Abstract
Numerous observations indicate a high flexibility of microbial communities in different biogas reactors during anaerobic digestion. Here, we describe the functional redundancy and structural changes of involved microbial communities in four lab-scale continuously stirred tank reactors (CSTRs, 39°C, 12 L volume) supplied with different mixtures of maize silage (MS) and sugar beet silage (SBS) over 80 days. Continuously stirred tank reactors were fed with mixtures of MS and SBS in volatile solid ratios of 1:0 (Continuous Fermenter (CF) 1), 6:1 (CF2), 3:1 (CF3), 1:3 (CF4) with equal organic loading rates (OLR 1.25 kgVS m−3 d−1) and showed similar biogas production rates in all reactors. The compositions of bacterial and archaeal communities were analysed by 454 amplicon sequencing approach based on 16S rRNA genes. Both bacterial and archaeal communities shifted with increasing amounts of SBS. Especially pronounced were changes in the archaeal composition towards Methanosarcina with increasing proportion of SBS, while Methanosaeta declined simultaneously. Compositional shifts within the microbial communities did not influence the respective biogas production rates indicating that these communities adapted to environmental conditions induced by different feedstock mixtures. The diverse microbial communities optimized their metabolism in a way that ensured efficient biogas production.
Introduction
In 2013, 7.6% of the end energy consumption in Germany was gained from biogas. 48% of the 7850 biogas plants in Germany use renewable raw materials as feedstocks, such as maize silage (MS, 73%) and grass silage (12%) (FNR, 2014).
Utilization in biogas reactors of these plant materials requires retention times of about 100 days even though fibre-rich feedstocks may not get fully degraded (Boe and Angelidaki, 2009; Procházka et al., 2012). In contrast, feedstocks containing easy-degradable sugars and alcohols, such as sugar beet silage (SBS) can be rapidly degraded leading to an acidification of the biogas sludge and consequently to an inhibition of methane production (Kryvoruchko et al., 2009). Furthermore, SBS is a difficult feedstock in terms of the availability of nutrients and buffering capacity (Demirel and Scherer, 2008). However, it is an interesting co-feedstock for biogas production (Weissbach and Strubelt, 2008) with a similar biogas yield compared with maize silage (Klang et al., 2008). Co-fermentation of SBS with other feedstocks can lead to a stable process and has been shown to enhance biogas production resulting in a more efficient biogas process (El-Mashad and Zhang, 2010). However, currently just 2% of the renewable raw materials used in biogas plants in Germany account for SBS (FNR, 2014).
Within the biogas process, methane is mainly produced from hydrogen and carbon dioxide and/or acetate by hydrogenotrophic or acetoclastic methanogenic Archaea (Liu and Whitman, 2008). In general, hydrogenotrophic methanogens are dominant in most agricultural biogas plants especially at high ammonia concentrations (Weiland, 2010). Two members of genera within the methanogenic Archaea, Methanosarcina and Methanosaeta, can use acetate as substrate for methane formation (Liu and Whitman, 2008). Since these two genera can degrade the same substrate using analogous enzymes, they are defined as functionally redundant in this study. Abundances of different acetoclastic methanogenic species are generally depended on their tolerance towards acetate (Ros et al., 2013).
However, an optimal biogas production process relies on a complex and efficient microbial community in order to deal with changing environmental conditions as well as altering substrate compositions. Although, similar archaeal and bacterial groups are present in different biogas reactors, compositions of each of the microbial communities are unique (Sundberg et al., 2013). Their identical end-products (CO2 and CH4) indicate that these microbial communities carry out similar functional processes, regardless of differences in their compositions (functional similarity) (Allison and Martiny, 2008).
In this study, the effect of different feedstock mixtures composed of MS and SBS (CF1, 1:0; CF2, 6:1; CF3, 3:1; CF4, 1:3) on the biogas process was assessed in four continuously fed 12 L lab-scale CSTRs. Therefore, we hypothesized that the biogas production from MS could be improved by co-fermentation of SBS in terms of biogas yield and process stability. Furthermore, in order to correlate possible changes in terms of an improvement of the biogas production, the archaeal and bacterial communities were investigated in relation to different feedstock mixtures by a 454-pyrosequencing approach based on 16S rRNA gene analysis and analysed by rdp (Ribosomal Database Project, Release 11.1; Cole et al., 2013).
Results
Process performance
All reactors performed well at high yields during the main experimental phase of 80 days at feedstock supply at 1.25 kgVS m−3 d−1. The cumulative specific biogas yields of all CSTRs (Table 1; Fig. S1) were higher compared with expected values calculated from KTBL data (KTBL 2013, Faustzahlen Biogas).
Table 1.
Biogas production characteristics of the studied CSTRs. Data recorded for 80 days at OLR of 1.25 kgVS m−3 d−1
| CSTR | CF1 | CF2 | CF3 | CF4 | |
|---|---|---|---|---|---|
| Feedstock mixture (MS : SBS) | 1:0 | 6:1 | 3:1 | 1:3 | |
| Average methane concentration (%) | 59 ± 4.3 | 59 ± 6.2 | 61 ± 3.9 | 60 ± 8.2 | (n = 80) |
| Cumulative specific biogas yield (lN kg−1VS) | 755 | 726 | 746 | 799 | |
| Expected specific biogas yield (lN kg−1VS) | 650 | 657 | 662 | 687 | |
| Avg. biogas production rate (lN h−1) | 0.39 ± 0.04 | 0.38 ± 0.06 | 0.36 ± 0.05 | 0.41 ± 0.09 | (n = 80) |
| pH | 7.8 ± 0.15 | 7.9 ± 0.10 | 7.9 ± 0.15 | 7.8 ± 0.10 | (n = 20) |
| VFA/TIC ratio | 0.06 ± 0.04 | 0.07 ± 0.04 | 0.06 ± 0.03 | 0.07 ± 0.04 | (n = 20) |
| C/N ratio | 11.1 ± 0.38 | 11.1 ± 0.24 | 11.3 ± 0.39 | 11.4 ± 0.77 | (n = 5) |
| NH4+−N [g l−1] | 2.7 ± 0.09 | 2.6 ± 0.010 | 2.5 ± 0.09 | 2.1 ± 0.19 | (n = 4) |
lN kg−1VS, norm litre per kilogram volatile solids; VFA/TIC ratio, volatile fatty acids/total inorganic carbon ratio; C/N ratio, carbon/nitrogen ratio.
Cumulative specific biogas yields of all reactors differed slightly (Table 1; Fig. S1), whereas the highest average methane concentration (61%) was determined for the 3:1 (MS : SBS) mixture (CF3). Regarding the process dynamics, no substantial differences between the individual mixtures were obvious, while a comparatively higher specific biogas yield (799 lN kg−1VS) has been found for CF4 containing the highest rate of SBS, which goes back to slightly higher biogas potential of SBS compared with MS.
Moreover, all reactors showed a stable process performance in terms of pH (7.5–7.9), VFA/TIC (0.04–0.18), C/N ratio (10.6–12.6) and ammonia concentrations (2.1–2.8 g l−1) (Table 1).
Overall phylogenetic analysis
454-amplicon sequencing of the five analysed biogas-producing microbial communities (inoculum and four CSTRs) resulted in a total of 399 258 archaeal and bacterial 16S rRNA gene sequences. The average sequence read length was 713 bp for bacterial and 559 bp for archaeal 16S rRNA gene sequences. The total numbers of sequences per sample obtained by the rdp workflow were given in Table S2 and used for further downstream analysis.
Taxonomic assignment of 16S rRNA gene sequences by the rdp classifier showed a high identification due to the read length of bacterial (760 bp) and archaeal sequences (550 bp). On average, 93% of the bacterial sequences were assigned at phylum level, 61% sequences on family level and 36% sequences at genus level. Furthermore, 99.7% of the archaeal sequences were assigned on phylum level, 89.8% sequences on family level and 87.8% sequences on genus level.
Species richness and biodiversity
At a sequence homology of 97%, the number of operational taxonomic units (OTUs) based on bacterial 16S rRNA gene sequences was on average 3474 and the Shannon index ranged from 4.0 to 7.4. Data analysis of archaeal 16S rRNA gene sequences resulted in average in 861 OTUs. The respective archaeal Shannon index ranged from 3.1 to 5.1. In general, the estimated numbers of archaeal OTUs as well as the diversity indices were lower compared with the corresponding values for Bacteria (Table S2).
Beta-diversity measurements (weighted UniFrac, Table S3) showed that there were high dissimilarities between bacterial communities (Fig. 1A). Compared with the inoculum (CF0), bacterial communities have shifted in similar way in respect to PCoA axis PC1. In comparison with reactor CF1, bacterial communities lined up along PCoA axis PC2 (Fig. 1A).
Figure 1.
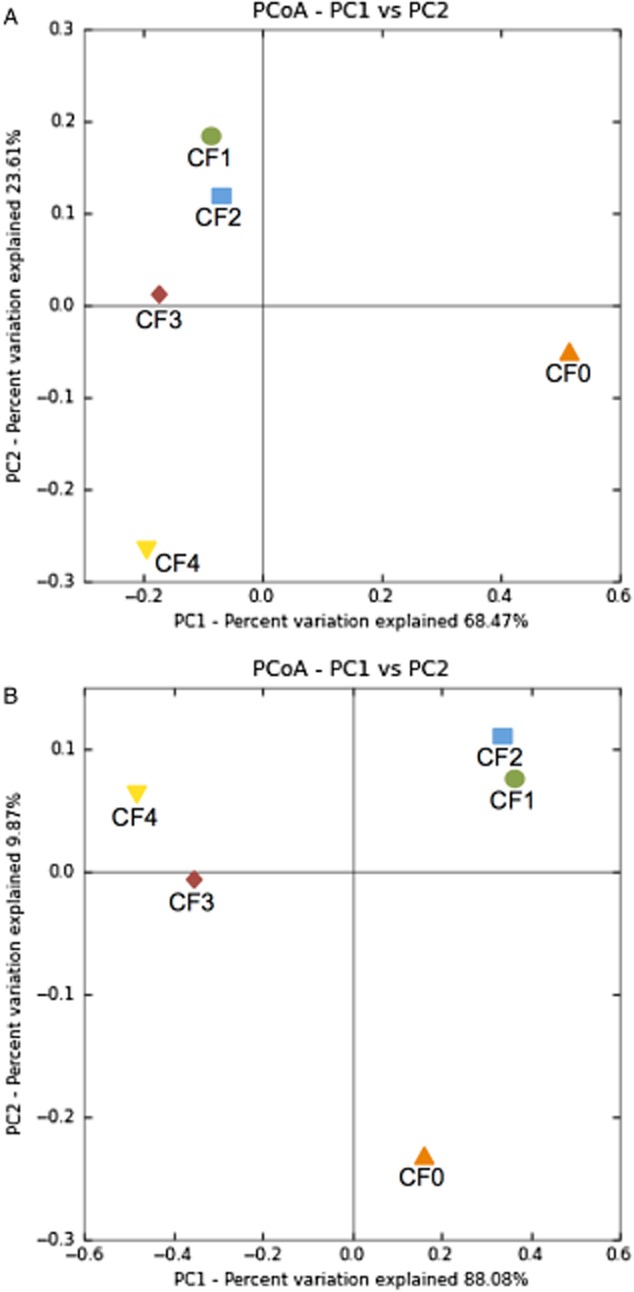
Two-dimensional PCoA of dissimilarities between (A) bacterial and (B) archaeal communities in biogas reactors based on weighted UniFrac analysis. Similar microbial communities are depicted near from each other. PC1 and PC2 explain (A) 92% and (B) 98% of the variation.
The archaeal community has also changed compared with the inoculum but at a relatively low extent as the PCoA axis PC2 explains only 9.9% of the total variance (Fig. 1B). The archaeal community in reactor CF1 with MS as mono-substrate was very similar to reactor CF2 with MS and only 14% SBS. Whereas, archaeal communities in reactors CF3 and CF4 with high amounts of SBS were highly dissimilar in comparison to reactors CF1 and CF2.
Bacterial communities
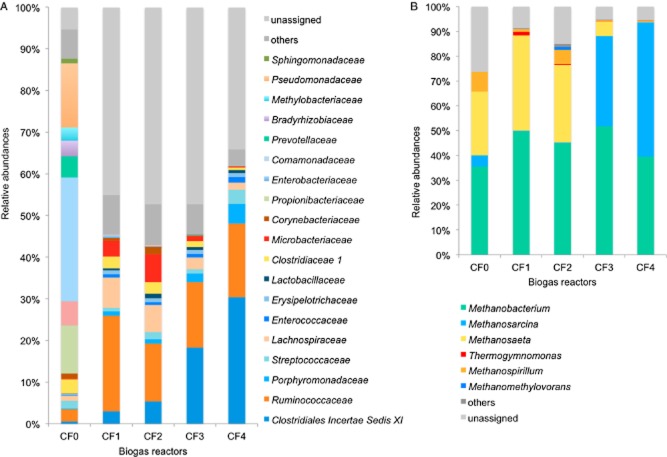
The bacterial population in the inoculum was dominated by the phyla Proteobacteria (63%) and Firmicutes (14%). In all CSTRs, the phylum Firmicutes (CF1, 77%; CF2, 70%; CF3, 80%; CF4, 78%) was prevalent. Furthermore, Bacteroidetes and Actinobacteria were present in the inoculum and all four CSTRs. Relative abundances of members of bacterial families comprising at least 1% of the communities were shown in Fig. 2A. At the start (CF0) of the experiment, the bacterial population was dominated by members of the families Comamonadaceae (30%), Pseudomonadaceae (15%), Enterobacteriaceae (5.8%) (Proteobacteria), Propionibacteriaceae (12%) (Actinobacteria) and Clostridiaceae 1 (3.4%) (Firmicutes). Only 6.9% of the sequences obtained from sample CF0 remained unassigned. With the addition of feedstock mixtures to the reactors CF1–CF4 over a period of 80 days, bacterial abundances were more unevenly distributed compared with the start (CF0), and the proportion of unassigned sequences was on average 43% (± 6). Furthermore, the bacterial community structure changed completely. Besides Clostridiaceae 1, the abundances of the mentioned families found in the inoculum (CF0) were smaller than 1% in all CSTRs (CF1-CF4).
Figure 2.
A. Bacterial community composition on family level and (B) archaeal community composition on genus level at the beginning of the experiment in the inoculum (CF0) and after 121 d AD in four lab-scale biogas reactors CF1–CF4 resulting from the 454 amplicon data analysis (confidence cut-off 80%).
The most dominant families of the domain Bacteria during monofermentation of MS (CF1) were classified as Ruminococcaceae (22.8%), Lachnospiraceae (7.2%), Clostridiaceae 1 (2.8%) within the phyla Firmicutes and Microbacteriaceae (3.8%) within the phylum Actinobacteria (Fig. 2A, marked in yellow-orange tones). With increasing amounts of SBS, the bacterial community shifted gradually, except for Ruminococcaceae. Bacteria belonging to the family Ruminococcaceae were present in high abundances in all reactors (CF2, 14%; CF3, 16%; CF4, 18%). Most notably, the abundances of members of Clostridiales Insertae Sedis XI increased considerably from 3% in reactor CF1 up to 30% in reactor CF4 (CF0, 0.6%; CF2, 5%; CF3, 18%) with the addition of SBS. Furthermore, relative abundances of the families Streptococcaceae (CF1, 0.9%; CF2, 1.6%; CF3, 1.1%; CF4, 3.4%), Enterococcaceae (CF1, 0.7%; CF2, 0.7%; CF3, 0.8%; CF4, 1.4%) and Porphyromonadaceae (CF1, 1%; CF2, 1.1%; CF3, 2.1%; CF4, 4.8%), (Fig. 2A, marked in blue tones) increased slightly with increasing amounts of SBS.
Archaeal communities
The Archaea detected in all samples belonged to the phyla Euryarchaeota (93.3%) and Crenarchaeota (6.4%). At the beginning of the experiment (CF0), the archaeal community showed a rather balanced composition between the four genera Methanobacterium (35.6%), Methanosaeta (25.8%), Methanospirillum (7.8%) and Methanosarcina (4.5%), while about 25% remained unclassified (Fig. 2B). After anaerobic digestion (AD) for 80 days, the abundances of the hydrogenotrophic Methanobacteria (CF1, 50%; CF2, 45%; CF3, 52% and CF4, 40%) in all samples (CF1–CF4) were similar to the start (CF0) independent from the amount of SBS fed to the four CSTRs (Fig. 2B). Furthermore, different members of acetoclastic Archaea were observed in relation to SBS. In reactors fed with MS mainly, Methanosaeta species (CF1, 38% and CF2, 32%) dominated. However, with increasing amounts of SBS the archaeal community shifted gradually. In reactors with high amounts of SBS, Methanosaeta proportion was as low as 6% in CF3 and almost absent in CF4 (0.4%), whereas members of the genus Methanosarcina were prevalent in reactors CF3 (36%) and CF4 (54%).
Discussion
Process performance
All four lab-scale CSTRs reached high-specific biogas yields (Table 1; Fig. S1). Biogas yields are significantly affected by system technology as well as feedstock quality (KTBL 2013; Mähnert and Linke, 2009). Moreover, high biogas yields are typical for reactors at low organic loading rates (OLRs; Mähnert and Linke, 2009). Biogas production rates and methane concentrations were similar in all CSTRs (Table 1). pH values between 7 and 8, ammonia concentrations below 5 g l−1 and low volatile fatty acid/total inorganic carbon (VFA/TIC) ratios (< 0.07 ± 0.04) indicate process stability (Drosg, 2013). Furthermore, C/N ratios were within the optimum range of 10–15 (Weiland, 2010). Consequently, there was no limitation in any of the reactors in terms of nutrients and process instability.
Feedstock characterization (Table S1) showed that SBS contains mainly ethanol, short chain fatty acids as well as sugar, which promote faster feedstock degradation (Weissbach, 2009; Böttcher et al., 2011). The comparatively higher specific cumulative biogas yield of reactor CF4 containing the highest rate of SBS (MS : SBS-1:3) goes back to slightly higher biogas potential of SBS (Table 1; Fig. S1). Consequently, the effect of SBS on AD of MS was not quite obvious in terms of biogas yields, methane concentrations and especially in terms of biogas production rates. Thus, the hypothesis of improving biogas production from MS with addition of SBS was rejected.
Species richness and biodiversity
Results showed a great species richness (OTUs) in all reactors (Table S2). Nelson and colleagues (2011) found similar numbers of archaeal (296 OTUs) and bacterial OTUs (5926) investigating multiple anaerobic digesters by a meta-analysis based on 16S rRNA gene sequencing. However, a high number of OTUs is not only caused by species richness but also by the number of sequence reads obtained by high-throughput sequencing (Roesch et al., 2007).
Regarding the total number of species/OTUs, the calculation of the Shannon Index showed that the biodiversity (H′) of the archaeal community was lower compared with bacterial biodiversity in biogas reactors (Table S2). This result is in line with other studies (Fernandez et al., 1999; Bengelsdorf et al., 2013; Gagliano et al., 2015). Differences in feedstock mixtures did not affect the biodiversity notably as indicated by similar values for H′.
Bacterial communities
CSTRs mainly fed with fibre-rich MS (i.e. CF1 and CF2; Fig. 2A) were dominated by cellulolytic Bacteria such as members of the families Ruminococcaceae, Lachnospiraceae, Clostridiaceae 1 and Microbacteriaceae. For example, members of the families Ruminococcaceae and Lachnospiraceae persist in fibrolytic communities (Brulc et al., 2009). They contain a wide range of glycoside hydrolases and carbohydrate-binding modules, enabling them to colonize complex plant material and to degrade recalcitrant polymers, such as cellulose and hemicellulose. Furthermore, they are able to cleave bonds of starch, cellobiose, cellodextrin and chitobiose (Biddle et al., 2013). Therefore, members of these families play a common role as active plant degraders (Biddle et al., 2013).
In contrast, CSTRs fed with high amounts of SBS were dominated by members of the families Clostridiales Insertae Sedis XI, Porphyromonadaceae, Streptococcaceae and Enterococcaceae (Fig. 2A). The family Clostridiales Insertae Sedis XI is a heterogeneous group that includes a wide range of different Bacteria (Pagnier et al., 2014), and any functional prediction for clostridia only by using phylogenetic information would be highly speculative due to their broad metabolic potential (Fernandez et al., 2000). Bacteria of the family Porphyromonadaceae are commonly found in mesophilic full-scale biogas plants (Liu et al., 2009; Bengelsdorf et al., 2013; Li et al., 2013; Pope et al., 2013). These Bacteria play a central role in glucose fermentation (Li et al., 2009). Members of the families Streptococcaceae and Enterococcaceae are facultative anaerobic, use homolactic acid fermentation (Fisher and Phillips, 2009; Whiley and Hardie, 2009) and were found in other mesophilic biogas reactors (Li et al., 2013). Moreover, Streptococcaceae-related microorganisms were dominant in biogas reactors fed with high amounts of glucose (Fernandez et al., 2000). An increased availability of monosaccharides in CSTRs with high amounts of SBS could have led to the increase of their relative abundances.
Altogether, the composition of each bacterial community was adapted to the supplied feedstock mixture. Thus, functional similarity of highly diverse bacterial communities led to similar biogas production rates in all reactors.
Archaeal communities
Methane production from acetate is a redundant function performed either by Methanosaeta and/or by Methanosarcina species (Liu and Whitman, 2008). Due to high affinity to acetate, Methanosaeta probably outcompeted Methanosarcina in reactors with low amounts of SBS (Fig. 2B). As indicated in Fig. 1B, this pronounced shift took place at the increasing share of SBS addition from 14% (CF2) to 25% (CF3). In reactors with high amounts of SBS, Methanosarcina became dominant most likely due to its tolerance to high acetate levels (Ros et al., 2013). Consequently, the diverse and functionally redundant members of archaeal communities are able to adapt to changing environmental conditions, and the most competent species consortium is prevailing in the metabolic functions.
Process performance linked to microbial community structures
Dynamics of microbial communities in biogas reactors depend on several factors for instance on hydraulic retention time, temperature, pH, OLR and ammonia concentration (Dollhopf et al., 2001; Ye et al., 2007; Rademacher et al., 2012; Hai et al., 2014; Werner et al., 2014). In this study, shifts in microbial communities (Fig. 2) were most likely induced by feedstock compositions varying in the proportions of MS and SBS. With respect to the hypothesis, we showed that these shifts clearly not affect the biogas production rates of CSTRs digesting complex feedstock mixtures (Table 1). A similar result was found by Fernandez and colleagues (2000). They indicated that the functional stability of AD was linked to a flexible microbial community. Moreover, Briones and Raskin (2003) pointed out the importance of diverse and flexible communities for a stable process performance.
In this study, microbial communities carried out functional processes at similar rates, regardless of differences in composition (Fig. S1; Fig. 2A). The individually composed bacterial communities in each CSTR had fulfilled similar functions (functional similarity), which finally resulted in similar biogas production rates in each reactor.
Even more important is the shift in the Archaea composition towards Methanosarcina with increasing proportion of SBS. Archaeal communities (Fig. 2B) newly established after SBS co-fermentation are functionally redundant, and thus biogas production rates and methane concentrations (Table 1) were not altered by compositional shifts (Allison and Martiny, 2008).
Functional redundancy is considered as insurance to maintain ecosystem functions under changing environmental conditions (McMahon et al., 2007). Moreover, stable ecosystem functions could be sustained by highly dynamic communities (Friedrich et al., 2003; Stamper et al., 2003; Wittebolle et al., 2008; Ayarza et al., 2010; Cabrol et al., 2012).
Conclusions
In this study, microbial communities stabilized the digestion process and led to an efficient biogas production from different feedstock compositions. Both bacterial and archaeal communities are responding simultaneously to the supply of easy-to-degrade feedstocks. This finding is unique for the AD research and explains the common observation from biogas practitioners that each biogas plant is an own specific ecosystem, hardly comparable to a second plant.
Experimental procedures
Experimental set-up
Anaerobic digestion was performed continuously in four mixed 12 L lab-scale CSTRs (operating volume 10 L, temperature 39°C).
The cylindrical, double-layered biogas reactors made of stainless steel were built in the mechanical workshop of the Ulm University under the supervision of the authors’ institute. Reactor temperature was maintained by passing water from additional hot water bath through copper pipes surrounding the reactors. The water inside the water bath was heated up to 42°C by a heating circulator (JULABO GmbH, Seelbach, Germany).
Feedstocks for biogas production were introduced into the reactors by screw conveyors placed on the bases of each feedstock storage tubes. The corresponding operating unit allowed the subdivision of each rotation into 12 impulses (1 impulse = 30°). Digital card under LabVIEW sent fixed number of impulses to the feeding system at given time interval. In the present experiment, the number of screw conveyor impulses was set individually for each mixture, and the feedstock input was realized automatically on hourly basis. Mixing of SBS with MS might produce high viscosity on the surface of the carrier. Thus, all surfaces were regularly cleaned for good performance as suggested by Scherer and colleagues (2009). The automatic feeding system was controlled daily by the operators of CSTRs. Homogeneous mixing inside CSTRs was achieved by stirring every 10 min for 5 min at 80 r.p.m.
The inoculum originated from a mesophilic (40°C) full-scale biogas plant (Ulm-Gögglingen, Germany) fed with MS (56%), pig manure (30%) and grass silage (14%). Inoculum was used after filtration and kept in the reactors at 40°C for 19 days in order to minimize its background methane production (Li et al., 2013; VDLUFA, 2011). Maize silage and SBS were obtained from the above-mentioned biogas plant (Gögglingen, Germany) and the Raiffeisenwarengenossenschaft Emsland-Süd (Lünne, Germany) respectively. Feedstock characteristics (Table S1) make obvious that SBS contains easily degradable components like alcohol, carboxylic acid and sugar, which leads to intensive formation of organic acid that causes low pH values (Böttcher et al., 2011). Furthermore, mono-fermentation of SBS requires larger volume of reactors or reduction of OLR and causes foam evolution (Kaiser et al., 2008; Böttcher et al., 2011).
The underlying aim of the study was to test the influence of different amounts of SBS on the reactor performance. Therefore, the lab-scale CSTRs were fed with different feedstock mixtures of MS and SBS in a ratio (based on volatile solids, VS) of 1:0 in CF1, 6:1 in CF2, 3:1 in CF3 and 1:3 in CF4 respectively. Anaerobic digestion in all CSTRS was started with a low OLR at 0.69 kgVS m−3 d−1 for a period of 41 days. After the process was stable in terms of biogas yield as well as VFA/TIC values (0.05 ± 0.01), OLR was increased to 1.25 kgVS m−3 d−1, and the feeding continued for further 80 days.
The experiment was designed for online monitoring of the biogas volume and methane concentrations in each reactor. The volume was assessed by Miligascounter (Ritter GmbH, Bochum, Germany) and given at standard condition (1.013 bar, 0°C and 0% RH). Methane concentrations were measured by infrared sensors (Bluesens GmbH, Herten, Germany, relative accuracy: ± 2% of the measured value). The sensors were calibrated with a 60% CH4 certified calibration gas.
Other process parameters like pH, the ratio of VFA/TIC, carbon/nitrogen (C/N) ratio and the amount of total solids (TS) and volatile solids (VS) were determined weekly. Total solids and VS were determined according to APHA methods 2540B and 2540E (APHA, 1999). The pH of the sludge was determined using Metrohm 605 (Filderstadt, Germany). The VFA/TIC ratio was measured by automatic titration (Dosimat 665, Metrohm, Hersau, Switzerland) with 1 M HCl to end-points of pH 5 and 4.3 (Voß et al., 2009). The analysis of C and N in the feedstocks and in the sludge was made by TrueSpec C/N analyser (LECO Instrumente GmbH, Mönchengladbach, Germany). Ammonia-nitrogen (NH4+−N) concentrations were determined with a gas-sensitive ammonia electrode (Type NH 500/2, WTW GmbH, Weilheim, Germany).
Ash-free neutral detergent fibre (NDFom), ash-free acid detergent fibre (ADFom), acid detergent lignin (ADL) and relevant characteristics (crude protein, crude fat, starch, sugar, crude fibre) were determined in the accredited Institute for Oil and Environment, LUFA Nord-West (Oldenburg, Germany). Cellulose, hemicellulose and lignin amounts were obtained from NDFom, ADFom and ADL.
Biogas yields, average methane concentrations, biogas and methane production rates were calculated on hourly basis respectively. Cumulative specific biogas yields were calculated based on cumulative biogas yields for total volatile solid inputs to each individual reactor. Methane concentrations as well as biogas production rates were calculated from average values per day (Table 1).
Sampling and deoxyribonucleic acid (DNA) extraction
For microbial community analysis, one sample was drawn from the inoculum (CF0) at the beginning of the experiment. Further samples were taken from each reactor (CF1, CF2, CF3 and CF4) after 80 days of AD. Samples were stored at −20°C before DNA was extracted by a modified protocol of Klocke and colleagues (2008). To remove polymerase chain reaction (PCR)-inhibiting substances, 4 ml of each sample were washed with 20 ml sodium phosphate buffer (Hugenholtz et al., 2002), 1 ml polyvinylpyrrolidon solution (35%) and 2 ml cetyl trimethylammonium bromide solution (10%) and centrifuged at 5000 r.p.m. for 20 min. The supernatants were removed, and the washing step was repeated 3–4x until the supernatants became clear. After washing, the respective pellets were suspended in 5 ml of a saline EDTA buffer (0.1 mol l−1 EDTA, 0.15 mol l−1 NaCl), and cells were lysed by Rybolyser (Hybaid, Middlesex, UK) for 3x 45 s at a speed of 6 ms−1. Total DNA of the samples was extracted based on phenol/chloroform DNA extraction as described in the protocol of Klocke and colleagues (2008). Deoxyribonucleic acid concentration of the extracted DNA was measured with the NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmingtion, USA) at 260 nm.
PCR amplification and sequencing
Barcode-tagged primers (Table S4) were used to amplify the V4-V6 region of archaeal 16S rRNA genes (555 bp) and the V3-V6 region of bacterial 16S rRNA genes (760 bp) from the extracted DNA of samples CF0, CF1, CF2, CF3 and CF4 by PCR. Approximately 100 ng of genomic DNA isolated from biogas reactors was used as template for each PCR reaction.
Archaeal 16S rRNA genes were amplified by ReproFast-DNA Polymerase (Genaxxon Bioscience GmbH, Ulm, Germany) under the following thermal cycling conditions: a prior denaturation step of 95°C for 5 min, a first loop of 15 cycles of 95°C for 45 s, 55°C for 60 s and 72°C for 60 s, and a second loop of 15 cycles of 95°C for 45 s, 57°C for 60 s and 72°C for 60 s, followed by a terminal elongation step of 72°C for 10 min. Amplification of bacterial 16S rRNA genes for the pyrosequencing approach was done with a DNA-free Taq DNA Polymerase (AppliChem GmbH, Darmstadt, Germany) under appropriate buffer conditions. Temperature steps of the PCR were: a prior denaturation step of 94°C for 5 min, a first loop of 10 cycles of 94°C for 60 s, 55°C for 30 s and 72°C for 60 s and a second loop of 20 cycles of 94°C for 30 s, 60°C for 30 s and 72°C for 60 s, followed by a terminal elongation step of 72°C for 10 min. 16S rRNA gene amplification and amplicon length were checked on a 2% agarose gel stained with ethidium bromide. For each sample, three independent PCR reactions were accomplished for either, archaeal and bacterial 16S rRNA genes. Merged PCR amplicons were purified by the purification kit DNA clean & concentrator-5 (ZYMO Research Europe GmbH, Freiburg, Germany). Deoxyribonucleic acid concentration of the purified PCR amplicons was measured with the NanoDrop 2000 Spectrophotometer at 260 nm. Finally, bar code-tagged DNA fragments of bacterial and archaeal 16S rRNA genes were equimolarly merged, and sequencing was performed by Roche GS FLX++ chemistry (MWG eurofins, Berlin, Germany) on 0.5 plate.
Data analysis
The analysis of 454 amplicon sequencing data was performed by Ribosomal Database Project (RDP). Following the RDP supervised workflow, archaeal and bacterial 16S rRNA gene sequences were sorted according to their bar codes and trimmed. Sequences with a deviation greater than ± 10% from the expected fragment lengths were discarded. Low-quality sequences (minimal quality score 25) were filtered out, and unspecific sequences were excluded by RDP’s sequence selection tool. Remaining sequences were checked for chimeras in the de novo mode by Usearch 6.0 (Edgar, 2010). After filtering processes, 55% of the sequences were discarded. Thus, 180 752 sequences remained for downstream analysis. These sequences were aligned by the infernal aligner (Nawrocki and Eddy, 2013). Based on the aligned sequences, complete linkage clustering (farthest neighbour method) was performed with a distance cut-off of 0.03.
The resulting cluster file was used to calculate rarefaction and alpha diversity (Shannon and Chao1 index). Furthermore, differences between microbial communities (ß-diversity) were measured by a weighted UniFrac analysis using qiime (Lozupone et al., 2007; Caporaso et al., 2010). Therefore, the depth of coverage was adapted to the lowest number of sequences within bacterial or archaeal samples for even sampling depth. A three-dimensional Principal Coordinates Analysis (PCoA) plot based on weighted UniFrac analysis was generated by Emperor for visualization of dissimilarities between microbial communities (Vazquez-Baeza et al., 2013).
Representative sequences were picked from the created clusters. Taxonomy was assigned to sequences by the RDP classifier with a confidence cut-off of 80 (Wang et al., 2007). Analysis was performed on RDP server infrastructure while data were uploaded. Bacterial and archaeal 16S rRNA gene sequences were deposed in the EMBL database under the study accession number PRJEB7938.
Acknowledgments
We are very thankful to Prof. P. Dürre at Ulm University for providing laboratory space. We gratefully acknowledge E. Wetzel for her support during fermentation experiments, and we like to thank Mr. T. Ströbele for providing inoculum from his biogas plant.
Conflict of interest
None declared.
Supporting Information
Fig. S1 Specific cumulative biogas yields of reactors CF1–CF4.
Table S1 Feedstock characteristics of maize and sugar beet silage supplied to CSTRs.
Table S2 Species richness and diversity estimated from archaeal (Arc) and bacterial (Bac) 16S rRNA gene sequences at 97% homology.
Table S3 Beta-diversity measurements for bacterial and archaeal communities. The distance of microbial communities was measured between pairs of samples by weighted UniFrac (0 = communities are identical).
Table S4 Primers for PCR amplification of archaeal and bacterial 16S rRNA genes.
References
- Allison SD. Martiny JBH. Resistance, resilience, and redundancy in microbial communities. Proc Natl Acad Sci USA. 2008;105:11512–11519. doi: 10.1073/pnas.0801925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHA. Standard methods for the examination of water and wastewater. Washington, DC, USA: American Public Health Association; 1999. Part 1000. [Google Scholar]
- Ayarza JM, Guerrero LD. Erijman L. Nonrandom assembly of bacterial populations in activated sludge flocs. Microb Ecol. 2010;59:436–444. doi: 10.1007/s00248-009-9581-1. [DOI] [PubMed] [Google Scholar]
- Bengelsdorf FR, Gerischer U, Langer S, Zak M. Kazda M. Stability of a biogas-producing bacterial, archaeal and fungal community degrading food residues. FEMS Microbiol Ecol. 2013;84:201–212. doi: 10.1111/1574-6941.12055. [DOI] [PubMed] [Google Scholar]
- Biddle A, Stewart L, Blanchard J. Leschine S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity. 2013;5:627–640. [Google Scholar]
- Boe K. Angelidaki I. Serial CSTR digester configuration for improving biogas production from manure. Water Res. 2009;43:166–172. doi: 10.1016/j.watres.2008.09.041. [DOI] [PubMed] [Google Scholar]
- Böttcher R, Stollberg C. Sakalauskas A. Efficient biogas production from beet. Agr Eng. 2011;43:16–22. [Google Scholar]
- Briones A. Raskin L. Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr Opin Biotechnol. 2003;14:270–276. doi: 10.1016/s0958-1669(03)00065-x. [DOI] [PubMed] [Google Scholar]
- Brulc JM, Antonopoulos DA, Berg-Miller ME, Wilson MK, Yannarell AC, Dinsdale EA, et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA. 2009;106:1948–1953. doi: 10.1073/pnas.0806191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrol L, Malhautier L, Poly F, Lepeuple AS. Fanlo JL. Bacterial dynamics in steady-state biofilters: beyond functional stability. FEMS Microbiol Ecol. 2012;79:260–271. doi: 10.1111/j.1574-6941.2011.01213.x. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2013;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirel B. Scherer P. Production of methane from sugar beet silage without manure addition by a single-stage anaerobic digestion process. Biomass Bioenergy. 2008;32:203–209. [Google Scholar]
- Dollhopf SL, Hashsham SA. Tiedje JM. Interpreting 16S rDNA T-RFLP data: application of self-organizing maps and principal component analysis to describe community dynamics and convergence. Microb Ecol. 2001;42:495–505. doi: 10.1007/s00248-001-0027-7. [DOI] [PubMed] [Google Scholar]
- Drosg B. 2013. IEA BioenergyProcess monitoring in biogas plants Task 37–Energy from biogas.
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- El-Mashad HM. Zhang R. Biogas production from co-digestion of dairy manure and food waste. Bioresour Technol. 2010;101:4021–4028. doi: 10.1016/j.biortech.2010.01.027. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Huang S, Seston S, Xing J, Hickey R, Criddle C. Tiedje J. How stable is stable? Function versus community composition. Appl Environ Microbiol. 1999;65:3697–3704. doi: 10.1128/aem.65.8.3697-3704.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AS, Hashsham SA, Dollhopf SL, Raskin L, Glagoleva O, Dazzo FB, et al. Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose. Appl Environ Microbiol. 2000;66:4058–4067. doi: 10.1128/aem.66.9.4058-4067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher K. Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155:1749–1757. doi: 10.1099/mic.0.026385-0. [DOI] [PubMed] [Google Scholar]
- FNR – Fachagentur für nachwachsende Rohstoffe e.V. (agency for renewable resources) 2014. Basisdaten Bioenergie Deutschland. [WWW document]. URL www.fnr.de/basisdaten/bioenergie/biogas.html.
- Friedrich U, Van Langenhove H, Altendorf K. Lipski A. Microbial community and physicochemical analysis of an industrial waste gas biofilter and design of 16S rRNA-targeting oligonucleotide probes. Environ Microbiol. 2003;5:183–201. doi: 10.1046/j.1462-2920.2003.00397.x. [DOI] [PubMed] [Google Scholar]
- Gagliano MC, Braguglia CM, Gallipoli A, Gianico A. Rossetti S. Microbial diversity in innovative mesophilic/thermophilic temperature-phased anaerobic digestion of sludge. Environ Sci Pollut Res Int. 2015;22:7339–7348. doi: 10.1007/s11356-014-3061-y. [DOI] [PubMed] [Google Scholar]
- Hai R, Wang Y, Wang X, Li Y. Du Z. Bacterial community dynamics and taxa-time relationships within two activated sludge bioreactors. PLoS ONE. 2014;9:e90175. doi: 10.1371/journal.pone.0090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Tyson G. Blackall L. Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridization. In: Rapley R, editor; de Muro M, editor. Gene Probes. Totowa, NJ: Humana Press; 2002. pp. 29–42. [DOI] [PubMed] [Google Scholar]
- Kaiser F, Metzner T, Effenberger M. Gronauer A. 2008. Sicherung der Prozessstabilität in landwirtschaftlichen Biogasanlagen. LfL-Information p. 9.
- Klang J, Theuerls S, Szewzyk U, Huth M, Tölle R. Klocke M. Dynamic variation of the microbial community structure during the long-time mono-fermentation of maize and sugar beet silage. Microb Biotechnol. 2008 doi: 10.1111/1751-7915.12263. doi: 10.1111/1751-7915.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klocke M, Nettmann E, Bergmann I, Mundt K, Souidi K, Mumme J. Linke B. Characterization of the methanogenic Archaea within two-phase biogas reactor systems operated with plant biomass. Syst Appl Microbiol. 2015;31:190–205. doi: 10.1016/j.syapm.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Kryvoruchko V, Machmüller A, Bodiroza V, Amon B. Amon T. Anaerobic digestion of by-products of sugar beet and starch potato processing. Biomass Bioenergy. 2009;33:620–627. [Google Scholar]
- Li T, Mazéas L, Sghir A, Leblon G. Bouchez T. Insights into networks of functional microbes catalysing methanization of cellulose under mesophilic conditions. Environ Microbiol. 2009;11:889–904. doi: 10.1111/j.1462-2920.2008.01810.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang R, Chen C, Liu G, He Y. Liu X. Biogas production from co-digestion of corn stover and chicken manure under anaerobic wet, hemi-solid, and solid state conditions. Bioresour Technol. 2013;149:406–412. doi: 10.1016/j.biortech.2013.09.091. [DOI] [PubMed] [Google Scholar]
- Liu FH, Wang SB, Zhang JS, Zhang J, Yan X, Zhou HK, et al. The structure of the bacterial and archaeal community in a biogas digester as revealed by denaturing gradient gel electrophoresis and 16S rDNA sequencing analysis. J Appl Microbiol. 2009;106:952–966. doi: 10.1111/j.1365-2672.2008.04064.x. [DOI] [PubMed] [Google Scholar]
- Liu Y. Whitman WB. Metabolic, phylogenetic, and ecological diversity of the methanogenic Archaea. Ann N Y Acad Sci. 2008;1125:171–189. doi: 10.1196/annals.1419.019. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST. Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KD, Martin HG. Hugenholtz P. Integrating ecology into biotechnology. Curr Opin Biotechnol. 2007;18:287–292. doi: 10.1016/j.copbio.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Mähnert P. Linke B. Kinetic study of biogas production from energy crops and animal waste slurry: Effect of organic loading rate and reactor size. Environ Technol. 2009;30:93–99. doi: 10.1080/09593330802246640. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP. Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MC, Morrison M. Yu Z. A meta-analysis of the microbial diversity observed in anaerobic digesters. Bioresour Technol. 2011;102:3730–3739. doi: 10.1016/j.biortech.2010.11.119. [DOI] [PubMed] [Google Scholar]
- Pagnier I, Croce O, Robert C, Raoult D. La Scola B. Non-contiguous finished genome sequence and description of Fenollaria massiliensis gen. nov., sp. nov., a new genus of anaerobic bacterium. Stand Genomic Sci. 2014;9:704–717. doi: 10.4056/sigs.3957647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope P, Vivekanand V, Eijsink VH. Horn S. Microbial community structure in a biogas digester utilizing the marine energy crop Saccharina latissima. Biotech. 2013;3:407–414. doi: 10.1007/s13205-012-0097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procházka J, Mrázek J, Strosová L, Fliegerová K, Zábranská J. Dohányos M. Enhanced biogas yield from energy crops with rumen anaerobic fungi. Eng Life Sci. 2012;12:343–351. [Google Scholar]
- Rademacher A, Nolte C, Schönberg M. Klocke M. Temperature increases from 55 to 75°C in a two-phase biogas reactor result in fundamental alterations within the bacterial and archaeal community structure. Appl Microbiol Biotechnol. 2012;96:565–576. doi: 10.1007/s00253-012-4348-x. [DOI] [PubMed] [Google Scholar]
- Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros M, Franke-Whittle IH, Morales AB, Insam H, Ayuso M. Pascual JA. Archaeal community dynamics and abiotic characteristics in a mesophilic anaerobic co-digestion process treating fruit and vegetable processing waste sludge with chopped fresh artichoke waste. Bioresour Technol. 2013;136:1–7. doi: 10.1016/j.biortech.2013.02.058. [DOI] [PubMed] [Google Scholar]
- Scherer P, Lehmann K, Schmidt O. Demirel B. Application of a fuzzy logic control system for continuous anaerobic digestion of low buffered, acidic energy crops as mono-substrate. Biotechnol Bioeng. 2009;102:736–748. doi: 10.1002/bit.22108. [DOI] [PubMed] [Google Scholar]
- Stamper DM, Walch M. Jacobs RN. Bacterial population changes in a membrane bioreactor for graywater treatment monitored by denaturing gradient gel electrophoretic analysis of 16S rRNA gene fragments. Appl Environ Microbiol. 2003;69:852–860. doi: 10.1128/AEM.69.2.852-860.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, et al. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol. 2013;85:612–626. doi: 10.1111/1574-6941.12148. [DOI] [PubMed] [Google Scholar]
- Vazquez-Baeza Y, Pirrung M, Gonzalez A. Knight R. Emperor: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2:1–4. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VDLUFA. 2011. Bestimmung der Biogas- und Methanausbeute in Gärtests. VDLUFA-Methodenvorschrift, Methodenbuch Band VII – Umweltanalytik, Speyer, 4. Auflage.
- Voß E, Weichgrebe D. Rosenwinkel K. FOS/TAC: Herleitung, Methodik, Anwendung und Aussagekraft. Int Wissenschaftstagung Biogas Sci. 2009;3:675–683. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM. Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland P. Biogas production: current state and perspectives. Applied Microbiol Biotechnol. 2010;85:849–860. doi: 10.1007/s00253-009-2246-7. [DOI] [PubMed] [Google Scholar]
- Weissbach F. Gas production potential of fresh and ensiled sugar beets in biogas production. Landtechnik. 2009;64:394–397. [Google Scholar]
- Weissbach F. Strubelt C. Die Korrektur des Trockensubstanzgehaltes von Maissilagen als Substrat für Biogasanlagen. Landtechnik. 2008;2:2–4. [Google Scholar]
- Werner JJ, Garcia ML, Perkins SD, Yarasheski KE, Smith SR, Muegge BD, et al. Microbial community dynamics and stability during an ammonia-induced shift to syntrophic acetate oxidation. Appl Environ Microbiol. 2014;80:3375–3383. doi: 10.1128/AEM.00166-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley RA. Hardie JM. Genus I. Streptococcus Rosenbach 1884, 22AL. In: Jones D, editor; De Vos P, Garrity GM, editors. Bergey’s Manual of Systematic Bacteriology The Firmicutes. Vol. 3. New York, USA: Springer; 2009. pp. 655–711. [Google Scholar]
- Wittebolle L, Vervaeren H, Verstraete W. Boon N. Quantifying community dynamics of nitrifiers in functionally stable reactors. Appl Environ Microbiol. 2008;74:286–293. doi: 10.1128/AEM.01006-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye NF, Lü F, Shao LM, Godon JJ. He PJ. Bacterial community dynamics and product distribution during pH-adjusted fermentation of vegetable wastes. J Appl Microbiol. 2007;103:1055–1065. doi: 10.1111/j.1365-2672.2007.03321.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Specific cumulative biogas yields of reactors CF1–CF4.
Table S1 Feedstock characteristics of maize and sugar beet silage supplied to CSTRs.
Table S2 Species richness and diversity estimated from archaeal (Arc) and bacterial (Bac) 16S rRNA gene sequences at 97% homology.
Table S3 Beta-diversity measurements for bacterial and archaeal communities. The distance of microbial communities was measured between pairs of samples by weighted UniFrac (0 = communities are identical).
Table S4 Primers for PCR amplification of archaeal and bacterial 16S rRNA genes.