopropyl β-d-1-thiogalactopyranoside (IPTG) at 30°C for 3 h. The clarified supernatant of bacterial lysates was loaded onto a nickel-ion affinity column (Qiagen, Hilden, Germany). After removal of the contaminant proteins with wash buffer containing 50 mmol/L imidazole, the 6x His-tagged protein of interest was eluted in elution buffer containing 150 mmol/L imidazole. The purified proteins were exchanged into 1X PBS buffer (pH 7.4) containing 10% glycerol, and visualized by 15% SDS-PAGE followed by staining with Coomassie Brilliant Blue R250 (Sigma, St. Louis, MO). Of note, the BioR1 is somewhat a weird protein, in that it easily precipitates during the process of purification, which is almost similar to scenarios seen with FabR proteins (Feng and Cronan 2011).
Liquid chromatography quadrupole time-of-flight mass spectrometry
The identity of two versions of P. denitrificans BioR proteins (BioR1 and BioR2) was verified using A Waters Q-Tof API-US Quad-ToF mass spectrometer connected to a Waters nano Acquity UPLC (Feng and Cronan 2011). As we described before (Feng and Cronan 2011), the protein band of interest was digested with Trypsin (G-Biosciences St. Louis, MO), and the resultant peptides were loaded on a Waters Atlantis C-18 column (0.03 mm particle, 0.075 × 150 mm). The dependently acquired data were further subjected to the ms/ms analyses.
Electrophoretic mobility shift assays
To test the functions of the predicted BioR-binding sites of P. denitrifican, gel shift assays were adopted as we described earlier (Feng and Cronan 2009b, 2010, 2011). In addition to the known probe bioBFDAZ_ at six more sets of DNA probes were prepared by annealing two complementary oligonucleotides (Table2). These probes included bioR1_pd probe, bioR2_pd probe, bioYB_pd1 probe, bioYB_pd2 probe, bioBFDAGC_pd1 probe, and bioBFDAGC_pd2 probe, respectively (Table2). In the gel shift experiments, the digoxigenin-labeled DNA probes (∼0.2 pmol) were incubated with the purified BioR protein (note: crude extract used for BioR1) in the binding buffer (Roche, Indianapolis, IN, USA) for 15 min at room temperature. When required, the cold probe (and/or biotin metabolites) was supplemented into the gel shift assays. The DNA–protein mixtures were separated with the native 7% PAGE, and transferred onto the nylon membrane via the direct contact gel transfer. Finally, the chemical-luminescence signals were captured through the exposure of the membrane to ECL films (Amersham, GE Healthcare, Piscataway, NJ, USA).
Table 2.
Primers used in this study
| Primers | Sequences (5′-3′) |
|---|---|
| bioR1pd-F (BamHI) | CG GGATCC ATG AAA CAC GCC CCT GAA GAG |
| bioR1pd-R (XhoI) | CCG CTCGAG TTA TCC GGG AAT CTC GTA AGT C |
| bioR2pd-F (BamHI) | CG GGATCC ATG AGC GCA GGT TCC GAA GAA |
| bioR2pd-R (SalI) | CCG GTCGAC TTA GCC GTG GAT GGC GAA GG |
| Pden1431rt-F | GGC GAC AAT GCC AGT ACC |
| Pden1431rt-R | AGG ATG ATC CGG TGA AAA TG |
| Pden1432rt-F | GCT ATC TGG CGG GCT ATC T |
| Pden1432rt-R | GAG GCC GAG GGC ATA GAC |
| Pden1433rt-F | AGCCTGCTCAGCATCAAGAC |
| Pden1433rt-R | GGATTGCGAGCAATAGCC |
| Pden2916rt-F | CTACAACCACAATATCGACACCTC |
| Pden2916rt-R | ATCCGGTCCTGGAAGGTC |
| Pden2917rt-F | CCTGGTGGTCCATGATGC |
| Pden2917rt-R | GGCATCGTTATGGGCAAA |
| Pden2918rt-F | GGCACCTGCTCTATTTGCAG |
| Pden2918rt-R | CGACAGCAGCGAATGGTT |
| Pden2919rt-F | GGGGCATGTGGTTCTATCAC |
| Pden2919rt-R | GCGATCTCGTCGAAAATCAG |
| Pden2922rt-F | TTCGGCGCCAGCCACGTCCCGGTGC |
| Pden2922rt-R | GTGCGGCGCGGCATGGCGCAGGG |
| Pden16Srt-F | AGGCCCTAGGGTTGTAAAGC |
| Pden16Srt-R | GGGGCTTCTTCTGCTGGTA |
| bioB_at probe-F | CTC TCT TGA GGA GGC AAA AAT TAT CTA TAA TTT GCC ATT TAA CGA CCT GC |
| bioB_at probe-R | GCA GGT CGT TAA ATG GCA AAT TAT AGA TAA TTT TTG CCT CCT CAA GAG AG |
| bioR1-probe-F1 | GGT GCA GCA TGA ATT ATC TAT AAT TCA TGA AAC ACG |
| bioR1-probe-R1 | CGT GTT TCA TGA ATT ATA GAT AAT TCA TGC TGC ACC |
| bioYB-probe1-F1 | GAT TCC CGG ATA ATT ATC TAT AAA CCT AAT TGC CAG |
| bioYB-probe1-R1 | CTG GCA ATT AGG TTT ATA GAT AAT TAT CCG GGA ATC |
| bioYB-probe2-F1 | CAA AGC CTT CGT AAT TAT AGA TAG ACT CGA TAC CTA TC |
| bioYB-probe2-R1 | GAT AGG TAT CGA GTC TAT CTA TAA TTA CGA AGG CTT TG |
| bioBFDAGC-probe-F2 | GGC GCT GAC CGT TTT ATA GAT ACT TCC ACA TGA GGC |
| bioBFDAGC-probe-R2 | GCC TCA TGT GGA AGT ATC TAT AAA ACG GTC AGC GCC |
The underlined sequences in italics are restriction sites, and the bold letters denote the predicted BioR-binding sites.
The genetic locus of genes (bioR1 and/or bioBY) is localized on Chromosome I.
The operon of bioBFDAGC is localized on Chromosome II.
β-Galactosidase assays
Bacterial samples stripped out of the MacConkey agar plates were suspended in Z-buffer and subjected to direct measurement of β-galactosidase activity (Miller 1992; Feng and Cronan 2009a,b). The data were recorded in triplicate more than three independent assays.
Real-time quantitative polymerase chain reaction
Cells were grown overnight in minimal media without biotin. This was used as an inoculum to inoculate 10 mL of fresh minimal media. Cells were grown upto 0.5 OD600 and pelleted and washed with minimal media. Cells were resuspended in 10 mL minimal media and divided into two 5 mL portions. A quantity of 100 nmol/L biotin was added into one portion. Cells were collected at 1/3 h for RNA isolation.
Quantitative real-time PCR was performed as previously described (Pfaffl 2001). Cells were harvested at different OD600, and RNA was extracted using RNeasy protect kit (Qiagen) according to the manufacturer’s recommendations. Total RNA was resuspended in PCR-grade nuclease-free water, and RNA quality and concentration were estimated by optical density measurement, using the Nanodrop 2000 (Thermo Fisher Scientific, San Jose, CA, USA). Each sample of 500 ng total RNA was reverse transcribed, using the First Strand cDNA Synthesis Kit (Fermentas, St. Leon-Rot, Germany). Real-time PCR reactions were carried out on a LightCycler 480 (Roche) using the SYBR Green detection format. Change in the expression was calculated relative to the expression of 16S rRNA. After each PCR run, a melting curve analysis was carried out to control for production of primer dimers and/or nonspecific PCR products. Fold change in mRNA expression during treatment was calculated using the crossing point (Cp) for each sample and the efficiency (Eff) of each transcript, using the formula (Efftarget gene)ΔCp/(Effhousekeeping gene)ΔCp. The fold change was estimated relative to 16SrRNA.
Bioinformatic analyses
The protein sequences of BioR regulators are derived from A. tumefaciens, B. melitensis, and P. denitrificans. The BioR-binding sites were all sampled from RegPrecise database (http://regprecise.lbl.gov/RegPrecise/regulon.jsp?regulon_id=53141). The multiple alignment of protein (and/or DNA) was performed with the program of ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html), and the final output of BLAST photography was given after being processed by the program ESPript 2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The sequence logo of the BioR-specific sites is generated using WebLogo (http://weblogo.berkeley.edu/logo.cgi). Transcription start sites of the bio operons were predicted using the method of Neutral Network Promoter Prediction (http://www.fruitfly.org/seq_tools/promoter.html).
Orthologs of BioB, BioR, and BioY proteins were identified by a procedure based on the analysis of phylogenetic trees for protein domains in MicrobesOnline (Dehal et al. 2010). Multiple protein alignments were done using MUSCLE tool (Edgar 2004a,b). Phylogenetic trees were constructed by the maximum-likelihood method with default parameters implemented in PhyML-3.0 (Guindon et al. 2010) and visualized using Dendroscope (Huson et al. 2007).
Results and Discussion
Complexity in biotin metabolism of P. denitrificans
The situation of genetic organization in P. denitrificans seemed to be unusual in that the gene duplication and/or redundancy is present in the context of biotin metabolism and regulation, which is far different from those of its two close-related cousins A. tumefaciens and B. melitensis (Fig.2 A–C). In addition to the megaplasmid (∼0.65 Mb), P. denitrificans also carries two chromosomes (designated as Ch-I (∼2.85 Mb) and Ch-II (∼1.73 Mb), Fig.2C). The bio operons in P. denitrificans included bioYB2 on Ch-1, bioBFDAGC on Ch-II, and bioMNY2 encoded by the megaplasmid, respectively (Fig.2C). Unlike the A. tumefaciens and B. melitensis both of which encode only one BioR repressor (Fig.2A and B), P. denitrificans has two BioR orthologs (Pden_1431 for BioR1, and Pden_2922 for BioR2) separately scattered on the two chromosomes (Fig.2C) (Rodionov and Gelfand 2006). Additionally, P. denitrificans also has two bioB homologs (one is located in the bioBFDAGC operon, the other is encoded by the bioYB2 operon) and two bioY paralogs (one is located in the bioYB2 operon, the other is encoded by the bioMNY2 operon) (Fig.2C) (Rodionov and Gelfand 2006). In much similarity to the scenario seen with B. melitensis bioR (Fig.2B) (Feng et al. 2013a), the two bioR homologs of P. denitrificans each has a putative BioR-specific palindrome in front of their coding sequences (Fig.2C and D), suggesting the possibility of autoregulation. No putative BioR-binding site was detected in the plasmid-borne bioMNY2 operon (Fig.2C), which is in much consistency with the scenario with the A. tumefaciens bioMNY (Fig.2A) (Feng et al. 2013b). In contrast, the other bioY-containing operon bioYB seemed likely to be controlled by the BioR regulator, in that it has two tandem BioR-recognizable sites (Fig.2C and D). As anticipated, the bioBFDAGC operon, a major gene cluster encoding the full de novo biotin synthesis pathway also has two tandem BioR-binding sites (Fig.2C), which is almost identical to the observation with B. melitensis bioBFDAZ (Fig.2B) (Feng et al. 2013a), but little bit different from that of the A. tumefaciens counterpart having only one palindrome for the BioR protein (Fig.2A) (Feng et al. 2013b). Of particular note, the bioZ gene is replaced with bioGC in this case (Fig.2A–C). Given the fact that two BioR homologs and 6 BioR-recognizable sites (representing 4 target genes/operons) coexist, we concluded that the BioR-mediated regulatory network in P. denitrifican is of unusual complexity (Fig.1E).
Tracing origins of bio operons/genes of P. denitrificans
Since the situation of bio operons/genes is pretty unusual in P. denitrificans, we are interested in tracing the origins of these genes esp. the duplicated cousins. The BLAST analyses revealed that the bio operons/genes of P. denitrificans can match no less than eight different species, including the plant pathogen Xylella fastidiosa and the marine bacteria Celeribacter indicus (Table3). Of being noteworthy, the P. denitrificans bioG is completely identical to the X. fastidiosa counterpart at the level of nucleotide acids (Table3). Systematic comparison of the GC contents showed that (1) bioR2 (pden_2922) with the GC percentage of 72.52% (but not bioR1 (pden_1431) with 66.36% of GC percentage) is significantly higher than that of the average GC% of the chromosome (66.7–66.8%); (2) bioY1 (pden_1431) with the 71.86% of GC percentage (but not bioY2 (pden_5033) with the GC percentage of 68.92%) is appreciably higher than that of the average GC% of the chromosome/megaplasmid (66.7–67.1%); (3) the group I BPL-encoding gene birA (pden_2230) exhibits the GC ratio of 72.6%, much higher than that of the Chromosome II (66.8%); (4) most of genes encoding the biotin synthesis pathway consistently showed higher GC% (74.37% for bioF, 74.06% for bioD, 71.18% for bioA, and 75.13% for bioC) than that of Chromosome II (66.8%), except that bioG presents 54.64%, the lowest GC% amongst the bio genes (Table3). Obviously, the above observations might indicate the possibility for HGT in the context of biotin metabolism-related gene clusters/operons. We anticipated that the heterogeneity (heterogeneous origins) somewhat is in part (if not all) why P. denitrificans evolves such kind of complicated machinery for biotin metabolism. However, the physiological/ecological advantage of this unusual mechanism requires further explorations.
Table 3.
GC% analyses of the Paracoccus denitrificans bio operon and exploration of their possible origins
| GC% | Origins matched1 | |||
|---|---|---|---|---|
| Ch-I | Ch-II | Plasmid | ||
| Ch-I | 66.7 | – | – | – |
| Ch-II | – | 66.8 | – | – |
| Plasmid | – | – | 67.1 | – |
| birA | – | 72.6 | – | Paracoccus aminophilus (76%) |
| bioR1 | 66.36 | – | – | Celeribacter indicus (84%) |
| bioR2 | – | 72.52 | – | Azorhizobium caulinodans (78%) |
| bioB1 | – | 66.77 | – | Rhodobacter capsulatus (81%) |
| bioB2 | 68.22 | – | – | Rhodobacter sphaeroides (90%) |
| bioY1 | 71.86 | – | – | Paracoccus denitrificans (100%) |
| bioY2 | – | – | 68.92 | P. denitrificans PD1222 plasmid 1 (100%) |
| bioM | – | – | 66.54 | P. denitrificans plasmid 1 (100%) |
| bioN | – | – | 70.23 | P. denitrificans plasmid 1 (100%) |
| bioF | – | 74.37 | – | P. denitrificans (100%) |
| bioD | – | 74.06 | – | P. aminophilus (70.9%) |
| bioA | – | 71.18 | – | R. capsulatus (76%) |
| bioG | – | 54.64 | – | Xylella fastidiosa (100%) |
| bioC | – | 75.13 | – | P. denitrificans (100%) |
–, not applicable; Ch, chromosome.
The nucleotide identity of the interested gene from P. denitrificans relative to its possible origins. The numbers in grey background denote the GC percentage of P. denitrificans Chromosome/plasmid.
For better understanding of origin of the duplicated genes, bioR (Fig.3A), bioB (Fig.3B), and bioY (Fig.3C), we analyzed their orthologs in genomes of Rhizobiales and Rhodobacterales. These phylogenetic analyses revealed that at least bioR1 gene (pden_1431) and bioYB2 operon (pden_1432-33) might be products of the horizontal transfer from Azorhizobium caulinodans or the related species (Fig.3).
Figure 3.
Phylogenetic trees for the biotin synthesis proteins duplicated in Paracoccus denitrificans and their orthologs in Rhodobacterales and Rhizobiales genomes. (A) Phylogenetic tree of the BioR homologs. (B) Phylogenetic tree of the BioB proteins. (C) Phylogenetic tree of the BioY transporters.
Characterization of two BioR homologs
Paracoccus denitrifIcans PD1222 contains two circular chromosomes: Ch-I (Accession no.: NC_008686.1) is 2.85 Mb long, while Ch II (Accession no.: NC_008687.1) is 1.73 Mb in length (http://www.ncbi.nlm.nih.gov/genome/?term=PD1222). Two bioR homologs separately are localized on the corresponding chromosome: bioR1 (pden_1431) on Ch-I encodes a 222 residues of polypeptide, whereas the bioR2 (pden2922) on Ch-II is a gene encoding a protein of 221 aa long (Figs.2C and 4A). Multiple sequence alignments of the two BioR proteins (BioR1 & BioR2) with the cousins of both A. tumefaciens and B. melitensis showed that they are appreciably conserved (Fig.4A). Given the very fact that BioR1 and BioR2 both share 76.1% identity and 66.7% similarity, respectively (not shown), we cannot figure out which one is the ancestor of the two duplicated bioR genes. Subsequent measurement for the GC contents of the two bioR genes ruled out the hypothesis that they are generated during the events of gene duplication in that the difference in GC% (66.36% for bioR1, and 72.52% for bioR2) raised the possibility that they are acquired by gene horizontal transfer (Table3). Further BLAST analyses indicated that bioR1 might be derived from C. indicus, whereas bioR2 can be traced to A. caulinodans (Table3).
Figure 4.

Characterization of the two BioR homologs of Paracoccus denitrificans. (A) Multiple sequence alignments of the the two BioR homologs of P. denitrificans with the paradigm members The multiple alignment of bacterial BioR homologs was performed using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html), and the final output was expressed after data processing by program ESPript 2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Identical residues are in white letters with red background, similar residues are in black letters in yellow background, and the varied residues are in gray letters. SDS-PAGE profile (B) and MS-identification (C) of the purified BioR1 (Pden_1431) protein SDS-PAGE profile (D) and MS-identification (E) of the purified BioR2 (Pden_2922) protein.
As predicted by Rodionov and Gelfand (2006), structural modeling suggested that the two BioR proteins are featuring with a conserved N-terminal DNA-binding motif with helix-turn-helix structure (not shown). To examine the putative function, we produced the two versions of recombinant BioR1 (and/or BioR2) protein in the E. coli expression system and purified them to homogeneity (Fig.4B and D). As anticipated, the two BioR proteins are weird (not easily tractable), in that most of them precipitates during the process of protein purification in vitro. The similar scenarios notorious in short survival time of protein were ever encountered in the cases of A. tumefaciens BioR (Feng et al. 2013b) and the counterpart of B. melitensis (Feng et al. 2013a). Subsequently, the two protein bands cut from the SDS-PAGE gel was subjected to the liquid chromatography mass spectrometry. The MS results of the resultant tryptic peptides showed that BioR1 (Fig.4B) and BioR2 (Fig.4D) we overexpressed in vitro well matched Pden_1431 with the coverage of 81% (Fig.4C), and Pden_2922 with the covering score of 72% (Fig.4E), respectively. Fortunately, we have luck to recover around 10% of soluble BioR2 protein, whereas we do not have any success to acquire trace amount of BioR1 protein even after a series of trials (that is why we have to fall back on the crude extract containing BioR1 protein for subsequent functional assays).
Binding of P. denitrificans BioR cognate genes
We performed an extensive bioinformatics analyses, using The Neutral Network Program of Promoter Prediction (http://www.fruitfly.org/seq_tools/promoter.html), which roughly illustrated the promoters of bio operons/genes (Fig. S1). Totally, six BioR-recognizable sites are assigned to four genes/operons: the two bioR each has one site, the two gene clusters (bioYB2, and bioBFDAGC) each has two discontinuous sites (Fig.2C and D). Prior to this study, we believed that the situation of biotin regulation in B. melitensis is quite complicated when compared with that of A. tumefaciens (Fig.1C and D). It seemed likely that the scenario is much more complex in the closely related organism P. denitrificans.
To test the functions of these predicted BioR sites (Fig.5A), electrophoresis mobility shift assay (EMSA) was conducted using the either the purified BioR2 protein (Fig.5B–F) or the crude extract containing BioR1 protein (Fig. S2). Gel shift assays confirmed that BioR2 protein effectively bind the probe of A. tumefaciens bioBFDAZ operon in a dose-dependent manner (Fig.5B), which is generally similar to our former observation with BioR proteins of both A. tumefaciens and B. melitensis (Feng et al. 2013a,b). The appreciable binding of the same bioBFDAZ_at probe to BioR1 protein was also confirmed (Fig. S2A). Obviously, it suggested that BioR homologs with a variety of origins are functionally exchangeable. The promoter of bioR1 interacted well with the BioR2 protein (Fig.5C) as well as the BioR1 protein (Fig. S2B), and vice versa (not shown). This implied that not only do the two regulators (BioR1 & BioR2) autoregulate themselves, but also they can crosstalk via direct DNA–protein interaction. As expected, the bioBFDAGC promoter of P. denitrificans exhibited strong binding to the BioR2 (Fig.5D) and BioR1 (Fig. S2C), validating the speculation by Rodionov and Gelfand (2006) that the biotin biosynthetic route is under the control by the BioR regulatory protein. Unlike the scenarios in A. tumefaciens (Feng et al. 2013b) and B. melitensis (Feng et al. 2013a), the situation in the case of P. denitrificans seemed unusual, in that two BioR (BioR1 and BioR2) transcription factors constitute a “double-safety locker” to guarantee the tight regulation exerted on the biotin synthesis pathway. In addition, the promoter region of the bioYB operon was found to bind both BioR2 (Fig.5E and F) and BioR1 (Fig. S2D). To the best of our knowledge, it might represent the second example of the BioR-regulated transport/scavenge of biotin in bacteria, in that the first paradigm was attributed to its close relative, the human pathogen B. melitensis (Feng et al. 2013a).
Figure 5.
Binding of Paracoccus denitrificans BioR2 (Pden_2922) to cognate target genes. (A) Comparative analyses for the BioR-recognizable sites. (B) Binding of Agrobacterium tumefaciens bioBFDAZ promoter to the P. denitrificans BioR2 (Pden_2922) protein. (C) Binding of P. denitrificans bioR1 (pden_1431) promoter to the P. denitrificans BioR2 (Pden_2922) protein. (D) P. denitrificans BioR2 (Pden_2922) protein interacts with the promoter of P. denitrificans bioBFDAGC operon Interplay between P. denitrificans BioR2 (Pden_2922) protein and the two putative sites of the bioYB operon, one of which is bioYB1 (E) and other is bioYB2 (F). at, Agrobacterium tumefaciens; bme, Brucella melitensis; pd, Paracoccus denitrificans..
Although the most straightforward model for BioR regulation referred to that BioR binding its cognate operators requires coexistence of either biotin or a biotin derivative such as biotinoyl-5′-adenylate, unfortunately we are still not aware of any direct evidence thus far. To address the long-term unresolved question, potential effectors/ligands for the DNA-BioR interplay, we systematically tested the precursor (pimeloyl-ACP), intermediates (KAPA, DAPA, and DTB), and the final product (biotin) of biotin synthesis pathway (Fig.6A) by employing EMSA approach. In much agreement with the scenarios seen with the BioR proteins of A. tumefaciens (Feng et al. 2013b) and B. melitensis (Feng et al. 2013a), we failed to visualize that the biotin-related metabolites we tested have obvious roles in interfering with the DNA-binding activity of BioR2 protein even after addition of excess metabolites (such as 500 pmol KAPA, DAPA, DTB, and biotin) (Fig.6B). As anticipated, we also noted that excess of cold DNA probe competitively impaired interplay of the DIG-labeled bioBFDAGC probe and BioR2 protein (Fig.6B), demonstrating that binding of BioR2 cognate DNA is a specific physical interaction.
Figure 6.

Probing possible roles of biotin-related metabolites in the specific interaction between BioR and cognate DNA. (A) Schematic diagram for the four-step pathway of bacterial biotin biosynthesis. (B) The binding of BioR cognate DIG-labeled DNA can be fully/specifically interfered with the excess of the relevant cold DNA probe, but no apparent roles of biotin metabolites (Pimeloyl-ACP, KAPA, DAPA, and DTB) are seen in such kinds of interaction. The minus sign denotes no addition of BioR2 protein (10–20 pmol). The protein samples were incubated with 0.2 pmol of DIG-labeled bioBFDAGC probe in a total volume of 15 μL. When required, the cold bioBFDAGC probe is supplemented at different levels (10, 20, and 50 pmol). A representative result from no less than 3 independent gel shift assays (7% native PAGE) is given. KAPA, 7-keto-8-aminopelargonic acid; DAPA, 7, 8-diaminopelargonic acid; DTB, dethiobiotin. BioF, 7-keto-8-amino pelargonic acid synthase; BioA, 7,8-diaminopelargonic acid aminotransferase; BioD, dethiobiotin synthase; BioB, biotin synthase; BirA, biotin protein ligase.
In vivo role of BioR regulatory protein
Very recently, we developed a reporter strain FYJ291, a ΔbioR mutant of A. tumefaciens engineered to carry the low-copy plasmid-borne PbioBat-lacZ transcriptional fusion (Table1) (Feng et al. 2013a,b). This reporter strain has been confirmed to work well in identifying the functional bioR ortholog in vivo (Feng et al. 2013a). In principle, growth of the reporter strain FYJ291 on a MacConkey agar plate with 0.2% lactose as a sole carbon source can give purple colonies, implying the robust β-gal activity by PbioBat-lacZ fusion is present upon removal of BioR repressor (Fig.7A). The introduction of both bioR1 and bioR2 into this reporter strain caused the formation of yellow colonies, suggesting that the expression of either BioR1 or BioR2 can downregulate β-gal activity of the PbioBat-lacZ fusion (Fig.7A). Indeed, such dramatic color alterations generally agreed with our former observations with BioR of A. tumefaceins (Feng et al. 2013b) and B. melitensis (Feng et al. 2013a) in this bioassay. Analyses for LacZ activities further showed that expressions of both bioR1 and bioR2 of P. denitrificans give a six to eightfold decrement of the bioB_at transcription level in comparison with that of FYJ291 indicator strain (Fig.7B). Therefore, both bioR1 and bioR2 of P. denitrificans encode a functional BioR ortholog having the in vivo role in modulating biotin metabolism.
Figure 7.
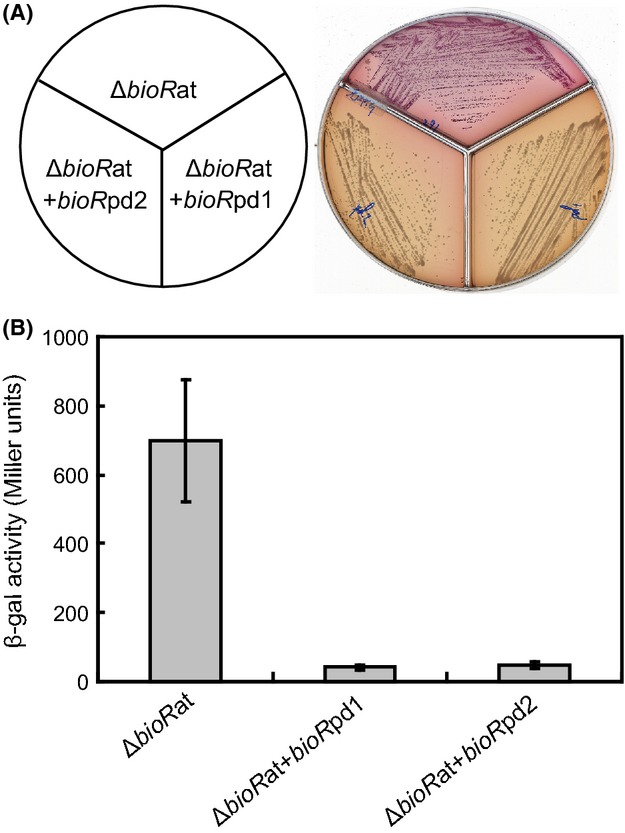
Functional analyses for two bioR homologs of the Paracoccus denitrificans using the Agrobacterium tumefaciens reporter system. (A) Maconkey plates-based assays for the activities of the two P. denitrificans BioR homologs (Pden_1431 and Pden_2922). (B) Measurment for β-gal activities of the Pden_1431 (and/or Pden_2922) promoter-driven LacZ reporter genes. Three strains used here include FYJ291 (NTL4, ΔbioR::Km, carrying pRG-PbioBat); FYJ376 (FYJ291 carrying pSRKGm-bioRpd1) and FYJ377 (FYJ291 carrying pSRKGm-bioRpd2), respectively.
Biotin sensing of P. denitrificans
We carried out qPCR assays to investigate the response of P. denitrificans to biotin by addressing the accumulated transcript level of the representative target genes that correspond to the two biotin-acquiring systems (bioY is for biotin uptake system, and bioB, bioF, bioD, and bioA are specific for biotin synthesis pathway, Fig.8). First, we observed that an addition of exogenous biotin (100 nmol/L) to cultures of the wild-type strain PD1222 gave more than 10-fold increment to transcription of the bioYB2 operon, but did not alter significantly the expression of bioR1 (Fig.8A). Somewhat, it seemed usual in that the increasing expression of BioB, an enzyme catalyzing the last committed step of biotin synthesis is energetically wasteful on the condition with the supply of exogenous biotin. Someone might conclude that it is not physiologically correct that the bioB2 is co-transcribed with bioY forming an operon of bioYB2. In contrast, we favored to believe it is possible. The reasons being in the following two points: (1) Although all the intermediates can enter E. coli at various efficiencies, the biotin transporter BioY might be helpful for uptake of DTB, a precursor for biotin (also the substrate of BioB biotin synthase); (2) when the DTB is available, expression of functional BioB is physiologically required to make biotin from DTB. Given the fact that no literature documented the above speculation thus far, it would be of much interest to test it. This hypothesis might be checked by seeing if DTB competes with biotin in a bioB strain. The criteria for this assay is described as follows: If DTB uses the same transporter, then the minimal amount of biotin will not be enough (this idea is mainly from personal communication with Prof. John Cronan, University of Illinois at Urbana-Champaign, Urbana, IL).
Figure 8.

Alteration of bioexpression by addition of biotin. (A) Induction of the bioY-containing operon bioYB2 by addition of exogenous biotin. (B) Repression of the biotin biosynthesis operon bioBFDA in the presence of excess of biotin The inside cartoon diagram illustrates the genetic organization of bio operons. 100 nmol/L biotin was added into the Paracoccus denitrificans grown in minimal medium, and real-time quantitative PCR is used to measure the relative expression of biotin-related genes.
Consequently, the supply of exogenous biotin (100 nmol/L) to bacterial cultures resulted in around fivefold decrement to expression of bioBFDAGC (note: the former four genes of this operon bioB, bioF, bioD, and bioA were checked), but no obvious change in bioR2 transcription. It demonstrated that the presence of biotin can effectively shut down the biotin synthesis pathway (Fig.8B). Together, the altered expression profile observed with P. denitrificans in responding to biotin is expected to be physiologically relevant.
Conclusions
The data shown here represented a first paradigm that the crosstalk between two functional BioR regulators is involved in modulating bacterial biotin metabolism. The BioR-mediated regulatory network for biotin metabolism is unprecedent, complicated/complex in that no less than four aspects are involved (Fig.1). Briefly, (1) The two BioR (BioR1 and BioR2) are autoregulators; (2) BioR1 and BioR2 can crossregulate each other; (3) BioR1 (and/or BioR2) can repress the bioY biotin transporter-containing bioYB2 operon; (4) BioR1 (and/or BioR2) negatively regulates the bioBFDAGC operon encoding the full biotin synthesis pathway (Fig.1). Given the fact that birA of P. denitrificans only encode a Group I BPL lacking the DNA-binding motif, it is reasonable that BioR, a novel GntR-like transcription factor, is evolved to compensate for the loss of regulatory function of BirA, a monofunctional BPL. As we earlier proposed (Feng et al. 2013a,b), we still favored a two-protein model of BirA and BioR, which represents an alternative mechanism for bacterial biotin sensing. The dramatic GC% difference of the two bioR homologs argues greatly the prediction that the event of bioR duplication exists in P. denitrificans (Table3). The fact that the number of BioR sites in P. denitrificans is most also determines in part the complexity in regulation of biotin metabolism by BioR.
Somewhat it seemed unexpected that the addition of exogenous biotin exerted an opposite effect on biotin biosynthesis operon bioBFDAGC and biotin transporter-containing operon bioYB in that it is quite different from the perspective of other biotin regulons in different bacterial species. For instance, BirA is a repressor of both biotin biosynthesis and transport genes in Bacillus sphaericus and other Firmicutes (Bower et al. 1995, 1996). Recently, BirA was found to act as a repressor of the novel biotin transporter yigM in E.coli (http://epub.uni-regensburg.de/15822/). BioQ in Actinobacteria also acts a repressor of both biotin biosynthesis and transporter operon (Brune et al. 2012; Tang et al. 2014). Moreover, even in yeasts, it has been shown that in addition to the transport genes, low biotin concentrations result in increased levels of transcription of the biosynthetic genes as well as the gene that encodes the BPL (Pirner and Stolz 2006; Beckett 2007).
We still have no success in identifying the possible ligands for BioR binding to cognate promoters (Fig.6). Crystallization of BioR alone and bound DNA might be a direct way to visualize if the ligand molecule is present or not. However, this approach seemed not easy in that the BioR protein is weird/hard tractable (precipitate at high level). In fact, we are frustrated by the in vitro performance of BioR proteins from three different organisms (A. tumefaciens, B. melitensis, and P. denitrificans) to some extent. The other possibility might be some unknown signaling pathway is linked to BioR-mediated regulation mechanism. While no evidence supports the above hypothesis right now. In summary, the existence of two BioR homologs in P. denitrificans defines a complex regulatory network, augmenting the diversity in the context of bacterial biotin metabolism.
Acknowledgments
This work was supported by the Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars (grant no. LR15H190001), and the start-up package from Zhejiang University (Y. F.). Feng is a recipient of the “Young 1000 Talents” Award.
Conflict of Interest
None declared.
Supporting Information
Figure S1. Molecular dissection for promoters of the bio operons from Paracoccus denitrificans. (A) The promoter of the bioR1 (Pden_1431) from P. denitrificans. (B) The promoter of the bioYB (Pden_1432) operon from P. denitrificans. (C) The promoter of the bioR2 (Pden_2922) from P. denitrificans. (D) The promoter of the bioBFDAGC (Pden_2916) operon from P. denitrificans. The predicted BioR site is given in cyan and underlined letter, and the possible ribosome binding site (RBS) is shown in purple and underlined type. The anticipated −10 and −35 regions are underlined in yellow. S, denotes transcription initiation site; M, denotes translation start site.
Figure S2. Binding of Paracoccus denitrificans BioR1 (Pden_1433) to cognate promoters. (A) Binding of the P. denitrificans BioR1 (Pden_1431) the Agrobacterium tumefaciens bioBFDAZ promoter. (B) Binding of the P. denitrificans BioR1 (Pden_1431) its own promoter. (C) Interplay between the P. denitrificans BioR1 (Pden_1431) protein with the promoter of the P. denitrificans bioBFDAGC operon. (D) The P. denitrificans BioR1 (Pden_1431) protein interact with the bioYB operon. The BioR1 protein seemed unusual in that it very easily precipitates during the process of prepration in vitro, thus the crude extract of Escherichia coli overexpressing the Pden_1431 protein is used in the EMSA assays.
References
- Baker SC, Ferguson SJ, Ludwig B, Page MD, Richter OM. van Spanning RJ. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol. Mol. Biol. Rev. 1998;62:1046–1078. doi: 10.1128/mmbr.62.4.1046-1078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DF. Campbell AM. The birA gene of Escherichia coli encodes a biotin holoenzyme synthetase. J. Mol. Biol. 1981a;146:451–467. doi: 10.1016/0022-2836(81)90042-5. [DOI] [PubMed] [Google Scholar]
- Barker DF. Campbell AM. Genetic and biochemical characterization of the birA gene and its product: evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J. Mol. Biol. 1981b;146:469–492. doi: 10.1016/0022-2836(81)90043-7. [DOI] [PubMed] [Google Scholar]
- Beckett D. The Escherichia coli biotin regulatory system: a transcriptional switch. J. Nutr. Biochem. 2005;16:411–415. doi: 10.1016/j.jnutbio.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Beckett D. Biotin sensing: universal influence of biotin status on transcription. Annu. Rev. Genet. 2007;41:443–464. doi: 10.1146/annurev.genet.41.042007.170450. [DOI] [PubMed] [Google Scholar]
- Beckett D. Biotin sensing at the molecular level. J. Nutr. 2009;139:167–170. doi: 10.3945/jn.108.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower S, Perkins J, Yocum RR, Serror P, Sorokin A, Rahaim P, et al. Cloning and characterization of the Bacillus subtilis birA gene encoding a repressor of the biotin operon. J. Bacteriol. 1995;177:2572–2575. doi: 10.1128/jb.177.9.2572-2575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower S, Perkins JB, Yocum RR, Howitt CL, Rahaim P. Pero J. Cloning, sequencing, and characterization of the Bacillus subtilis biotin biosynthetic operon. J. Bacteriol. 1996;178:4122–4130. doi: 10.1128/jb.178.14.4122-4130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PH, Cronan JE, Grotli M. Beckett D. The biotin repressor: modulation of allostery by corepressor analogs. J. Mol. Biol. 2004;337:857–869. doi: 10.1016/j.jmb.2004.01.041. [DOI] [PubMed] [Google Scholar]
- Brune I, Gotker S, Schneider J, Rodionov DA. Tauch A. Negative transcriptional control of biotin metabolism genes by the TetR-type regulator BioQ in biotin-auxotrophic Corynebacterium glutamicum ATCC 13032. J. Biotechnol. 2012;159:225–234. doi: 10.1016/j.jbiotec.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Chakravartty V. Cronan JE. Altered regulation of Escherichia coli biotin biosynthesis in birA superrepressor mutant strains. J. Bacteriol. 2012;194:1113–1126. doi: 10.1128/JB.06549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman-Smith A. Cronan JE., Jr The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. Trends Biochem. Sci. 1999;24:359–363. doi: 10.1016/s0968-0004(99)01438-3. [DOI] [PubMed] [Google Scholar]
- Cronan JE., Jr The E. coli bio operon: transcriptional repression by an essential protein modification enzyme. Cell. 1989;58:427–429. doi: 10.1016/0092-8674(89)90421-2. [DOI] [PubMed] [Google Scholar]
- Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, et al. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 2010;38:D396–D400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dillewijn P, Soto MJ, Villadas PJ. Toro N. Construction and environmental release of a Sinorhizobium meliloti strain genetically modified to be more competitive for alfalfa nodulation. Appl. Environ. Microbiol. 2001;67:3860–3865. doi: 10.1128/AEM.67.9.3860-3865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinfor. 2004a;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004b;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Cronan JE. Escherichia coli unsaturated fatty acid synthesis: complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J. Biol. Chem. 2009a;284:29526–29535. doi: 10.1074/jbc.M109.023440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Cronan JE. A new member of the Escherichia coli fad regulon: transcriptional regulation of fadMybaW. J. Bacteriol. 2009b;191:6320–6328. doi: 10.1128/JB.00835-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Cronan JE. Overlapping repressor binding sites result in additive regulation of Escherichia coli FadH by FadR and ArcA. J. Bacteriol. 2010;192:4289–4299. doi: 10.1128/JB.00516-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Cronan JE. Complex binding of the FabR repressor of bacterial unsaturated fatty acid biosynthesis to its cognate promoters. Mol. Microbiol. 2011;80:195–218. doi: 10.1111/j.1365-2958.2011.07564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Xu J, Zhang H, Chen Z. Srinivas S. Brucella BioR regulator defines a complex regulatory mechanism for bacterial biotin metabolism. J. Bacteriol. 2013a;195:3451–3467. doi: 10.1128/JB.00378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Zhang H. Cronan JE. Profligate biotin synthesis in α-Proteobacteria – A develoing or degenerating regulatory system? Mol. Microbiol. 2013b;88:77–92. doi: 10.1111/mmi.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillen-Navarro K, Encarnacion S. Dunn MF. Biotin biosynthesis, transport and utilization in Rhizobia. FEMS Microbiol. Lett. 2005;246:159–165. doi: 10.1016/j.femsle.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W. Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hebbeln P, Rodionov DA, Alfandega A. Eitinger T. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc. Natl. Acad. Sci. USA. 2007;104:2909–2914. doi: 10.1073/pnas.0609905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke SK. Cronan JE. Successful conversion of the Bacillus subtilis BirA Group II biotin protein ligase into a Group I ligase. PLoS ONE. 2014;9:e96757. doi: 10.1371/journal.pone.0096757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Richter DC, Rausch C, Dezulian T, Franz M. Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinfor. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John P. Whatley FR. Paracoccus denitrificans and the evolutionary origin of the mitochondrion. Nature. 1975;254:495–498. doi: 10.1038/254495a0. [DOI] [PubMed] [Google Scholar]
- Ke Y, Yuan X, Wang Y, Bai Y, Xu J, Song H, et al. Genome sequences of Brucella melitensis 16M and its two derivatives 16M1w and 16M13w, which evolved in vivo. J. Bacteriol. 2012;194:5489. doi: 10.1128/JB.01293-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Zhao S, Vetting MW, Wood BM, Sakai A, Cho K, et al. Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine. MBio. 2014;5:e00933-13. doi: 10.1128/mBio.00933-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Mittenhuber G. Friedrich CG. Transfer of Thiosphaera pantotropha to Paracoccus denitrificans. Int. J. Syst. Bacteriol. 1993;43:363–367. doi: 10.1099/00207713-43-2-363. [DOI] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, et al. RegPrecise 3.0–a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genom. 2013;14:745. doi: 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirner HM. Stolz J. Biotin sensing in Saccharomyces cerevisiae is mediated by a conserved DNA element and requires the activity of biotin-protein ligase. J. Biol. Chem. 2006;281:12381–12389. doi: 10.1074/jbc.M511075200. [DOI] [PubMed] [Google Scholar]
- Rainey FA, Kelly DP, Stackebrandt E, Burghardt J, Hiraishi A, Katayama Y, et al. A re-evaluation of the taxonomy of Paracoccus denitrificans and a proposal for the combination Paracoccus pantotrophus comb. nov. Int. J. Syst. Bacteriol. 1999;49(Pt. 2):645–651. doi: 10.1099/00207713-49-2-645. [DOI] [PubMed] [Google Scholar]
- Rodionov DA. Gelfand MS. Computational identification of BioR, a transcriptional regulator of biotin metabolism in Alphaproteobacteria, and of its binding signal. FEMS Microbiol. Lett. 2006;255:102–107. doi: 10.1111/j.1574-6968.2005.00070.x. [DOI] [PubMed] [Google Scholar]
- Rodionov DA, Mironov AA. Gelfand MS. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 2002;12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddavattam D, Karegoudar TB, Mudde SK, Kumar N, Baddam R, Avasthi TS, et al. Genome of a novel isolate of Paracoccus denitrificans capable of degrading N, N-dimethylformamide. J. Bacteriol. 2011;193:5598–5599. doi: 10.1128/JB.05667-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit WR. Entcheva P. Biotin in microbes, the genes involved in its biosynthesis, its biochemical role and perspectives for biotechnological production. Appl. Microbiol. Biotechnol. 2003;61:21–31. doi: 10.1007/s00253-002-1186-2. [DOI] [PubMed] [Google Scholar]
- Tang Q, Li X, Zou T, Zhang H, Wang Y, Gao R, et al. Mycobacterium smegmatis BioQ defines a new regulatory network for biotin metabolism. Mol. Microbiol. 2014;94:1006–1023. doi: 10.1111/mmi.12817. [DOI] [PubMed] [Google Scholar]
- Van den Eede G, Deblaere R, Goethals K, Van Montagu M. Holsters M. Broad host range and promoter selection vectors for bacteria that interact with plants. Mol. Plant Microbe Interact. 1992;5:228–234. doi: 10.1094/mpmi-5-228. [DOI] [PubMed] [Google Scholar]
- Zhao S, Kumar R, Sakai A, Vetting MW, Wood BM, Brown S, et al. Discovery of new enzymes and metabolic pathways by using structure and genome context. Nature. 2013;502:698–702. doi: 10.1038/nature12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Molecular dissection for promoters of the bio operons from Paracoccus denitrificans. (A) The promoter of the bioR1 (Pden_1431) from P. denitrificans. (B) The promoter of the bioYB (Pden_1432) operon from P. denitrificans. (C) The promoter of the bioR2 (Pden_2922) from P. denitrificans. (D) The promoter of the bioBFDAGC (Pden_2916) operon from P. denitrificans. The predicted BioR site is given in cyan and underlined letter, and the possible ribosome binding site (RBS) is shown in purple and underlined type. The anticipated −10 and −35 regions are underlined in yellow. S, denotes transcription initiation site; M, denotes translation start site.
Figure S2. Binding of Paracoccus denitrificans BioR1 (Pden_1433) to cognate promoters. (A) Binding of the P. denitrificans BioR1 (Pden_1431) the Agrobacterium tumefaciens bioBFDAZ promoter. (B) Binding of the P. denitrificans BioR1 (Pden_1431) its own promoter. (C) Interplay between the P. denitrificans BioR1 (Pden_1431) protein with the promoter of the P. denitrificans bioBFDAGC operon. (D) The P. denitrificans BioR1 (Pden_1431) protein interact with the bioYB operon. The BioR1 protein seemed unusual in that it very easily precipitates during the process of prepration in vitro, thus the crude extract of Escherichia coli overexpressing the Pden_1431 protein is used in the EMSA assays.






