Abstract
The diagnostic techniques based on polymerase chain reaction (PCR) for the detection of Schistosoma spp. DNA in stool, serum, plasma and urine has shown high sensitivity and specificity solving the problems for the low worm burdens and low transmission rates facing the routine microscopic diagnosis. Since PCR assays require efficient unbiased procedures of extraction and purification of nucleic acids. This study compared the efficiencies of simple, manual and feasible DNA extraction methods; a salting out and resin method, phenol/chloroform method to a commercial extraction kit through PCR analysis of human urine and serum samples spiked with known amounts of adult Schistosoma mansoni DNA confirmed by the application on real samples from patients. In artificially spiked urine gradient, the best mean diagnostic performance was that of salting out and resin then phenol/chloroform and last for the commercial kit. All three methods gave positive results in all tested urine samples which insures comparable high efficiency for DNA detection. In artificially spiked serum gradient, the highest mean diagnostic performance was obtained by the kit then salting out and resin and last by phenol chloroform. In patients’ urine samples the phenol/chloroform method showed the highest mean diagnostic performance followed by the resin and then the kit. Using patients’ serum samples the resin method showed equal mean diagnostic performance with the phenol/chloroform method which was higher compared to the kit. As regards sensitivity from urine samples the resin and phenol/chloroform showed equal results using artificial gradients and patients’ samples. In serum samples the resin and phenol/chloroform showed equal results using artificial gradients while the resin showed better results in patients’ samples. It is recommended to extract DNA from urine samples and to use the salting out and resin as a manual DNA extraction method from patients’ samples for the molecular diagnosis of Schistosoma mansoni infection.
Keywords: Schistosoma mansoni, Salting out resin, Phenol/chloroform, DNA extraction, Urine, Serum, PCR
Introduction
Schistosomiasis is from a global public health perspective one of the most important water-borne parasitosis and a major neglected tropical disease, with more than 200 million people infected and close to 800 million at risk (Steinmann et al. 2006). Schistosoma mansoni is endemic in 54 countries and territories in South America, the Caribbean, Africa and the Eastern Mediterranean regions including Egypt (Chitsulo et al. 2000). Although substantial progress has been made in the control of schistosomiasis mansoni in Egypt and Latin America by reducing morbidity and prevalence, transmission continues, and the disease has spread to previously non-endemic areas (Hotez et al. 2008). Although the Kato-Katz parasitological technique is currently the recommended method for diagnosis because it is a quantitative, relatively inexpensive and simple one (Katz et al. 1972; Doenhoff et al. 1993), it lacks the sensitivity to identify reliably positive cases (Kongs et al. 2001). Consequently, the prevalence is significantly underestimated hampering further progress of the control efforts (Enk et al. 2008).
As an alternative, serological tests can be applied together with or without coproscopic techniques (Doenhoff et al. 2004). Antibody detection assays are more sensitive than parasitological examination but available tests present either low sensitivity, cross-reactivity with other helminth infections or cannot distinguish between active and past infections, which are particularly important in endemic areas (Gryseels et al. 2006; Bergquist et al. 2009). The detection of circulating antigens is highly specific, but is not more sensitive than detection of eggs in areas of low endemicity (Van Lieshout et al. 1994; Dias Neto et al. 1996). New candidates for serological diagnosis of schistosomiasis are still required (Bligh et al. 2010; Zhong et al. 2010). Moreover, taking into consideration the yearly rising numbers of schistosomiasis infected travelers, who in general show very low infection intensity in its early stage, more sensitive methods for diagnosis are urgently needed (Rabello and Enk 2005; Bergquist et al. 2009).
The diagnostic techniques based on polymerase chain reaction (PCR) for the detection of Schistosoma spp. DNA in stool, serum (Pontes et al. 2002, 2003; Ten Hove et al. 2008; Gomes et al. 2009, 2010), plasma (Wichmann et al. 2009) and urine samples (Sandoval et al. 2006a) has shown high sensitivity and specificity giving a promising solution. PCR methods are used to improve the direct detection of Schistosoma. These tests involve the extraction of DNA from eggs prior to PCR amplification. Unfortunately, only a small volume of sample can be processed for DNA extraction, and is dependent on chance whether the processed sample contains eggs or not (Sandoval et al. 2006b; Pontes et al. 2003). In schistosomiasis, there is a huge parasite turnover, liberating DNA from decaying parasites which would reach the blood and be partially excreted into urine (Umansky and Tomei 2006). These nucleic acids could be used for development of clinical diagnostic PCR-based assays (Melkonyan et al. 2008).
Multicellular parasites like Schistosoma contain DNA copies in excess over parasite count. So, it might be possible to find cell-free DNA circulating in the blood which can be used to diagnose schistosomiasis. In contrast to eggs in stool or urine, the circulating DNA would be equally distributed throughout blood of the patient, resolving the issue of random sampling that spoils clinical sensitivity of classical detection methods (Wichmann et al. 2009). Hamburger et al. (1991) described a 121-base pair (bp) tandem repeat DNA sequence present in 12 % of S. mansoni genome. This sequence has been successfully used in the diagnosis of human infection using fecal (Pontes et al. 2002), serum (Sadek et al. 2008), plasma (Wichmann et al. 2009) and urine samples (Enk et al. 2010, 2012).
An efficient unbiased method of DNA extraction that yields pure, high quality DNA is crucial for the success of PCR and sequencing reactions, and the subsequent treatment of disease (McOrist et al. 2002). Many DNA extraction methods have been described in literature. The salting out and resin method for DNA extraction from urine samples is a simple and inexpensive method (Enk et al. 2010, 2012). Also, the phenol/chloroform extraction procedure for DNA which minimizes the inhibition of nucleic acid amplification by PCR in clinical samples (Bergallo et al. 2006), has been traditionally used to obtain nucleic acids from cells and microorganisms in addition to separation and purification from contaminating materials (Wiedbrauk et al. 1995).
These extraction methods utilize easily urine and serum samples as DNA source, in combination with the high sensitivity of PCR technique, constituting a promising diagnostic tool to overcome the difficulties of detecting schistosomiasis in areas of low transmission or in individual cases with low worm burden (Bergallo et al. 2006).
In Egypt there is an urge for evaluating the different DNA extraction procedures not only because there are limited economic and diagnostic resources, but also the use of non toxic materials during the extraction process and its easy handling, may, in future, make these techniques easily applicable in survey of poor areas in comparison to the expensive kits.
The aim of this study was to compare simple, manual and feasible DNA extraction methods; a salting out and resin method versus a phenol/chloroform extraction compared to a commercial extraction kit through PCR analysis of urine and serum samples spiked with known amounts of adult S. mansoni DNA and to compare the applicability on real samples from patients.
Materials and methods
DNA extraction from adult Schistosoma mansoni
Adult worms (obtained from Tudor Bilharz Research Institute, Giza, Egypt) were individually vacuum-dried for 15 min in a Speedvac evaporator. Next, 20 μl of NaOH (250 mM) was added. After a 15 min incubation period at 25 °C, the tubes were heated at 99 °C for 2 min. Then, 10 μl HCl (250 mM), 5 μl of Tris–HCl (500 mM) and 5 μl Triton X-100 (2 %) were added and a second heat shock at 99 °C for 2 min was performed. The products were stored at −20 °C (Beltran et al. 2008). Commercial DNA extraction was done using Pure link genomic DNA extraction kit for purification of genomic DNA (Invitrogen life technologies—Germany) according to the manufacturer’s instructions. Briefly, cells were resuspended in 180 μl Pure Link Genomic digestion buffer, 20 μl proteinase K was added, mixed well by brief vortexing then incubated at 55 °C for 45 min. 20 μl RNAse was added to the lysate, mixed well by brief vortexing and incubated at room temperature for 2 min, then 200 μl Pure Link Genomic lysis/binding buffer was added and briefly vortexed to obtain homogenous solution. 200 μl of absolute ethanol was added to the lysate and vortexed for 5 s. About 640 μl lysate was added to the Pure Link Spin column and centrifuged for 1 min at room temperature. Washing was done twice to eliminate unwanted materials. The DNA was eluted with 100 μl Pure Link Genomic elution buffer and stored at −20 °C till use.
Preparation of artificially spiked urine and serum samples
Fifty milliliters of fresh urine from a healthy individual were treated with EDTA to a final concentration of 40 mM (Milde et al. 1999). Serum samples negative for schistosomiasis by indirect haemagglutination test (IHAT) were obtained from healthy volunteers and each 1 ml of serum was diluted in 1 ml of PBS. 30 ml of the urine sample and diluted serum samples were artificially spiked with 120 ng of adult S. mansoni DNA (Enk et al. 2010, 2012). Artificially spiked samples were adjusted to six consecutive DNA dilutions to form a gradient of; 4, 800, 160, 32, 6.40 and 1.28 pg/ml to have a final of six artificially infected urine samples and six artificially infected serum samples which were processed to evaluate each extraction method’s efficiency by conventional PCR.
Collection of samples from patients
Twenty patients were selected from the Internal Medicine Department, Ain Shams University hospitals with given history of infection with S. mansoni. They were subjected to: stool examination by Kato-Katz method (Katz et al. 1972); urine examination; antibody detection in serum for schistosomiasis by the IHAT (Bilharzios Fumouze Diagnostics/SERFIB, France). All patients were suffering active intestinal schistosomiasis showing S. mansoni eggs in their stools. Informed consent was obtained from the study subjects using standard guidelines. The study protocol was approved by the Scientific-Ethics Committee of Faculty of Medicine Ain Shams University. All participants provided one morning fresh urine sample. 5 ml of blood were collected from each subject and centrifuged at 760×g at 4 °C for 10 min and the obtained serum samples were stored at −70 °C until use.
Methods of DNA extraction from artificially spiked and patients’ urine and serum samples
Salting out and resin method
All aliquots (the gradients and patients samples) were heated at 100 °C, in a water bath for 10 min. After that, 5 M NaCl, in a volume of 1/10 of the sample volume was added to each tube. The tubes were shaken vigorously for 15 s, placed on ice for 1 h and centrifuged for 10 min at 4,000 rpm. The supernatant was transferred to another tube, shaken vigorously for 15 s and centrifuged for 10 min at 4,000 rpm. Again the supernatant was transferred to another clean tube, and twice the sample volume of absolute ethanol was added. The DNA was then precipitated at −20 °C for at least 2 h. The DNA strand was removed with a pipette, transferred to a 0.5 mL microcentrifuge tube and washed in 200 μl 70 % ethanol. The tubes were centrifuged again for 20 min at 14,000 rpm. The pellets were dried and resuspended in 100 μl of DNAse free water and 100 μl of InstaGene matrix® resin (BioRad). Samples were incubated at 56 °C for 30 min then at 100 °C for 8 min, vortexed at high speed for 10 s, and centrifuged at 14,000 rpm for 3 min. The supernatant transferred to a new tube and used as template in PCR reactions (Enk et al. 2010, 2012).
Phenol/chloroform method
A home-made procedure based on the classical phenol/chloroform extraction method was applied (Bergallo et al. 2006). A total of 250 μl of each sample was added to 250 μl of buffer-saturated phenol/chloroform/isoamilalcool (pH 7.5–7.8) and 25 μl of 3 M sodium acetate pH 5, vortexed for 1 min, incubated in ice for 15 min, and centrifuged for 10 min at 12,000 rpm at room temperature. The aqueous layer was transferred to a 1.5 ml screw-cap plastic tube containing 250 μl of isopropanol and incubated at −20 °C for 5 min. The sample was centrifuged (5 min, 12000 rpm at 4 °C) in order to precipitate DNA. The supernatant was removed, and the pellet resuspended in 500 μl of 70 % ethanol, vortexed and centrifuged for 2 min at 13,000 rpm at room temperature. The supernatant was removed again and the remaining ethanol eliminated by a quick centrifuge spin. The pellet was air-dried in a half-open tube for at least 30 min, suspended in 20 μl of TE (10 mM TrisCl, 1 mM EDTA pH 8) by vortexing, and stored at −20 °C prior to use.
Commercial DNA extraction
DNA extraction was done using Pure link genomic DNA extraction kit for purification of genomic DNA (Invitrogen life technologies—Germany).
Polymerase chain reaction
PCR was applied on all the samples from the gradients and patients. PCR was carried out using a pair of primers, directed to a 121 bp repetitive fragment designed by Pontes et al. (2002), GoTaq DNA polymerase and STR 10× buffer (Promega). Into each reaction tube were added 1 μl of 10× buffer, 0.1 μg/μl of 1× BSA, 0.8 U of Taq DNA polymerase, 0.5 pmol of each forward and reverse primers, 4 μl of DNA template and enough water to a final volume of 10 μl. As PCR positive controls of 1 ng of S. mansoni adult worm DNA was used as template. PCR negative control containing all elements of the reaction mixture and water instead of DNA were also included in each PCR assay as surveillance for contamination. A total of 40 cycles of amplification were conducted using a 30 s denaturing step at 95 °C, 30 s annealing at 65 °C and 30 s extension at 72 °C. A 20 μl sample of each amplification product was electrophoresed on 8 % PAGE. The amplification products were quantified using a digital imaging system (Quantity One 1-D Analysis Software; BIO-RAD, USA). Each of the three DNA extraction methods and PCR was applied twice for each sample.
Statistical analysis
All results are presented as mean and standard deviation (SD) values. Categorical results are presented as numbers of cases and percentages. Continuous variables were compared between independent groups using Mann/Whitney U/− test, depending on the distribution of raw data. In cases in which the data was paired, the Wilcoxon signed rank or paired t test was used. Categorical variables were compared using the fisher exact test. Significance level of P < 0.05 was used in all tests. All statistical procedures were carried out using SPSS version15 for Windows (SPSS Inc, Chicago, IL, USA).
Results
In artificial urine gradient, the mean diagnostic performance of salting out and resin was 149.8 ± 53.9, for phenol/chloroform was 142.8 ± 42.6 and for commercial kit was 132 ± 24.9. All three methods gave positive results in all tested urine samples (Table 1).
Table 1.
The diagnostic performance of the three methods of extraction using urine versus serum artificially spiked gradients
| Parameter | Resin | Phenol chloroform | Kit | ||||
|---|---|---|---|---|---|---|---|
| Urine | Serum | Urine | Serum | Urine | Serum | ||
| Mean density of PCR product ± SD | 149.8 ± 53.9 | 74.7 ± 58 | 142.8 ± 42.6 | 55.2 ± 60.5 | 132 ± 24.9 | 93.3 ± 17.2 | |
| Samples (n = 6) | |||||||
| Positive | 6 | 5 | 6 | 5 | 6 | 6 | |
| Negative | 0 | 1 | 0 | 1 | 0 | 0 | |
| % of positivity among samples | 100 | 83.4 | 100 | 83.4 | 100 | 100 | |
| P value | *0.042 | *0.018 | *0.011 | ||||
Paired t test
* P < 0.05 Significant (S)
In artificial serum gradient, the mean diagnostic performance of salting out and resin was 74.7 ± 58.0, for phenol chloroform was 55.2 ± 60.5 and for commercial kit was 93.3 ± 17.2. Results showed that salting out and resin and phenol chloroform were positive in 83.4 % of serum samples, while commercial kit showed 100 % positivity (Table 1).
Paired t test showed that there was a significant difference (P < 0.05) between artificial urine and serum gradient by the three methods as regards the mean diagnostic performance. In each of the three tested methods of extraction, the mean diagnostic performance of urine samples was significantly higher compared to that of serum samples (Table 1).
In patients’ urine samples the phenol chloroform method showed the highest mean diagnostic performance results (128.1 ± 68.4) in comparison to the resin (105.3 ± 54.3) and the kit (100.9 ± 45.4) (Table 2). Using Wilcoxon signed rank, the urine samples showed a highly significant difference (P < 0.01) between the resin and phenol chloroform while a non significant difference was found between each of the resin and the kit, and the phenol chloroform and the kit (Table 3).
Table 2.
The diagnostic performance among the three methods of extraction in patients’ urine versus serum samples
| Extraction method | Sample | P value | |
|---|---|---|---|
| Urine (No = 20) | Serum (No = 20) | ||
| Mean ± SD | Mean ± SD | ||
| Resin | 105.3 ± 54.3 | 61.1 ± 51.2 | **0.0001 |
| Phenol chloroform | 128.1 ± 68.4 | 61.0 ± 51.2 | **0.0001 |
| Kit | 100.9 ± 45.4 | 58.4 ± 49.2 | **0.003 |
Mann–Whitney test
** P < 0.01 Highly significant (HS)
Table 3.
Comparison between the three extraction methods on patients’ urine samples
| Extraction method | Mean ± SD (No = 20) | P value |
|---|---|---|
| Resin | 105.3 ± 54.3 | **0.001 |
| Phenol chloroform | 128.1 ± 68.4 | |
| Resin | 105.3 ± 54.3 | 1.00 |
| Kit | 100.9 ± 45.4 | |
| Phenol chloroform | 128.1 ± 68.4 | 0.108 |
| Kit | 100.9 ± 45.4 |
Wilcoxon signed rank. P > 0.05 Non significant (NS)
** P < 0.01 Highly significant (HS)
Using patients ‘serum samples the resin method showed equal mean diagnostic performance with the phenol chloroform method (61.1 ± 51.2) while the kit showed lower mean diagnostic performance (58.4 ± 49.2) (Table 4). Using Wilcoxon signed rank, there was a significant difference (P < 0.05) between phenol chloroform and the kit while a non significant difference was obtained in comparing resin and phenol chloroform (Table 4).
Table 4.
Comparison between the three extraction methods on patients’ serum samples
| Extraction method | Mean ± SD (No = 20) | P value |
|---|---|---|
| Resin | 61.1 ± 51.2 | 0.541 |
| Phenol chloroform | 61.00 ± 51.2 | |
| Resin | 61.1 ± 51.2 | 0.054 |
| Kit | 58.4 ± 49.2 | |
| Phenol chloroform | 61.00 ± 51.2 | *0.046 |
| Kit | 58.4 ± 49.2 |
Wilcoxon signed rank. P > 0.05 Non significant (NS)
* P < 0.05 Significant (S)
Fisher exact test revealed a significant difference between urine and serum samples using resin method and a highly significant difference using phenol chloroform method as regard positivity (Table 5). In each of the three tested methods, patient’s urine samples gave significantly higher results compared to serum samples as regards mean diagnostic performance (Table 2).
Table 5.
Comparison between patients’ urine and serum among the three extraction methods
| Extraction method | Sample | P value | ||||
|---|---|---|---|---|---|---|
| Urine (No = 20) | Serum (No = 20) | |||||
| No | % | No | % | |||
| Resin | ||||||
| Negative | 0 | 0 | 5 | 25 | *0.047 | |
| Positive | 20 | 100 | 15 | 75 | ||
| Phenol chloroform | ||||||
| Negative | 0 | 0 | 8 | 40 | **0.003 | |
| Positive | 20 | 100 | 12 | 60 | ||
| Kit | ||||||
| Negative | 0 | 0 | 0 | 0 | ||
| Positive | 20 | 100 | 20 | 100 | ||
Fisher exact test
* P < 0.05 Significant (S)
** P < 0.01 Highly significant (HS)
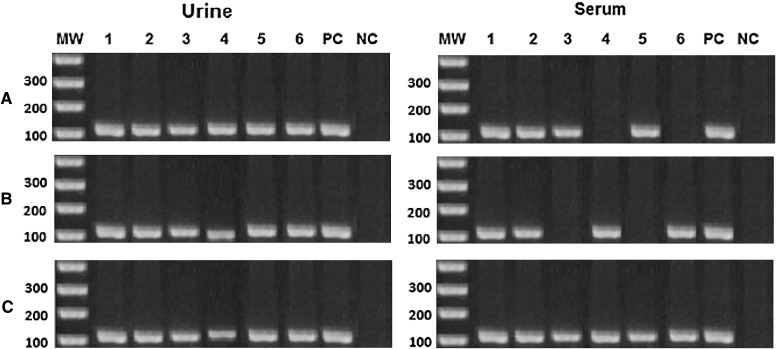
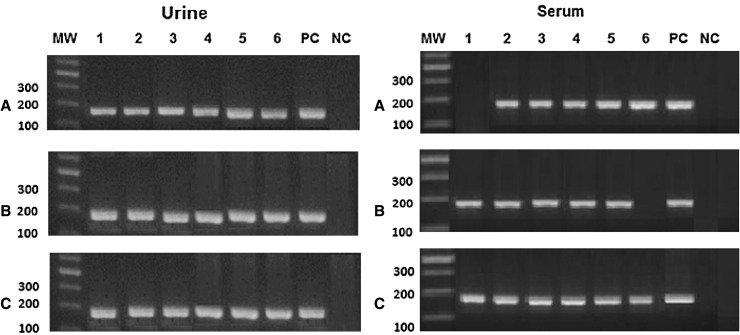
Applying the 3 methods of extraction on artificially spiked gradients, the sensitivity among urine samples was 100 % while among serum samples it was 83.4 % for resin and phenol chloroform methods and 100 % for the kit (Table 1). Using patients′ urine samples the sensitivity was also 100 % from the three methods while serum samples showed highest outcome from the kit (100 %), then the resin method (75 %) and last the phenol chloroform (60 %) (Table 5, Fig. 2). The reproducibility of results was confirmed by repeating the three methods of extraction twice giving same outcome. Concerning the artificially spiked serum samples the resin showed good results at low concentrations only starting from 800 pg/ml but no results were obtained at the higher concentration of 4 ng/ml. Also, no outcome was obtained from the phenol/chloroform extraction method at the least (1.28 pg) concentration of the samples (Fig. 1).
Fig. 2.
PCR product from urine and serum samples of six patients (lanes 1–6), PC positive control, NC negative control, A salting out and resin method, B phenol/chloroform method and C kit
Fig. 1.
PCR product from urine and serum gradients. MW molecular weight marker, lanes 1–6 six consecutive DNA dilutions of; 4, 800, 160, 32, 6.40 and 1.28 pg/ml. PC positive control, NC negative control, A salting out and resin method, B phenol/chloroform method and C kit
Discussion
The underestimated schistosomiasis mansoni in low transmission areas, emphasizes the urgent need of an accurate diagnostic tool capable of detecting infections in the early phase which would be of great value in permitting early treatment that could prevent the pathology associated with chronic infections. S. mansoni DNA in urine of mice had been detected by PCR very early after infection (Sandoval et al. 2006a, Oliveira et al. 2010).
Given these findings, it is probable that the PCR can detect infection in humans during the larval stage or at least before parasite eggs can be found in the stool. Pontes et al. (2002) reported high sensitivity of PCR in the detection of free circulating S. mansoni DNA in serum of two infected patients with active intestinal schistosomiasis. Moreover, Sadek et al. (2008), Xia et al. (2009) and Kato-Hayashi et al. (2010) proved that PCR was highly valuable in the diagnosis of early experimentally active intestinal schistosomiasis with increase in the density of the specific band as the infection progressed.
DNA fragments from cells that have died throughout the body appear in the urine, presenting relatively low molecular size fragments (100–200 bp) which should be considered when deciding methods of DNA extraction and PCR design. When PCR is used for amplification and detection of DNA sequences, as in our case of S. mansoni, the application of a small sized amplicon significantly enhances sensitivity. Therefore, a pair of primers targeting a sequence of 121 bp, which has been previously applied extensively in S. mansoni diagnostics (Pontes et al. 2002; Gomes et al. 2009, 2010), was used in the present study.
The method for detection of S. mansoni in human feces and serum (Pontes et al. 2002) was based on amplification of a highly repeated DNA sequence (Hamburger et al. 1998). The high sensitivity of this assay is certainly due to an unusual high copy number (600,000/cell) of target sequence, which comprises at least 12 % of S. mansoni genome (Hamburger et al. 1991). This enables the detection of fractions of a single S. mansoni individual cell, instead of the egg needed for microscopic identification.
In our study we aim to detect the DNA regardless to the amount of eggs shed from the patient as the bigger future goal of the work is to detect the early prepatent infection using the best simple extraction method were no eggs may be present in endemic area of low prevalence. Gradients were used to control the amount of DNA in the sample and assess the extraction efficiency and sensitivity. Patients were chosen to pass eggs here to ensure the presence of DNA for the mere qualification of the three methods of extraction.
Although many different extraction methods for PCR have been described, there have been relatively few studies comparing them. In our study, three methods of extraction were chosen on basis of being sensitive, specific, reproducible, rapid, and simple allowing nucleic acids purification from urine and serum samples in a routine setting. A technique recently described based on a salting-out and resin procedure, was applied for its high analytical sensitivity, simplicity and low cost. It proved to be sufficient for following molecular analysis of urine samples from an endemic area (Enk et al. 2010, 2012). Also, the phenol/chloroform method was chosen based on being the traditional extraction procedure of DNA from plasmids or clinical samples and widely used in many research and diagnostic laboratories (Sambrook et al. 1989).
The reagent cost for both methods is much lower compared to commercial nucleic acid extraction kits. Also, these methods are expected to reduce the concentration of residual proteins and membrane components potentially inhibiting the enzymatic activity of Taq polymerase in comparison to other manual extraction procedures (Bergallo et al. 2006; Enk et al. 2010, 2012). In our study, the commercially available kit was included as a reliable reference.
The artificially spiked samples were used to qualify the sensitivity of each extraction method used, which may later on be applied in endemic areas where the diagnosis should cover any stage of infection and the DNA appears from the turnover and tegument shedding from the dwelling adults. The application of the methods on samples from patients well known to suffer infection and shedding eggs was applied to qualify the methods on a real basis of infection.
Using artificial urine gradient, PCR results showed that the commercial kit was able to detect the DNA in all the different concentrations and that was used as reference method for extraction. Also an outcome yield of DNA covering the whole gradient concentrations was obtained from the other two extraction methods, thus revealing comparable levels of efficiency. The resin extraction procedure showed the highest diagnostic performance in detecting the maximal DNA content from the different gradient concentrations, considering the mean density of the PCR product, followed by the phenol/chloroform then the kit. Our results goes with Enk et al. (2008) who proved that the salting out and resin extraction procedure to be reproducible up to 100 % and sensitive in detection up to a concentration of 1.28 pg DNA/ml in artificially spiked urine samples. A different outcome was obtained from urine samples of the patients; the phenol/chloroform showed the highest diagnostic performance then the resin and the kit. This goes with Bergallo et al. (2006) who demonstrated that the phenol/chloroform extraction method was the most efficient one compared to many other extraction methods used from human urine samples. Comparing the three methods of extraction; the resin, phenol chloroform and kit showed no difference as regards positivity in urine samples of the patients.
Concerning the artificially spiked serum samples, the kit showed the highest diagnostic performance followed by the resin then the phenol chloroform. The kit showed the highest efficiency compared to the other two extraction methods in covering the whole gradients. The resin showed good results at low concentrations only starting from 800 pg/ml but no results were obtained at the higher concentrations of 4 ng/ml. The resin beads if resembled like a sieve may be hindered by the high concentration of DNA which may actually cause its blockage (Klein et al. 1997). Also, no outcome was obtained from the phenol/chloroform extraction method at the least (1.28 pg) concentration of the samples which may highlight its lesser efficiency. Again a different outcome was obtained using serum samples of the patients, the resin and phenol/chloroform methods showed equal diagnostic performance compared to a lower diagnostic performance from the kit. This may add a value for the two methods in diagnosis using serum samples from patients. The different outcome from serum samples as regard sensitivity obtained by the two manual methods may be attributed to the different DNA content in the patients′ serum.
Presence of inhibitory factors in patients’ serum may hinder the PCR outcome which may explain the different results obtained in comparison to the gradient as regard sensitivity. The artificial gradients are considered a preparatory step for showing the role and efficacy of extraction methods in diagnosis. Also, it can be used to quantify DNA level in a patient which may reflect the stage of infection if spotted on artificial gradients standard curves.
In the three tested extraction methods using artificially spiked and real samples from patients, urine outstand serum as regards diagnostic performance and positivity. This fact was supported by Enk et al. (2012) who approved the operational advantage of urine sample to be a promising alternative for the diagnosis of schistosomiasis mansoni .
It is recommended to extract DNA from urine samples and to use the salting out and resin as a manual DNA extraction method from patients’ samples for the molecular diagnosis of S. mansoni infection. There is still room for cost reduction by optimizing the extraction and amplification procedures of the system for poor endemic areas. Further studies are needed to verify the diagnosis of early schistosomiasis mansoni infection using the resin as a manual method of extraction.
References
- Beltran S, Galinier R, Allienne JF, Boissier J. Cheap, rapid and efficient DNA extraction method to perform multilocus microsatellite genotyping on all Schistosoma mansoni stages. Mem Inst Oswaldo Cruz. 2008;103:501–503. doi: 10.1590/S0074-02762008000500017. [DOI] [PubMed] [Google Scholar]
- Bergallo M, Costa C, Gribaudo G, Tarallo S, Baro S, Negro Ponzi A, Cavallo R. Evaluation of six methods of extraction and purification of viral DNA from urine and serum samples. New Microbiol. 2006;29:111–119. [PubMed] [Google Scholar]
- Bergquist R, Johansen MV, Utzinger J. Diagnostic dilemmas in helminthology: what tools to use and when. Trends Parasitol. 2009;25:151–156. doi: 10.1016/j.pt.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Bligh J, Schramm G, Chiodini PL, Doenhoff MJ. Serological analysis of the outcome of treatment of Schistosoma mansoni infections with praziquantel. Ann Trop Med Parasitol. 2010;104:511–520. doi: 10.1179/136485910X12786389891245. [DOI] [PubMed] [Google Scholar]
- Chitsulo L, Engels D, Montresor A, Savioli L. The global status of schistosomiasis and its control. Acta Trop. 2000;77:41–51. doi: 10.1016/S0001-706X(00)00122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias Neto E, Harrop R, Correia-Oliveira R, Pena SD, Wilson RA, Simpson AJ. The Schistosome genome project: RNA arbitrarily primed PCR allows the accelerated generation of expressed sequence tags. Mem Inst Oswaldo Cruz. 1996;91:655–657. doi: 10.1590/S0074-02761996000500020. [DOI] [PubMed] [Google Scholar]
- Doenhoff MJ, Butterworth AE, Hayes RJ, Sturrock RF, Ouma JH, Koech D, Prentice M, Bain J. Seroepidemiology and serodiagnosis of schistosomiasis in Kenya using crude and purified egg antigens of Schistosoma mansoni in ELISA. Trans R Soc Trop Med Hyg. 1993;87:42–48. doi: 10.1016/0035-9203(93)90415-M. [DOI] [PubMed] [Google Scholar]
- Doenhoff MJ, Chiodini PL, Hamilton JV. Specific and sensitive diagnosis of Schistosome infection: can it be done with antibodies. Trends Parasitol. 2004;20:35–39. doi: 10.1016/j.pt.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Enk MJ, Lima AC, Drummond SC, Schall VT, Coelho PM. The effect of the number of stool samples on the observed prevalence and the infection intensity with Schistosoma mansoni among a population in an area of low transmission. Acta Trop. 2008;108:222–228. doi: 10.1016/j.actatropica.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Enk MJ, Oliveira e Silva G, Rodrigues NB. A salting out and resin procedure for extracting Schistosoma mansoni DNA from human urine samples. BMC Res Notes. 2010;3:115. doi: 10.1186/1756-0500-3-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enk MJ, Oliveira e Silva G, Rodrigues NB. Diagnostic accuracy and applicability of a PCR system for the detection of Schistosoma mansoni DNA in human urine samples from an endemic area. PLoS One. 2012;7(6):e38947. doi: 10.1371/journal.pone.0038947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LI, Marques LH, Enk MJ, Coelho PM, Rabello A. Further evaluation of an updated PCR assay for the detection of Schistosoma mansoni DNA in human stool samples. Mem Inst Oswaldo Cruz. 2009;104:1194–1196. doi: 10.1590/S0074-02762009000800021. [DOI] [PubMed] [Google Scholar]
- Gomes LI, Marques LH, Enk MJ, De Oliveira MC, Coelho PM, et al. Development and evaluation of a sensitive PCR-ELISA system for detection of Schistosoma infection in feces. PLoS Negl Trop Dis. 2010;4:e664. doi: 10.1371/journal.pntd.0000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet. 2006;368:1106–1118. doi: 10.1016/S0140-6736(06)69440-3. [DOI] [PubMed] [Google Scholar]
- Hamburger J, Turetski T, Kapelle I, Deresiewicz R. Highly repeated short DNA sequences in the genome of Schistosoma mansoni recognized by a species-specific probe. Mol Biochem Parasitol. 1991;44:73–80. doi: 10.1016/0166-6851(91)90222-R. [DOI] [PubMed] [Google Scholar]
- Hamburger J, Xin XY, Ramzy RM, Jourdane J, Ruppel A. A polymerase chain reaction for detection snails infected with bilharzia parasites (Schistosoma mansoni) from very early prepatency. Am J Trop Med Hyg. 1998;59(6):872–876. doi: 10.4269/ajtmh.1998.59.872. [DOI] [PubMed] [Google Scholar]
- Hotez PJ, Bottazz ME, Franco-Paredes C, Ault SK, Periago MR. The neglected tropical diseases of Latin America and the Caribbean: a review of disease burden and distribution and a roadmap for control and elimination. PLoS Negl Trop Dis. 2008;2:e300. doi: 10.1371/journal.pntd.0000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Hayashi N, Kirinoki M, Iwamura Y, Kanazawa T, Kitikoon V, Matsuda H, Chigusa Y. Identification and differentiation of human schistosomes by polymerase chain reaction. Exp Parasitol. 2010;124(3):325–329. doi: 10.1016/j.exppara.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Katz N, Chaves A, Pellegrino J. A simple device for quantitative stool thick smear technique in Schistosomiasis mansoni. Rev Inst Med Trop. 1972;14:337–340. [PubMed] [Google Scholar]
- Klein A, Barsuk R, Dagan S, Nusbaum O, Shouval D, Galun E. Comparison of methods for extraction of nucleic acid from haemolytic serum for PCR amplification of hepatitis B virus DNA sequences. J Clin Microbiol. 1997;35:1897–1899. doi: 10.1128/jcm.35.7.1897-1899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongs A, Marks G, Verle P, Van der Stuyf P. The unreliability of the Kato-Katz technique limits its usefulness for evaluating S. mansoni infections. Trop Med Int Health. 2001;6:163–169. doi: 10.1046/j.1365-3156.2001.00687.x. [DOI] [PubMed] [Google Scholar]
- McOrist AL, Jackson M, Bird AR. A comparison of five methods for extraction of bacterial DNA from human faecal samples. J Microbiol Methods. 2002;50:131–139. doi: 10.1016/S0167-7012(02)00018-0. [DOI] [PubMed] [Google Scholar]
- Melkonyan HS, Feaver WJ, Meyer E, Scheinke V, Shekhtman EM, et al. Transrenal nucleic acids: from proof of principle to clinical tests. Ann NY Acad Sci. 2008;1137:73–81. doi: 10.1196/annals.1448.015. [DOI] [PubMed] [Google Scholar]
- Milde A, Haas-Rochholz H, Kaatsch HJ. Improved DNA typing of human urine by adding EDTA. Int J Legal Med. 1999;112:209–210. doi: 10.1007/s004140050237. [DOI] [PubMed] [Google Scholar]
- Oliveira E, Silva G, Enk MJ, Rodrigues NB (2010) The detection of schistosoma mansoni transrenal DNA in urine samples using the murine model. Annais of the 12th international symposium on schistosomiasis
- Pontes LA, Dias-Neto E, Rabello A. Detection by polymerase chain reaction of Schistosoma mansoni DNA in human serum and feces. Am J Trop Med Hyg. 2002;66:157–162. doi: 10.4269/ajtmh.2002.66.157. [DOI] [PubMed] [Google Scholar]
- Pontes LA, Oliveira MC, Katz N, Dias-Neto E, Rabello A. Comparison of a polymerase chain reaction and the Kato-Katz technique for diagnosing infection with Schistosoma mansoni. Am J Trop Med Hyg. 2003;68:652–656. [PubMed] [Google Scholar]
- Rabello AL, Enk MJ (2005) Progress towards the detection of schistosomiasis. Report of the scientific working group on schistosomiasis 67–71
- Sadek SM, El Missiry AM, El-Tonsy MM, Hussein HM, El Asmar MF, Ahmed SA. Detection of Schistosoma mansoni DNA in human blood as a diagnostic method for schistosomiasis. PUJ. 2008;1(1):31–36. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sandoval N, Siles-Lucas M, Lopez AJ, Perez-Arellano JL, Garate T, et al. Schistosoma mansoni: a diagnostic approach to detect acute schistosomiasis infection in a murine model by PCR. Exp Parasitol. 2006;114:84–88. doi: 10.1016/j.exppara.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Sandoval N, Siles-Lucas M, Perez-Arellano JL, Carranza C, Puente S, et al. A new PCR-based approach for the specific amplification of DNA from different Schistosoma species applicable to human urine samples. Parasitology. 2006;133:581–587. doi: 10.1017/S0031182006000898. [DOI] [PubMed] [Google Scholar]
- Steinmann P, Keiser J, Bos R, Tanner M, Utzinger J. Schistosomiasis and water resources development: systematic review, meta-analysis, and estimates of people at risk. Lancet Infect Dis. 2006;6:411–425. doi: 10.1016/S1473-3099(06)70521-7. [DOI] [PubMed] [Google Scholar]
- Ten Hove RJ, Verweij JJ, Vereecken K, Polman K, Dieye L, et al. Multiplex real-time PCR for the detection and quantification of Schistosomamansoni and S. haematobium infection in stool samples collected in northern Senegal. Trans R Soc Trop Med Hyg. 2008;102:179–185. doi: 10.1016/j.trstmh.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Umansky SR, Tomei LD. Transrenal DNA testing: progress and perspectives. Expert Rev Mol Diagn. 2006;6:153–163. doi: 10.1586/14737159.6.2.153. [DOI] [PubMed] [Google Scholar]
- Van Lieshout L, De Jonge N, El-Masry N, Mansour MM, Bassily S, Krijger FW, Deelder AM. Monitoring the efficacy of different doses of praziquantel by quantification of circulating antigens in serum and urine of schistosomiasis patients. Parasitology. 1994;108:519–526. doi: 10.1017/S0031182000077386. [DOI] [PubMed] [Google Scholar]
- Wichmann D, Panning M, Quack T, Kramme S, Burchard GD, et al. Diagnosing schistosomiasis by detection of cell-free parasite DNA in human plasma. PLoS Negl Trop Dis. 2009;3:e422. doi: 10.1371/journal.pntd.0000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedbrauk DL, Werner JC, Drevon AM. Inhibition of PCR by aqueous and vitreous fluids. J Clin Microbiol. 1995;33:2643–2646. doi: 10.1128/jcm.33.10.2643-2646.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia CM, Rong R, Lu ZX, Shi CJ, Xu J, Zhang HQ, Gong W, Luo W. Schistosomajaponicum: a PCR assay for the early detection and evaluation of treatment in a rabbit model. Exp Parasitol. 2009;121(2):175–179. doi: 10.1016/j.exppara.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Zhong ZR, Zhou HB, Li XY, Luo QL, Song XR, et al. Serological proteome-oriented screening and application of antigens for the diagnosis of Schistosomiasis japonica. Acta Trop. 2010;116:1–8. doi: 10.1016/j.actatropica.2010.04.014. [DOI] [PubMed] [Google Scholar]