Abstract
Objective
To elucidate the molecular mechanisms involved in renal inflammation during the progression, remission and relapse of nephritis in murine lupus models using transcriptome analysis.
Methods
Kidneys from NZB/W F1 and NZM2410 mice were harvested at intervals during their disease course or after remission induction. Genome wide expression profiles were obtained from microarray analysis of perfused kidneys. Real time PCR analysis for selected genes was used to validate the microarray data. Comparisons between groups using SAM, and unbiased analysis of the entire dataset using singular value decomposition and self-organizing map were performed.
Results
Few changes in the renal molecular profile were detected in pre-nephritic kidneys but a significant shift in gene expression, reflecting inflammatory cell infiltration and complement activation occurred at proteinuria onset. Subsequent changes in gene expression predominantly affected mitochondrial dysfunction and metabolic stress pathways. Endothelial cell activation, tissue remodeling and tubular damage were the major pathways associated with loss of renal function. Remission induction reversed most, but not all of the inflammatory changes and progression towards relapse was associated with recurrence of inflammation, mitochondrial dysfunction and metabolic stress signatures.
Conclusion
Immune cell infiltration and activation is associated with proteinuria onset and reverses with immunosuppressive therapy but disease progression is associated with renal hypoxia and metabolic stress. Optimal therapy of SLE nephritis may therefore need to target both immune and non-immune disease mechanisms. In addition, the overlap of a substantial subset of molecular markers with those expressed in human lupus kidneys suggests potential new biomarkers and therapeutic targets.
Lupus nephritis affects 30-70% of SLE patients and its treatment remains insufficiently effective and excessively toxic (1-4). Although biomarkers for nephritis are being identified (5-6) there is still no reliable way of predicting an impending renal flare or determining which patients will respond to therapy. Because human renal tissue cannot be obtained sequentially during remission and relapse, animal models are often used to study progression of lupus nephritis.
Complete histologic remission of proliferative glomerulonephritis is induced in 80-90% of NZB/W F1 mice (7-8) by a combination of cyclophosphamide and the costimulatory antagonists CTLA4Ig and anti-CD40L (triple therapy) (9), or a combination of CTLA4Ig with a BAFF antagonist ((10) and Davidson, unpublished). Similarly, BAFF-R-Ig induces long-lasting remission of nephritis in NZM2410 mice that ordinarily develop glomerulosclerosis with rapid progression to renal failure (11). The induced remission does not abrogate renal immune complex deposition but is associated with decreased renal inflammation and preservation of the glomerular filtration barrier (11-12). Evidence from multiple murine models confirms that there is a checkpoint in disease progression between immune complex deposition and the development of renal impairment (13-15).
In this study, we profiled NZB/W kidneys to define the molecular characteristics of nephritis onset and progression, complete remission and progression towards relapse. Surprisingly, only few changes in the renal gene expression profile were detected after immune complex deposition but a major shift in transcripts occurred at proteinuria onset, reflecting rapid onset of inflammation. Subsequent changes in gene expression reflected mitochondrial dysfunction and metabolic stress. Remission induction reversed much of the inflammatory gene expression pattern but during late remission mitochondrial dysfunction and metabolic stress signatures recurred before proteinuria onset. In NZM2410 mice, a limited inflammatory signature arose during remission but was not associated with renal decline. In both strains, the presence of proteinuria was closely associated with renal tubular dysfunction. Our findings suggest that optimal therapy of SLE nephritis may need to target both immune and non-immune disease mechanisms.
MATERIALS AND METHODS
Mouse models and treatment protocols
NZB/NZW F1 females (Jackson Laboratories, Bar Harbor ME) were followed clinically as previously described (9, 16-17). Mice with proteinuria of >300 mg/dl on 2 occasions 24-48 hours apart were treated with a single dose of CTX 50mg/kg and 6 doses of CTLA4Ig 100μg and anti-CD154 250μg (9). Remission was defined as ≤ 30mg/dl proteinuria on ≥ 2 occasions. An early remission group was sacrificed 3-4 weeks after remission induction and had complete histologic remission by light microscopy as defined by composite glomerular and tubulointerstitial scores ≤ 2.5, similar to young controls. The late remission group was sacrificed >5-14 weeks after remission induction and had composite glomerular and tubulointerstitial scores ≥ 3.0 (Supplementary Figure 1) (12). We also induced remission in 30 week old mice with 100-300mg/dl of proteinuria using a single dose of adenovirus expressing TACI-Ig, together with two weeks of CTLA4Ig (10). These mice were sacrificed 15 weeks after remission induction therapy; their renal scores were similar to those of mice in late remission.
22 week old NZM2410 mice (Taconic, Albany, NY) were treated with adenovirus expressing BAFF-R-Ig and sacrificed at 30-35 or at 55 weeks of age as previously described (11).
Control mice were sacrificed at sequential disease stages as previously described (16-17). The ages, and mean renal scores of each group are shown in Supplementary Table 1.
Analysis of renal tissues
H & E stained kidney sections were scored for glomerular and interstitial damage using a semi-quantitative scale from 0-4 (18). Renal RNA extraction, cDNA synthesis, hybridization, microarray processing, data normalization and filtering were performed as previously described (16-17). Significantly regulated genes were analyzed using Genomatix Pathway System (GePS - www.genomatix.de) and Ingenuity Pathway Analysis (IPA - www.ingenuity.com) software. Gene expression datasets are available at GEO #GSE49898, GSE32583 and GSE32591.
Identifying coherent expression modules using self organizing map (SOM)
To identify the biological processes driving coherent expression modules, we used a self-organizing map (SOM) to extract the fundamental patterns of gene expression inherent in the serial data sets from NZB/W mice (19). The average expression values across multiple samples were taken for each gene for each disease stage. To capture the dynamic trend across different stages, we centered the expression pattern for each gene across all stages at zero and then applied SOM to identify the coherent expression modules (19). We used Ingenuity Pathway Analysis software to identify significantly represented biological processes.
Identifying major expression patterns using Singular Value Decomposition (SVD)
To detect major expression responses during disease progression, we applied singular value decomposition (SVD) on the time-course data. SVD is a mainstream technique for analysis of multivariate time-course data to identify orthogonal patterns. It allows us to transform genome-wide expression data from genes × arrays space to diagonalized “eigengenes” × “eigenassays” space (20). The eigengenes (also referred to as patterns) represent orthogonal expression patterns that may be hidden in the original data (20-22). The top 600 genes in the dominant pattern for each comparison were further analyzed.
Quantitative Real Time PCR (qPCR)
Quantitative real time PCR analysis of 166 genes was performed as previously described (12, 16, 23).
Statistical Analysis
The TIGR MultiExperiment Viewer (TMEV) application was used for statistical analysis of microarray data as previously described (16-17). Genes undergoing further analyses met the following criteria: q-value ≤ 0.05 and absolute value of log2 fold change ≥ 0.26. Genes that met the above criteria and whose mean fold expression changed by ≥1.5 fold between nephritic and remission stages were considered to be reversed with remission. qPCR data were scaled to the mean of the young mice in each strain, given a value of 1. Genes with q-value (false discovery rate) <0.05 were considered significant (16-17). As all pathways in Genomatix Pathway System are derived from Homo Sapiens, the mouse gene IDs were converted to the corresponding human orthologs using the NCBI homolog (Build 67) for all GePS and IPA analyses.
Comparisons for Supplementary Table 1 were performed using Mann-Whitney test. p values ≤ 0.05 were considered significant.
RESULTS
Gene expression profile associated with disease onset
Only 180 changes in gene expression were found between non-proteinuric 16w and 23w old mice. The major signature was for immunoglobulin genes and B cells (Supplementary Table 2A). Indeed, small lymphoid aggregates containing plasma cells are found in the renal pelvis of 23w old NZB/W mice (not shown). Other pathways included nitrogen metabolism and fatty acid synthesis.
Similarly, only 109 genes were regulated between 23w and 30w old non-proteinuric mice reflecting innate immune cell activation, complement protein production, early tubular involvement and collagen gene expression (Supplementary Table 2B). By contrast, major shifts in gene expression were observed when 23w old non-proteinuric mice were compared with 23w old mice with new onset proteinuria (1342 genes - Supplementary Table 2C). Ingenuity Pathway analysis indicated that immune activation accounted for the top 10 pathways (Table 1). Tubular damage was exemplified by increased expression of lipocalin 2 (LCN2), fibronectin, and vimentin. Further upregulation of several collagen genes indicated early interstitial fibrosis.
Table 1.
Top significantly regulated canonical pathways and molecules in each pathway, as assessed by IPA (Ingenuity Pathway Analysis). p-values are indicated in parentheses. In square brackets are the numbers of genes regulated / number of genes in the pathway.
|
Top 10 pathways from the 1342 genes regulated in 23 weeks old mice with new onset of proteinuria vs. 23 wks old mice without proteinuria (corresponding to 1194 human gene IDs). |
| 1. Dendritic Cell Maturation (4.37E-09) [35/207] |
| B2M, ICAM1, NFKBIE, HLA-DQA1, LTB, HLA-DRB1, CD83, HLA-DMB, HLA-DQB1, FCGR1A, EP300, COL1A2, PLCE1, PIK3CG, HLA-B, COL18A1, STAT1, PLCD4, FCGR3A, HLA-DMA, TYROBP, FCGR2A, RELB, IKBKE, DDR1, TLR2, IL33, COL1A1, TLR4, IL18, FCER1G, CD86, IL1B, IRF8, COL3A1 |
| 2. Altered T Cell and B Cell Signaling in Rheumatoid Arthritis (3.89E-07) [21/92] |
| TLR1, HLA-DMA, SPP1, CD79B, RELB, HLA-DQA1, HLA-DRB1, LTB, HLA-DMB, HLA-DQB1, TNFRSF17, IL33, TLR2, TLR4, IL18, CXCL13, TGFB1, FCER1G, CD86, IL1B, TNFSF13B |
| 3. Antigen Presentation Pathway (3.63E-06) [11/40] |
| B2M, PSMB9, HLA-DMA, HLA-DQA1, HLA-B, HLA-DRB1, HLA-DMB, PSMB8, CD74, TAP1, TAP2 |
| 4. Atherosclerosis Signaling (7.08E-06) [24/136] |
| APOE, VCAM1, ICAM1, CXCR4, PLA2G7, F3, TNFRSF12A, SELPG, COL1A2, IL33, PLA2G4A, COL1A1, ITGB2, IL18, CCL2, APOC3, TGFB1, IL1B, SERPINA1, S100A8, COL18A1, CCR2, ITGA4, COL3A1 |
| 5. TREM1 Signaling (1.00E-05) [15/71] |
| ITGB1, TLR1, ICAM1, TYROBP, CD83, STAT3, TLR2, TLR4, CXCL3, IL18, CCL2, CASP1, CD86, IL1B, ITGAX |
| 6. Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes (1.10E-05) [21/102] |
| FYN, ARPC1B, FCGR2A, ACTB, ACTA2, ARPC5, FCGR1A, PRKCZ, PLD4, HMOX1, ARF6, NCF1, ACTR3, PIK3CG, ARPC2, HCK, VAMP3, LYN, ARPC3, FCGR3A, PRKCB |
| 7. Communication between Innate and Adaptive Immune Cells (1.10E-05) [17/109] |
| B2M, TLR1, HLA-DRB1, CD83, CCL5, TNFRSF17, IL33, CXCL10, TLR2, TLR4, IL18, FCER1G, HLA-B, CD86, CCL3L1/CCL3L3, IL1B, TNFSF13B |
| 8. Complement System (1.23E-05) [10/35] |
| C1R, SERPING1, C3, C1S, CFI, C1QC, C1QA, C1QB, CFH, C3AR1 |
| 9. CD28 Signaling in T Helper Cells (1.41E-05) [23/132] |
| HLA-DMA, FYN, PTPN6, ARPC1B, NFATC3, NFKBIE, MAP3K1, ARPC5, HLA-DQA1, HLA-DRB1, PTPRC, CD3G, FOS, JUN, ACTR3, PIK3CG, ARPC2, FCER1G, CD86, ARPC3 |
| 10. Cell Cycle Control of Chromosomal Replication (1.86E-05) [10/31] |
| MCM5, MCM3, MCM6, MCM2, CDT1, ORC6, CHECK2, MCM4, DBF4, MCM7 |
|
Top 5 pathways from the 321 genes regulated in 36 weeks old mice with established proteinuria vs. 23 wks old mice at onset of proteinuria (corresponding to 311 human gene IDs). |
| 1. TCA Cycle II (Eukaryotic) (1.48E-10) [9/41] |
| SUCLA2, CS, SUCLG1, SDHD, IDH3A, MDH1, MDH2, ACO1, IDH3B |
| 2. Mitochondrial Dysfunction (4.90E-06) [13/186] |
| COX6B1, NDUFA9, COX6A1, COX11, SOD2, ATP5B, COX5A, NDUFB6, SDHD, CYC1, ATP5G3, NDUFA8, COX15 |
| 3. Inosine-5′-phosphate Biosynthesis II (1.26E-03) [2/16] |
| ADSL, PAICS |
| 4. Arsenate Detoxification I (Glutaredoxin) (2.51E-03) [2/20] |
| AS3MT, GSTO1 |
| 5. Aspartate Degradation II (8.32E-03) [2/14] |
| MDH1, MDH2 |
Gene expression profile associated with disease progression
321 gene differences were noted between the kidneys of 23w old mice with new onset proteinuria and those of older mice with established proteinuria of >2 weeks duration; these, surprisingly, were all downregulated (Supplementary Table 2D). Ingenuity analysis indicated that the top 5 pathways involve metabolic alterations and mitochondrial dysfunction (Table 1).
Major changes in gene expression were observed between 30w old non-proteinuric mice and 36w old mice with established proteinuria (Supplementary Table 2E). 427 upregulated genes reflected inflammation, similar to the pattern observed at disease onset (Supplementary Table 3A). 1356 downregulated genes, reflected metabolic and mitochondrial dysfunction, as observed during progression of nephritis from onset to established disease (Supplementary Table 3B). Gene expression changes during disease onset and progression are summarized in Figure 1A.
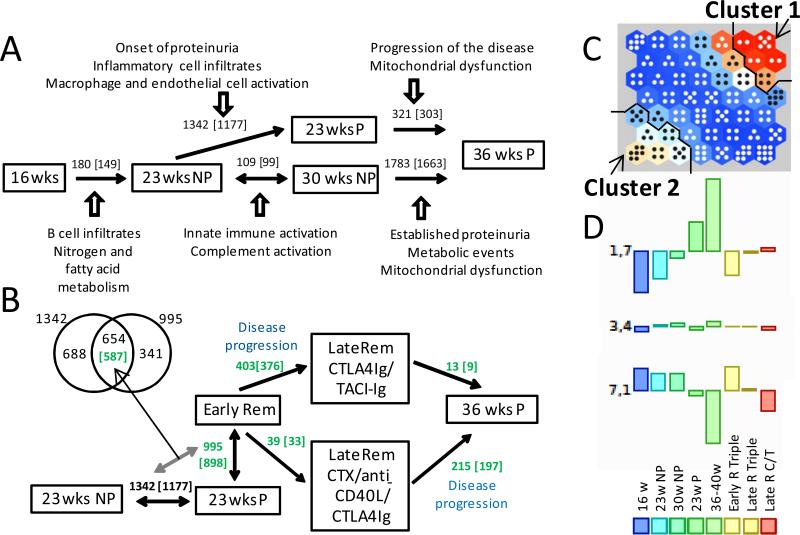
Figure 1. Patterns of gene expression in NZB/W mice.
A. Numbers of regulated genes (number of human orthologs) between the indicated stages of disease progression. Dominant pathologic processes identified by IPA are shown for each disease stage. B. Numbers of regulated genes between the indicated stages before (black) and after (green) remission induction. The Venn-diagram shows the 654 genes regulated in both during proteinuria onset and remission. NP: non-proteinuric; P: proteinuric. C. Self-organizing map of datasets from each of the indicated disease stages. Number of dots in each module reflects the number of regulated genes and the color reflects the degree of change - either up (red – Cluster 1) or down (white – Cluster 2) or not changing (blue). D. Average gene expression for each data set in three representative modules.
Transcriptional indicators for disease remission
To identify mechanisms for remission and transcriptional indicators of renal clinical status we identified genes whose expression changed at proteinuria onset and reversed upon remission. Of the 1342 genes that were upregulated at proteinuria onset, 654 (49.1%) reversed, 603 by >1.5 fold, during complete remission (Supplementary Table 2F, Figure 1B). These genes reflected inflammatory cell infiltration, complement activity, endothelial cell activation and extracellular matrix formation (Table 2, Supplementary Figure 2A). To determine the relevance to human disease we interrogated datasets of genes upregulated in glomeruli and tubulointerstitium of SLE nephritis biopsies compared with normal control biopsies (17). A striking level of concordance was observed. Of the 654 mouse marker genes (corresponding to 587 human genes), 175 were concordantly regulated in both glomeruli and tubules of SLE nephritis biopsies (Supplementary Table 4A) and a further 145 were concordantly regulated in one of the two compartments (Supplementary Table 4B). Genomatix analysis of the 175 shared genes revealed several nodes of pathogenic significance (Supplementary Figure 2B) including CD14 and CD18 (mononuclear cell infiltration/activation), CD44 (cell migration), Fn1 (extracellular matrix and fibrosis), CXCL10, CCL5, IL18 (chemokines/cytokines) and Stat1 (type 1 interferon pathway). Stat1 was the most represented transcription factor; 54 of the 175 genes had a Stat1 binding site in their promoter (Supplementary Table 4C). Ig transcripts, CXCL13 and ITGAM were not completely downregulated during remission, consistent with the persistence of small inflammatory infiltrates.
Table 2.
Top 10 significantly regulated canonical pathways and molecules in each pathway, from the 654 genes reversed during complete remission (corresponding to 587 human gene IDs), as assessed by IPA (Ingenuity Pathway Analysis). p-values are indicated in parentheses. In square brackets are the numbers of genes regulated / number of genes in the pathway.
| 1. Dendritic Cell Maturation (6.31E-08) [22/185] |
| B2M, ICAM1, TYROBP, FCG2RA, RELB, NFKBIE, HLADQA1, IKBKE, CD83, FCGR1A, COL1A2, IL33, TLR2, TLR4, IL18, PIK3CG, HLA-DRA, CD86, IL1B, IRF8, STAT1, HLA-C |
| 2. Communication between Innate and Adaptive Immune Cells (1.38E-07) [15/109] |
| B2M, CD83, CCL5, CCL9, IL33, CXCL10, TLR2, TLR4, IL18, HLA-DRA, IL1B, TLR13, CD86, TNFSF13B, HLA-C |
| 3. Cell Cycle Control of Chromosomal Replication (9.77E-07) [9/31] |
| MCM5, MCM3, MCM6, MCM2, CDT1, ORC6, MCM4, DBF4, MCM7 |
| 4. TREM1 Signaling (6.03E-06) [11/66] |
| TLR2, TLR4, IL18, ICAM1, CCL2, TYROBP, CASP1, TLR3, CD86, IL1B, CD83 |
| 5. Hepatic Fibrosis / Hepatic Stellate Cell Activation (2.29E-05) [18/147] |
| CCR5, VCAM1, ICAM1, FN1, CTGF, MMP2, CCL5, COL1A2, MYL9, CXCL3, TLR4, LY96, CCL2, IL10RA, CD14, IL1B, STAT1, PDGFRB |
| 6. MSP-RON Signaling Pathway (7.41E-05) [9/51] |
| TLR2, CSF2RB, ITGB2, TLR4, KLK3, CCL2, PIK3CG, ACTB, CCR2 |
| 7. Complement System (7.76E-05) [7/35] |
| C1R, SERPING1, C3, C1S, C1QA, C1QB, CFH |
| 8. Altered T Cell and B Cell Signaling in Rheumatoid Arthritis (1.17E-04) [12/92] |
| IL33, TLR2, TLR4, IL18, SPP1, RELB, HLA-DRA, HLA-DQA1, TLR13, CD86, IL1B, TNFSF13B |
| 9. Role of Hypercytokinemia/hyperchemokinemia in the Pathogenesis of Influenza (1.32E-04) [7/44] |
| IL33, CXCL10, IL18, CCR5, CCL2, IL1B, CCL5 |
| 10. Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses (1.58E-04) [13/106] |
| C3, C1QA, C1QB, CCL5, TLR2, IFIH1, TLR4, CLEC7A, IRF7, PI3CG, CASP1, IL1B, CLEC6A |
Gene expression profile associated with progression towards relapse
Only 39 differences were observed between early and late remission after triple therapy (Supplementary Table 2G). By contrast, 403 gene differences were noted between early remission and late remission after CTLA4-Ig/TACI-Ig treatment, all being downregulated and involving metabolic pathways (Supplementary Table 2H). Compared with mice studied late after triple therapy, mice with established proteinuria had 174 upregulated genes and 41 downregulated genes (Supplementary Table 2I). Only few differences were observed between mice with established proteinuria and CTLA4-Ig/TACI-Ig mice, confirming that these mice represent a later stage in the progressive relapse process. Gene expression changes during nephritis and early and late remission are summarized in Figure 1B.
Confirmation using Self-Organizing Map (SOM)
To identify genes that changed coherently throughout disease progression and remission we applied Self Organizing Map, an unbiased approach to cluster genes based on the similarity between their sequential expression profiles. Figure 1C displays the SOM for this study. In the SOM panel, the top right hand set of modules (cluster 1, containing modules 1,6/1,7/2,6/2,7/3,7) includes mostly pro-inflammatory genes with coherent expression throughout the study. By contrast, the bottom left hand set of modules (cluster 2, containing modules 5,1/6,1/6,2/7,1/7,2/7,3) is enriched for genes associated with metabolic stress and mitochondrial dysfunction (Supplementary Table 5). Representative modules are shown in Figure 1D. IPA analysis of clusters 1 and 2 is shown in Table 3. Remarkably, 426 of the 1098 mitochondrial genes in the mouse Mitocarta database were represented in Cluster 2. 98 of these (23%) were significantly regulated in the kidneys of mice in late remission after CTLA4Ig and TACI-Ig compared with mice in early remission (Figure 2, top panel). In addition, of 197 selected genes representing mouse hypoxia, cellular stress and toxicity and oxidative stress (SABiosciences), 34 were represented in Cluster 2 (Figure 2, lower panel); of these 8 (23.5%) were significantly regulated in the kidneys of mice in late remission after combination CTLA4Ig and TACI-Ig compared with mice in early remission.
Table 3.
Top 5 canonical pathways significantly regulated (p-value<0.05) from the genes in each module of the SOM analysis, as assessed by IPA (Ingenuity Pathway Analysis). p-values are indicated in parentheses.
| Module | Canonical pathways (p-value) |
|---|---|
| Cluster 1 (915 genes) | |
| 1_6 | IL-12 Signaling and Production in Macrophages (1.58E-06), Prolactin Signaling (7.59E-05), PKCθ Signaling in T Lymphocytes (8.32E-05), CTLA4 Signaling in Cytotoxic T Lymphocytes (1.17E-04) |
| 1_6 / 1_7 | Granulocyte Adhesion and Diapedesis (9.33E-05 / 1.58E-12) |
| 1_7 | Altered T Cell and B Cell Signaling in Rheumatoid Arthritis (1.26E-13), Dendritic Cell Maturation (2.00E-13), Agranulocyte Adhesion and Diapedesis (3.09E-10), Communication between Innate and Adaptive Immune Cells (4.90E-10) |
| 2_6 | Semaphorin Signaling in Neurons (7.59E-06), Toll-like Receptor Signaling (8.51E-06), Tec Kinase Signaling (8.91E-06), LXR/RXR Activation (6.76E-05), CD28 Signaling in T Helper Cells (6.76E-05) |
| 2_7 | Cell Cycle Control of Chromosomal Replication (5.37E-05), Role of IL-17A in Psoriasis (2.29E-03), Estrogen-mediated S-phase Entry (1.62E-02), p53 Signaling (3.39E-02), Adenine and Adenosine Salvage III (3.98E-02) |
| 3_7 | Mitotic Roles of Polo-Like Kinase (2.24E-04), G Beta Gamma Signaling (1.41E-03), Chemokine Signaling (2.34E-03), HGF Signaling (2.51E-03), fMLP Signaling in Neutrophils (3.39E-03) |
| Cluster 2 (1728 genes) | |
| 5_1 | Catecholamine Biosynthesis (2.88E-05), Mineralocorticoid Biosynthesis (1.26E-02), Noradrenaline and Adrenaline Degradation (1.51E-02), Glucocorticoid Biosynthesis (1.55E-02), VDR/RXR Activation (1.55E-02) |
| 6_1 | Flavin Biosynthesis IV (Mammalian) (1.58E-04), Tryptophan Degradation X (Mammalian, via Tryptamine) (9.55E-04), Glutathione-mediated Detoxification (2.88E-03), Glycogen Degradation II (4.17E-03) |
| 6_1 / 7_1 | Ethanol Degradation II (4.07E-03 / 3.98E-07) |
| 6_2 | Heme Biosynthesis II (2.34E-04), Tetrapyrrole Biosynthesis II (2.04E-03), Sphingomyelin Metabolism (4.17E-03), Ubiquinol-10 Biosynthesis (Eukaryotic) (1.05E-02) |
| 6_2 / 7_2 / 7_3 | Mitochondrial Dysfunction (1.38E-02 / 1.26E-13 / 5.01E-15) |
| 7_1 | Glycine Betaine Degradation (9.77E-08), Nicotine Degradation II (3.24E-07), Superpathway of Methionine Degradation (1.35E-06), Serotonin Degradation (1.74E-06) |
| 7_2 | TCA Cycle II (Eukaryotic) (1.48E-08), Valine Degradation I (1.26E-04), Branched-chain α-keto acid Dehydrogenase Complex (1.32E-03) |
| 7_2 / 7_3 | Fatty Acid β-oxidation I (8.51E-04 / 1.86E-03) |
| 7_3 | L-glutamine Biosynthesis II (tRNA-dependent) (3.39E-04), Coenzyme A Biosynthesis (1.00E-03), Stearate Biosynthesis I (Animals) (3.02E-03) |
Figure 2. Heatmaps of genes representing the mitochondrial signature (top panel) and the hypoxia, stress/toxicity, oxidative stress signature (lower panel) that are present in SOM cluster 2 and are regulated during late remission.
The heatmaps were generated using the SOM module in MultiExperiment Viewer Application (MeV) (http://www.tm4.org/). The “SOM vector” bar represents an average of the gene expression for the corresponding sample.
Unbiased identification of biomarkers for disease onset, progression and remission
A second unbiased approach to biomarker identification was to apply a singular value decomposition method to the sequential NZB/W datasets. This analysis identifies genes that were most tightly regulated during changes in disease stage and was performed without consideration of actual fold change in gene expression. Of the top 300 positively and 300 negatively regulated genes within the dominant pattern (Supplementary Figure 3A), 554 with human homologs were differentially expressed between 16w pre-nephritic and 36w proteinuric mice. These 554 genes were then overlaid on the sets of genes differentially expressed in human SLE biopsies (16-17). 201 genes were concordantly regulated in glomeruli and 132 in tubulointerstitium of human SLE biopsies with 97 (31 upregulated and 66 downregulated) concordantly regulated in both compartments (Supplementary Table 6A). The most highly upregulated of these 97 genes were associated with myeloid and lymphoid cell infiltration and activation, antigen presentation and innate immune processes including ubiquitination, nucleic acid exonuclease activity and type 1 interferon production. Ubd (Fat10) was the most highly upregulated non-immune gene.
Because the genes thus identified were dominated by those associated with established nephritis, we repeated the analysis using only the first 3 groups of the time series and the two remission groups (Supplementary Figure 3B). Of the top 600 genes in the dominant pattern, 121 were concordantly regulated in human glomeruli and 119 in interstitium with 81 concordantly regulated in both compartments. Of these 81 genes, 40 (30 upregulated and 10 downregulated) were within the 603 gene subset that was >1.5 fold regulated both during disease onset and remission. The most highly upregulated of these 40 genes in the human biopsies included genes involved in innate immunity such as IFIH1, CXCL10, and PLAC8 (Onzin), a regulator of pro-inflammatory responses. Other upregulated genes include VCAM1, NMI, an IFNγ and IL2 induced signaling molecule, CKLF, a chemoattractant, FGL2 a prothrombinase induced during inflammation and SNAI2, a mediator of epithelial to mesenchymal transition (Supplementary Table 6B).
Confirmation by real time PCR
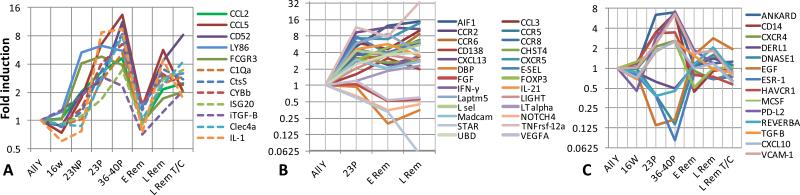
Results from real-time PCR analyses are shown in Supplementary Table 7 and Supplementary Figure 4. There was a high level of concordance with those genes regulated by microarray but the PCR was more sensitive and therefore detected expression of some genes not detected by microarray analysis. Of 166 genes, only 4 (CCL2, CCL5, CD52 and Ly86) were ≥1.5 fold regulated before nephritis onset, reversed with remission induction, became abnormal again during late remission and were also expressed in both human glomeruli and tubulointerstitium. FCGR, with a similar expression pattern, was upregulated in human glomeruli (Figure 3A).
Figure 3. Gene expression by PCR over time of selected genes in NZB/W kidneys.
A. Genes that were upregulated prior to (thick lines) or at (dashed lines) proteinuria onset, reverted during remission and became abnormal again during relapse and were also expressed in human SLE kidney biopsies. B. Genes that were upregulated at proteinuria onset and did not revert during remission. C. Genes that were upregulated at proteinuria onset, reverted at remission and remained normal during late remission.
A subset of genes displayed a pattern of regulation at proteinuria onset, reversal with complete remission induction and re-expression during late remission; many of these were associated with macrophage activation. C1qA, ISG20, CTSS, TGFBI (a component of extracellular matrix) and CYBB (NADPH oxidase 2) were robust markers of progression towards relapse and also highly expressed in human biopsies (Figure 3A). By contrast, TNFRSF12, Serpina3g, Fpr2, Clec4e and Grem-1 were highly associated with disease stage in the mouse tissues but were not detected in human biopsies. Genes that were regulated before proteinuria onset were significantly less likely to revert to normal during remission than genes that were regulated after proteinuria onset (p=0.005, data not shown).
Upregulated genes whose expression did not completely revert to normal at remission included a subset of chemokines, cytokines and adhesion molecules, probably reflecting the incomplete disappearance of lymphoid aggregates, as well as Ubd (FAT10), a marker of tubular epithelial dysfunction. In addition, the downregulation of VEGFA and FGF2 did not reverse completely with remission induction, suggesting a continuing defect in angiogenesis (Figure 3B).
We confirmed that several genes related to mitochondrial dysfunction or metabolic stress, including NOX4, and COX15, became abnormal late after treatment with CTLA4Ig/TACI-Ig. In addition, LCN2 and TIMP1, genes that were upregulated at proteinuria onset and reversed with remission induction, were modestly upregulated late after treatment with CTLA4Ig/TACI-Ig before the onset of overt proteinuria.
Finally, we identified a subset of genes whose expression remained at remission levels even during late remission. Upregulated genes with this pattern included CD14, ANKRD1 (podocyte injury), TGFβ, CXCL9, 10, 11, 16 and CCL9 (chemokines), ICAM1 and VCAM-1 (endothelial activation), HAVCR1 and C3 (tubular injury), whereas downregulated genes included, EGF, CLYBL, DNASE1 and several genes involved in drug detoxification (Figure 3C).
Application of these data to a second murine model
Of 3759 genes that were regulated in nephritic vs. young NZM2410 mice (17), 2364 (62.9%) reversed after BAFF inhibition (Supplementary Table 8), reflecting improvements in inflammation, endothelial cell activation, tubular dysfunction, metabolic stress and mitochondrial dysfunction. Reversal of nephrin loss suggests that podocyte repair may occur. Of these genes, 399 were concordantly regulated in both glomeruli and tubulointerstitium of SLE nephritis biopsies; this 399 geneset was quite similar to that identified in NZB/W mice (Supplementary Table 9). 1395 genes did not reverse after BAFF inhibition including both pro-inflammatory genes and genes related to mitochondrial dysfunction (data not shown), suggesting that, as in NZB/W mice, immune modulation may not completely reverse processes that could contribute to ongoing renal damage.
qRT-PCR analyses showed increased expression of a subset of pro-inflammatory cytokines and chemokines in the remission mice, (not shown) but this was not associated with clinical relapse. As in NZB/W mice, a cluster of genes was identified whose expression reversed after remission induction and remained at remission levels even 33 weeks after BAFF-R-Ig therapy (Supplementary Table 10 and Supplementary Figure 5).
DISCUSSION
Our study identifies sequential molecular events in the progression and remission of SLE nephritis in informative mouse models. We show limited expression of inflammatory mediators during the pre-nephritic stage in NZB/W mice. By contrast, a major signature of inflammatory cell infiltration and activation occurs at the onset of proteinuria. . This is accompanied by increased expression of a variety of collagen genes and evidence of endothelial cell activation, and tubular damage. These changes are associated with glomerular hypertrophy and proliferative changes, and progressive accumulation of periglomerular and perivascular lymphoid aggregates (9).
As glomerular inflammation progresses, compromise of the downstream peritubular blood flow (reviewed (24)) is associated with a signature of oxidative and metabolic stress, mitochondrial dysfunction and tubular damage. In addition, activation of the peritubular endothelium can induce inflammatory cell infiltration, particularly of macrophages, into the tubulointerstitium. We show that disease progression is characterized by continued recruitment of pro-inflammatory mediators and expression of genes involved in macrophage activation, alternative complement activation, prothrombotic tendency, and fibrosis. Importantly, an unbiased analysis of the sequential microarray datasets from NZB/W mice confirms that there are two major gene clusters that are dynamically regulated during disease onset and remission,, one, occurring at proteinuria onset, that identifies the inflammatory component of nephritis and the other, occurring during established disease that reflects the metabolic disturbances that are associated with hypoxia.
Previous studies have described the gene expression profiles in NZB/W (25) and MRL/lpr kidneys (26), including the effect of preventive treatments (Rapamycin and Prednisone respectively). Despite the different methods used (e.g. glomeruli laser microdissection in the case of the MRL/lpr study), a significant gene overlap (67% and 57% respectively) was confirmed between our 16w vs. 36-40w dataset and the genes that were regulated in established nephritis compared to young mice in those studies.
We further show here that remission induced by costimulatory blockade or BAFF inhibition (9, 11) is associated with reversal of much of the inflammatory signature that accompanies proteinuria onset. Importantly however, in NZB/W mice, the mitochondrial and metabolic signature recurs before the reappearance of proteinuria, perhaps reflecting an increased propensity to hypoxia conferred by the initial tissue insult. In NZM2410 mice reversal of this signature is incomplete despite histologic remission. In these mice a subset of inflammatory markers and inflammatory cells accumulates during long-lasting remission following BAFF inhibition but this is not sufficient for clinical relapse. A similar checkpoint has been described in BAFF deficient NZM2328 mice and in mice congenic for a NZM derived gene locus that develop glomerular pathology and lymphoid infiltrates but do not progress to renal failure (15, 27). These data show that renal decline does not inevitably follow renal inflammatory cell infiltration. Rather, our molecular data suggests that podocyte damage, renal tubular dysfunction, endothelial cell activation and tissue remodeling are the functional features associated with clinical onset of proteinuria and consequent renal failure.
Our studies gave us an opportunity to identify transcriptional profiles characteristic of each specific disease stage, with the potential to serve as biomarker candidates. Using PCR we identified CCL2, CCL5, Ly86, CD52 and FCGR3 as genes that were upregulated in prenephritic kidneys, fluctuated with early and late remission, and were also highly expressed in human SLE nephritis biopsies. Of these, increasing urinary CCL2 (MCP1) is currently the most promising biomarker for disease flare (28-29), whereas failure to normalize urinary CCL5 during remission is associated with an increased risk for subsequent flare (30). Ly86 is expressed by plasma cells and renal macrophages; interestingly, serum levels of soluble Ly86 correlate with disease progression in MRL/lpr mice (31). CD52 and FCGR3 are markers for infiltrating renal cells. Surprisingly, TNFRSF12 (TWEAK receptor), that is upregulated in pre-nephritic mice and is an excellent biomarker for remission and progression, was not detected in our human lupus biopsies. Nevertheless a previous study using real-time PCR detected TNFRSF12 mRNA in lupus glomeruli and interstitium (32).
Genes that are regulated at proteinuria onset, reverse with remission induction and become abnormal again during late remission may be useful biomarkers of impending flare or of failure to respond to therapy. We identified several such genes that are associated with macrophage activation. Of those that were also identified in human kidneys, C1q components are of interest because these are produced mostly by tissue macrophages and may therefore reflect macrophage/DC infiltration and/or activation. Importantly, antibodies to C1q are associated with lupus nephritis, and may amplify tissue damage (33). How C1q modulates local macrophage function in the setting of inflammation is not currently known.
To identify genes whose expression most tightly correlated with disease stage we used an unbiased approach. Using a NZB/W dataset that included the pre-nephritic stage, new onset proteinuria and remission we identified 41 such genes that were also expressed in human SLE biopsies. These include the procoagulant Fgl2 and PLAC8 (onzin), a molecule required for antibacterial and inflammatory responses (34). Two additional genes, CXCL10 and VCAM-1, are soluble proteins that have already been identified as potential urinary biomarkers for human lupus nephritis (5, 35-36). In the human biopsies, the expression of CXCL10 was coupled with expression of its inducing protein IFIH1 (MDA5), an interferon inducible gene that is highly expressed in the glomeruli of SLE nephritis patients (37). Urinary levels of VCAM-1 are strongly associated with renal disease activity in mouse and human lupus nephritis in cross-sectional studies (reviewed (35)). Fgl2 and PLAC8 were regulated at proteinuria onset, reverted with remission induction and were expressed again during late remission whereas IFIH1, VCAM1 and CXCL10 remained normal during late remission.
When the whole series of NZB/W samples was included in an unbiased analysis of genes regulated with disease stage, 97 genes were identified that were also expressed in both compartments of human SLE biopsies. These include genes involved in innate immunity many of which are IFN inducible, confirming the sensitivity of genes involved in this pathway to changes in disease activity. Notably, this geneset included LCN2, a marker of proximal tubular damage. While initial longitudinal studies suggested that increasing urine levels of LCN2 could predict a flare of SLE nephritis especially in children (38-39), a recent large longitudinal study in adults failed to reproduce these results (40). Our data suggests that LCN2 is a relatively late marker of SLE nephritis that is highly responsive to remission induction. Indeed, LCN2 was not expressed in the human lupus biopsies, most of which were harvested from patients who have already begun glucocorticoid therapy (17). We also found among this geneset genes whose expression was detected only in proteinuric mice and remained normal even during late remission. Although such genes may not be sensitive markers of an impending renal flare, they may be of potential use to follow a therapeutic response.
Future studies will determine whether the genes we have identified herein can serve as biomarkers for flares or therapeutic responses or whether they will help distinguish active disease from chronic changes. While not all the gene products of our biomarker gene set are soluble, methodology for performing expression, proteomic or metabolic analysis of urine cells is constantly improving and could be used to test sequential expression of some of the other highly expressed genes or pathways.
Given the current lack of sufficiently effective therapy for lupus nephritis and the unacceptably high rate of disease relapse, one important goal of these studies was to identify pathways and molecules that suggest potential therapeutic targets. While lupus nephritis is classically treated with immunosuppressive therapies, less attention has been paid to correction of metabolic inefficiency and cellular stress. Multiple strategies to achieve this have been tested in animal models (41-42). In addition our studies have identified several understudied molecules that are tightly regulated with disease stage, highly expressed in the human biopsies and whose role can be further explored in the appropriate murine models. These include Laptm5, a positive regulator of macrophage pro-inflammatory cytokine production (43), Plac8 (onzin) and ubiquitin D (Fat10) a proteasome binding protein that promotes tubulointerstitial inflammation in kidney diseases via NFkB activation (44-45).
In conclusion, our studies show that limited renal chemokine and cytokine production occurs during the pre-nephritic phase whereas inflammatory events escalate at or just before proteinuria onset. This is followed by hypoxia and metabolic stress and finally by tubular and endothelial dysfunction. Regimens that include costimulatory blockade and BAFF inhibition reverse inflammatory events and but progression towards relapse is associated with recurrence of inflammation, cellular stress and mitochondrial dysfunction. Not all molecular abnormalities are reversed by remission induction therapies, perhaps increasing the sensitivity of the kidney to hypoxia during flare and contributing to progressive renal damage even during periods of quiescence. By contrast, some molecules maintain their normal expression profile even during the late stages of remission. We show in sum, that there are several checkpoints along the path to renal failure in lupus nephritis that are associated with renal molecular changes and expression of stage specific biomarkers, some of which can now be tested in humans.
Supplementary Material
REFERENCES
- 1.Davidson A, Aranow C. Lupus nephritis: lessons from murine models. Nat Rev Rheumatol. 2010;6(1):13–20. doi: 10.1038/nrrheum.2009.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sprangers B, Monahan M, Appel GB. Diagnosis and treatment of lupus nephritis flares--an update. Nat Rev Nephrol. 2012;8(12):709–17. doi: 10.1038/nrneph.2012.220. [DOI] [PubMed] [Google Scholar]
- 3.Costenbader KH, Solomon DH, Winkelmayer W, Brookhart MA. Incidence of End-Stage Renal Disease Due to Lupus Nephritis in the U.S.,1995-2004. Arthritis and Rheumatism. 2008;(Supplementary) doi: 10.1002/art.30293. Abstract 1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward MM. Changes in the incidence of endstage renal disease due to lupus nephritis in the United States, 1996-2004. J Rheumatol. 2009;36(1):63–7. doi: 10.3899/jrheum.080625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mok CC. Biomarkers for lupus nephritis: a critical appraisal. J Biomed Biotechnol. 2010;2010:638413. doi: 10.1155/2010/638413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Fang X, Li QZ. Biomarker profiling for lupus nephritis. Genomics Proteomics Bioinformatics. 2013;11(3):158–65. doi: 10.1016/j.gpb.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perry D, Sang A, Yin Y, Zheng YY, Morel L. Murine models of systemic lupus erythematosus. J Biomed Biotechnol. 2011;2011:271694. doi: 10.1155/2011/271694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theofilopoulos AN, Dixon FJ. Murine models of systemic lupus erythematosus. Adv Immunol. 1985;37(1):269–390. doi: 10.1016/s0065-2776(08)60342-9. [DOI] [PubMed] [Google Scholar]
- 9.Schiffer L, Sinha J, Wang X, Huang W, von Gersdorff G, Schiffer M, et al. Short term administration of costimulatory blockade and cyclophosphamide induces remission of systemic lupus erythematosus nephritis in NZB/W F1 mice by a mechanism downstream of renal immune complex deposition. J Immunol. 2003;171(1):489–97. doi: 10.4049/jimmunol.171.1.489. [DOI] [PubMed] [Google Scholar]
- 10.Ramanujam M, Wang X, Huang W, Schiffer L, Grimaldi C, Akkerman A, et al. Mechanism of action of transmembrane activator and calcium modulator ligand interactor-Ig in murine systemic lupus erythematosus. J Immunol. 2004;173(5):3524–34. doi: 10.4049/jimmunol.173.5.3524. [DOI] [PubMed] [Google Scholar]
- 11.Ramanujam M, Bethunaickan R, Huang W, Tao H, Madaio MP, Davidson A. Selective blockade of BAFF for the prevention and treatment of systemic lupus erythematosus nephritis in NZM2410 mice. Arthritis Rheum. 2010;62(5):1457–68. doi: 10.1002/art.27368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schiffer L, Bethunaickan R, Ramanujam M, Huang W, Schiffer M, Tao H, et al. Activated renal macrophages are markers of disease onset and disease remission in lupus nephritis. J Immunol. 2008;180(3):1938–47. doi: 10.4049/jimmunol.180.3.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clynes R, Dumitru C, Ravetch JV. Uncoupling of immune complex formation and kidney damage in autoimmune glomerulonephritis. Science. 1998;279(5353):1052–4. doi: 10.1126/science.279.5353.1052. [DOI] [PubMed] [Google Scholar]
- 14.Bagavant H, Fu SM. New insights from murine lupus: disassociation of autoimmunity and end organ damage and the role of T cells. Curr Opin Rheumatol. 2005;17(5):523–8. doi: 10.1097/01.bor.0000169361.23325.1e. [DOI] [PubMed] [Google Scholar]
- 15.Jacob CO, Pricop L, Putterman C, Koss MN, Liu Y, Kollaros M, et al. Paucity of clinical disease despite serological autoimmunity and kidney pathology in lupus-prone New Zealand mixed 2328 mice deficient in BAFF. J Immunol. 2006;177(4):2671–80. doi: 10.4049/jimmunol.177.4.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bethunaickan R, Berthier CC, Ramanujam M, Sahu R, Zhang W, Sun Y, et al. A unique hybrid renal mononuclear phagocyte activation phenotype in murine systemic lupus erythematosus nephritis. J Immunol. 2011;186(8):4994–5003. doi: 10.4049/jimmunol.1003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berthier CC, Bethunaickan R, Gonzalez-Rivera T, Nair V, Ramanujam M, Zhang W, et al. Cross-species transcriptional network analysis defines shared inflammatory responses in murine and human lupus nephritis. J Immunol. 2012;189(2):988–1001. doi: 10.4049/jimmunol.1103031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan O, Madaio MP, Shlomchik MJ. The roles of B cells in MRL/lpr murine lupus. Ann N Y Acad Sci. 1997;815:75–87. doi: 10.1111/j.1749-6632.1997.tb52046.x. [DOI] [PubMed] [Google Scholar]
- 19.Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, et al. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci U S A. 1999;96(6):2907–12. doi: 10.1073/pnas.96.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wall M, Rechtsteiner A, Rocha L. Singular value decomposition and principal component analysis. In: Berrar D, Dubitzky W, Granzow M, editors. A Practical Approach to Microarray Data Analysis. Kluwer Academic Publishers; Norwell, MA: 2002. [Google Scholar]
- 21.Alter O, Brown PO, Botstein D. Generalized singular value decomposition for comparative analysis of genome-scale expression data sets of two different organisms. Proc Natl Acad Sci U S A. 2003;100(6):3351–6. doi: 10.1073/pnas.0530258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holter NS, Mitra M, Maritan A, Cieplak M, Banavar JR, Fedoroff NV. Fundamental patterns underlying gene expression profiles: simplicity from complexity. Proc Natl Acad Sci U S A. 2000;97(15):8409–14. doi: 10.1073/pnas.150242097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bethunaickan R, Berthier CC, Zhang W, Kretzler M, Davidson A. Comparative Transcriptional Profiling of 3 Murine Models of SLE Nephritis Reveals Both Unique and Shared Regulatory Networks. PLoS One. 2013;8(10):e77489. doi: 10.1371/journal.pone.0077489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson A, Berthier C, Kretzler M. Pathogenetic mechanisms in lupus nephritis. In: Hahn B, Wallace D, editors. Dubois SLE. 8 ed Elsevier; 2012. [Google Scholar]
- 25.Reddy PS, Legault HM, Sypek JP, Collins MJ, Goad E, Goldman SJ, et al. Mapping similarities in mTOR pathway perturbations in mouse lupus nephritis models and human lupus nephritis. Arthritis Res Ther. 2008;10(6):R127. doi: 10.1186/ar2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teramoto K, Negoro N, Kitamoto K, Iwai T, Iwao H, Okamura M, et al. Microarray analysis of glomerular gene expression in murine lupus nephritis. J Pharmacol Sci. 2008;106(1):56–67. doi: 10.1254/jphs.fp0071337. [DOI] [PubMed] [Google Scholar]
- 27.Ge Y, Jiang C, Sung SS, Bagavant H, Dai C, Wang H, et al. Cgnz1 allele confers kidney resistance to damage preventing progression of immune complex-mediated acute lupus glomerulonephritis. J Exp Med. 2013;210(11):2387–401. doi: 10.1084/jem.20130731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunner HI, Bennett MR, Mina R, Suzuki M, Petri M, Kiani AN, et al. Association of noninvasively measured renal protein biomarkers with histologic features of lupus nephritis. Arthritis Rheum. 2012;64(8):2687–97. doi: 10.1002/art.34426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rovin BH, Song H, Birmingham DJ, Hebert LA, Yu CY, Nagaraja HN. Urine chemokines as biomarkers of human systemic lupus erythematosus activity. J Am Soc Nephrol. 2005;16(2):467–73. doi: 10.1681/ASN.2004080658. [DOI] [PubMed] [Google Scholar]
- 30.Tian S, Li J, Wang L, Liu T, Liu H, Cheng G, et al. Urinary levels of RANTES and M-CSF are predictors of lupus nephritis flare. Inflamm Res. 2007;56(7):304–10. doi: 10.1007/s00011-007-6147-x. [DOI] [PubMed] [Google Scholar]
- 31.Sasaki S, Nagai Y, Yanagibashi T, Watanabe Y, Ikutani M, Kariyone A, et al. Serum soluble MD-1 levels increase with disease progression in autoimmune prone MRL(lpr/lpr) mice. Mol Immunol. 2012;49(4):611–20. doi: 10.1016/j.molimm.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 32.Michaelson JS, Wisniacki N, Burkly LC, Putterman C. Role of TWEAK in lupus nephritis: a bench-to-bedside review. J Autoimmun. 2012;39(3):130–42. doi: 10.1016/j.jaut.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsirogianni A, Pipi E, Soufleros K. Relevance of anti-C1q autoantibodies to lupus nephritis. Ann N Y Acad Sci. 2009;1173:243–51. doi: 10.1111/j.1749-6632.2009.04750.x. [DOI] [PubMed] [Google Scholar]
- 34.Johnson RM, Kerr MS, Slaven JE. Plac8-dependent and inducible NO synthase-dependent mechanisms clear Chlamydia muridarum infections from the genital tract. J Immunol. 2012;188(4):1896–904. doi: 10.4049/jimmunol.1102764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reyes-Thomas J, Blanco I, Putterman C. Urinary biomarkers in lupus nephritis. Clin Rev Allergy Immunol. 2011;40(3):138–50. doi: 10.1007/s12016-010-8197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avihingsanon Y, Benjachat T, Tassanarong A, Sodsai P, Kittikovit V, Hirankarn N. Decreased renal expression of vascular endothelial growth factor in lupus nephritis is associated with worse prognosis. Kidney Int. 2009 doi: 10.1038/ki.2009.75. [DOI] [PubMed] [Google Scholar]
- 37.Imaizumi T, Aizawa-Yashiro T, Tsuruga K, Tanaka H, Matsumiya T, Yoshida H, et al. Melanoma differentiation-associated gene 5 regulates the expression of a chemokine CXCL10 in human mesangial cells: implications for chronic inflammatory renal diseases. Tohoku J Exp Med. 2012;228(1):17–26. doi: 10.1620/tjem.228.17. [DOI] [PubMed] [Google Scholar]
- 38.Rubinstein T, Pitashny M, Levine B, Schwartz N, Schwartzman J, Weinstein E, et al. Urinary neutrophil gelatinase-associated lipocalin as a novel biomarker for disease activity in lupus nephritis. Rheumatology (Oxford) 2010;49(5):960–71. doi: 10.1093/rheumatology/kep468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hinze CH, Suzuki M, Klein-Gitelman M, Passo MH, Olson J, Singer NG, et al. Neutrophil gelatinase-associated lipocalin is a predictor of the course of global and renal childhood-onset systemic lupus erythematosus disease activity. Arthritis Rheum. 2009;60(9):2772–81. doi: 10.1002/art.24751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiani AN, Wu T, Fang H, Zhou XJ, Ahn CW, Magder LS, et al. Urinary vascular cell adhesion molecule, but not neutrophil gelatinase-associated lipocalin, is associated with lupus nephritis. J Rheumatol. 2012;39(6):1231–7. doi: 10.3899/jrheum.111470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Small DM, Coombes JS, Bennett N, Johnson DW, Gobe GC. Oxidative stress, anti-oxidant therapies and chronic kidney disease. Nephrology (Carlton) 2012;17(4):311–21. doi: 10.1111/j.1440-1797.2012.01572.x. [DOI] [PubMed] [Google Scholar]
- 42.Tabas I, Glass CK. Anti-inflammatory therapy in chronic disease: challenges and opportunities. Science. 2013;339(6116):166–72. doi: 10.1126/science.1230720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glowacka WK, Alberts P, Ouchida R, Wang JY, Rotin D. LAPTM5 protein is a positive regulator of proinflammatory signaling pathways in macrophages. J Biol Chem. 2012;287(33):27691–702. doi: 10.1074/jbc.M112.355917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong P, Canaan A, Wang B, Leventhal J, Snyder A, Nair V, et al. The ubiquitin-like protein FAT10 mediates NF-kappaB activation. J Am Soc Nephrol. 2010;21(2):316–26. doi: 10.1681/ASN.2009050479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren J, Wang Y, Gao Y, Mehta SB, Lee CG. FAT10 mediates the effect of TNF-alpha in inducing chromosomal instability. J Cell Sci. 2011;124(Pt 21):3665–75. doi: 10.1242/jcs.087403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.