Abstract
Pancreatic cancer remains a lethal disease with limited treatment options. At the time of diagnosis, approximately 80% of these patients present with unresectable tumors caused by either locally advanced lesions or progressive metastatic growth. Therefore, development of novel treatment strategies and new therapeutics are needed. Xanthohumol (XN) has emerged as a potential compound that inhibits various types of cancer, but the molecular mechanism underlying the effects of XN remain unclear. In the present study, we have assessed the efficacy of XN on pancreatic cancer cell lines (AsPC-1, PANC-1, L3.6pl, MiaPaCa-2, 512, and 651) against cell growth in real time and using colony forming assays. Treatment with XN resulted in reduction in cellular proliferation in a dose and time dependent manner. The growth suppression effect of XN in pancreatic cancer cell lines is due to increased apoptosis via the inhibition of the Notch1 signaling pathway, as evidenced by reduction in Notch1, HES-1, and survivin both at mRNA as well as protein levels. Notch1 promoter reporter analysis after XN treatment indicated that XN down regulates Notch promoter activity. Importantly, overexpression of active Notch1 in XN-treated pancreatic cancer cells resulted in negation of growth suppression. Taken together, these findings demonstrate, for the first time, that the growth suppressive effect of XN in pancreatic cancer cells is mainly mediated by Notch1 reduction.
Keywords: Notch signaling, xanthohumol, apoptosis, pancreatic cancer
Introduction
Pancreatic cancer is the 8th leading cause of cancer death worldwide and it is the 4th leading cause of cancer deaths in the United States (US). The incidence and death rates for pancreatic cancer are rising. It is estimated that pancreatic cancer incidence and mortality will be approximately 46,420 and 39,590 respectively in 2014 (1). It is projected that by 2030, pancreatic cancer will likely be the 2nd leading cause of cancer-related death in the USA (2). The prognosis for pancreatic cancer is extremely poor.(3-5). Combination cytotoxic therapies, including FOLFIRINIX and gemcitabine with nab-paclitaxel have showed clinically meaningful improvements over the single agent gemcitabine which was the prior standard of care, however the median survival benefit is an additional 4-6 months [5-8]. Regardless of current therapies, less than 5% of patients with pancreatic cancer survive for 5 years due to the aggressiveness of the disease and the lack of effective therapies (3-5). Therefore, development of new therapeutic compounds is needed.
. . Natural compounds and novel synthetic drugs have been shown to induce the process of apoptosis by altering various signaling pathways. Xanthohumol (XN), a natural flavonoid isolated from the cones of the hop plant (Humulus lupulus L.), has been shown to inhibit cancer cell proliferation in vitro in various solid organ human malignancies such as breast, colon, prostate, ovarian, hepatocellular, and medullary thyroid cancers (6-12). XN has also been shown to reduce growth by inducing apoptosis, both caspase dependent and independent, in cancer cells (13, 14). In vivo, oral administration of XN showed delayed advanced tumor progression and also reduced cell growth of poorly differentiated prostrate carcinoma [16]. In addition, XN exerts a broad range of biological activities such as antioxidant, anti-inflammatory, antimicrobial, immune modulatory activity, and may also have therapeutic potential for metabolic diseases including type 2 diabetes, a risk factor for the development of pancreatic cancer (15-21). However, the biological effects of XN in pancreatic cancer are not known. In the present study, we investigated the antiproliferative effects of XN on established human pancreatic cancer cell lines and cells derived from human pancreatic cancer tissues. We found that XN inhibited cellular growth in a dose and time dependent manner. Treatment of pancreatic cancer cell lines with XN also induced apoptosis, demonstrated growth suppression which was associated with reduction in Notch pathway proteins and mRNA, and resulted in reduction in Notch1 promoter activity. Importantly, overexpression of active Notch1 negated the growth suppressive effect of XN in pancreatic cancer cell lines. These findings suggested that growth suppression of pancreatic cancer cells by XN may be mediated by Notch1 reduction.
Materials and Methods
Cell lines and culture conditions
The human pancreatic cancer cell lines (AsPC-1, PANC-1, and MiaPaCa-2) and human normal fetal lung fibroblast, WI-38, were purchased in 2012 from American Type Culture Collection (ATCC, Rockville, MD, USA) and expanded and frozen several vials after 3rd generation. 4-10 generations cells were used for the entire experiments. L3.6pl was a kind gift from Dr. Jose G. Trevino, University of Florida-Gainesville and received in 2013. Unique patient derived pancreatic cancer cells (512 and 651) were obtained during 2013 from the Medical College of Wisconsin Surgical Oncology Biorepository in 2013. Pancreatic cancer cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen) whereas WI-38 was maintained in Modified Eagle Medium (MEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% penicillin/streptomycin (Invitrogen) at 37°C in a humidified atmosphere with 5% CO2. Patient derived pancreatic cancer cells 512 and 651 were cultured in DMEM-F12 (1:1, Invitrogen) supplemented with 10%FBS, recombinant TNF-α, EGF, insulin growth factor I (IGF) and basic fibroblast growth factor (bFGF) (Invitrogen). The culture media was replaced every 2-3 days. The confluent cells were subcultured by splitting them at 1:5 ratios. Authenticity of ATCC cell lines were done by ATCC before purchase by the standard short tandem repeat DNA typing methodology. Pancreatic cancer cells (512 and 651) from MCW were not independently authenticated. L3.6pl was authenticated by STR analysis by Dr. Jose Trevino at University of Florida. No authentication of these cell lines were done by the authors. However, these cells were expanded and frozen in multiple vials within three weeks of receipt for this study. Experiments were carried out with in the 15-20 passages of each thawing of cells.
Reagents
Xanthohumol (XN) and 1-chloro-2,4-dimethylthylthiazol-2yl-2,5-dipheynltetrazolium bromide (MTT), dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St.Louis, MO, USA). The XN was solubilized in DMSO at stock concentrations of 50mM and diluted in the media when used for treatment. Antibodies against survivin, glyceraldehyde phosphate dehydrogenase (GAPDH), Notch1, HES-1, and caspase-3 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and cleaved caspase-3 and cleaved PARP were obtained from Cell Signaling Technology Inc. (Denvers, MA, USA) respectively.
Cell proliferation, viability, and colony forming ability assays
Cells were seeded in 24-well plates and were treated with different concentrations of XN for up to 96 hours. Cell proliferation/viability was measured by using colorimetric assay with MTT (22). Cell viability was also assessed by trypan blue staining. The results represent the average of three experiments each conducted in quadruplicates. Values were calculated to percent inhibition relative to vehicle control (0.1% DMSO). The effect of XN on colony forming ability was determined by the measurement of the colonogenic cell survival as previously described (22).
Non- invasive cellular proliferation assay in real time
Using IncuCyte Live-Cell Imaging systems (Essen Bioscience, Ann Arbor, MI, USA), cellular proliferation of pancreatic cancer cell lines was measured as previously described (23). Briefly, MiaPaCa-2 (1000 cells/well), AsPC-1 (2000 cells/well), and PANC-1 (2000 cells/well) were plated onto 96-well plate and incubated in an XL-3 incubation chamber maintained at 37°C. After 12 hours, the cells were treated with varying concentrations of XN (0-50μM) for up to 4 days. The cells were photographed every two hours using a 10X objective for the entire duration of the incubation. Cell confluence was calculated using IncuCyte 2011A software and importantly the IncuCyte Analyzer provides real-time cellular confluence data based on segmentation of high-definition phase-contrast images. The cell proliferation was expressed as an increase in percentage of confluence in 12 hours intervals.
Western blot analysis
Western blot analyses were conducted as previously described (22). Cells were lysed in radioimmunoprecipitation assay (RIPA, Thermo Fisher) buffer supplemented with protease inhibitor cocktail (Sigma) and phenylmethylsulfonylfluoride (Sigma). Equal amount of proteins were quantified by bicinchonic method (BCA, Thermo Fisher) and analyzed by SDS-PAGE (Bio-Rad Laboratories, Hercules, CA, USA). Then the proteins were transferred to nitrocellulose membranes (Bio-Rad) using a Trans-Blot (Bio-Rad) and analyzed by specific antibodies as indicated in the experiments. The detection of immune complexes was conducted using chemiluminescence with an HRP antibody detection kit and then images were taken using Molecular ImagerTM ChemiDoc XRS+ imager with image lab software (Bio-Rad).
Caspase-3 and -7 activities
Caspase-Glo 3/7 Assay (Promega, Madison, WI, USA) kit was used to measure the cleaved caspase-3 and -7 activities from the lysates of cells treated with XN. Ten to fifteen μg of protein samples in 25 μL total volume was mixed with equal volume (25 μL) of Caspase-Glo reagent and incubated at room temperature in a white 96-well plate for 30 minutes. Then the luminescence was measured using Infinite M200PRO Microplate reader (TECAN, San Jose, CA).
Real-time reverse transcriptase polymerase chain reaction (RT-PCR)
RT-PCR was conducted to quantify gene expression using mRNA from control or XN treated cells. Total RNA (1μg) was reverse transcribed using either iScript cDNA Synthesis Kit (Bio-Rad) or Reverse Transcriptase 2X Mastermix (MidSci, St. Louis, MO, USA) and PCR was carried out using SsoAdvanced SYBR Green Supermix (Bio-Rad) or Evagreen qPCR Mastermix (MidSci) in a master cycler (CFX96; Bio-Rad) as per the manufacturer’s instruction. Sequences for each PCR primers pairs were listed in Table 1. Data were analyzed as per to the comparative CT method and normalized for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression in each sample.
Table 1.
Sequence of primers used for real-time PCR
| Primer | Sequence (5'to3') |
|---|---|
| Notch1 forward primer | TCCACCAGTTTGAATGGTCA |
| Notch1 reverse primer | AGCTCATCATCTGGGACAGG |
| HES1 forward primer | TGAAGAAAGATAGCTCGCGG |
| HES1 reverse primer | GGTACTTCCCCAGCACACTT |
| HEY1 forward primer | TGGATCACCTGAAAATGCTG |
| HEY1 reverse primer | CGAAATCCCAAACTCCGATA |
| GAPDH forward primer | GAGTCAACGGATTTGGTCGT |
| GAPDH reverse primer | TTGATTTTGGAGGGATCTCG |
| Survivin forward primer | GACCACCGCATCTCTACATTC |
| Survivin reverse primer | AAGTCT GGCTCGTTCTCAGTG |
Luciferase reporter assay
To determine the effect of XN on Notch1 promoter, pancreatic cancer cells were transfected with luciferase reporter gene under the control of Notch1 promoter as previously described (24). Notch1 promoter and promoterless control plasmids were a generous gift from Dr. Yugawa, National Cancer Center Research Institute, Tokyo, Japan. Briefly, plasmid containing Notch1 promoter or promoterless vector was cotransfected with simian virus β-galactosidase (SV-β-gal; Promega, Madison, WI, USA) in to pancreatic cancer cells using Lipofectamine 2000 (Invitrogen). Next day, cells were treated with or without XN at indicated concentrations and incubated for another 24-48 hours. Cell lysates were prepared and luciferase and β-galactosidase assays (Promega) were carried out as per the manufacturer’s instructions.
Statistical analysis
Analysis of variance (ANOVA) with Bonferroni post hoc testing was performed using a statistical analysis software package (IBM SPSS Statistics version 22, New York, NY, USA). A P value of < 0.05 was considered significant. Data were represented as ± standard error.
Results
Effects of Xanthohumol on pancreatic cancer cells proliferation
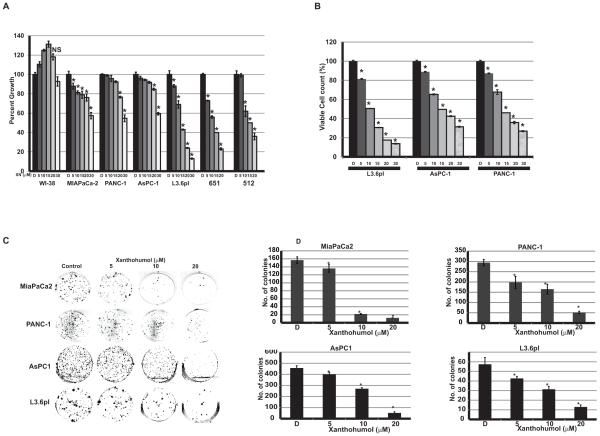
Effect of Xanthohumol (XN) on cellular proliferation/viability of established human pancreatic cancer cells (MIAPaCa-2, PANC-1, AsPC-1, and L3.6pl) and patient-derived pancreatic cancer cells (512 and 651) was evaluated by MTT assay. As shown in Fig.1A treatment of MIAPaCa-2, PANC-1, AsPC-1, and L3.6pl cells with XN (5-30 μM) resulted in a dose-dependent reduction in cell viability compared to vehicle (DMSO) controls after 2 days of treatment. Pancreatic cancer patient derived cells 512 and 651 were also showed growth reduction in a dose-dependent manner. However, in contrast, WI-38 cells showed no significant growth reduction even at 30 μM of XN suggesting that normal cells may not be affected by XN (Fig. 1A). To determine whether the effect of XN was persistant, cell viability was measured by harvesting cells after 4 days of XN treatment and stained with trypan blue and counted the viable cells. The percentages of viable cells were significantly reduced in a dose-dependent manner in PANC-1, AsPC-1, and L3.6pl cells (Fig. 1B). This was further confirmed by assessing the colony forming ability of these cell lines after XN treatment. As shown in Fig. 1C and 1D, the colonogenic potential in XN treated cells was significantly decreased. Increasing concentrations of XN demonstrated a decrease in cellular proliferation represented by decrease in confluence in MiaPaCa-2, AsPC-1, and PANC-1 in a time and dose-dependent manner as measured by the cell confluence real time kinetics (Fig. 2).
Figure 1.
Effect of XN on pancreatic cancer cellular proliferation. A. Human pancreatic cancer cell lines and human normal lung fibroblast cells (WI-38) were treated with XN at indicated doses for 24 hours and cytotoxicity was measured using MTT assay (n=3; p=0.05 at 20 μM concentrations for all cancer cell lines except WI-38; not significant compared to control). B. Viable cells were counted after 48hr of XN treatment using trypan blue staining. (n=3; p<0.05 for all cancer cell lines). C. Cells were treated with XN up to 20μM for 4 days and cell viability was measured by colony formation assay. D. Bar graph shows the number of colonies formed after XN treatment (n=3; *p>0.05 in all treatment; NS, not significant).
Figure 2.
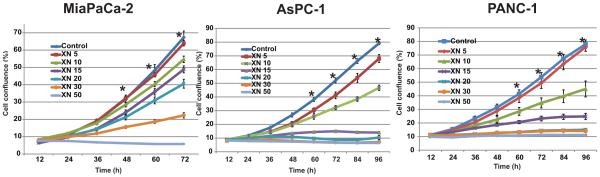
Effects of XN on pancreatic cancer cell proliferation in real-time. Cells were treated with indicated concentrations of XN and cell proliferation was monitored in real time with the continuous presence of XN. The cells were photographed and the cell confluence was calculated using IncuCyte 2011A software. The changes in cell confluence are used as a surrogate marker of cell proliferation. Statistically significant (* p<0.05) growth suppressions were observed at or above 10 μM XN compared to control.
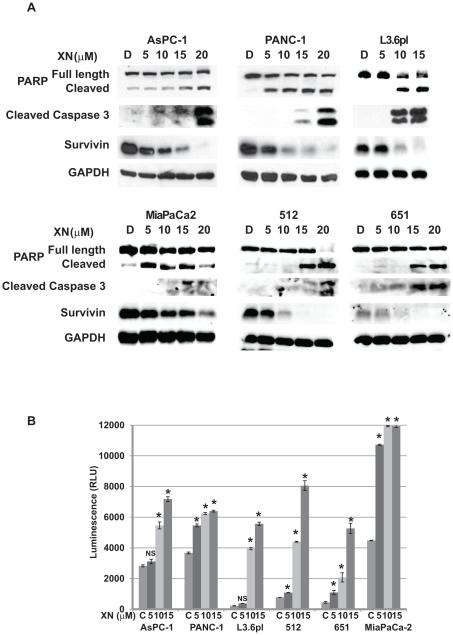
Xanthohumol induces apoptosis in pancreatic cancer cells
To determine if the increased antiproliferative effect of XN maybe be due to apoptosis, we analyzed apoptotic markers by Western analysis in four pancreatic cancer cell lines after treatment with XN. As shown in Fig. 3A, XN treatment induces the cleavage of caspase-3 as well as the full length poly (ADP) ribose polymerase (PARP) in AsPC-1, PANC-1 and L3.6pl in a dose-dependent manner. These results were confirmed by luminescence assay that measures caspase-3 and -7 activities. As shown in Fig. 3B, there was an increase in caspase activity, as evidenced by an increase in luminescence with increased concentrations of XN treatment. Furthermore, XN treatment also reduced the levels of survivin in all tested cell lines. These findings suggest that XN blocks proliferation and/or survival mechanism in pancreatic cancer cells.
Figure 3.
Mechanism of action of XN in pancreatic cancer cell lines. A. Levels of cleaved PARP, and cleaved caspase-3 (markers of apoptosis) were analyzed from lysates after 48hrs of XN treatment by Western. A level of anti-apoptotic protein survivin was also determined. GAPDH was used as loading control. B. Caspase-3 and -7 activities were measured by caspase-Glo3/7 assay. (n=3; * p<0.05 compared to control in all cell lines tested, NS, not significant).
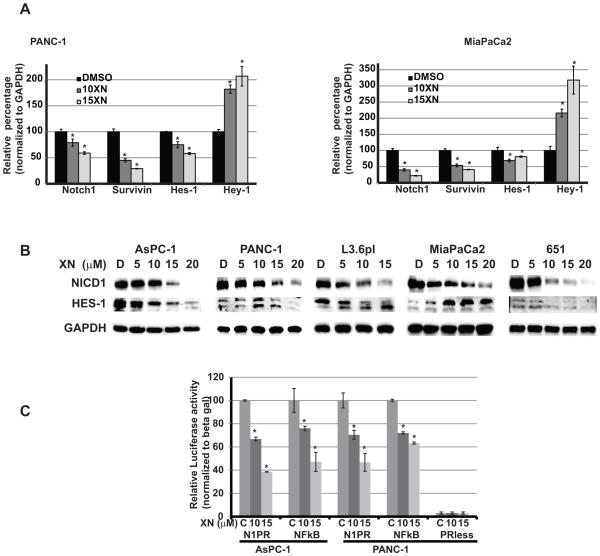
Effects of xanthohumol on Notch1 signaling in pancreatic cancer cells
XN has been shown to reduce the levels of Notch1 at both transcriptional and translational process in ovarian cancer (25). However, until now there has been no direct association between Notch reduction and growth in XN treated pancreatic cancer cells. Therefore, we analyzed the mechanisms by which XN inhibits pancreatic cancer cell growth and the role of Notch1 in growth suppression. We analyzed the Notch1 pathway mRNAs such as Notch1, Hes-1 and Hey-1 in MiaPaCa-2 and PANC-1 cells after XN treatment using real time quantitative-PCR (qPCR). The expression levels of Notch1 pathway genes were altered with treatment in a dose-dependent manner (Figure 4A). In contrast, we have observed a robust increase in Hey-1 mRNA levels with XN treatment. To verify that the reduction in mRNA expressions also lead to reduction in protein expression, we measured the Notch1 and Hes-1 protein levels after XN treatment by Western analysis. As shown in Fig. 4B, the level of Notch1 is markedly decreased with increasing concentrations of XN treated AsPC-1, PANC-1, L3.6pl, and MiaPaCa-2 cells. However, HES-1 was reduced in three cell lines whereas it was increased in MiaPaCa-2 cells. The reason for the increase in HES-1 protein after XN treatment is not clear, but the HES-1 regulation by the Notch1 signaling pathway may be cell-line specific. Furthermore, HES-1 protein contains both DNA-binding and protein-protein interaction domains that are important for its function as a transcriptional regulator of various genes including negative regulation of its own transcription (26-29). Therefore, we speculate that the reduction in mRNA level may be due to the negative regulation of its transcription by increase in HES-1 protein. Collectively, results from the qPCR and western analysis experiments on XN treated cells, indicated that XN Inhibits Notch signaling pathway as evidenced by the reduction in Notch1 with concomitant reduction in HES-1, an important downstream target of Notch pathway.
Figure 4.
XN inhibits Notch1 signaling pathway. A. mRNA levels of Notch1 signaling pathway members were measured by quantitative real-time polymerase chain reaction normalized to GAPDH for each cell lines and presented in bar graph, after XN treatment. The data were presented relative to control treatment (n=3; p=0.05 in all cell lines compared to control treatment). B. Protein levels of NICD and HES-1, a downstream target of Notch1 signaling, were analyzed by western blot after XN-treatment. Equal loading was confirmed by GAPDH. C. Promoter reporter analyses after XN treatment. Promoterless (PRless) plasmid was used as experimental control. Luciferase activity was normalized to β-galactosidase activity (n=3; * p<0.05 in both cell lines tested).
Xanthohumol suppresses Notch1 promoter activity in pancreatic cancer cells
XN treatment leds to a significant reduction in Notch1 mRNA in pancreatic cancer cells suggesting that it may directly or indirectly regulate the Notch1 promoter activity. To assess the effect on notch1 promoter activity, a plasmid containing luciferase gene under the control of Notch1 promoter or promoterless plasmid were transfected in to pancreatic cancer cells and then treated with XN at indicated concentrations. There is very minimal luciferase activity in the lysate derived from cells with promoterless plasmid compared to Notch1 promoter containing plasmid (Fig 4C). However, increased concentrations of XN caused decrease in luciferase activity in plasmid containing Notch1 promoter suggesting that XN affects the Notch1 promoter activity (Fig. 4C) which leads to the decrease in the levels of Notch1 mRNA and protein levels.
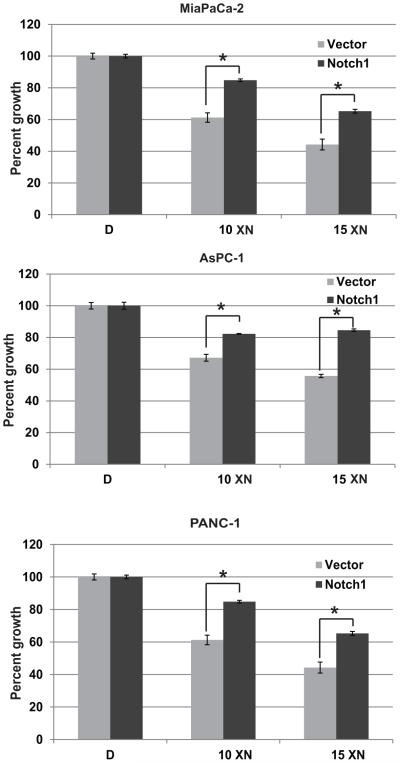
Forced over expression of active Notch1 reverse the growth suppression effect of xanthohumol in pancreatic cancer cells
Multiple studies have demonstrated the anti-proliferative effects of XN on variety of cancer types, but the mechanism of action remains unclear. The observed impairment of cell proliferation and the associated reduction of Notch1 mRNA and protein level suggest that Notch pathway inhibition may be important in XN treatment. To examine whether Notch1 pathway inhibition directly mediates growth suppression by XN, active Notch1 (NICD1) was over expressed in MiaPaCa-2, AsPC-1, and PANC-1 cells, treated with XN for 2 days, and then cell viability was measured. Notch1 overexpressed XN treated MiaPaCa-2 and AsPC-1 cells showed less growth suppression as compared to cells transfected with empty vector (Fig. 5). These results indicated that inhibition of Notch1 is important for the growth suppression effect of XN.
Figure 5.
Effect of Notch1 overexpression in XN treatment. Cells were transfected with Notch1plasmid and treated with or without XN for 2 days. Percent of growth was measured by MTT assay (n=3; * p=0.05 compared to vector with XN10 in all three cell lines tested).
Discussion
Human pancreatic cancer remains a major challenge for treatment because of the development of inevitable chemoresistance. Various studies have suggested the involvement of multiple signaling pathways in the development, progression and metastasis of pancreatic cancer (30-37). One such pathway, Notch1 signaling, a highly conserved pathway throughout the animal kingdom, plays an important role in cellular differentiation, proliferation, and survival. Increased expression of Notch receptors and their ligands has been detected in human pancreatic cancer tissues and cell lines (38-40). Importantly, it has been reported that Notch mediates tumor initiating effect and required for early stages of pancreatic neoplasia that leads to pancreatic tumorigenesis (40). Inhibition of Notch1 or the Notch signaling pathway by Notch1 siRNA in pancreatic cancer cells enhanced chemosensitivity to gemcitabine (41). Furthermore, natural products either alone or in combination with chemotherapeutic drugs have shown to reduce pancreatic cancer through targeting signaling molecules (30, 42-47). Unfortunately, clinical trials utilizing Notch pathway inhibitors in patients with solid tumors resulted in significant side effects. However, several clinical trials are underway based on the inhibition of the Notch pathway via antibody therapy or by gamma secretase inhibitors (48, 49). In addition to it’s role in pancreatic cancer pathogenesis, Notch1 may be an important factor in the development of chemotherapeutic resistance. One cause of resistance to drug treatment in pancreatic cancer is an increase in nuclear transcription factor kB (NF-kB) promoter activity by Notch1 (50).
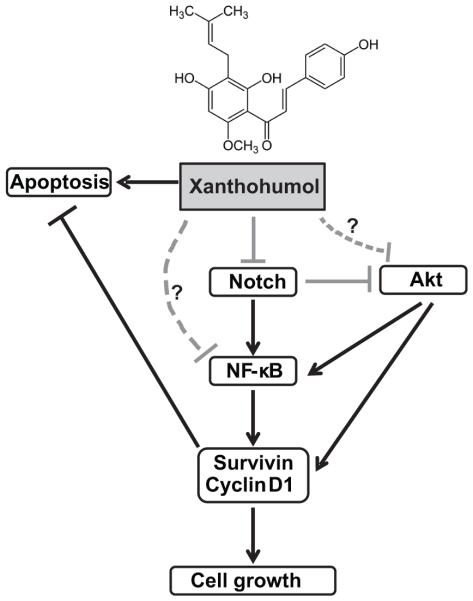
Xanthohumol (XN), a flavonoid compound isolated from hop plant, humulus lupulus, has significant antitumor activities against breast, prostate, ovarian, thyroid, colon, and hepatocellular cancer cells (6-8, 12, 14, 25, 51-54). Therefore, in recent years, XN has received much attention for its biological effects. Pharmacological aspects of XN and its effects are described (55). XN has been shown to inhibit cancer cell proliferation by association with reduction in several major targets such as NF-kB activation, drug efflux transporters expression, angiogenesis, AKT/NF-kB, and Notch1 (11, 12, 25, 56-60). However, the underlying molecular mechanism by which XN inhibits growth is still not clear. It has also been shown to induce caspase dependent as well as caspase independent apoptosis, and inhibit cell invasion (8, 9, 13, 14). We have previously shown that XN treatment in medullary thyroid cancer cells resulted in suppression of transcription factor achaete scute complex-like1 (ASCL1) with concomitant induction of phosphorylation of ERK1/2 and reduction in growth (6). There is a single report on XN reduces Notch1 at both mRNA and protein level in ovarian cancer cells in vitro (25). However, the findings in their report, in our opinion, lack functional analysis to confirm the role of Notch1 in XN treated cells. Though similar growth suppression effect is observed in variety of cancers, the associated effects are correlative. In the present study, we have shown that XN inhibits growth of human pancreatic cancer cell lines in vitro with a concomitant induction of apoptosis in a dose-dependent manner. We observed a significant reduction in Notch1 signaling pathway members such as Notch1, HES-1, basic helix-loop-helix transcription factor, and survivin both at mRNA and protein levels when treated with XN. Importantly, down regulation of Notch1 signaling pathway appears to be the mechanism of growth suppression because over expression of Notch1 negated the growth suppression effect of XN. Therefore, we believe that our research findings are significant for demonstrating that inhibition of Notch signaling pathway is most likely the predominant mechanism of action of XN for the following reasons: 1) overexpression of Notch1 rescued XN-induced cell growth suppression, and 2) inhibition of Notch1 reduced Notch1 promoter activity and its downstream targets. Furthermore, our results indicated that XN treatment reduced Notch expression that may lead to a reduction in NF-kB activity and therefore, providing a new strategies for pancreatic cancer. Based on our findings and other reports, we propose a mechanism by which XN induces apoptosis in pancreatic cancer cells demonstrating possible cross talk between Notch, PI3K-Akt pathway, and the NF-kB pathway reported in other cancer types (Fig. 6).
Figure 6.
Proposed mechanism of XN in pancreatic cancer. Schematic representation of XN induced apoptosis through Notch1 signaling pathway.
In vivo, XN has been shown to reduce the growth of poorly differentiated prostate tumors without adverse side effects (12) suggesting that XN may be a novel agent for the management of solid organ tumor such as prostate cancer. Recently, XN metabolism and pharmacokinetics parameters were tested (Clinical trial# NCT01367431)[27] and single-dose pharmacokinetic study conducted in healthy humans following oral consumption of XN making XN an viable addition to anti-tumor therapies for pancreatic cancer. We believe that the concentrations of XN we used to suppress the growth in pancreatic cancer cells could be achieved in humans with fewer side effects. However phase 1 dose-escalation studies are needed to find out the maximum tolerable dose and side effects in human. In summary, results from human and animal pharmacodynamics studies, as well as antiproliferative studies on various cancers including the present study on pancreatic cancer, suggested for the use of XN in future clinical studies not only aimed at improving pancreatic cancer treatment but also have implications beyond pancreatic cancer that are expressing Notch.
Acknowledgments
Financial support: This study was supported in part by the grants from National Institute of Health R03 CA 155691 (M. Kunnimalaiyaan), Institutional Research Grant # 86-004-26 from the American Cancer Society (M. Kunnimalaiyaan and T.C.Gamblin), MCW Cancer Center grant (M. Kunnimalaiyaan and T.C.Gamblin), The Medical College of Wisconsin Dean’s Program Development (T.C.Gamblin and M. Kunnimalaiyaan), and Froedtert Hospital Foundation (M. Kunnimalaiyaan and T.C.Gamblin)
Footnotes
Conflict of interest: No conflict of interest
REFERENCES
- 1.American Cancer Society Cancer Facts and Figures 2014. 2014 [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Gysin S, Lee SH, Dean NM, McMahon M. Pharmacologic inhibition of RAF-->MEK-->ERK signaling elicits pancreatic cancer cell cycle arrest through induced expression of p27Kip1. Cancer Res. 2005;65:4870–80. doi: 10.1158/0008-5472.CAN-04-2848. [DOI] [PubMed] [Google Scholar]
- 4.Michl P, Gress TM. Current concepts and novel targets in advanced pancreatic cancer. Gut. 2013;62:317–26. doi: 10.1136/gutjnl-2012-303588. [DOI] [PubMed] [Google Scholar]
- 5.Saleem M, Kaur S, Kweon MH, Adhami VM, Afaq F, Mukhtar H. Lupeol, a fruit and vegetable based triterpene, induces apoptotic death of human pancreatic adenocarcinoma cells via inhibition of Ras signaling pathway. Carcinogenesis. 2005;26:1956–64. doi: 10.1093/carcin/bgi157. [DOI] [PubMed] [Google Scholar]
- 6.Cook MR, Luo J, Ndiaye M, Chen H, Kunnimalaiyaan M. Xanthohumol inhibits the neuroendocrine transcription factor achaete-scute complex-like 1, suppresses proliferation, and induces phosphorylated ERK1/2 in medullary thyroid cancer. American journal of surgery. 2010;199:315–8. doi: 10.1016/j.amjsurg.2009.08.034. discussion 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorn C, Weiss TS, Heilmann J, Hellerbrand C. Xanthohumol, a prenylated chalcone derived from hops, inhibits proliferation, migration and interleukin-8 expression of hepatocellular carcinoma cells. International journal of oncology. 2010;36:435–41. [PubMed] [Google Scholar]
- 8.Gerhauser C, Alt A, Heiss E, Gamal-Eldeen A, Klimo K, Knauft J, et al. Cancer chemopreventive activity of Xanthohumol, a natural product derived from hop. Molecular cancer therapeutics. 2002;1:959–69. [PubMed] [Google Scholar]
- 9.Lust S, Vanhoecke B, Janssens A, Philippe J, Bracke M, Offner F. Xanthohumol kills B-chronic lymphocytic leukemia cells by an apoptotic mechanism. Molecular nutrition & food research. 2005;49:844–50. doi: 10.1002/mnfr.200500045. [DOI] [PubMed] [Google Scholar]
- 10.Szliszka E, Czuba ZP, Mazur B, Sedek L, Paradysz A, Krol W. Chalcones Enhance TRAIL-Induced Apoptosis in Prostate Cancer Cells. IntJMolSci. 2009;11:1–13. doi: 10.3390/ijms11010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanhoecke B, Derycke L, Van Marck V, Depypere H, De Keukeleire D, Bracke M. Antiinvasive effect of xanthohumol, a prenylated chalcone present in hops (Humulus lupulus L.) and beer. International journal of cancer Journal international du cancer. 2005;117:889–95. doi: 10.1002/ijc.21249. [DOI] [PubMed] [Google Scholar]
- 12.Vene R, Benelli R, Minghelli S, Astigiano S, Tosetti F, Ferrari N. Xanthohumol impairs human prostate cancer cell growth and invasion and diminishes the incidence and progression of advanced tumors in TRAMP mice. Molecular medicine. 2012;18:1292–302. doi: 10.2119/molmed.2012.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delmulle L, Vanden Berghe T, De Keukeleire D, Vandenabeele P. Treatment of PC-3 and DU145 prostate cancer cells by prenylflavonoids from hop (Humulus lupulus L.) induces a caspase-independent form of cell death. Phytother Res. 2008;22:197–203. doi: 10.1002/ptr.2286. [DOI] [PubMed] [Google Scholar]
- 14.Pan L, Becker H, Gerhauser C. Xanthohumol induces apoptosis in cultured 40-16 human colon cancer cells by activation of the death receptor- and mitochondrial pathway. Molecular nutrition & food research. 2005;49:837–43. doi: 10.1002/mnfr.200500065. [DOI] [PubMed] [Google Scholar]
- 15.Gerhauser C. Broad spectrum anti-infective potential of xanthohumol from hop (Humulus lupulus L.) in comparison with activities of other hop constituents and xanthohumol metabolites. Molecular nutrition & food research. 2005;49:827–31. doi: 10.1002/mnfr.200500091. [DOI] [PubMed] [Google Scholar]
- 16.Legette LL, Luna AY, Reed RL, Miranda CL, Bobe G, Proteau RR, et al. Xanthohumol lowers body weight and fasting plasma glucose in obese male Zucker fa/fa rats. Phytochemistry. 2013;91:236–41. doi: 10.1016/j.phytochem.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 17.Lupinacci E, Meijerink J, Vincken JP, Gabriele B, Gruppen H, Witkamp RF. Xanthohumol from hop (Humulus lupulus L.) is an efficient inhibitor of monocyte chemoattractant protein-1 and tumor necrosis factor-alpha release in LPS-stimulated RAW 264.7 mouse macrophages and U937 human monocytes. Journal of agricultural and food chemistry. 2009;57:7274–81. doi: 10.1021/jf901244k. [DOI] [PubMed] [Google Scholar]
- 18.Miranda CL, Stevens JF, Ivanov V, McCall M, Frei B, Deinzer ML, et al. Antioxidant and prooxidant actions of prenylated and nonprenylated chalcones and flavanones in vitro. JAgricFood Chem. 2000;48:3876–84. doi: 10.1021/jf0002995. [DOI] [PubMed] [Google Scholar]
- 19.Nozawa H. Xanthohumol, the chalcone from beer hops (Humulus lupulus L.), is the ligand for farnesoid X receptor and ameliorates lipid and glucose metabolism in KK-A(y) mice. Biochemical and biophysical research communications. 2005;336:754–61. doi: 10.1016/j.bbrc.2005.08.159. [DOI] [PubMed] [Google Scholar]
- 20.Peluso MR, Miranda CL, Hobbs DJ, Proteau RR, Stevens JF. Xanthohumol and related prenylated flavonoids inhibit inflammatory cytokine production in LPS-activated THP-1 monocytes: structure-activity relationships and in silico binding to myeloid differentiation protein-2 (MD-2) Planta medica. 2010;76:1536–43. doi: 10.1055/s-0029-1241013. [DOI] [PubMed] [Google Scholar]
- 21.Xuan NT, Shumilina E, Gulbins E, Gu S, Gotz F, Lang F. Triggering of dendritic cell apoptosis by xanthohumol. Molecular nutrition & food research. 2010;54(Suppl 2):S214–24. doi: 10.1002/mnfr.200900324. [DOI] [PubMed] [Google Scholar]
- 22.Carter YM, Kunnimalaiyaan S, Chen H, Gamblin TC, Kunnimalaiyaan M. Specific glycogen synthase kinase-3 inhibition reduces neuroendocrine markers and suppresses neuroblastoma cell growth. Cancer biology & therapy. 2014;15:510–5. doi: 10.4161/cbt.28015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng G, Zielonka J, McAllister D, Tsai S, Dwinell MB, Kalyanaraman B. Profiling and targeting of cellular bioenergetics: inhibition of pancreatic cancer cell proliferation. British journal of cancer. 2014;111:85–93. doi: 10.1038/bjc.2014.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yugawa T, Handa K, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol Cell Biol. 2007;27:3732–42. doi: 10.1128/MCB.02119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drenzek JG, Seiler NL, Jaskula-Sztul R, Rausch MM, Rose SL. Xanthohumol decreases Notch1 expression and cell growth by cell cycle arrest and induction of apoptosis in epithelial ovarian cancer cell lines. Gynecologic oncology. 2011;122:396–401. doi: 10.1016/j.ygyno.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 26.Coglievina M, Guarnaccia C, Pintar A, Pongor S. Different degrees of structural order in distinct regions of the transcriptional repressor HES-1. Biochimica et biophysica acta. 2010;1804:2153–61. doi: 10.1016/j.bbapap.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. Journal of cellular physiology. 2003;194:237–55. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 28.Takebayashi K, Akazawa C, Nakanishi S, Kageyama R. Structure and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-5. Identification of the neural precursor cell-specific promoter element. The Journal of biological chemistry. 1995;270:1342–9. doi: 10.1074/jbc.270.3.1342. [DOI] [PubMed] [Google Scholar]
- 29.Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. The Journal of biological chemistry. 1994;269:5150–6. [PubMed] [Google Scholar]
- 30.Shi P, Yin T, Zhou F, Cui P, Gou S, Wang C. Valproic acid sensitizes pancreatic cancer cells to natural killer cell-mediated lysis by upregulating MICA and MICB via the PI3K/Akt signaling pathway. BMC cancer. 2014;14:370. doi: 10.1186/1471-2407-14-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Zhang J, Xu K, Xiao Z, Sun J, Xu J, et al. PTEN/PI3K/mTOR/B7-H1 signaling pathway regulates cell progression and immuno-resistance in pancreatic cancer. Hepato-gastroenterology. 2013;60:1766–72. [PubMed] [Google Scholar]
- 32.Onishi H, Katano M. Hedgehog signaling pathway as a new therapeutic target in pancreatic cancer. World journal of gastroenterology : WJG. 2014;20:2335–42. doi: 10.3748/wjg.v20.i9.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma J, Xia J, Miele L, Sarkar FH, Wang Z. Notch Signaling Pathway in Pancreatic Cancer Progression. Pancreatic disorders & therapy. 2013;3 [PMC free article] [PubMed] [Google Scholar]
- 34.Avila JL, Kissil JL. Notch signaling in pancreatic cancer: oncogene or tumor suppressor? Trends in molecular medicine. 2013;19:320–7. doi: 10.1016/j.molmed.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams TM, Flecha AR, Keller P, Ram A, Karnak D, Galban S, et al. Cotargeting MAPK and PI3K signaling with concurrent radiotherapy as a strategy for the treatment of pancreatic cancer. Molecular cancer therapeutics. 2012;11:1193–202. doi: 10.1158/1535-7163.MCT-12-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma D, Wang J, Zhao Y, Lee WN, Xiao J, Go VL, et al. Inhibition of glycogen phosphorylation induces changes in cellular proteome and signaling pathways in MIA pancreatic cancer cells. Pancreas. 2012;41:397–408. doi: 10.1097/MPA.0b013e318236f022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preis M, Korc M. Signaling pathways in pancreatic cancer. Critical reviews in eukaryotic gene expression. 2011;21:115–29. doi: 10.1615/critreveukargeneexpr.v21.i2.20. [DOI] [PubMed] [Google Scholar]
- 38.Du X, Zhao YP, Zhang TP, Zhou L, Chen G, Cui QC, et al. Notch1 contributes to chemoresistance to gemcitabine and serves as an unfavorable prognostic indicator in pancreatic cancer. World journal of surgery. 2013;37:1688–94. doi: 10.1007/s00268-013-2010-0. [DOI] [PubMed] [Google Scholar]
- 39.Fukushima N, Sato N, Prasad N, Leach SD, Hruban RH, Goggins M. Characterization of gene expression in mucinous cystic neoplasms of the pancreas using oligonucleotide microarrays. Oncogene. 2004;23:9042–51. doi: 10.1038/sj.onc.1208117. [DOI] [PubMed] [Google Scholar]
- 40.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–76. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 41.Du X, Wang YH, Wang ZQ, Cheng Z, Li Y, Hu JK, et al. [Down-regulation of Notch1 by small interfering RNA enhances chemosensitivity to gemcitabine in pancreatic cancer cells through activating apoptosis activity] Zhejiang da xue xue bao Yi xue ban = Journal of Zhejiang University Medical sciences. 2014;43:313–8. doi: 10.3785/j.issn.1008-9292.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Zhou L, Qi L, Jiang L, Zhou P, Ma J, Xu X, et al. Antitumor activity of gemcitabine can be potentiated in pancreatic cancer through modulation of TLR4/NF-kappaB signaling by 6-shogaol. The AAPS journal. 2014;16:246–57. doi: 10.1208/s12248-013-9558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xin Y, Shen XD, Cheng L, Hong DF, Chen B. Perifosine inhibits S6K1-Gli1 signaling and enhances gemcitabine-induced anti-pancreatic cancer efficiency. Cancer chemotherapy and pharmacology. 2014;73:711–9. doi: 10.1007/s00280-014-2397-9. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Zhou Y, Jia G, Han B, Liu J, Teng Y, et al. Shikonin suppresses tumor growth and synergizes with gemcitabine in a pancreatic cancer xenograft model: Involvement of NF-kappaB signaling pathway. Biochemical pharmacology. 2014;88:322–33. doi: 10.1016/j.bcp.2014.01.041. [DOI] [PubMed] [Google Scholar]
- 45.Deeb D, Gao X, Liu YB, Pindolia K, Gautam SC. Pristimerin, a quinonemethide triterpenoid, induces apoptosis in pancreatic cancer cells through the inhibition of pro-survival Akt/NF-kappaB/mTOR signaling proteins and anti-apoptotic Bcl-2. International journal of oncology. 2014;44:1707–15. doi: 10.3892/ijo.2014.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balic A, Sorensen MD, Trabulo SM, Sainz B, Jr., Cioffi M, Vieira CR, et al. Chloroquine targets pancreatic cancer stem cells via inhibition of CXCR4 and hedgehog signaling. Molecular cancer therapeutics. 2014;13:1758–71. doi: 10.1158/1535-7163.MCT-13-0948. [DOI] [PubMed] [Google Scholar]
- 47.Sun XD, Liu XE, Huang DS. Curcumin reverses the epithelial-mesenchymal transition of pancreatic cancer cells by inhibiting the Hedgehog signaling pathway. Oncol Rep. 2013;29:2401–7. doi: 10.3892/or.2013.2385. [DOI] [PubMed] [Google Scholar]
- 48.Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling--are we there yet? Nature reviews Drug discovery. 2014;13:357–78. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]
- 49.Espinoza I, Miele L. Notch inhibitors for cancer treatment. Pharmacology & therapeutics. 2013;139:95–110. doi: 10.1016/j.pharmthera.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oswald F, Liptay S, Adler G, Schmid RM. NF-kappaB2 is a putative target gene of activated Notch-1 via RBP-Jkappa. Mol Cell Biol. 1998;18:2077–88. doi: 10.1128/mcb.18.4.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim SY, Lee IS, Moon A. 2-Hydroxychalcone and xanthohumol inhibit invasion of triple negative breast cancer cells. Chemico-biological interactions. 2013;203:565–72. doi: 10.1016/j.cbi.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 52.Kang Y, Park MA, Heo SW, Park SY, Kang KW, Park PH, et al. The radio-sensitizing effect of xanthohumol is mediated by STAT3 and EGFR suppression in doxorubicin-resistant MCF-7 human breast cancer cells. Biochimica et biophysica acta. 2013;1830:2638–48. doi: 10.1016/j.bbagen.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 53.Deeb D, Gao X, Jiang H, Arbab AS, Dulchavsky SA, Gautam SC. Growth inhibitory and apoptosis-inducing effects of xanthohumol, a prenylated chalone present in hops, in human prostate cancer cells. Anticancer research. 2010;30:3333–9. [PMC free article] [PubMed] [Google Scholar]
- 54.Monteiro R, Calhau C, Silva AO, Pinheiro-Silva S, Guerreiro S, Gartner F, et al. Xanthohumol inhibits inflammatory factor production and angiogenesis in breast cancer xenografts. Journal of cellular biochemistry. 2008;104:1699–707. doi: 10.1002/jcb.21738. [DOI] [PubMed] [Google Scholar]
- 55.Liu M, Hansen PE, Wang G, Qiu L, Dong J, Yin H, et al. Pharmacological profile of xanthohumol, a prenylated flavonoid from hops (Humulus lupulus) Molecules. 2015;20:754–79. doi: 10.3390/molecules20010754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Albini A, Dell'Eva R, Vene R, Ferrari N, Buhler DR, Noonan DM, et al. Mechanisms of the antiangiogenic activity by the hop flavonoid xanthohumol: NF-kappaB and Akt as targets. FASEB J. 2006;20:527–9. doi: 10.1096/fj.05-5128fje. [DOI] [PubMed] [Google Scholar]
- 57.Colgate EC, Miranda CL, Stevens JF, Bray TM, Ho E. Xanthohumol, a prenylflavonoid derived from hops induces apoptosis and inhibits NF-kappaB activation in prostate epithelial cells. Cancer letters. 2007;246:201–9. doi: 10.1016/j.canlet.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 58.Dell'Eva R, Ambrosini C, Vannini N, Piaggio G, Albini A, Ferrari N. AKT/NF-kappaB inhibitor xanthohumol targets cell growth and angiogenesis in hematologic malignancies. Cancer. 2007;110:2007–11. doi: 10.1002/cncr.23017. [DOI] [PubMed] [Google Scholar]
- 59.Lee SH, Kim HJ, Lee JS, Lee IS, Kang BY. Inhibition of topoisomerase I activity and efflux drug transporters' expression by xanthohumol. from hops. Archives of pharmacal research. 2007;30:1435–9. doi: 10.1007/BF02977368. [DOI] [PubMed] [Google Scholar]
- 60.Monteghirfo S, Tosetti F, Ambrosini C, Stigliani S, Pozzi S, Frassoni F, et al. Antileukemia effects of xanthohumol in Bcr/Abl-transformed cells involve nuclear factor-kappaB and p53 modulation. Molecular cancer therapeutics. 2008;7:2692–702. doi: 10.1158/1535-7163.MCT-08-0132. [DOI] [PubMed] [Google Scholar]