Abstract
Membrane biofilm development was evaluated to improve psychrophilic (15°C) anaerobic membrane bioreactor (AnMBR) treatment of domestic wastewater. An AnMBR containing three replicate submerged membrane housings with separate permeate collection was operated at three levels of membrane fouling by independently controlling biogas sparging for each membrane unit. High membrane fouling significantly improved permeate quality, but resulted in dissolved methane in the permeate at a concentration two to three times the equilibrium concentration predicted by Henry’s law. Illumina sequencing of 16S rRNA targeting Bacteria and Archaea and reverse transcription-quantitative polymerase chain reaction targeting the methyl coenzyme-M reductase (mcrA) gene in methanogens indicated that the membrane biofilm was enriched in highly active methanogens and syntrophic bacteria. Restoring fouled membranes to a transmembrane pressure (TMP) near zero by increasing biogas sparging did not disrupt the biofilm’s treatment performance, suggesting that microbes in the foulant layer were tightly adhered and did not significantly contribute to TMP. Dissolved methane oversaturation persisted without high TMP, implying that methanogenesis in the biofilm, rather than high TMP, was the primary driving force in methane oversaturation. The results describe an attractive operational strategy to improve treatment performance in low-temperature AnMBR by supporting syntrophy and methanogenesis in the membrane biofilm through controlled membrane fouling.
Introduction
Anaerobic membrane bioreactor (AnMBR) treatment allows for the direct recovery of energy from wastewater in the form of methane-rich biogas. In AnMBRs, methane is produced during the anaerobic microbial degradation of the organic compounds present in wastewater in a bioreactor containing microbial biomass in suspension. This suspended biomass is separated from the treated wastewater using membrane filtration to produce a particle-free wastewater effluent (permeate). The recent recognition of the potential benefits of AnMBR treatment of domestic wastewater compared with conventional activated sludge treatment has resulted in a surge in AnMBR research activity (e.g. Yoo et al., 2012; Ma et al., 2013; Smith et al., 2015) and pilot-scale evaluations (Dagnew et al., 2011; Gimenez et al., 2011; Martinez-Sosa et al., 2011; Robles et al., 2013; Shin et al., 2014; Gouveia et al., 2015). As the pumping energy demand needed for membrane filtration increases during the development of a membrane fouling layer, membrane fouling has received considerable attention in AnMBR research (Gao et al., 2010; Huang et al., 2011; Yang et al., 2011; Chen et al., 2012; Kola et al., 2014). The consensus in the water quality engineering field has been to operate membrane-filtration systems, including AnMBRs, with minimal membrane fouling (Yang et al., 2006), which is accomplished using gas sparging, backflushing and chemical cleaning. As a result, almost no research has been performed on the potential benefits of membrane fouling. The membrane fouling layer contains considerable microbial biomass and can thus be considered a membrane biofilm, which has the potential to improve effluent quality by providing additional biodegradation not accomplished by the suspended biomass (Smith et al., 2015).
Anaerobic microbial communities in a membrane biofilm could have an advantage over suspended microbial communities because of reduced mass-transfer limitations. Mass-transfer phenomena likely have a substantial effect on substrate utilization when substrate concentrations are low, such as during domestic wastewater treatment (Gonzalez-Gil et al., 2001), at low temperatures (Wu et al., 1995) and when mass transport is influenced by advective forces such as liquid flow through a membrane biofilm. In addition, biofilms may facilitate interspecies hydrogen transfer (Ishii et al., 2005; 2006) or direct interspecies electron transfer (DIET; Summers et al., 2010; Morita et al., 2011) between methanogens and their syntrophic partners, and thus provide enhanced microbial activity relative to the suspended biomass activity.
The complexity of anaerobic microbial communities and reported differences in suspended and biofilm AnMBR community structure (Gao et al., 2010; Yu et al., 2012; Ma et al., 2013; Smith et al., 2013; 2015) suggest that careful monitoring of community structure during development of AnMBR operational strategies is important. RNA-based approaches targeting either 16S rRNA (e.g. Eichler et al., 2006; Foesel et al., 2013; Hunt et al., 2013; Männistö et al., 2013) or transcripts of functional genes [e.g. the methyl coenzyme-M reductase (mcrA) gene in methanogens; Freitag and Prosser, 2009] may be more useful than DNA-based approaches in characterizing microbial community function in AnMBRs. The need for RNA-based approaches is particularly important given the slow growth rates and low biomass yields of anaerobic microbes, high biomass retention provided by membrane separation, and short operational periods commonly studied in AnMBRs especially relative to the long solids retention time (SRT) in these systems.
This study elucidated the contribution of the membrane biofilm in AnMBR treatment of synthetic domestic wastewater using a bench-scale AnMBR operated at 15°C equipped with three submerged membrane housings, designated P1, P2 and P3, with separate permeate collection and independent biogas sparging control. The three membrane housings were operated to allow for three different levels of membrane fouling and membrane biofilm development. Illumina sequencing of 16S rRNA genes (rDNA) and 16S rRNA and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) targeting the mcrA gene transcripts were applied to compare microbial community structure and activity dynamics in the suspended biomass and in the membrane biofilms.
Results and discussion
Slow start-up after inoculating the psychrophilic AnMBR with mesophilic sludge
The AnMBR with three membrane housings was initially operated for 99 days (Phase 1; Fig. 1) under low fouling (LF) conditions by maintaining a high biogas sparging flow rate to prevent biofilm development for each of the membrane units. COD removal during the first 99 days of operation (Phase 1) was limited, averaging 57% ± 12% (Fig. S1). The majority of the permeate chemical oxygen demand (COD) was comprised of acetate (average 70 ± 19 mg l−1) and propionate (average 52 ± 18 mg l−1) (Fig. S2). Further information regarding start-up is presented in Appendix S6.
Figure 1.
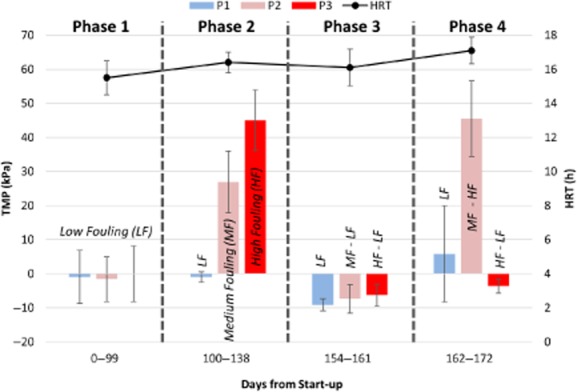
Average transmembrane pressure (TMP) for each of the membranes P1, P2 and P3 (left y-axis) and bioreactor hydraulic retention time (HRT; right y-axis) from days 0 to 172. This time period is divided in four phases defined by the degree of membrane fouling or biofilm development. Data from days 139 to 153 are not reported due to poor AnMBR performance. Error bars for HRT represent the standard deviation of daily flow rate measurements. Error bars for TMP represent the standard deviation of pressure data recorded every minute of operation.
Biofilm development improves effluent quality but results in dissolved methane oversaturation
To improve permeate quality, a controlled membrane fouling experiment was conducted to encourage biofilm development on P2 and P3 by independently reducing the biogas sparging flow rates (Phase 2). Three different levels of membrane fouling were targeted – low fouling (LF; P1), medium fouling (MF; P2) and high fouling (HF; P3) – resulting in the need to operate with different transmembrane pressures (TMPs) to maintain similar fluxes. During Phase 2, P1, P2 and P3 TMP averaged −0.96 ± 1.5, 27 ± 9.0 and 45 ± 8.9 kPa, respectively, indicating the targeted fouling levels were achieved (Fig. 1). Hereafter, P1, P2 and P3 are referred to based on their fouling level (LF, MF and HF respectively).
Differences in permeate COD concentrations were observed throughout Phase 2 and corresponded to the level of membrane fouling (Fig. 2A). The HF permeate consistently had the lowest COD with a concentration of 22 mg l−1 at the end of Phase 2. Permeate volatile fatty acid (VFA) levels showed a similar trend (Fig. 2B and C). The VFA concentrations in the bioreactor and LF permeate were similar throughout Phase 2, indicating minimal biological activity across the LF membrane. These observations indicate that controlled membrane fouling can substantially improve effluent quality in AnMBR, and further suggest that the activity of syntrophic propionate oxidizing populations and their methanogenic partners can be promoted through membrane biofilm development (see below).
Figure 2.
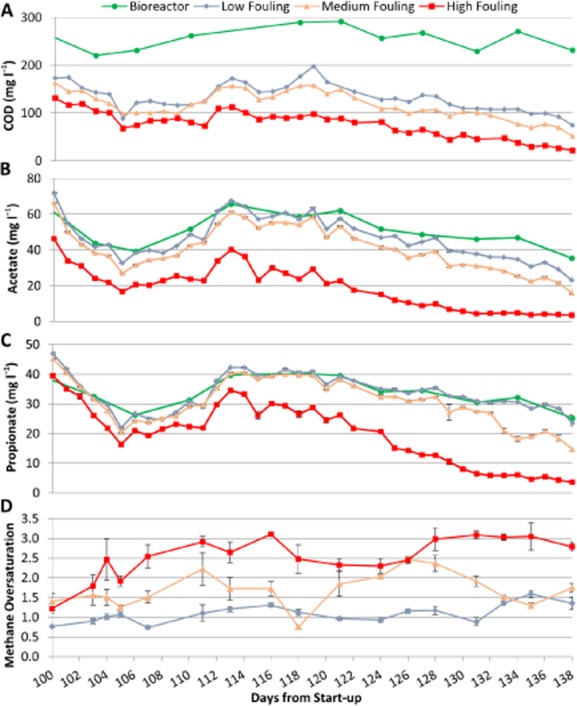
Effect of different degrees of biofilm development (low fouling, medium fouling and high fouling) on permeate quality during Phase 2 of AnMBR operation.A. Bioreactor (soluble) and permeate COD concentrations. Influent COD was 410 ± 46 mg l−1.B. Bioreactor and permeate acetate concentrations. Error bars represent the standard deviation of triplicate IC injections.C. Bioreactor and permeate propionate concentrations. Error bars represent the standard deviation of triplicate IC injections.D. Dissolved methane oversaturation in the permeate calculated assuming a Henry’s law constant of 34 300 atm at 15°C (Tchobanoglous et al., 2003), and measured methane partial pressure in the biogas and dissolved methane concentration in the permeate. The methane content of the biogas was approximately 90%, with the balance being carbon dioxide. A high methane content is expected given the low organic loading rate and differences in methane and carbon dioxide solubility at this temperature. Error bars represent the standard deviation of duplicate dissolved methane extractions and triplicate GC injections of each dissolved methane extract.
Consistent with this, dissolved methane concentrations in MF and HF permeates indicated significant oversaturation of methane (Fig. 2D), suggesting that methanogenesis occurred in the biofilm and that methane produced in the biofilm left the system in the dissolved form. From days 107 to 138, dissolved methane concentrations in LF, MF and HF permeate averaged 1.1 ± 0.22, 1.7 ± 0.44 and 2.6 ± 0.30 times the concentrations predicted by Henry’s law respectively. The dissolved methane concentration in the bioreactor, which could not be measured, was believed to be near saturation due to vigorous biogas sparging. The observation that the dissolved methane concentration in LF permeate was close to saturation during Phase 2, as it was during Phase 1 for all three permeates, provided further evidence of minimal biological activity during LF conditions. Dissolved methane recovery downstream of anaerobic treatment has been attempted using membrane degasification, but substantial energy was required (Bandara et al., 2011) and methane content of the off-gas may be insufficient for cogeneration (Cookney et al., 2012). If released to the atmosphere, this ‘lost’ energy source could constitute a potent greenhouse gas emission (Smith et al., 2014). Biological oxidation of dissolved methane using aerobic methanotrophy or nitrite-dependent anaerobic methane oxidation (n-damo) could prevent greenhouse gas emissions but would require an additional unit process and energy input and would not recover the methane for electricity production.
Biofilm development leads to a specialized microbial community enriched in active methanogens
High-throughput sequencing of both 16S rDNA and 16S rRNA was employed to study controlled membrane fouling during Phase 2, and the terms ‘relative abundance’ and ‘relative activity’, respectively, are used to report the results of these sequencing efforts. The relative abundance and relative activity levels of populations differed greatly in suspended and biofilm biomass (Fig. 3). The 16S rDNA sequence data indicated that the suspended and biofilm community comprised < 10% methanogens. The hydrogenotrophic methanogens were more abundant than the acetoclastic methanogens in the suspended biomass 26 days after start-up and in the biofilm biomass at the end of Phase 2 (Fig. S5A), suggesting the hydrogen utilization pathway became more important after biomass adaptation to the psychrophilic temperature. A shift towards hydrogenotrophic methanogenesis has also been observed previously in other anaerobic systems when transitioning from mesophilic to psychrophilic conditions using DNA-based analyses and specific methanogenic activity assays (McHugh et al., 2013; Collins et al., 2006; Connaughton et al., 2006), and has been explained by increased hydrogen solubility and thus increased substrate availability for hydrogenotrophic metabolisms (Lettinga et al., 2001). In contrast to 16S rDNA sequence data, 16S rRNA sequencing indicated that the relative activity of acetoclastic and hydrogenotrophic methanogens was similar in our system (Fig. S5B). MF and HF biofilms also had substantially greater activity of methanogens relative to the suspended biomass and LF biofilm on day 138. Specifically, methanogens represented 33% and 34% of the combined bacterial and archaeal activity in MF and HF biofilms, respectively, in comparison to only 15% in the suspended biomass and LF biofilm (Fig. 3). These observations correlated with the low levels of acetate and propionate and high dissolved methane oversaturation by the end of Phase 2 (Fig. 2), and suggest that a high level of methanogenesis occurred in the MF and HF biofilms.
Figure 3.
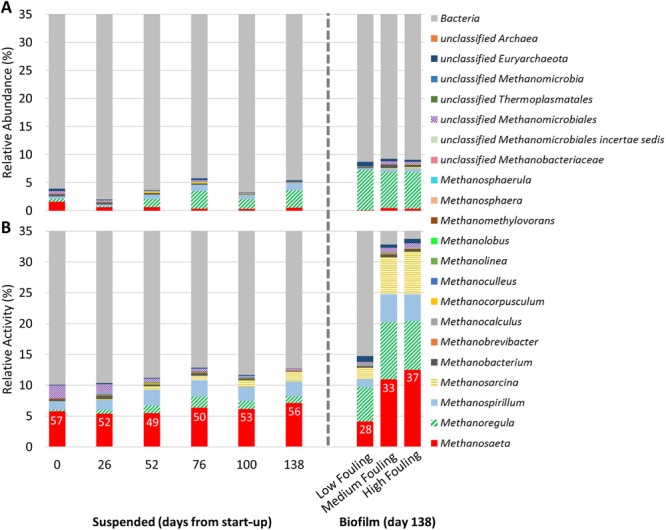
(A) Relative abundance of methanogens identified at the genus level based on 16S rDNA sequencing and (B) relative activity of methanogens identified at the genus level based on 16S rRNA sequencing. Data were obtained for suspended biomass samples collected from start-up to the end of Phase 2 and in biofilm samples obtained at the end of Phase 2. Data are expressed as percentages and were normalized using the total number of 16S rDNA sequences (A) and 16S rRNA sequences (B) (including Bacteria and Archaea). Numbers within bars in (B) represent the relative activity of Methanosaeta spp. based on 16S rRNA sequence data normalized to the 16S rRNA sequences of all Archaea. A truncated y-axis (0–35%) is shown to accentuate changes in methanogen abundance and activity.
Methanosaeta is believed to be the only methanogen that exclusively produces methane through the acetoclastic pathway (Smith and Ingram-Smith, 2007). In the suspended biomass, the relative abundance of Methanosaeta spp. decreased from 1.6% on day 0 to 0.55% on day 138 (Fig. 3A). However, their relative activity was fairly stable (Fig. 3B), resulting in an increase in the activity/abundance ratio over time. The consistently low relative abundance and high relative activity of Methanosaeta spp. in the suspended biomass suggest that growth was negligible, possibly due to the psychrophilic temperature. Further, the observation of long-term (over 138 days) stable suspended biomass concentrations (Fig. S4) supports this notion.
Methanosarcina produces methane from acetate, hydrogen and C1 compounds (Mladenovska and Ahring, 1997; Welander and Metcalf, 2005) and has thus been categorized as mixotrophic. The relative activity of Methanosarcina spp. increased over time in the suspended biomass and comprised 18% and 21% of relative methanogenic activity in MF and HF biofilm biomass respectively (Fig. S5). Methanosarcina spp. were either not detected or detected at ≤ 0.23% of the combined bacterial and archaeal abundance (Fig. 3A). Similarly, Methanosarcina spp. were not detected via 16S rDNA sequencing in another psychrophilic AnMBR study (Bandara et al., 2012) and were detected at < 0.50% of the archaeal community in our previous AnMBR work at 15°C (Smith et al., 2013). Methanosarcina has a greater maximum growth rate and half-saturation coefficient than Methanosaeta, which often leads to the dominance of Methanosaeta when acetate concentrations are low, such as in continuously fed anaerobic digestion (Conklin et al., 2006) or during low-strength wastewater treatment. It is unclear why the activity of Methanosarcina was high in this study, particularly in the MF and HF biofilms, as the acetate concentration was below or within the reported range of threshold levels at which Methanosarcina spp. are typically inactive (11–71 mg l−1 acetate; Jetten et al., 1992) (Fig. 2B). Psychrotolerant Methanosarcina spp. have been observed in the environment (Simankova et al., 2001; von Klein et al., 2002), but a specific mechanism for low-temperature adaptation that would give a competitive advantage over Methanosaeta or other methanogens in psychrophilic AnMBR has not been reported. However, Methanosarcina spp. have a unique surface structure (Francoleon et al., 2009; De Vrieze et al., 2012), which may aid in cell attachment to surfaces (Robinson et al., 2012; De Vrieze et al., 2012). We hypothesize that the metabolic flexibility and unique surface structure of Methanosarcina spp. offered a competitive advantage in the biofilm relative to other methanogens.
Methanoregula spp. and Methanospirillum spp. were the dominant active hydrogenotrophic methanogens classified in suspended and biofilm biomass and comprised 24% and 14% of methanogenic activity in the HF biofilm biomass respectively (Fig. 3B). Methanoregula spp., mesophilic hydrogenotrophic methanogens, were only recently cultivated from a full-scale upflow anaerobic sludge blanket reactor (Yashiro et al., 2011) and an acidic peat bog (Bräuer et al., 2011). Growth for both of these Methanoregula spp. was demonstrated at temperatures as low as 10°C, suggesting tolerance to psychrophilic temperature. The activity/abundance ratio of Methanoregula spp. was 0.41 and 0.33 in MF and HF biofilm biomass, respectively, whereas the ratio for Methanospirillum spp. was 2.5 and 2.0 in MF and HF biofilm biomass, respectively, suggesting that the activity per cell for Methanospirillum spp. was significantly greater than for Methanoregula spp. in the biofilm biomass.
It is important to note the limitations of our approach to infer microbial abundance and activity. 16S rRNA operon number varies from 1 to 15 copies per genome (Klappenbach et al., 2000), which can lead to over or under-representation of specific phylogenies if a constant operon number is assumed across all phylogenies (Větrovský and Baldrian, 2013). Normalization of sequencing results to operon number is challenging since the operon number is not available for all methanogens (Lee et al., 2014), and variations in operon number exist at the phylogenetic resolution provided by our sequencing data (species level). Variations in 16S rRNA abundance between phylogenies based on cell size and other factors are another concern with our approach. 16S rRNA abundance is also not directly linked to a specific cellular function (e.g. methanogenesis) and does not always correlate well with activity even for pure cultures under steady-state conditions (Blazewicz et al., 2013). Further, inactive or dormant microorganisms sometimes contain high amounts of rRNA (Sukenik et al., 2012). Another limitation is the lack of absolute abundance or gene expression data, which can only be obtained when accurate quantitative nucleic acid extraction is possible. Quantitative DNA and RNA extractions are particularly challenging when working with biomass from environmental systems with different characteristics, such as suspended and biofilm biomass in this study. For example, elevated concentrations of extracellular polymeric substances in biofilm biomass (Smith et al., 2013) may reduce extraction efficiency and quality of nucleic acids extracted. Such matrix effects may influence microbial characterization due to biases in nucleic acids extraction, PCR, reverse transcription or other steps (Martin-Laurent et al., 2001). These concerns were motivation for validating our approach by monitoring changes in the expression of the mcrA gene in methanogens (Thauer, 1998).
The RT-qPCR results quantifying mcrA transcripts correlated well with performance observations and 16S rRNA sequence data, indicating significantly higher methanogenic activity in MF and HF biofilm biomass relative to suspended or LF biofilm biomass (Fig. 4). Taken together, our results provide strong evidence that the membrane biofilm was enriched in highly active acetoclastic and hydrogenotrophic methanogens.
Figure 4.
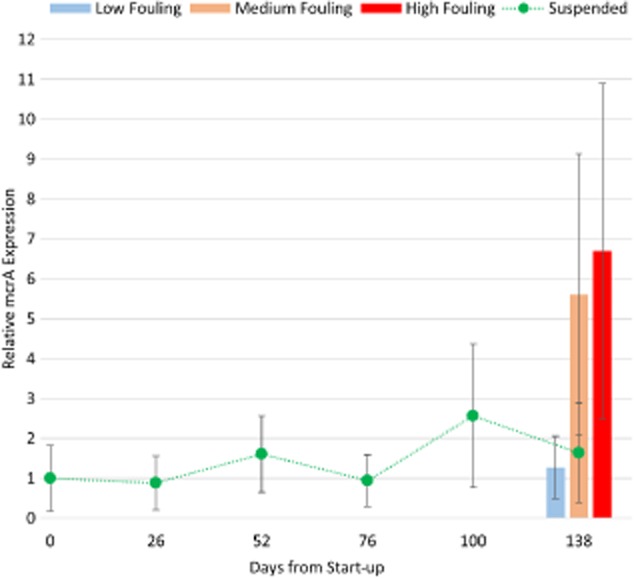
Relative expression of mcrA in suspended and biofilm biomass. Copies of mcrA transcripts in each biomass sample were first normalized to 16S rRNA copies. Next, relative mcrA expression was calculated by normalizing the ratio of mcrA transcript copies to 16S rRNA copies in each sample to the corresponding ratio in the suspended biomass sample collected on day 0. Error bars represent the standard deviation of the ratio of triplicate qPCR reactions at serial dilutions of cDNA template concentration.
Phylogenetically distinct syntrophic bacterial operational taxonomic unit (OTU) was highly active in the biofilm
The relative activity of fatty acid-oxidizing obligate syntrophic bacteria correlated well with the differing VFA concentrations in the permeates (Fig. 5). On day 0, a high level of syntrophic activity was observed in the suspended biomass, which quickly fell to 0.33% activity of total bacteria by day 26, likely due to the introduction of a mesophilic inoculum into a system with a psychrophilic operational temperature. In addition, the calculated average velocity gradient (g) due to biogas sparging in our system of 410 s−1 is much higher than recommended g values for effective mixing in anaerobic digestion (50–80 s−1; Tchobanoglous et al., 2003), and raises the question of how this high shear affected the microbial community and syntrophic associations in particular. Research has suggested that high shear can be detrimental to anaerobic digester performance under high loading rates due to increased hydrolysis and fermentation resulting in acidification (Stroot et al., 2001; Vavilin and Angelidaki, 2005; Padmasiri et al., 2007). Hoffmann and colleagues (2008) demonstrated that even when loading rates were low, high shear conditions had a detrimental effect on digester performance. None of these studies conclusively determined how high shear conditions may impact anaerobic microbial communities, and the effects of shear in AnMBRs operated at low loading rates, such as the system in this study, have not been studied. The relative activity levels of syntrophic bacteria in the suspended biomass gradually increased over time and showed differences in the three biofilm samples collected on day 138, consistent with AnMBR performance observations and methanogenic activity (Fig. 5). Known syntrophic acetate-oxidizing bacteria (Hattori, 2008) were not detected in the biofilm, suggesting that acetate removal was primarily via acetoclastic methanogenesis.
Figure 5.
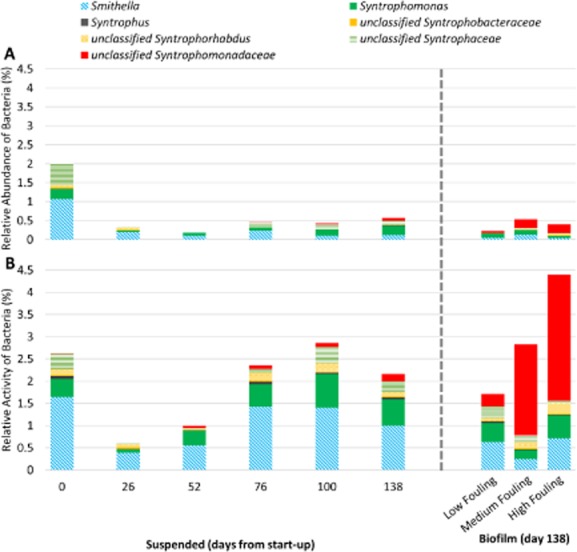
(A) Relative abundance of known fatty acid-oxidizing obligate syntrophic bacteria identified at the genus level or family level based on 16S rDNA sequencing and (B) relative activity of syntrophic bacteria identified at the genus level or family level based on 16S rRNA sequencing. Data were obtained for suspended biomass samples collected from start-up to the end of Phase 2 and in biofilm samples obtained at the end of Phase 2. Data are expressed as percentages and were normalized using the total number of 16S rDNA bacterial sequences (A) and 16S rRNA bacterial sequences (B). A truncated y-axis (0–4.5%) is shown to accentuate changes in abundance and activity.
An OTU unclassified at the genus level according to the Ribosomal Database Project (RDP) belonging to the family Syntrophomonadaceae comprised a significant proportion of the relative activity of all identified syntrophs in MF and HF biofilm biomass (i.e. 72% and 64% respectively), but accounted for only 7.7% of the relative activity of syntrophs in the suspended biomass on day 138. A representative sequence from this OTU exhibited 95% identity with Syntrophomonas zehnderi, an obligate syntrophic microorganism (Sousa et al., 2007). Interestingly, genera in Syntrophomonadaceae have only been observed to syntrophically oxidize C4 compounds (e.g. butyrate) and higher order organics (Stams et al., 2012). In our system, butyrate concentrations were below the detection limit, and thus it is surprising that a butyrate oxidizing syntroph would have such high activity in the biofilm particularly relative to propionate oxidizing syntrophs (e.g. Smithella) given the significant propionate removal in the biofilm. The unclassified OTU may be a yet to be described species of Syntrophomonadaceae capable of C3 oxidation. Alternatively, Gan and colleagues (2012) using DNA-based stable isotope probing proposed a novel pathway in which Smithella spp. first dismutate propionate to acetate and butyrate followed by butyrate oxidation by Syntrophomonas spp. via a trophic interaction. Based on the significant removal of propionate in the biofilm, non-detectable levels of butyrate in the bioreactor, and activity of both Smithella spp. and the unclassified OTU belonging to Syntrophomonadaceae, it is possible that this novel pathway occurred here. In this scenario, butyrate may have remained non-detectable acting as a transient metabolite. This scenario would require cooperation between two syntrophic bacteria and a hydrogenotrophic methanogen, and thus may benefit from the increased spatial organization afforded to the biofilm community relative to the suspended biomass. The unclassified OTU may also be more active in the biofilm than in the suspended biomass due to differential preferences in growth mode (i.e. attached versus suspended).
The AnMBR biofilm may support other syntrophic interactions. The sulfate reducer Desulfovibrio vulgaris has been identified as capable of growing syntrophically with a hydrogenotrophic methanogen on lactate in the absence of sulfate (Scholten et al., 2007). The majority of sulfate reduction in our system occurred in the suspended biomass with sulfate concentrations < 1 mg l−1 in the bioreactor at the end of Phase 2 (prior to biofilm biomass sampling). The relative activity of some sulfate reducers correlated well with performance data. For example, Desulfobulbus spp. had a relative activity of 2.2% in the suspended biomass but only 0.25% in HF biofilm biomass (Fig. S6). However, the relative activity of Desulfovibrio spp. was greater in the biofilm biomass, 7.2% versus 4.3% in HF biofilm and suspended biomass, respectively, despite limited sulfate reduction across the biofilm. It is possible that, in the absence of sulfate, Desulfovibrio spp. transition to syntrophic metabolisms potentially enhanced in the biofilm. The biofilm may also provide an environment conducive to DIET (Summers et al., 2010; Morita et al., 2011). However, Geobacter spp., a genus with members likely to participate in DIET (Summers et al., 2010), had lower relative activity in the biofilm than in the suspended biomass. The highly active unclassified Syntrophomonadaceae OTU could potentially be involved in DIET, although genome sequencing of Syntrophomonas wolfei did not identify the outer membrane cytochromes necessary for DIET (Sieber et al., 2010). The potential role of DIET in AnMBR domestic wastewater treatment has yet to be explored and deserves further investigation.
We hypothesize that reduced mass-transfer limitations, increased substrate availability and spatial organization of the biofilm community may all play a role in the high microbial activity observed in the biofilm. It is important to note that microbial activity in the biofilm could also be influenced by elevated temperatures due to exothermic reactions occurring there. To estimate the maximum potential temperature differential inside the biofilm relative to the suspended biomass, we considered hydrogenotrophic methanogenesis, an exothermic reaction, in accordance with the measured permeate dissolved methane oversaturation and reaction enthalpies as discussed by Westermann (1994). The calculated maximum heat differential was < 0.2°C, suggesting a negligible difference between biofilm and suspended biomass temperature. Further, other reactions occurring in the biofilm (i.e. acetoclastic methanogenesis and propionate oxidation) are endothermic. Future research should evaluate the underlying mechanism(s) responsible for the high microbial activity in the biofilm.
Biofilm treatment performance is maintained in the absence of TMP
During Phases 3 and 4, biogas sparging on HF was increased to evaluate if biological treatment in the biofilm could be maintained without high TMP (i.e. HF operation was switched to LF operation, and is indicated in Fig. 1 as HF–LF). The HF–LF permeate COD during Phases 3 and 4 averaged 24 ± 7.1 mg l−1 (Fig. S7), a similar effluent quality to that obtained at the end of Phase 2. Further, HF–LF permeate propionate concentration was below detection by the start of Phase 4 (Fig. S8), implying that the activity of syntrophic propionate oxidizers improved during this time period, despite low TMP. Dissolved methane oversaturation remained high, averaging 2.2 ± 0.49, and was thus primarily driven by biological activity in the biofilm rather than high TMP or a combination of the two. One concern with our comparative evaluation is that pump slippage from high TMP resulted in reduced flux for HF, which increased substrate contact time in the biofilm and could have affected our observations. However, HF flux was restored after returning TMP to near zero, suggesting that the potentially higher substrate contact time did not impact our comparison. These results demonstrate that biofilm activity can be maintained in the absence of TMP and suggest that the active microbial community in the biofilm is tightly adhered to the membrane surface. The active community is either distinct from the layer of foulants contributing to high TMP or has sufficient biological activity to maintain treatment under LF conditions (i.e. less biomass).
After restoring the TMP in the MF membrane unit to near zero during Phase 3 (indicated as MF–LF in Fig. 1), fouling for this membrane was increased during Phase 4 in an attempt to replicate the performance obtained previously with HF (indicated as MF–HF in Fig. 1). The MF–HF permeate COD during Phase 4 was 37 ± 7.0 mg l−1, approaching an effluent quality similar to that of the HF permeate in Phase 2. Dissolved methane oversaturation in the MF–HF permeate increased, averaging 2.6 ± 0.68. These observations provide evidence that biofilm promotion via reduced biogas sparging to enhance treatment performance is replicable.
Because we were able to maintain biological activity after returning to near zero TMP, biofilm promotion strategies may only require an inoculation period in which the membrane is colonized from the suspended biomass and can then be operated at low TMP. Long-term operation with substantial membrane fouling is undesirable from an operation’s standpoint, and thus our demonstration of biofilm activity at low TMP is encouraging. The industry’s current reliance on aggressive chemical cleaning in membrane installations disrupts the active biofilm community and may have prevented similar observations in the past. Since we were able to return to a low TMP after extended periods of fouling by solely adjusting biogas sparging flow rate without chemical cleaning, this fouling control method or an alternative strategy could be explored in full-scale systems to sustain biological activity in the biofilm. The energy demands of a higher biogas sparging flow rate to do so need to be weighed against the benefits of operation without chemical cleaning.
Biofilm development is an attractive operational strategy for low-temperature AnMBR
We have shown that effluent quality in AnMBR domestic wastewater treatment can be improved by rethinking common perceptions of membrane fouling. Multiple lines of evidence (i.e. 16S rRNA sequencing, RT-qPCR targeting the mcrA gene and performance observations) show that controlled membrane fouling leads to the development of a membrane biofilm enriched in highly active acetoclastic and hydrogenotrophic methanogens and syntrophic bacteria. This active biofilm may also have additional unexplored benefits in AnMBR, such as removal of antibiotic resistance genes (e.g. as demonstrated in aerobic membrane bioreactors; Riquelme Breazeal et al., 2012) or other micropollutants. Future research should evaluate the underlying mechanisms behind the biofilm community’s high biological activity (e.g. reduced mass transfer limitations, lower intercellular distances or other factors), the impact of biofilm promotion strategies on long-term membrane fouling and the biofilm’s response to chemical membrane cleaning. It is important to note that this research was performed using synthetic domestic wastewater and that evaluation using real domestic wastewater is necessary. Future research is also required to develop low-energy dissolved methane recovery technologies to prevent greenhouse gas emissions, particularly when biofilm activity results in significant oversaturation.
Experimental procedures
AnMBR operation and chemical assays
A bench-scale AnMBR described previously (Smith et al., 2013) was redesigned to incorporate three submerged flat-sheet membrane housings with microfiltration polyethersulfone membranes (GE Osmonics, Greenville, SC) at a pore size of 0.2 μm and a total effective membrane area of 924 cm2. The system was operated at 15°C with a synthetic domestic wastewater (Smith et al., 2013). Reactor temperature was controlled using a water jacket connected to a Polystat 6-L re-circulating water bath (Cole-Parmer, Vernon Hills, IL). Water bath temperature was adjusted based on temperature measurement of a submerged probe located in close proximity to the membrane surface. Three mini diaphragm pumps (KNF Neuberger, Trenton, NJ) re-circulated headspace biogas and dispersed it directly below each membrane housing by horizontally placed sparging tubes for fouling control. Biogas sparging flow rates were independently controlled for each membrane housing using in-line flow metres. The liquid volume of the reactor was 4 l. The AnMBR was inoculated with sludge from a mesophilic (32°C) wastewater treatment plant anaerobic sludge digester (Northfield Wastewater Treatment Plant, Whitmore Lake, MI) at an initial volatile suspended solid concentration of approximately 8000 mg l−1.
The system was operated at a target hydraulic retention time (HRT) of 16 h, which corresponded to an organic loading rate of 670 mg COD/l●d. Biomass was only removed from the AnMBR for sampling purposes, which resulted in an SRT of approximately 300 days. From days 1 through 99 (Phase 1), a membrane flux of 2.7 l m−2 h−1 (LMH) was targeted for each membrane housing. This relatively low membrane flux ensured operation with minimal membrane fouling could be maintained without chemical cleaning and provided good operational control. The high biogas sparging flow rate selected for Phase 1 (3.0 l biogas min−1 for each membrane housing or 5.8 m3 biogas h−1 m−2 membrane surface area) helped prevent the formation of a membrane biofilm. Backflushing was performed for 30 s every 10 min of bioreactor operation. Due to pump slippage at high TMP (Phases 2 and 4), the flux for P1 was increased as necessary to maintain an HRT of 16 h. Data from days 139 to 151 are not presented due to a brief exposure of the system to air during biofilm sampling on day 138 (described below), which resulted in poor system performance. Chemical assays were performed as described in Appendix S1.
Biomass samples and nucleic acids extraction
Suspended biomass samples from the AnMBR were taken on days 0, 26, 52, 76, 100 and 138 of operation, pelletized by centrifugation at 5000× g for 5 min at 4°C, and immediately stored at −80°C after decanting the supernatant. Biofilm biomass samples were gently scraped from the membrane surface of P1 (LF), P2 (MF) and P3 (HF) on day 138 using sterile lazy-l spreaders, pelletized, decanted and immediately stored at −80°C. P1 (LF) had limited biofilm biomass, which was loosely associated with the membrane, consistent with the LF condition. Biomass samples for RNA extraction were prepared similarly except for the addition of RNAlater (Qiagen, Valencia, CA) prior to storage. DNA and RNA extractions were accomplished as described in Appendix S2.
RT-qPCR
Primers targeting the mcrA gene were designed via an in silico analysis described in Appendix S3. Universal primers targeting the V4 region of the 16S rDNA (Caporaso et al., 2011) were used to quantify 16S rRNA for normalization of mcrA transcript quantification. Coverage of 16S rRNA primers was verified using TestPrime 1.0 (Klindworth et al., 2012) (Tables S1 and S2).
Reverse transcription to generate single-stranded complementary DNA (cDNA) from RNA extracts was performed using the SuperScript VILO cDNA Synthesis Kit according to manufacturer’s instruction (Life Technologies, Grand Island, NY). Two-step RT-qPCR, as opposed to one-step in which cDNA synthesis and qPCR occur sequentially in one reaction, was done to allow for sequencing of synthesized cDNA (described below).
Standards for RT-qPCR were prepared as described in Appendix S4. Reverse transcription-qPCR was conducted on a Mastercycler realplex ep (Eppendorf, Hamburg, Germany) with a total reaction volume of 20 μl as described in Appendix S4. The R2 and efficiencies for mcrA and 16S rRNA standard curves were 0.991 and 0.997 and 75% and 71% respectively.
16S rDNA and rRNA sequencing
Polymerase chain reaction, sample multiplexing and Illumina MiSeq sequencing of 16S rDNA and rRNA were performed by the Host Microbiome Initiative (University of Michigan, Ann Arbor, MI; further information provided in Appendix S5). After quality filtering and subsampling, 16 587 paired-end reads (2 × 250 bp) per sample were generated. The resulting sequences were processed with mothur (Schloss et al., 2009) following the Schloss MiSeq SOP and classified using the Ribosomal Database Project (Maidak et al., 2013) and Basic Local Alignment Search Tool (blast; NCBI, Bethesda, MD). These sequence data have been submitted to the DDBJ/EMBL/GenBank databases under Accession Number SRP056737.
Acknowledgments
The authors wish to thank Julie Pierce and Qaboos Imran for their help in the lab, and Judy Opp for help with Illumina sequencing.
Conflict of Interest
None declared.
Supporting Information
Fig. S1. Influent (total and soluble), bioreactor (soluble) and permeate COD during days 1–100.
Fig. S2. P1 VFA concentrations (concentrations are expressed as the actual compound, not as COD), theoretical COD contribution from measured VFAs and measured COD during days 1–100. Total as COD is the calculated theoretical COD contribution from measured VFAs. Results for P2 and P3 were very similar (data not reported). Error bars represent standard deviations of triplicate IC injections.
Fig. S3. COD mass balance for days 100–138. Total CODout is the summation of measured permeate COD, measured dissolved methane, measured gaseous methane, theoretical COD removal from measured sulfate reduction and theoretical COD from measured biomass wasting.
Fig. S4. Total suspended solids (TSS) and volatile suspended solids (VSS) in the bioreactor during days 1–173 (primary y-axis) and inverse Simpson index in suspended biomass based on 16S rDNA sequencing (secondary y-axis). Error bars represent standard deviation of triplicate sample analysis.
Fig. S5. (A) Relative abundance of methanogens identified at the genus level based on 16S rDNA sequencing and (B) relative activity of methanogens identified at the genus level based on 16S rRNA sequencing in suspended biomass from start-up to the end of Phase 2 and in biofilms at the end of Phase 2. Data are expressed as a percentage and were normalized using the total archaeal 16S rDNA sequences (A) and 16S rRNA sequences (B).
Fig. S6. (A) Relative abundance of sulfate-reducing bacteria identified at the genus level based on 16S rDNA sequencing and (B) relative activity of sulfate-reducing bacteria identified at the genus level based on 16S rRNA sequencing in suspended biomass from start-up to the end of Phase 2 and in biofilms at the end of Phase 2. Data are expressed as a percentage and were normalized using the total 16S rDNA sequences (A) and 16S rRNA sequences (B) (including archaeal and bacterial sequences). A truncated y-axis (0–8%) is shown to accentuate changes in abundance and activity.
Fig. S7. Influent (total and soluble), bioreactor (soluble) and permeate COD during Phases 3 and 4.
Fig. S8. P1, P2 and P3 permeate VFA concentrations (concentrations are expressed as the actual compound, not as COD), theoretical COD contribution from measured VFAs, and measured COD during Phases 3 and 4. Total as COD is the calculated theoretical COD contribution from measured VFAs. Error bars represent standard deviations of triplicate IC injections.
Table S1. Primer coverage of Archaea for 16S rDNA primers F515 (GTGCCAGCMGCCGCGGTAA) and R806 (GGACTACHVGGGTWTCTAAT) targeting the V4 region (Caporaso et al., 2011) according to TestPrime 1.0. TestPrime 1.0 evaluates the coverage of primer pairs by running an in silico PCR using the SILVA databases. Zero primer mismatches were allowed.
Table S2. Primer coverage of Bacteria for 16S rDNA primers targeting the V4 region according to TestPrime 1.0 (see Table S1 legend for additional details). The coverage of taxa with known fatty acid-oxidizing syntrophic bacteria is specified down to the genus or family levels.
Appendix S1. Chemical assays for process monitoring.
Appendix S2. DNA and RNA extractions.
Appendix S3. Primer design for mcrA gene.
Appendix S4. RT-qPCR.
Appendix S5. Illumina sequencing.
Appendix S6. Start-up of psychrophilic AnMBR.
References
- Bandara WM, Kindaichi T, Satoh H, Sasakawa M, Nakahara Y, Takahashi M. Okabe S. Anaerobic treatment of municipal wastewater at ambient temperature: analysis of archaeal community structure and recovery of dissolved methane. Water Res. 2012;46:5756–5764. doi: 10.1016/j.watres.2012.07.061. [DOI] [PubMed] [Google Scholar]
- Bandara WMKRTW, Satoh H, Sasakawa M, Nakahara Y, Takahashi M. Okabe S. Removal of residual dissolved methane gas in an upflow anaerobic sludge blanket reactor treating low-strength wastewater at low temperature with degassing membrane. Water Res. 2011;45:3533–3540. doi: 10.1016/j.watres.2011.04.030. [DOI] [PubMed] [Google Scholar]
- Blazewicz SJ, Barnard RL, Daly RA. Firestone MK. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. The ISME J. 2013;7:2061–2068. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräuer SL, Cadillo-Quiroz H, Ward RJ, Yavitt JB. Zinder SH. Methanoregula boonei gen. nov., sp. nov., an acidophilic methanogen isolated from an acidic peat bog. Int J Syst Evol Microbiol. 2011;61:45–52. doi: 10.1099/ijs.0.021782-0. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang M, Wang A, Lin H, Hong H. Lu X. Osmotic pressure effect on membrane fouling in a submerged anaerobic membrane bioreactor and its experimental verification. Bioresour Technol. 2012;125:97–101. doi: 10.1016/j.biortech.2012.08.038. [DOI] [PubMed] [Google Scholar]
- Collins G, McHugh S, Connaughton S, Enright AM, Kearney A, Scully C, et al. New low-temperature applications of anaerobic wastewater treatment. J Environ Sci Health Part A Tox Hazard Subst Environ Eng. 2006;41:881–895. doi: 10.1080/10934520600614504. [DOI] [PubMed] [Google Scholar]
- Conklin A, Stensel HD. Ferguson J. Growth kinetics and competition between Methanosarcina and Methanosaeta in mesophilic anaerobic digestion. Water Environ Res. 2006;78:486–496. doi: 10.2175/106143006x95393. [DOI] [PubMed] [Google Scholar]
- Connaughton S, Collins G. O’Flaherty V. Development of microbial community structure and activity in a high-rate anaerobic bioreactor at 18 degrees C. Water Res. 2006;40:1009–1017. doi: 10.1016/j.watres.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Cookney J, Cartmell E, Jefferson B. McAdam E. Recovery of methane from anaerobic process effluent using poly-di-methyl-siloxane membrane contactors. Water Sci Technol. 2012;65:604–610. doi: 10.2166/wst.2012.897. [DOI] [PubMed] [Google Scholar]
- Dagnew M, Parker W, Seto P, Waldner K, Hong Y, Bayly R. Cumin J. 2011. Pilot testing of an AnMBR for municipal wastewater treatment. 84th Annual Water Environment Federation Technical Exhibition and Conference, Los Angeles, CA, USA.
- De Vrieze J, Hennebel T, Boon N. Verstraete W. Methanosarcina: the rediscovered methanogen for heavy duty biomethanation. Bioresour Technol. 2012;112:1–9. doi: 10.1016/j.biortech.2012.02.079. [DOI] [PubMed] [Google Scholar]
- Eichler S, Christen R, Höltje C, Westphal P, Bötel J, Brettar I, et al. Composition and dynamics of bacterial communities of a drinking water supply system as assessed by RNA-and DNA-based 16S rRNA gene fingerprinting. Appl Environ Microbiol. 2006;72:1858–1872. doi: 10.1128/AEM.72.3.1858-1872.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foesel BU, Nägele V, Naether A, Wüst PK, Weinert J, Bonkowski M, et al. Determinants of Acidobacteria activity inferred from the relative abundances of 16S rRNA transcripts in German grassland and forest soils. Environ Microbiol. 2013;16:658–675. doi: 10.1111/1462-2920.12162. [DOI] [PubMed] [Google Scholar]
- Francoleon DR, Boontheung P, Yang Y, Kim U, Ytterberg AJ, Denny PA, et al. S-layer, surface-accessible, and concanavalin a binding proteins of Methanosarcina acetivorans and Methanosarcina mazei. J Proteome Res. 2009;8:1972–1982. doi: 10.1021/pr800923e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag TE. Prosser JI. Correlation of methane production and functional gene transcriptional activity in a peat soil. Appl Environ Microbiol. 2009;75:6679–6687. doi: 10.1128/AEM.01021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Qiu Q, Liu P, Rui J. Lu Y. Syntrophic oxidation of propionate in rice field soil at 15 and 30 C under methanogenic conditions. Appl Environ Microbiol. 2012;78:4923–4932. doi: 10.1128/AEM.00688-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D-W, Zhang T, Tang C-YY, Wu W-M, Wong C-Y, Lee YH, et al. Membrane fouling in an anaerobic membrane bioreactor: differences in relative abundance of bacterial species in the membrane foulant layer and in suspension. J Memb Sci. 2010;364:331–338. [Google Scholar]
- Gimenez JB, Robles A, Carretero L, Duran F, Ruano MV, Gatti MN, et al. Experimental study of the anaerobic urban wastewater treatment in a submerged hollow-fibre membrane bioreactor at pilot scale. Bioresour Technol. 2011;102:8799–8806. doi: 10.1016/j.biortech.2011.07.014. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Gil G, Seghezzo L, Lettinga G. Kleerebezem R. Kinetics and mass-transfer phenomena in anaerobic granular sludge. Biotechnol Bioeng. 2001;73:125–134. doi: 10.1002/bit.1044. [DOI] [PubMed] [Google Scholar]
- Gouveia J, Plaza F, Garralon G, Fdz-Polanco F. Peña M. Long-term operation of a pilot scale anaerobic membrane bioreactor (AnMBR) for the treatment of municipal wastewater under psychrophilic conditions. Bioresour Technol. 2015;185:225–233. doi: 10.1016/j.biortech.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Hattori S. Syntrophic acetate-oxidizing microbes in methanogenic environments. Microbes Environ. 2008;23:118–127. doi: 10.1264/jsme2.23.118. [DOI] [PubMed] [Google Scholar]
- Hoffmann RA, Garcia ML, Veskivar M, Karim K, Al-Dahhan MH. Angenent LT. Effect of shear on performance and microbial ecology of continuously stirred anaerobic digesters treating animal manure. Biotechnol Bioeng. 2008;100:38–48. doi: 10.1002/bit.21730. [DOI] [PubMed] [Google Scholar]
- Huang Z, Ong SL. Ng HY. Submerged anaerobic membrane bioreactor for low-strength wastewater treatment: effect of HRT and SRT on treatment performance and membrane fouling. Water Res. 2011;45:705–713. doi: 10.1016/j.watres.2010.08.035. [DOI] [PubMed] [Google Scholar]
- Hunt DE, Lin Y, Church MJ, Karl DM, Tringe SG, Izzo LK. Johnson ZI. Relationship between abundance and specific activity of bacterioplankton in open ocean surface waters. Appl Environ Microbiol. 2013;79:177–184. doi: 10.1128/AEM.02155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii SI, Kosaka T, Hori K, Hotta Y. Watanabe K. Coaggregation facilitates interspecies hydrogen transfer between Pelotomaculum thermopropionicum and Methanothermobacter thermautotrophicus. Appl Environ Microbiol. 2005;71:7838–7845. doi: 10.1128/AEM.71.12.7838-7845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii SI, Kosaka T, Hotta Y. Watanabe K. Simulating the contribution of coaggregation to interspecies hydrogen fluxes in syntrophic methanogenic consortia. Appl Environ Microbiol. 2006;72:5093–5096. doi: 10.1128/AEM.00333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten MS, Stams AJ. Zehnder AJ. Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol Lett. 1992;88:181–197. [Google Scholar]
- Klappenbach JA, Dunbar JM. Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Klein D, Arab H, Völker H. Thomm M. Methanosarcina baltica, sp. nov., a novel methanogen isolated from the Gotland Deep of the Baltic Sea. Extremophiles. 2002;6:103–110. doi: 10.1007/s007920100234. [DOI] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M. Glöckner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012;41:1–11. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kola A, Ye Y, Le-Clech P. Chen V. Transverse vibration as novel membrane fouling mitigation strategy in anaerobic membrane bioreactor applications. J Memb Sci. 2014;455:320–329. [Google Scholar]
- Lee ZM-P, Bussema C. Schmidt TM. rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res. 2009;37:D489–D493. doi: 10.1093/nar/gkn689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettinga G, Rebac S. Zeeman G. Challenge of psychrophilic anaerobic wastewater treatment. Trends Biotechnol. 2001;19:363–370. doi: 10.1016/s0167-7799(01)01701-2. [DOI] [PubMed] [Google Scholar]
- Ma J, Wang Z, Zou X, Feng J. Wu Z. Microbial communities in an anaerobic dynamic membrane bioreactor (AnDMBR) for municipal wastewater treatment: comparison of bulk sludge and cake layer. Process Biochem. 48:510–516. [Google Scholar]
- Maidak BL, Olsen GJ, Larsen N, Overbeek R, McCaughey MJ. Woese CR. The RDP (Ribosomal Database Project) Nucleic Acids Research. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh S, Carton M, Collins G. O’Flaherty V. Reactor performance and microbial community dynamics during anaerobic biological treatment of wastewaters at 16–37 C. FEMS Microbiol Ecol. 2004;48:369–378. doi: 10.1016/j.femsec.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Martin-Laurent F, Philippot L, Hallet S, Chaussod R, Germon J, Soulas G. Catroux G. DNA extraction from soils: old bias for new microbial diversity analysis methods. Appl Environ Microbiol. 2001;67:2354–2359. doi: 10.1128/AEM.67.5.2354-2359.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sosa D, Helmreich B, Netter T, Paris S, Bischof F. Horn H. Pilot-scale anaerobic submerged membrane bioreactor (AnSMBR) treating municipal wastewater: the fouling phenomenon and long-term operation. Water Sci Technol. 2011;64:1804–1811. doi: 10.2166/wst.2011.745. [DOI] [PubMed] [Google Scholar]
- Männistö MK, Kurhela E, Tiirola M. Häggblom MM. Acidobacteria dominate the active bacterial communities of Arctic tundra with widely divergent winter-time snow accumulation and soil temperatures. FEMS Microbiol Ecol. 2013;84:47–59. doi: 10.1111/1574-6941.12035. [DOI] [PubMed] [Google Scholar]
- Mladenovska Z. Ahring BK. Mixotrophic growth of two thermophilic Methanosarcina strains, Methanosarcina thermophila TM-1 and Methanosarcina sp. SO-2P, on methanol and hydrogen/carbon dioxide. Appl Microbiol Biotechnol. 1997;48:385–388. [Google Scholar]
- Morita M, Malvankar NS, Franks AE, Summers ZM, Giloteaux L, Rotaru AE, et al. Potential for direct interspecies electron transfer in methanogenic wastewater digester aggregates. MBio. 2011;2:e159–e111. doi: 10.1128/mBio.00159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmasiri SI, Zhang J, Fitch M, Norddahl B, Morgenroth E. Raskin L. Methanogenic population dynamics and performance of an anaerobic membrane bioreactor (AnMBR) treating swine manure under high shear conditions. Water Res. 2007;41:134–144. doi: 10.1016/j.watres.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Riquelme Breazeal MV, Novak JT, Vikesland PJ. Pruden A. Effect of wastewater colloids on membrane removal of antibiotic resistance genes. Water Res. 2012;47:130–140. doi: 10.1016/j.watres.2012.09.044. [DOI] [PubMed] [Google Scholar]
- Robinson RW, Aldrich H, Hurst SF. Bleiweis A. Role of the cell surface of Methanosarcina mazei in cell aggregation. Appl Environ Microbiol. 1985;49:321–327. doi: 10.1128/aem.49.2.321-327.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles A, Ruano M, Ribes J. Ferrer J. Performance of industrial scale hollow-fibre membranes in a submerged anaerobic MBR (HF-SAnMBR) system at mesophilic and psychrophilic conditions. Separation and Purification Technology. 2013;104:290–296. [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholten JC, Culley DE, Brockman FJ, Wu G. Zhang W. Evolution of the syntrophic interaction between Desulfovibrio vulgaris and Methanosarcina barkeri: involvement of an ancient horizontal gene transfer. Biochem Biophys Res Commun. 2007;352:48–54. doi: 10.1016/j.bbrc.2006.10.164. [DOI] [PubMed] [Google Scholar]
- Shin C, McCarty PL, Kim J. Bae J. Pilot-scale temperate-climate treatment of domestic wastewater with a staged anaerobic fluidized membrane bioreactor (SAF-MBR) Bioresour Technol. 2014;159:95–103. doi: 10.1016/j.biortech.2014.02.060. [DOI] [PubMed] [Google Scholar]
- Sieber JR, Sims DR, Han C, Kim E, Lykidis A, Lapidus AL, et al. The genome of Syntrophomonas wolfei: new insights into syntrophic metabolism and biohydrogen production. Environ Microbiol. 2010;12:2289–2301. doi: 10.1111/j.1462-2920.2010.02237.x. [DOI] [PubMed] [Google Scholar]
- Simankova MV, Parshina SN, Tourova TP, Kolganova TV, Zehnder AJ. Nozhevnikova AN. Methanosarcina lacustris sp. nov., a new psychrotolerant methanogenic archaeon from anoxic lake sediments. Syst Appl Microbiol. 2001;24:362–367. doi: 10.1078/0723-2020-00058. [DOI] [PubMed] [Google Scholar]
- Smith A, Skerlos S. Raskin L. Anaerobic membrane bioreactor treatment of domestic wastewater at psychrophilic temperatures ranging from 15°C to 3°C. Environ Sci: Water Res Technol. 2015;1:56–64. [Google Scholar]
- Smith AL, Skerlos SJ. Raskin L. Psychrophilic anaerobic membrane bioreactor treatment of domestic wastewater. Water Res. 2013;47:1655–1665. doi: 10.1016/j.watres.2012.12.028. [DOI] [PubMed] [Google Scholar]
- Smith AL, Stadler LB, Cao L, Love NG, Raskin L. Skerlos SJ. Navigating wastewater energy recovery strategies: a life cycle comparison of anaerobic membrane bioreactor and conventional treatment systems with anaerobic digestion. Environ Sci Technol. 2014;48:5972–5981. doi: 10.1021/es5006169. [DOI] [PubMed] [Google Scholar]
- Smith KS. Ingram-Smith C. Methanosaeta, the forgotten methanogen? Trends Microbiol. 2007;15:150–155. doi: 10.1016/j.tim.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Sousa DZ, Smidt H, Alves MM. Stams AJ. Syntrophomonas zehnderi sp. nov., an anaerobe that degrades long-chain fatty acids in co-culture with Methanobacterium formicicum. Int J Syst Evol Microbiol. 2007;57:609–615. doi: 10.1099/ijs.0.64734-0. [DOI] [PubMed] [Google Scholar]
- Stams AJ, Sousa DZ, Kleerebezem R. Plugge CM. Role of syntrophic microbial communities in high-rate methanogenic bioreactors. Water Sci Technol. 2012;66:352–362. doi: 10.2166/wst.2012.192. [DOI] [PubMed] [Google Scholar]
- Stroot PG, McMahon KD, Mackie RI. Raskin L. Anaerobic codigestion of municipal solid waste and biosolids under various mixing conditions – I. Digester performance. Water Res. 2001;35:1804–1816. doi: 10.1016/s0043-1354(00)00439-5. [DOI] [PubMed] [Google Scholar]
- Sukenik A, Kaplan-Levy RN, Welch JM. Post AF. Massive multiplication of genome and ribosomes in dormant cells (akinetes) of Aphanizomenon ovalisporum (Cyanobacteria) The ISME Journal. 2012;6:670–679. doi: 10.1038/ismej.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers ZM, Fogarty HE, Leang C, Franks AE, Malvankar NS. Lovley DR. Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science. 2010;330:1413–1415. doi: 10.1126/science.1196526. [DOI] [PubMed] [Google Scholar]
- Tchobanoglous G, Burton FL. Stensel HD. Wastewater Engineering: Treatment and Reuse. New York, NY, USA: McGraw-Hill; 2003. [Google Scholar]
- Thauer RK. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology-UK. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- Vavilin V. Angelidaki I. Anaerobic degradation of solid material: importance of initiation centers for methanogenesis, mixing intensity, and 2D distributed model. Biotechnol Bioeng. 2005;89:113–122. doi: 10.1002/bit.20323. [DOI] [PubMed] [Google Scholar]
- Větrovský T. Baldrian P. The variability of the 16S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS ONE. 2013;8:e57923. doi: 10.1371/journal.pone.0057923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welander PV. Metcalf WW. Loss of the mtr operon in Methanosarcina blocks growth on methanol, but not methanogenesis, and reveals an unknown methanogenic pathway. Proc Natl Acad Sci USA. 2005;102:10664–10669. doi: 10.1073/pnas.0502623102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann P. The effect of incubation temperature on steady-state concentrations of hydrogen and volatile fatty acids during anaerobic degradation in slurries from wetland sediments. FEMS Microbiol Ecol. 1994;13:295–302. [Google Scholar]
- Wu MM, Criddle CS. Hickey RF. Mass transfer and temperature effects on substrate utilization in brewery granules. Biotechnol Bioeng. 1995;46:465–475. doi: 10.1002/bit.260460511. [DOI] [PubMed] [Google Scholar]
- Yang J, Spanjers H. van Lier JB. Pulse shear stress for anaerobic membrane bioreactor fouling control. Water Sci Technol. 2011;64:355–360. doi: 10.2166/wst.2011.727. [DOI] [PubMed] [Google Scholar]
- Yang W, Cicek N. Ilg J. State-of-the-art of membrane bioreactors: worldwide research and commercial applications in North America. J Memb Sci. 2006;270:201–211. [Google Scholar]
- Yashiro Y, Sakai S, Ehara M, Miyazaki M, Yamaguchi T. Imachi H. Methanoregula formicica sp. nov., a methane-producing archaeon isolated from methanogenic sludge. Int J Syst Evol Microbiol. 2011;61:53–59. doi: 10.1099/ijs.0.014811-0. [DOI] [PubMed] [Google Scholar]
- Yoo R, Kim J, McCarty PL. Bae J. Anaerobic treatment of municipal wastewater with a staged anaerobic fluidized membrane bioreactor (SAF-MBR) system. Bioresour Technol. 2012;120:133–139. doi: 10.1016/j.biortech.2012.06.028. [DOI] [PubMed] [Google Scholar]
- Yu Z, Wen X, Xu M. Huang X. Characteristics of extracellular polymeric substances and bacterial communities in an anaerobic membrane bioreactor coupled with online ultrasound equipment. Bioresour Technol. 2012;117:333–340. doi: 10.1016/j.biortech.2012.04.075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Influent (total and soluble), bioreactor (soluble) and permeate COD during days 1–100.
Fig. S2. P1 VFA concentrations (concentrations are expressed as the actual compound, not as COD), theoretical COD contribution from measured VFAs and measured COD during days 1–100. Total as COD is the calculated theoretical COD contribution from measured VFAs. Results for P2 and P3 were very similar (data not reported). Error bars represent standard deviations of triplicate IC injections.
Fig. S3. COD mass balance for days 100–138. Total CODout is the summation of measured permeate COD, measured dissolved methane, measured gaseous methane, theoretical COD removal from measured sulfate reduction and theoretical COD from measured biomass wasting.
Fig. S4. Total suspended solids (TSS) and volatile suspended solids (VSS) in the bioreactor during days 1–173 (primary y-axis) and inverse Simpson index in suspended biomass based on 16S rDNA sequencing (secondary y-axis). Error bars represent standard deviation of triplicate sample analysis.
Fig. S5. (A) Relative abundance of methanogens identified at the genus level based on 16S rDNA sequencing and (B) relative activity of methanogens identified at the genus level based on 16S rRNA sequencing in suspended biomass from start-up to the end of Phase 2 and in biofilms at the end of Phase 2. Data are expressed as a percentage and were normalized using the total archaeal 16S rDNA sequences (A) and 16S rRNA sequences (B).
Fig. S6. (A) Relative abundance of sulfate-reducing bacteria identified at the genus level based on 16S rDNA sequencing and (B) relative activity of sulfate-reducing bacteria identified at the genus level based on 16S rRNA sequencing in suspended biomass from start-up to the end of Phase 2 and in biofilms at the end of Phase 2. Data are expressed as a percentage and were normalized using the total 16S rDNA sequences (A) and 16S rRNA sequences (B) (including archaeal and bacterial sequences). A truncated y-axis (0–8%) is shown to accentuate changes in abundance and activity.
Fig. S7. Influent (total and soluble), bioreactor (soluble) and permeate COD during Phases 3 and 4.
Fig. S8. P1, P2 and P3 permeate VFA concentrations (concentrations are expressed as the actual compound, not as COD), theoretical COD contribution from measured VFAs, and measured COD during Phases 3 and 4. Total as COD is the calculated theoretical COD contribution from measured VFAs. Error bars represent standard deviations of triplicate IC injections.
Table S1. Primer coverage of Archaea for 16S rDNA primers F515 (GTGCCAGCMGCCGCGGTAA) and R806 (GGACTACHVGGGTWTCTAAT) targeting the V4 region (Caporaso et al., 2011) according to TestPrime 1.0. TestPrime 1.0 evaluates the coverage of primer pairs by running an in silico PCR using the SILVA databases. Zero primer mismatches were allowed.
Table S2. Primer coverage of Bacteria for 16S rDNA primers targeting the V4 region according to TestPrime 1.0 (see Table S1 legend for additional details). The coverage of taxa with known fatty acid-oxidizing syntrophic bacteria is specified down to the genus or family levels.
Appendix S1. Chemical assays for process monitoring.
Appendix S2. DNA and RNA extractions.
Appendix S3. Primer design for mcrA gene.
Appendix S4. RT-qPCR.
Appendix S5. Illumina sequencing.
Appendix S6. Start-up of psychrophilic AnMBR.


