Abstract
Beta-tubulin is an important multifunctional protein of eukaryotes abundant in the cytoskeleton and responsible for the formation of tubulin, structures responsible for cell morphology and which aid in motility and intracellular transportation. It has been used as a genotypic marker for studying the evolutionary history and phylogenetic relationships between eukaryotic organisms. Internal transcribed spacers of the ribosomal RNA genes have been widely used for typing inter-species and intra-species variation. An Indian isolate of Eimeria tenella was genotyped following the cloning and sequencing of beta-tubulin and internal transcribed spacer-2 (ITS-2) and compared with other reference isolates of E. tenella. The β-tubulin has 99 % intra-species similarity at the gene level and the functional homology of the protein is very high with more than 95 % amino-acid similarity across the Apicomplexa. The ITS-2 sequence had a similar pattern of nucleotide base arrangement with 99 % homology to Houghton and Nippon strains of E. tenella.
Keywords: Eimeria tenella, Beta tubulin, ITS-2, Genotyping, India
Introduction
Poultry coccidiosis is a disease of immense economic importance to the poultry industry, worldwide. The disease is caused by seven well known species of the genus Eimeria, which vary in their pathogenicity and hence the severity of disease. Eimeria tenella, the causative agent of caecal coccidiosis is one of the two most pathogenic species, the other being Eimeria necatrix. Vaccines and anti-coccidial drugs are available in the world market for control of coccidiosis and the economic burden of chemotherapy further worsened by cases of fast developing anti-coccidial resistance, has paved the way to search for new drug targets. Anti-coccidial vaccines are an important measure for immunoprophylactic control of chicken coccidiosis, but have limitations as they are live vaccines composed of oocysts of Eimeria species derived from local isolates/strains.
The poultry industry, along with the associated axillary/supporting industries make the largest contribution to the economy within the livestock sector in India. Studies have been carried out in India on chemotherapeutic resistance against anticoccidials (Panda et al. 1973; Gill and Bajwa 1979; Yadav and Gupta 2001; Gautam and Gupta 2004). Genotyping studies are an essential approach for development of suitable vaccines and drug trials and currently very little information is available on genetic diversity within Eimeria species (Beck et al. 2009). Internal transcribed spacer-1 (ITS-1) based genotyping studies for species identification and intra-species variation have been carried out in India (Bhaskaran et al. 2010; Kumar 2012).
Genetic characterization of strains of parasite species from a geographical area should include housekeeping genes, as well as genes related to the host-parasite relationship. Βeta-tubulin an important nucleotide binding protein forms the conspicuous constituent of the eukaryotic cytoskeletal protein. It is highly conserved across several genera of algae, fungi and protists (Barabona et al. 1988; Conner et al. 1989; Gurr et al. 1988; Trivinos-Lagos et al. 1993). The β-tubulin is a polypeptide monomer of microtubules, the other component being α-tubulin. The polymerization and de-polymerization of β-tubulin monomers is a dynamic process contributing to cell locomotion (flagellar and cilial motility), intracellular transport and cell morphology. Zhu and McDougald (1995) localized the protein in the cytoskeleton, apical complex and sub-pellicular region of the asexual stages (sporozoites and merozoites) of E. tenella by confocal microscopy. Beta-tubulin serves as an important drug target for many nematodes, fungi, moulds, parasitic protozoa, but has not been explored against Eimeria. Despite this it is an important factor for genotyping studies.
Internal transcribed spacers (ITS) are regions of unknown function located between the 18S, 5.8S and 28S ribosomal RNA (r RNA) genes located on the same stretch of DNA within the eukaryotic chromosome. Sequencing of ITS is an important tool for understanding congeneric populations, within genus homology (inter-species differences), differences between highly related genera and intra-species differences. ITS has been used in species specific detection of parasites. Kawahara et al. (2008) used real time PCR to detect five important Eimeria species of domestic chicken. ITS-2 based species status study of Otodectes cynotis from different host species and geographical areas was carried out by Lohse et al. (2002).
The present study has been carried out as a part of multiple locus sequence typing (MLST) of Eimeria tenella Indian isolate1. Βeta-tubulin and ITS-2 of Indian isolate 1 of E. tenella were partially sequenced, characterized, annotated and compared with the standard reference laboratory strains of E. tenella, as well as with other apicomplexan parasites at the level of translated polypeptide.
Materials and methods
DNA extraction
The parasite oocysts used in the study were derived from Indian isolate 1 of Eimeria tenella (origin Kaithal district, Haryana). E. tenella oocysts, 1.5 × 105, were mixed with equal volume of previously autoclaved 0.3–0.5 mm glass beads (Sigma, USA) and a small volume of lysis buffer supplied by the manufacturer (Qiagen) and smashed by vigorous vortexing in order to release the sporocysts and sporozoites. Vortexing was done till 80 % of the oocysts were ruptured and sporozoites were released. QIAmp stool DNA minikit (Qiagen, Germany) was used for extraction of DNA as per the manufacturer’s protocol, excepting that the purified DNA was eluted in 100 μl volume of elution buffer.
PCR and cloning
Eimeria tenella beta-tubulin gene and ITS-2 (both partial fragments) were amplified from genomic DNA using specific primers (Table 1). Amplifications were carried out under thermal cycling conditions of initial denaturation at 95 °C for 3 min, 35 cycles of denaturation at 95 °C for 30 s, annealing at 56 °C for 30 s, extension at 72 °C for 2/1 min(s) and final extension at 72 °C for 15 min in a S1000 thermal cycler (Biorad, USA). The reaction mixture of 25 μl volume consisted of 2 μl DNA, 2.5 μl of 10× High Fidelity enzyme mix buffer with 1.5 mM·MgCl2, 0.5 μl of 25 mM MgCl2 (to obtain a final concentration of 2 mM MgCl2), 0.5 U of High Fidelity enzyme mix (Thermo Scientific, USA), 0.5 μl 10 mM dNTP mix, 1 μl each of forward and reverse primers (400 nM) and nuclease free water to make up the volume. Electrophoresis of PCR products was carried out at constant voltage of 5 V/cm in a Biorad electrophoresis assembly, in 1 % agarose gel in 1× Tris acetate EDTA (TAE) buffer. Products were purified from other reactants in the PCR mixture using Minelute PCR purification kit (Qiagen, Germany).
Table 1.
Details of primers used for amplification and cloning of E. tenella beta-tubulin
| Primers | Sequence | Approximate product size | Annealing temperature used | |
|---|---|---|---|---|
| CIDte21f | Βeta-tubulin | CCATGAAATGAAAGCGGGTCAC | 2,007 bp | Ta = 56 °C |
| CIDte21r | GGGGTATTCTTCGCGGACTTTG | |||
| CIDte26f | ITS-2 | TTTAGTCCATCGCAACCCTTGA | 923 bp | |
| CIDte26r | TCCGAATGAAAGGAGCATACGA |
Purified PCR products were quantified and ligated into TA cloning vector, pTZ57R/T (provided with the InsTA cloning kit, Thermo Scientific, USA). The ligation mixture of 20 μl volume containing PCR product 8 μl (β-tubulin)/5 μl (ITS-2), vector 1 μl, T4 DNA ligase 1 μl (5 Weiss U/μl), 5× ligase buffer 4 μl and nuclease free water 6 μl, was incubated overnight at 4 °C.
Each ligation mixture was used to transform Escherichia coli DH5α cells using the chemicals supplied with the InsTA cloning kit and the transformed cells were poured on to ampicillin and X-gal added Luria–Bertani agar (LB agar) pate and incubated overnight at 37 °C. From the population of blue-white selection of the colonies on each plate, four white colonies were picked using a bacteriological loop and streaked on a fresh LB agar plate containing 1.2 mg/ml ampicillin and incubated at 37 °C. Colony PCR was performed to confirm the presence of the desired insert. In short the 4–6 white colonies from the LB agar plate were streaked onto a fresh ampicillin added LB agar plate and incubated overnight at 37 °C. PCR reaction mixture of 25 μl was prepared as per the composition mentioned earlier except the template DNA. A single colony from the streak was touched with a sterile tip (10 μl volume) and transferred to the respective PCR reaction mix. The PCR reaction was carried out under the same conditions as above with 30 cycles of amplification.
Plasmid extraction and restriction digestion
Two positive colonies were inoculated into 5 ml of LB broth containing ampicillin and cultured overnight at 37 °C in a shaking water bath. Plasmid extraction was performed using plasmid extraction kit (Invitrogen, USA). Plasmid quantity was estimated by UV spectrophotometry. Restriction double digestion of 500–600 ng recombinant plasmids (beta-tubulin and ITS-2) with restriction enzymes, EcoRI and HindIII was carried out in a total 20 μl volumes including1 μl each of the two Fast digest enzymes, 2 μl Fast Digest green buffer and nuclease free water, incubated at 37 °C for 15 min.
Sequencing of the clones
Cultured broth was used for preparation of stab cultures in 2 ml micro centrifuge tubes, containing 1 ml volume of LB agar. The stab cultures of the confirmed recombinant clones were forwarded to the DNA sequencing facility in the Department of Biochemistry, University of Delhi. The sequences received were aligned with the known sequences of beta-tubulin of other E. tenella isolates. Multiple alignments were carried out using the Megalin programme of the DNAStar Lasergene software version 6, pairwise alignment report and phylogenetic tree construction was done using Mega 5.1.
Results
DNA extraction, PCR and cloning
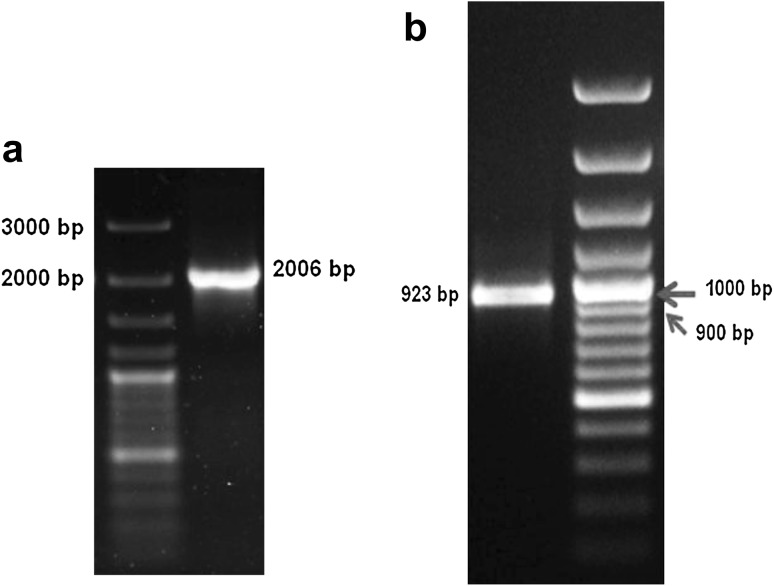
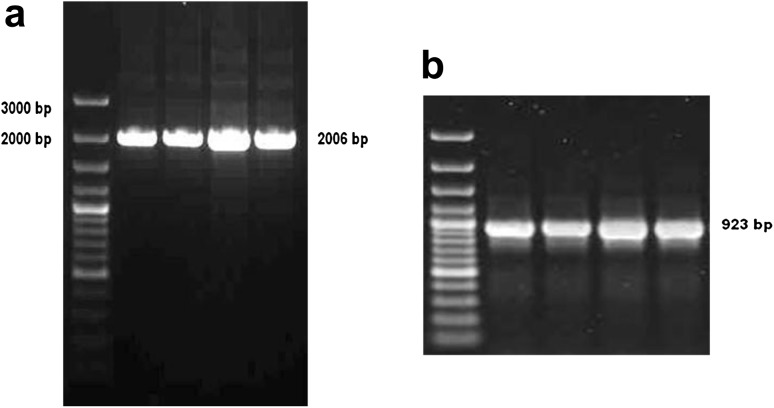
400–500 ng genomic DNA was extracted from 105 oocysts, with absorbance ratio 260:280 nm, >1.7. A single PCR product of approximately 2.0 kb was amplified during the reaction (Fig. 1a) with primers CIDte21f and CIDte21r; and a single PCR product of approximately 0.9 kb was amplified with CIDte26f and CIDte26r (Fig. 1b). Colony PCR amplified similar sized products (Fig. 2a, b).
Fig. 1.
a Beta-tublin amplicon, 2,006 bp, b E. tenella ITS-2 amplicon, 923 bp
Fig. 2.
a Beta-tublin colony PCR b E. tenella ITS-2 colony PCR, 923 bp
Plasmid extraction and restriction digestion
Plasmid yield from the broth culture of positive recombinant colonies was 70 ng/μl. Restriction digestion of the beta-tubulin recombinant plasmid with EcoRI and HindIII generated 1,200 bp and 900 bp fragments (Fig. 3). ITS-2 recombinant plasmid on restriction digestion with the same enzyme pair generated 923 bp fragment (figure not shown).
Fig. 3.

Characterisation of E. tenella beta-tublin by EcoRI-Hindlll double digest: release of two products because of the presence of internal Hindlll restriction site (a) against an uncut plasmid (b)
Sequence analysis of beta-tubulin
A 2,006 bp contiguous sequence was generated during the Sanger sequencing and the BLAST search in NCBI gave hits with the E. tenella beta-tubulin. The sequence was submitted to the NCBI database via Sequin, after annotation of introns, exons, coding sequence (cds), 5′ UTR (un-translated region) based on the reference sequence (accession numbers U19609.1 and ETU19609). The detailed annotation of the beta-tubulin sequence is presented in Table 2.
Table 2.
Detailed annotation of E. tenella India 1, beta-tubulin
| Gene fragment | Location | Size (base pairs) | Remarks |
|---|---|---|---|
| Intron1; intron2 | 1–894; 1,082–1,622 | 894; 541 | 5′ end partial |
| Exon1; exon2 | 895–1,081; 1,623–2,006 | 187; 384 | 3′ end partial |
| 5′ UTR | 895–985 | 91 | |
| Coding sequence (cds) | 986–1,081; 1,623–2,006 | 96; 384 | 3′ end partial |
| Point mutations when aligned with reference Houghton strain | Deletion at 955 | Point mutation in 5′UTR | |
| Substitution at 489 | Point mutation in intron 1 (A substituted by C) | ||
| Substitution at 1,372 | Point mutation in intron 2 (C substituted by G) |
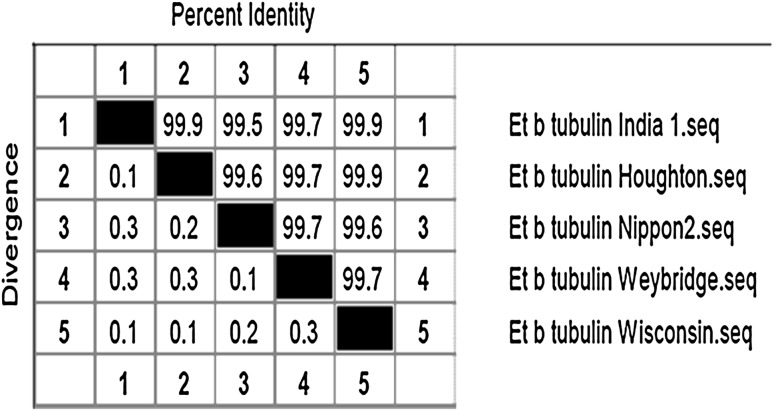
Intraspecies distance relationship was analyzed with CLUSTAL W algorithm of the Megalin programme in Lasergene DNA Star ver 6. Figure 4 depicts 99 % intraspecies similarity of beta-tubulin.
Fig. 4.
Intraspecies homology: percent similarity between beta-tubulin
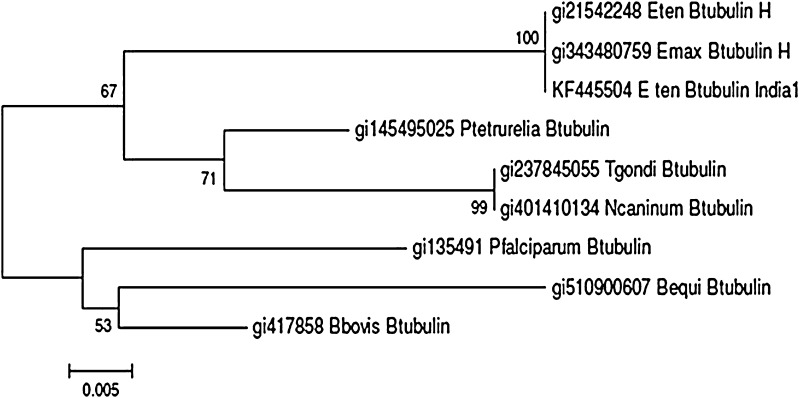
The translated coding sequence (predicted amino acid sequence of the cds) when applied to BLAST homology search, provided several hits with >90 % homology, among protozoa. Pairwise alignment with MEGA5.1 programme showed high similarity with other apicomplexans (Table 3; Fig. 5).
Table 3.
Translated coding sequence of β-tubulin: relative pairwise distance between Indian isolate1, other apicomplexans and ciliate Paramaecium tetraurelia with >90 % similarity
| Query sequence | Subject sequence | Distance |
|---|---|---|
| E. tenella β-tubulin India1 | gi21542248 E. tenella β-tubulin | 0.0000 |
| E. tenella β-tubulin India1 | gi343480759 E. maxima β-tubulin Houghton strain | 0.0000 |
| E. tenella β-tubulin India1 | gi237845055 Toxoplasma gondi β-tubulin | 0.0645 |
| E. tenella β-tubulin India1 | gi401410134 Neospora caninum β-tubulin | 0.0645 |
| E. tenella β-tubulin India1 | gi135491 Plasmodium falciparum β-tubulin | 0.0847 |
| E. tenella β-tubulin India1 | gi510900607 Babesia equi β-tubulin | 0.0780 |
| E. tenella β-tubulin India1 | gi417858 Babesia bovis β-tubulin | 0.0645 |
| E. tenella β-tubulin India1 | gi145495025 Paramaecium tetraurelia β-tubulin | 0.0513 |
Fig. 5.
Beta-tublin of E. tenella: Phylogenetic evaluation with apicomplexans and Paramaecium tetraurelia (NJ tree)
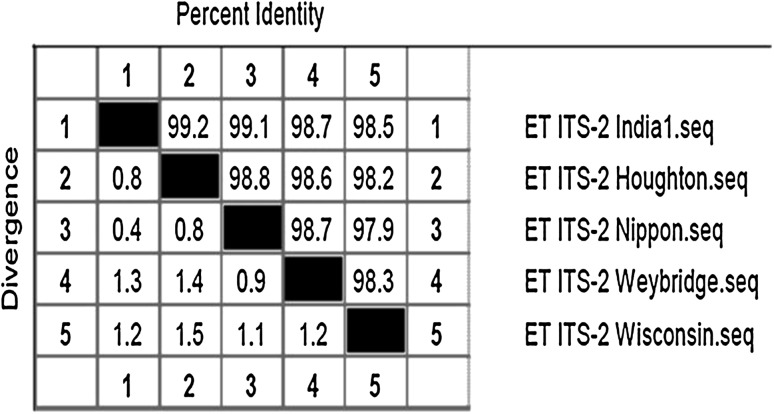
Sequence analysis of ITS-2
The partial ITS-2 sequence of 923 bp size showed high sequence homology with the other reference strains of E. tenella. The similarity analyses by CLUSTALW algorithm of Megalin programme is depicted in Fig. 6. The phylogeny of the isolates were obtained by neighbor joining tree method (N-J method), the India isolate grouped with the Houghton isolate in the same clade. The sequence was submitted to the NCBI gene bank.
Fig. 6.
Intraspecies homology: percent similarity between ITS-2
Discussion
Zhu and Keithly (1996), first reported the detailed sequence of E. tenella β-tubulin from the E. tenella Wisconsin strain. They reported a 2,225 bp gene sequence with 1,550 bp open reading frame (orf), a 146 bp 5′ untranslated region (UTR), a 315 bp 3′ UTR and 146 bases of poly A tail. In the study, a 91 bp 5′UTR was annotated after aligning with the known sequence of β-tubulin from the Houghton strain. Since the gene was partially sequenced, it covers only two introns (the first intron being partial from the 5′ end) and two exons. Exon1 is complete and exon2 is partial from the 3′ end. The pairwise alignment results revealed no differential variation from the β-tubulin sequence of known strains, namely the Houghton, Wisconsin, Weybridge and Nippon2.The point mutations were present in the introns, one substitution in intron1, second substitution and single deletion in intron2. No change in exon indicates the strict conservation of structure and function of the polypeptide sequence, which is of immense importance for survival of the unicellular organisms.
The orf sequence was GC rich (60 %), similar to Cryptosporidium parvum (62 % GC content, Caccio et al. 1997) contrary to the AT rich sequence of another apicomplexan, Plasmodium falciparum (Delves et al. 1989). In E. tenella the gene contains three intron sequences (Zhu and Keithly 1996) similar to Toxoplasma gondi (Nagel and Boothroyd 1988). In P. falciparum and C. parvum there are two and one intron/s, respectively (Delves et al. 1989 and Caccio et al. 1997). Within Protista, in general, β-tubulin exists as a multigene family, attributable to various cellular functions. Beta tubulin of apicomplexan parasites, in sharp contrast is a single gene product. This is quite similar to ciliates like Paramaecium tetraurelia. The phylogenetic tree constructed by the neighbor joining method (N-J method) follows the trend of pair wise alignment distances between species. This predicts a strong homology at the structural level within the evolutionary set up. More than 93 % conserved sequence is in the C-terminal. The ciliate, P. tetraurelia β-tubulin is closely related to E. tenella and E. maxima than any other apicomplexan parasite, compared in this study. Fast et al. (2002) reported cladal clustering of alveolates (dinoflagellates, ciliates and apicomplexans) based on phylogenetic tree of β-tubulin, Hsp70 (heat shock protein 70) and Hsp90. However they claimed that the β-tubulin phylogenetic tree provides rather less conclusive results on the evolutionary aspect of alveolates.
Heijden et al. (2006) used ITS-1 sequence C-profiling as an epidemiological tool to identify a new isolate of Histomonas meleagridis from the outbreaks of clinical histomonosis in poultry farms.The isolate was morphologically dissimilar from the other known isolates. ITS-2 was used as a marker to study the genetic heterogeneity within the congeneric species of Fasciola of mainland China by Huang et al. (2004). Encephalitozoon hellem, microsporidian parasite of human beings have been ITS genotyped and intra-species variants have been confirmed (Haro et al. 2003). ITS-2 sequencing data has immense potential in field epidemiology, for studying the spatial distribution of pathogens and their spread. The Further studies on multiple genes related to antigens and housekeeping factors will help in analysis of the intra-species and interspecies position of Indian isolate 1 of E. tenella.
Acknowledgement
Authors are thankful to the Indian Council of Agriculture Research, New Delhi and the Director, IndianVeterinary Research Institute (IVRI), Izatnagar for providing necessary facilities. The financial assistance provided by DFID and BBSRC, UK in the form of CIDLID project BB/H009337(Anticoccidial vaccine development: the importance of genetic diversity and delivery strategy) and IVRI for providing institute senior research fellowship to Ph.D scholar Dr. Krishnendu Kundu.
References
- Barabona I, Soares H, Cyrne L, Penque D, Denoulet P, Rodrigues PC. Sequence of one α and two β tubulin genes of Tetrahymena pyriformis. Structural and functional relationships with the eukaryotic tubulin genes. J Mol Biol. 1988;202:365–382. doi: 10.1016/0022-2836(88)90271-9. [DOI] [PubMed] [Google Scholar]
- Beck HP, Blake D, Darde ML, Felger I, Pedraza Diaz S, Regidor-Cerrillo J, Gomez-Bautista M, Ortega-Mora ML, Putignani L, Shiels B, Tait A, Weir W. Molecular approaches to diversity of subpopulations of apicomplexan parasites. Intl J Parasitol. 2009;39:175–189. doi: 10.1016/j.ijpara.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Bhaskaran MS, Venkatesan L, Aadimoolam R, Jayagopal HT, Sriraman R. Sequence diversity of internal transcribed spacer-1 (ITS-1) of Eimeria infecting chicken and its relevance in species identification from indian field samples. Parasitol Res. 2010;106:513–521. doi: 10.1007/s00436-009-1696-2. [DOI] [PubMed] [Google Scholar]
- Caccio S, LaRosa G, Pozio E. The β-tubulin gene of Cryptosporidium parvum. Mol Biochem Parasitol. 1997;89:307–311. doi: 10.1016/S0166-6851(97)00122-9. [DOI] [PubMed] [Google Scholar]
- Conner TW, Thomson MD, Silflow CD. Structure of three β-tubulin genes of unicellular alga. Polytomelia agilis. Gene. 1989;84:345–351. doi: 10.1016/0378-1119(89)90509-X. [DOI] [PubMed] [Google Scholar]
- Delves CJ, Ridley RG, Goman M, Holloway SP, Hyde JE, Scaife JG. Cloning of a β-tubulin gene from Plasmodium falciparum. Mol Microbiol. 1989;3:1511–1519. doi: 10.1111/j.1365-2958.1989.tb00137.x. [DOI] [PubMed] [Google Scholar]
- Fast NM, Xue L, Bingham S, Keeling PJ. Re-examining alveolate evolution using multiple protein molecular phylogenies. J Eukaryot Microbiol. 2002;49:30–37. doi: 10.1111/j.1550-7408.2002.tb00336.x. [DOI] [PubMed] [Google Scholar]
- Gautam RK, Gupta SK. Drug resistance in field isolates of Eimeria tenella in poultry. Ind J Poult Sci. 2004;39:43–48. [Google Scholar]
- Gill BS, Bajwa RS. Drug resistance in field isolates of chicken from Punjab state. Ind J Parasitol. 1979;3:132–134. [Google Scholar]
- Gurr SJ, Unkles SE, Kaghorn JR. The structure and organization of nuclear genes of filamentous fungi. In: Kinghorn JR, editor. Gene structure in Eukaryotic microbes. Oxford: IRL Press; 1988. pp. 93–139. [Google Scholar]
- Haro M, del Aguila C, Fenoy S, Henriques-Gil N. Intraspecies genotypic variability of microsporidian parasite Encephlitozoon hellem. J Clin Microbiol. 2003;41(9):4166–4171. doi: 10.1128/JCM.41.9.4166-4171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WY, He B, Wang CR, Zhu XQ. Characterization of Fasciola species from mainland China by ITS-2 ribosomal DNA sequence. Vet Parasitol. 2004;120:75–83. doi: 10.1016/j.vetpar.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Kawahara A, Taira K, Nagai S, Onaga H, Onuma M, Nunoya T. Detection of five avian Eimeria species by species-specific real-time polymerase chain reaction assay. Avian Dis. 2008;52:652–656. doi: 10.1637/8351-050908-Reg.1. [DOI] [PubMed] [Google Scholar]
- Kumar S (2012) Molecular identification and prevalence of Eimeria spp. of chicken. M.V.Sc. thesis submitted in Veterinary Parasitology to the Deemed University, Indian Veterinary research Institute, Bareilly
- Lohse J, Rinder H, Gothe R, Zahler M. Validity of species status of the parasitic mite Otodectescynotis. Med Vet Entomol. 2002;16:133–138. doi: 10.1046/j.1365-2915.2002.00355.x. [DOI] [PubMed] [Google Scholar]
- Nagel SD, Boothroyd JC. The α and β-tubulins of Toxoplasma gondii are encoded by single copy genes containing multiple introns. Mol Biochem Parasitol. 1988;29:261–273. doi: 10.1016/0166-6851(88)90081-3. [DOI] [PubMed] [Google Scholar]
- Panda NC, Pradhan HK, Mitra A, Parichha SN. Drug resistance of coccidia exposed to a single type of coccidiostat. Indian Vet J. 1973;50:22–26. [Google Scholar]
- Trivinos-Lagos L, Ohmachi T, Albrightson C, Burns RG, Ennis HL, Chishom RL. The structure and organization of nuclear genes from Dictyostelium discoidium are encoded by single genes. J Cell Sci. 1993;105:903–911. doi: 10.1242/jcs.105.4.903. [DOI] [PubMed] [Google Scholar]
- Van der Heijden MJF, Landman WJM, Greve S, Peek R. Genotyping of Histomonas meleagridis isolates based on internal transcribed spacer-1 sequences. Avian Pathol. 2006;35:330–334. doi: 10.1080/03079450600815499. [DOI] [PubMed] [Google Scholar]
- Yadav A, Gupta SK. Study of resistance against some ionophores in Eimeria tenella field isolates. Vet Parasitol. 2001;102:69–75. doi: 10.1016/S0304-4017(01)00512-X. [DOI] [PubMed] [Google Scholar]
- Zhu G, Keithly JS. The beta tubulin gene of Eimeria tenella. Mol Biochem Parasitol. 1996;76:315–319. doi: 10.1016/0166-6851(95)02536-7. [DOI] [PubMed] [Google Scholar]
- Zhu G, McDougald LR. Confocal laser scanning microscopy of β tubulin and α actinin asexual stages of Eimeriatenella (Cocccidia, Apicomplexa) in vitro. Arch Protistenk. 1995;145:112–118. doi: 10.1016/S0003-9365(11)80305-2. [DOI] [Google Scholar]