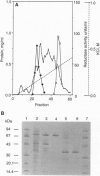
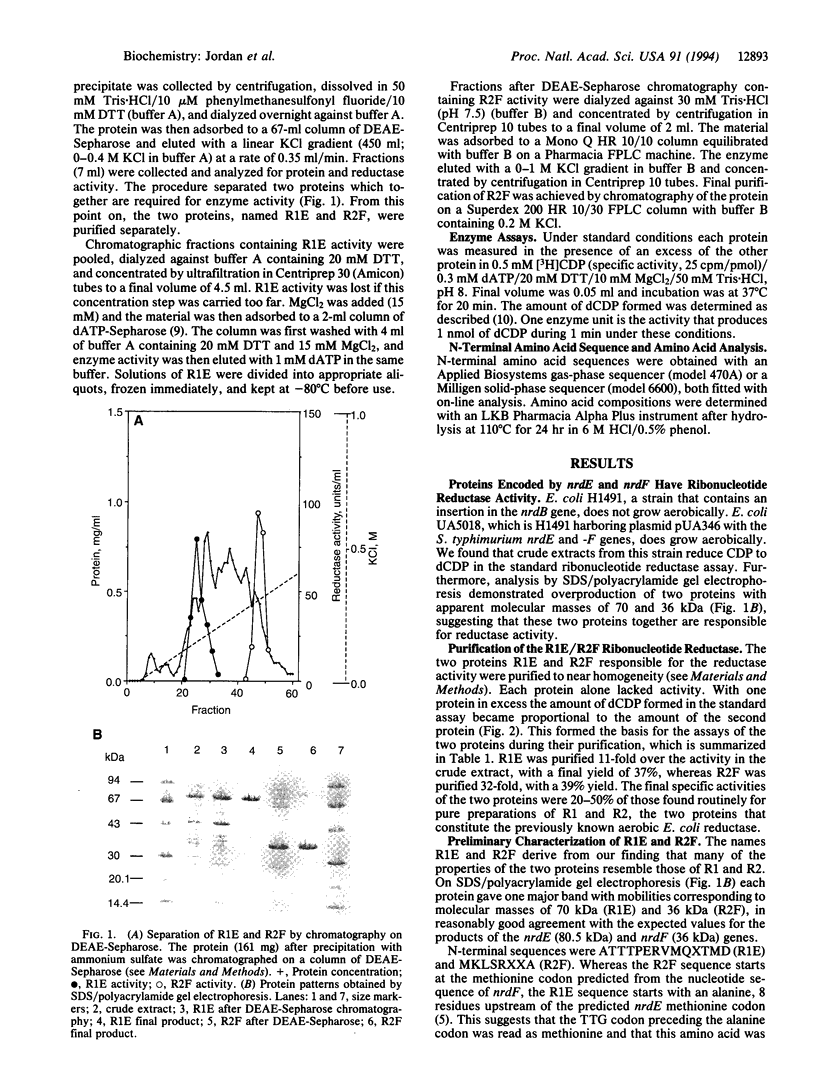
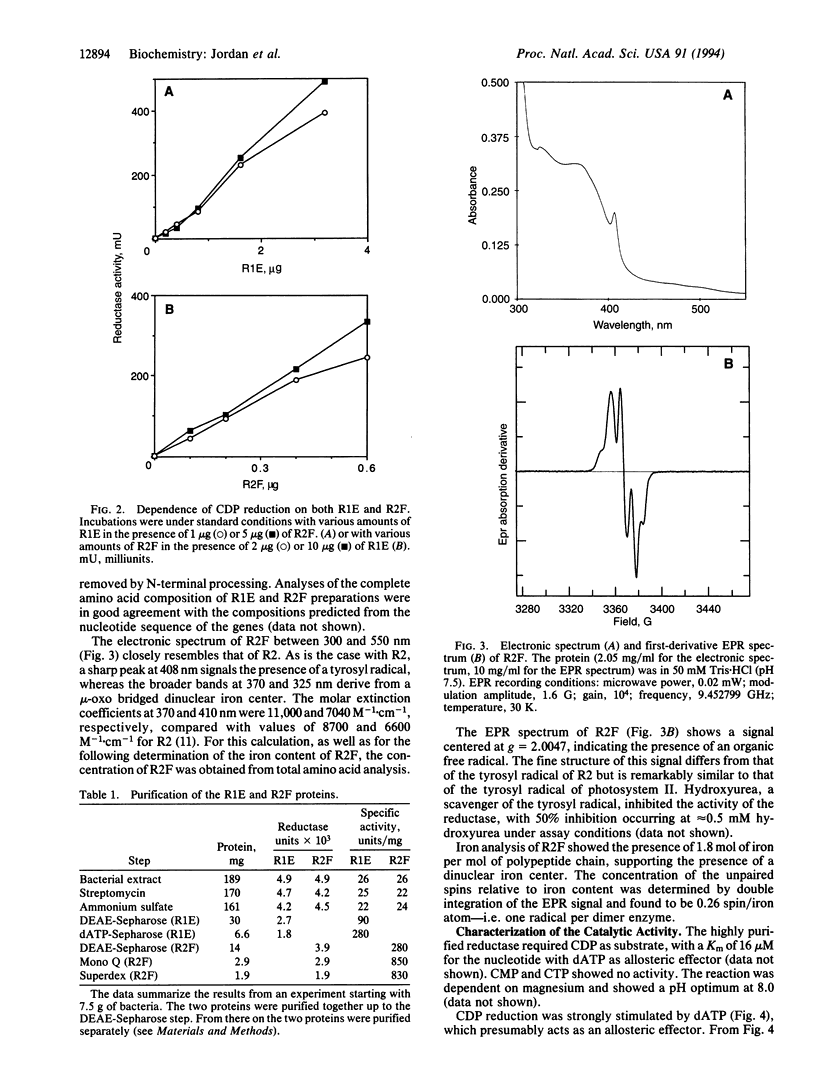
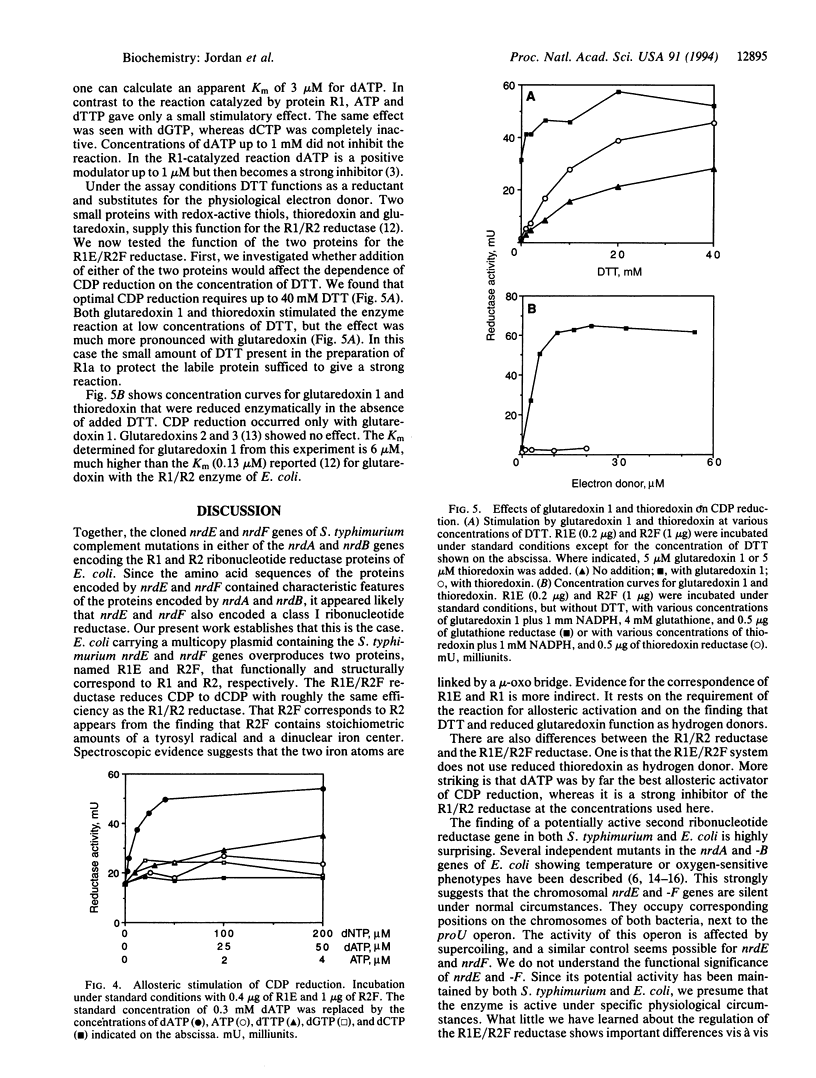
Abstract
The nrdA and nrdB genes of Escherichia coli and Salmonella typhimurium encode the R1 and R2 proteins that together form an active class I ribonucleotide reductase. Both organisms contain two additional chromosomal genes, nrdE and nrdF, whose corresponding protein sequences show some homology to the products of the genes nrdA and nrdB. When present on a plasmid, nrdE and nrdF together complement mutations in nrdA or nrdB. We have now obtained in nearly homogeneous form the two proteins encoded by the S. typhimurium nrdE and nrdF genes (R1E and R2F). They correspond to the R1 and R2 proteins. Each protein is a homodimer. Together they catalyze the reduction of CDP to dCDP, using dithiothreitol or reduced glutaredoxin, but not thioredoxin, as an electron donor. CDP reduction is strongly stimulated by low concentrations of dATP, presumably acting as an allosteric effector. Protein R2F contains an antiferromagnetically coupled dinuclear iron center and a tyrosyl free radical. The E. coli and S. typhimurium chromosome thus have maintained the information for a potentially active additional class I ribonucleotide reductase, whose role in vivo is as yet unknown. The allosteric regulation of this enzyme differs from that of the normally expressed reductase.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aslund F., Ehn B., Miranda-Vizuete A., Pueyo C., Holmgren A. Two additional glutaredoxins exist in Escherichia coli: glutaredoxin 3 is a hydrogen donor for ribonucleotide reductase in a thioredoxin/glutaredoxin 1 double mutant. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):9813–9817. doi: 10.1073/pnas.91.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund O., Eckstein F. Synthesis of ATP- and dATP-substituted sepharoses and their application in the purification of phage-T4-induced ribonucleotide reductase. Eur J Biochem. 1972 Aug 4;28(4):492–496. doi: 10.1111/j.1432-1033.1972.tb01936.x. [DOI] [PubMed] [Google Scholar]
- Berglund O. Ribonucleoside diphosphate reductase induced by bacteriophage T4. II. Allosteric regulation of substrate sepecificity and catalytic activity. J Biol Chem. 1972 Nov 25;247(22):7276–7281. [PubMed] [Google Scholar]
- Brown N. C., Reichard P. Role of effector binding in allosteric control of ribonucleoside diphosphate reductase. J Mol Biol. 1969 Nov 28;46(1):39–55. doi: 10.1016/0022-2836(69)90056-4. [DOI] [PubMed] [Google Scholar]
- Eliasson R., Pontis E., Fontecave M., Gerez C., Harder J., Jörnvall H., Krook M., Reichard P. Characterization of components of the anaerobic ribonucleotide reductase system from Escherichia coli. J Biol Chem. 1992 Dec 15;267(35):25541–25547. [PubMed] [Google Scholar]
- Fontecave M., Nordlund P., Eklund H., Reichard P. The redox centers of ribonucleotide reductase of Escherichia coli. Adv Enzymol Relat Areas Mol Biol. 1992;65:147–183. doi: 10.1002/9780470123119.ch4. [DOI] [PubMed] [Google Scholar]
- Fuchs J. A., Karlström H. O., Warner H. R., Reichard P. Defective gene product in dnaF mutant of Escherichia coli. Nat New Biol. 1972 Jul 19;238(81):69–71. doi: 10.1038/newbio238069a0. [DOI] [PubMed] [Google Scholar]
- Hantke K. Characterization of an iron sensitive Mud1 mutant in E. coli lacking the ribonucleotide reductase subunit B2. Arch Microbiol. 1988;149(4):344–349. doi: 10.1007/BF00411654. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989 Aug 25;264(24):13963–13966. [PubMed] [Google Scholar]
- Jamison C. S., Adler H. I. Mutations in Escherichia coli that effect sensitivity to oxygen. J Bacteriol. 1987 Nov;169(11):5087–5094. doi: 10.1128/jb.169.11.5087-5094.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling M. G., Holmes R. K. Construction of vectors with the p15a replicon, kanamycin resistance, inducible lacZ alpha and pUC18 or pUC19 multiple cloning sites. Nucleic Acids Res. 1990 Sep 11;18(17):5315–5316. doi: 10.1093/nar/18.17.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A., Gibert I., Barbé J. Cloning and sequencing of the genes from Salmonella typhimurium encoding a new bacterial ribonucleotide reductase. J Bacteriol. 1994 Jun;176(11):3420–3427. doi: 10.1128/jb.176.11.3420-3427.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A., Sjöberg B. M. Identification of the stable free radical tyrosine residue in ribonucleotide reductase. EMBO J. 1986 Aug;5(8):2037–2040. doi: 10.1002/j.1460-2075.1986.tb04461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson L., Gräslund A., Ehrenberg A., Sjöberg B. M., Reichard P. The iron center in ribonucleotide reductase from Escherichia coli. J Biol Chem. 1980 Jul 25;255(14):6706–6712. [PubMed] [Google Scholar]
- Reichard P. From RNA to DNA, why so many ribonucleotide reductases? Science. 1993 Jun 18;260(5115):1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- Taschner P. E., Verest J. G., Woldringh C. L. Genetic and morphological characterization of ftsB and nrdB mutants of Escherichia coli. J Bacteriol. 1987 Jan;169(1):19–25. doi: 10.1128/jb.169.1.19-25.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander L., Sjöberg B. R., Eriksson S. Ribonucleoside diphosphate reductase (Escherichia coli). Methods Enzymol. 1978;51:227–237. doi: 10.1016/s0076-6879(78)51032-x. [DOI] [PubMed] [Google Scholar]