Abstract
ACE inhibitory and antioxidative peptides identified by LCMS/MS, from mixed milk (Bubalus bubalis and Bos taurus) tryptic whey protein hydrolysate, were compared with the in silico predictions. α la and ß lg sequences, both from Bubalus bubalis and Bos taurus, were used for in silico study. SWISS-PROT and BIOPEP protein libraries were accessed for prediction of peptide generation. Study observed gaps in the prediction versus actual results, which remain unaddressed in the literature. Many peptides obtained in vitro, were not reflected in in silico predictions. Differences in identified peptides in separate libraries were observed too. In in silico prediction, peptides with known biological activities were also not reflected. Predictions, towards generation of bioactive peptides, based upon in silico release of proteins and amino acid sequences from different sources and thereupon validation in relation to actual results has often been reported in research literature. Given that computer aided simulation for prediction purposes is an effective research direction, regular updating of protein libraries and an effectual integration, for more precise results, is critical. The gaps addressed between these two techniques of research, have not found any address in literature. Inclusion of more flexibility with the variables, within the tools being used for prediction, and a hierarchy based database with search options for various peptides, will further enhance the scope and strength of research.
Electronic supplementary material
The online version of this article (doi:10.1007/s13197-014-1669-z) contains supplementary material, which is available to authorized users.
Keywords: In silico, In vitro, Antihypertensive, Antioxidant, Whey protein
Introduction
Mostly all proteins may be considered as the precursors of bioactive peptides that reside in dormant state within the parent sequence and may confer variety of biological activities after being defragmented by proteolysis (Dziuba et al. 2004). Subsequently, proteolytic digestion of milk and whey proteins for generation of peptides with specific physiologically beneficial activity, have attracted thick research since late. Often the term Peptidome has been associated with such kind of study where peptides present in an organism are described. The identified peptides then can be well recommended as a component for functional foods. Antibacterial, immunomodulatory, anticancerous, antioxidative, mineral binding, hypotensive etc. peptides have been found to address the corresponding issues. It is imperative to state that the health benefits enticed by these peptides encompass multi facets depending upon what sequence has been freed, which again rests on parent sequence and release process subjected to synchronization by many other factors. Hypotensive competency of bioactive peptides remains a potent study scope in relation to the risen stress levels and time crunches amongst the current populace. This has primarily been related to the prevention of conversion of angiotensin I to angiotensin II by way of impeding the angiotensin I converting enzyme (ACE) resulting a depression in blood pressure and thereby hypertension. The whey protein hydrolysates are well capable of fostering antioxidative benefits by means of halting oxidation intervened by free radicals too (Kamau and Lu 2010). The function of free radicals against ails as aging, cancer, inflammation etc. is quite on records (Ames et al. 1993). Association and application of computers have imperatively become an integral part of research. Protein libraries and databases like BIOPEP and SWISS-PROT have been utilized for various simulated and comparative research activities related to proteins and peptides. Tools like PeptideCutter, PeptideMass, Peptide Builder, PepPepSearch available at Bioinformatics Resource Portal, ExPASy, assist predictions related to research with peptides through a database search. Researchers like Dziuba et al. (2004), Vermeirssen et al. (2004), Pripp (2005), Chang and Alli (2012) have effectively applied the in silico technique for prediction and comparison of results. The paper attempts to compare and corroborate results from an in vitro and in silico study and also addresses the identified gaps related to the integration of these two methodologies.
Materials and methods
Materials
Amino acid sequences
Amino acid sequences of α la from Bubalus bubalis (Entry – Q9TSN6, Entry Name – LALBA_BUBBU, Length – 142 AA, Mass - 16,275 Da), Bos taurus (Entry – P00711, Entry Name – LALBA_BOVIN, Length – 142 AA, Mass - 16,247 Da), ß lg sequence from Bubalus bubalis (Entry – P02755, Entry Name – LACB_BUBBU, Length – 180 AA, Mass - 19,883 Da) and Bos taurus, (Entry – P02754, Entry Name – LACB_BOVIN, Length – 178 AA, Mass – 19,883 Da) were obtained in FASTA format from the Protein Knowledgebase (UniProtKB) database (SWISS-PROT and TrEMBL) available at www.uniprot.org.
Whey protein concentrate (WPC) and enzyme
WPC 70 was procured from Modern Dairies Pvt. Ltd. Karnal, Haryana, India, and stored in airtight container till further use. The enzyme trypsin (EC 3.4.21.4) was obtained from M/s Sigma (USA) and stored at 4 °C.
Chemicals
Analytical grade reagents and chemicals from reputed companies were used for chemical analysis.
Methods
In vitro analyses of experimental hydrolysate
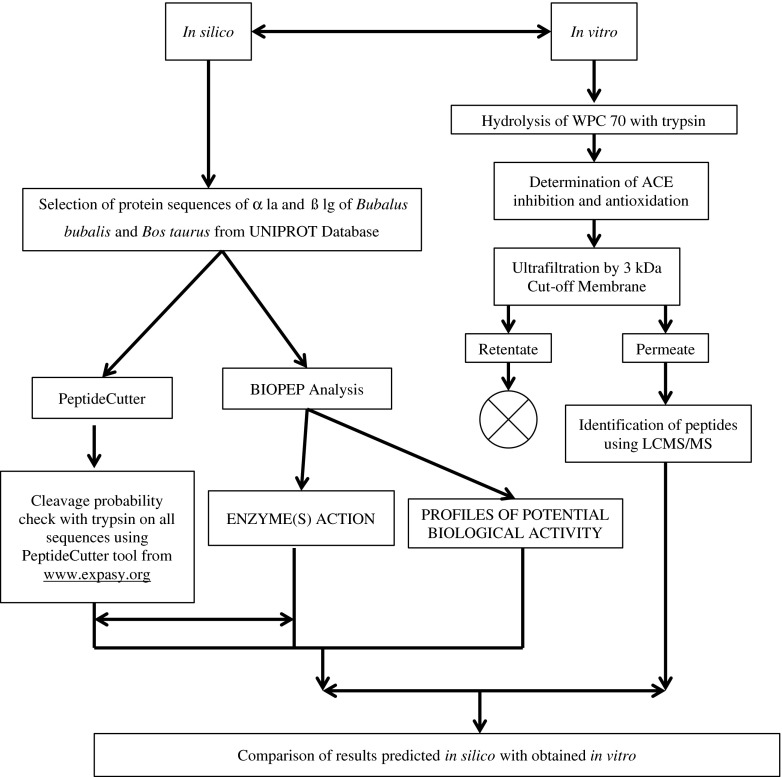
The complete experimental design has been presented at Fig. 1. WPC 70, prepared from mixed milk of Bubalus bubalis and Bos taurus, was procured. The determination of optimum hydrolysis conditions for most hypotensive and antioxidation effect was done by the study carried out employing Central Composite Rotatable Design model under Response Surface Methodology using DesignExpertTM software with pH, time and temperature as process variables over different range (complete optimization study not published yet). The pH, time and temperature ranges were taken as 5–8, 37–50 °C and 2–8 h respectively and the initial static conditions were kept as 65 °C pre-heat temperature, 0.01 enzyme-substrate ratio and 90 °C /10 min enzyme inactivation temperature-time combination. Based upon the results from this study, hydrolysis of 5 % protein solution of whey protein concentrate of 70 % protein at pH of 7.97 (~ 8.00) and temperature of 37 °C for 8 hours was performed that resulted in 76.09 % ACE inhibition and 51.83 % antioxidation property at 13.22 % degree of hydrolysis (work not published yet). ACE inhibition was calculated in vitro as per method given by Cushman and Cheung (1971), degree of hydrolysis by pH stat method (Adler-Nissen 1986) and antioxidative activity by DPPH (2, 2 diphenyl 1 picryl hydrazyl) method (Shimada et al. 1992). The selection of 13 % as keeping particular percentage for lowest cleavage probability was based on Degree of Hydrolysis obtained from this optimization study of tryptic whey protein hydrolysate for maximum generation of ACE inhibitory and antioxidative peptides. The flow for the preparation of hydrolysate is provided in Fig. 2. The hydrolysate obtained was fractionated using ultrafiltration system (Amicon) in which the hydrolysate was filtered through 3,000 Dalton molecular weight cut off membrane, at 7,000 g. Permeate and retentate obtained was separately collected and the permeate was subjected to freeze drying as preparation of samples for peptide identification by LCMS/MS. The identification of these peptides was done using LCMS/MS. The samples were sequenced at Proteomics International Pvt. Ltd, Western Australia through TechnoConcept Pvt Ltd., New Delhi. The detailed results are presented at Table 1.
Fig. 1.
Experimental design
Fig. 2.
Skeleton for the preparation of whey protein hydrolysate using trypsin
Table 1.
Mass-to-charge ratio with calculated and observed masses of peptide sequences
| Ion (m/z) | Calculated mass | Observed mass | Sequence |
|---|---|---|---|
| 932.54 | 932.5365 | 467.2753 | A. IIVTQTMK.G |
| 2030.30 | 2010.10 | 753.4499 | Y. SLAMAASDISLLDAQSAPLR.S |
| 1626.68 | 2045.0463 | 1023.9800 | A. ASDISLLDAQSAPLR.V |
| 855.47 | 856.4403 | 856.4782 | L. DAQSAPLR.V |
| 878.55 | 878.4749 | 440.2825 | R. VYVEELK.P |
| 1452.87 | 1451.7871 | 726.9620 | K. PTPEGDLEILLQK.W |
| 545.36 | 545.3213 | 546.3274 | K. IPAVF.K |
| 560.31 | 560.3322 | 561.3227 | I. PAVFK.I |
| 673.47 | 545.3213 | 546.3762 | K. IPAVFK.I |
| 914.60 | 915.4661 | 916.6319 | K. IDALNENK.V |
| 1064.66 | 1064.5754 | 533.2992 | K. VLVLDTDYK.K |
| 852.48 | 852.4229 | 853.4700 | L. VLDTDYK.K |
| 1244.65 | 1244.5772 | 1245.6582 | R. TPEVDDEALEK.F |
| 1046.59 | 1046.4768 | 524.3031 | P. EVDDEALEK.F |
| 836.53 | 836.4691 | 419.2351 | K. ALPMHIR.L |
| 1164.77 | 1164.6801 | 583.3948 | K. ALKALPMHIR.L |
Peptide release using peptidecutter
The complete approach to prediction and validation is summarized in Fig. 1. Protein sequences of α la and ß lg were selected from the database at www.uniprot.com, the details of which are provided under Materials section. Each sequence was then subjected to in silico release of peptides by enzyme trypsin, using the tool PeptideCutter available at www.expasy.org. Thereafter, each individual sequence in FASTA format was entered in the tool and the cutting of the sequence was performed by selecting sophisticated model for trypsin, mapping of cleavage site (60 amino acids per block), table of sites sorted alphabetically by enzyme and chemical name; and table of sites sorted sequentially by amino acid number. The lowest cleavage probability to be displayed was kept at 13 %. After the cutting of the sequence was performed, a list of probable peptides with cleavage sites, length of peptides, peptide mass and cleavage probability was obtained.
BIOPEP analyses
Parallel to PeptideCutter, a two way BIOPEP analyses was performed at http://www.uwm.edu.pl/biochemia/index.php/pl/biopep. Here, firstly individual sequences in FASTA format was used for prediction of release of peptides using enzyme trypsin. This analysis was carried out by selecting the option ‘ENZYME(S) ACTION’ at the above portal. However, factors such as percentage cleavage probability, enzyme/substrate ratio, time, temperature and pH for the lysis could not be taken into account due to non availability of any such possibility. Thereafter, from the above stated portal, another available option ‘PROFILES OF POTENTIAL BIOLOGICAL ACTIVITY’ was selected and ACE inhibition activity (Code from the available Drop Down Menu – ACE inhibitor | ah) and anitoxidative property (Code from the available Drop Down Menu – antioxidative | ao) were sequentially selected and the above stated sequences under ‘Materials’ section were used one by one for prediction of the peptides with the sought properties.
Results and discussion
Results and discussion
Since, throughout the in silico experiments, the results from α la sequence of Bubalus bubalis and α la sequence of Bos taurus were truly identical, only results from α la sequence of Bubalus bubalis have been discussed, and presented wherever required. Peptides from prepared hydrolysate, as identified through LCMS/MS and other details as obtained are provided in Table 2. All the obtained peptides were fractions of β-lg. Several studies have attempted linking the inhibitory activity with the primary structure of the peptides. The favorability of hydrophobic amino acids at each of the three C-terminal positions has been suggested by Li et al. (2004), Lopez-Fandino et al. (2006), Otte et al. (2007). Cheung et al. (1980) found that amino acids, namely Pro, Trp, Tyr and Phe, to be utmost effective at the ultimate C-terminal. They further advocated the exceptional better binding of Pro to ACE. This is even seconded by QSAR models studies by Pripp et al. (2006) thus concluding that hypotensive property as expressed by small peptides are strongly and progressively correlated to the positioning of hydrophobic amino acid at the ultimate C-terminal. However, cleavage specificity of an enzyme also plays an equal determining role in proteolytic peptide release. Since the enzyme used here was trypsin, all the cleavage points are either at AA residue Arginine (R) or Lysine (K) thus conforming to the specificity of tryptic action (Table 2). This is also corroborated by the results from in silico analysis where in Fig. 3; starting from 1st AA and moving to 142nd; and in Figs. 4 and 5; starting from 1st AA and moving to 180th and 178th respectively; the cleavage points and percentage probability of cleavage for α la, ß-lg from Bubalus bubalis and Bos taurus sequences are presented and all the cleavage points are either at AA residue ‘R’ or ‘K’. It must be stated that differences in the molecular weights were observed between the obtained peptides and reported peptides in the literature. This may be due to the difference in the amino acid composition of the protein.
Table 2.
[In-vitro] - Peptides from prepared hydrolysate
| Sl. No. | Protein fragments | Peptide sequence identified | Molecular weight (Da) | Reported biological activity of identified peptides | Reference |
|---|---|---|---|---|---|
| 1 | β-lg f (1–8) | IIVTQTMK | 932.54 | ACE-inhibitory | (Chobert et al. 2005) |
| 2 | β-lg f (21–40) | SLAMAASDISLLDAQSAPLR | 2030.30 | ACE-inhibitory, Immunomodulatory | (Chobert et al. 2005; Rodriguez-carrio et al. 2014) |
| 3 | β-lg f (25–40) | AASDISLLDAQSAPLR | 1626.68 | Bactericidal | (Pellegrini et al. 2001) |
| 4 | β-lg f (33–40) | DAQSAPLR | 855.47 | ACE-inhibitory | (Pihlanto-leppala et al. 2000) |
| 5 | β-lg f (41–47) | VYVEELK | 878.55 | Antioxidant | (Hernandez-ledesma et al. 2005) |
| 6 | β-lg f (48–60) | PTPEGDLEILLQK | 1452.87 | - | - |
| 7 | β-lg f (78–82) | IPAVF | 545.36 | ACE-inhibitory | (Konard et al. 2014) |
| 8 | β-lg f (79–83) | PAVFK | 560.31 | ACE-inhibitory | (Dalsgaard et al. 2008) |
| 9 | β-lg f (78–83) | IPAVFK | 673.47 | ACE-inhibitory, Antimicrobial | (Chobert et al. 2005; Pellegrini et al. 2001) |
| 10 | β-lg f (84–91) | IDALNENK | 914.60 | ACE-inhibitory, Immunomodulatory | (Chobert et al. 2005; Rodriguez-carrio et al. 2014) |
| 11 | β-lg f (92–100) | VLVLDTDYK | 1064.66 | ACE-inhibitory, Antimicrobial | (Pihlanto-leppala et al. 2000) |
| 12 | β-lg f (94–100) | VLDTDYK | 852.48 | ACE-inhibitory | (Pihlanto-leppala et al. 2000) |
| 13 | β-lg f (125–135) | TPEVDDEALEK | 1244.65 | ACE inhibitory | (Roufik et al. 2006) |
| 14 | β-lg f (127–135) | EVDDEALEK | 1046.59 | - | - |
| 15 | β-lg f (139–148) | ALKALPMHIR | 1164.77 | Antioxidant | (Hernandez-ledesma et al. 2005) |
| 16 | β-lg f (142–148) | ALPMHIR | 836.53 | ACE-inhibitory, Antioxidant | (Mullally et al. 1997) |
Fig. 3.
Cleavage sites in α-la sequence from Bubalus bubalis
Fig. 4.
Cleavage sites in ß-lg sequence from Bubalus bubalis
Fig. 5.
Cleavage sites in ß-lg sequence from Bos taurus
PeptideCutter software has been used previously by Pripp (2005) for evaluating the proteolytic effect by selected enzymes, over milk proteins. The guidelines of the research was further utilized by Otte et al. (2007) for similar work where they studied the effect of substrate, enzyme and time of hydrolysis on ACE inhibitory activity of milk proteins. With adherence to similar guidelines with the same software, results obtained are discussed hereunder. Table 3 shows the action of enzyme trypsin on the α la sequence of Bubalus bubalis and the possible peptides generated thereof, along with length of peptide and molecular weight. Throughout the study, the results of all in silico experimentation on α la sequence from Bubalus bubalis and on that of the sequence from Bos taurus were exactly identical since the parent sequence, as selected from the UNIPROT database, was a true match.
Table 3.
[In-silico] - Peptides after cutting of α la sequence from Bubalus bubalis
| Sl. No. | Resulting peptide sequence | Peptide length [aa] | Peptide mass [Da] |
|---|---|---|---|
| 1 | MMSFVSLLLVGILFHATQAEQLTK | 24 | 2678.243 |
| 2 | CEVFR | 5 | 652.766 |
| 3 | ELK | 3 | 388.464 |
| 4 | DLK | 3 | 374.437 |
| 5 | DYGGVSLPEWVCTAFHTSGYDTQAI VQNNDSTEYGLFQINNK | 42 | 4684.040 |
| 6 | IWCK | 4 | 548.701 |
| 7 | DDQNPHSSNICNISCDK | 17 | 1889.986 |
| 8 | FLDDDLTDDIMCVK | 14 | 1642.856 |
| 9 | K | 1 | 146.189 |
| 10 | ILDK | 4 | 487.597 |
| 11 | VGINYWLAHK | 10 | 1200.406 |
| 12 | ALCSEK | 6 | 649.760 |
| 13 | LDQWLCEK | 8 | 1034.195 |
| 14 | L | 1 | 131.175 |
Similarly, Table 4 and Table 5 enlist the peptides from ß lg sequence of Bubalus bubalis and Bos taurus, respectively, with cleavage points and percentage probability of cleavage being presented at Figs. 4 and 5, correspondingly. The presented sequence ß lg from Bubalus bubalis has AAs GLALACAAQAI at location {9–19} whereas from Bos taurus, has AAs ALTCGAQAL at location {9–17}. Also, the latter has AA ‘I’ at location {178–178} whereas the former has AA ‘V’ at location {180–180}. The difference in the length of sequence is drawn from the difference at position {9–19}, as the sequence from Bos taurus has 02 AA less as compared to Bubalus bubalis, as discussed above.
Table 4.
[In-silico] - Peptides after cutting of ß lg sequence from Bubalus bubalis
| Sl. No. | Resulting peptide sequence | Peptide length [aa] | Peptide mass [Da] |
|---|---|---|---|
| 1 | MK | 2 | 277.382 |
| 2 | CLLLALGLALACGAQAIIVTQTMK | 24 | 2416.037 |
| 3 | GLDIQK | 6 | 672.779 |
| 4 | VAGTWYSLAMAASDISLLDAQSAPLR | 26 | 2708.081 |
| 5 | VYVEELK | 7 | 879.021 |
| 6 | PTPEGDLEILLQK | 13 | 1452.668 |
| 7 | WENGECAQK | 9 | 1064.138 |
| 8 | K | 1 | 146.189 |
| 9 | IIAEK | 5 | 572.702 |
| 10 | TK | 2 | 247.294 |
| 11 | IPAVFK | 6 | 673.853 |
| 12 | IDALNENK | 8 | 915.999 |
| 13 | VLVLDTDYK | 9 | 1065.232 |
| 14 | K | 1 | 146.189 |
| 15 | YLLFCMENSAEPEQSLACQCLVR | 23 | 2648.077 |
| 16 | TPEVDDEALEK | 11 | 1245.306 |
| 17 | FDK | 3 | 408.455 |
| 18 | ALK | 3 | 330.428 |
| 19 | ALPMHIR | 7 | 837.051 |
| 20 | LSFNPTQLEEQCHV | 14 | 1644.819 |
Table 5.
[In-silico] - Peptides after cutting of ß lg sequence from Bos taurus
| Sl. No. | Resulting peptide sequence | Peptide length [aa] | Peptide mass [Da] |
|---|---|---|---|
| 1 | MK | 2 | 277.382 |
| 2 | CLLLALALTCGAQALIVTQTMK | 22 | 2275.852 |
| 3 | GLDIQK | 6 | 672.779 |
| 4 | VAGTWYSLAMAASDISLLDAQSAPLR | 26 | 2708.081 |
| 5 | VYVEELK | 7 | 879.021 |
| 6 | PTPEGDLEILLQK | 13 | 1452.668 |
| 7 | WENGECAQK | 9 | 1064.138 |
| 8 | K | 1 | 146.189 |
| 9 | IIAEK | 5 | 572.702 |
| 10 | TK | 2 | 247.294 |
| 11 | IPAVFK | 6 | 673.853 |
| 12 | IDALNENK | 8 | 915.999 |
| 13 | VLVLDTDYK | 9 | 1065.232 |
| 14 | K | 1 | 146.189 |
| 15 | YLLFCMENSAEPEQSLACQCLVR | 23 | 2648.077 |
| 16 | TPEVDDEALEK | 11 | 1245.306 |
| 17 | FDK | 3 | 408.455 |
| 18 | ALK | 3 | 330.428 |
| 19 | ALPMHIR | 7 | 837.051 |
| 20 | LSFNPTQLEEQCHI | 14 | 1658.846 |
The analysis was simultaneously performed at BIOPEP. α la sequence when subjected to enzyme action of trypsin, returned fractions that were a replica of Table 3. However, amongst 13 peptides, only 02 peptides, namely ALCSEK at location {128–133} and VGINYWLAHK at location {118–125} were found to be the only peptides to have been reported biologically active, in the database, with antibacterial and ACE inhibitory activity, respectively. In literature, the reference for this peptide and its’ biological activity, can be drawn from the works by Pellegrini et al. (1999), Dziuba et al. (2009) and Madureira et al. (2010).
Likewise, both the sequences of ß lg (i.e. from Bubalus bubalis and Bos taurus) were also subjected to enzyme action by trypsin and they returned sequences, that were corroboration of Tables 4 and 5, correspondingly. From these, the identified active peptides with reported biological activity were exactly similar. ALPMHIR and GLDIQK were 02 peptides with ACE inhibitory property whereas other 02 peptides i.e. IPAVFK and VLVLDTDYK had antibacterial property, as reported in the library.
List of peptides with ACE inhibiting property and antioxidant property; individually; from α la sequence of Bubalus bubalis and ß lg sequence of Bubalus bubalis and Bos taurus, was obtained from BIOPEP analyses. These results were compared with those obtained from the whey protein hydrolysate prepared by trypsin. The LCMS/MS identification returned 16 peptides out of which 14 peptides have been previously reported in the literature with biological activity(ies), 01 peptide, that although matches with the in silico prediction, the biological activity of it is yet unreported, subjected to any, and 01 peptide did not find any match with regard to library or literature. These peptides have been enlisted in Table 2. It shows 06 peptides with ACE inhibitory property, 02 peptides with ACE inhibitory and antimicrobial property, 01 peptide with ACE inhibitory and antioxidant property, 02 peptides with antioxidative property, 01 peptide with bactericidal property, 02 peptides with ACE inhibitory and immunomodulatory property and 02 unreported peptide. When these obtained peptides were tallied with the results from in silico experiment by PeptideCutter, 07 peptides (from Table 2; S1. No. 5, 6, 9, 10, 11, 13 and 16) out of 19 (Table 4 and Table 5; S1. No. 5, 6, 11, 12, 13, 16 and 19; each) were observed to be coinciding. However, all the peptides were from ß lg fraction of whey protein. Also, all the 07 peptides were in the ß lg fraction of both Bubalus bubalis (Table 4) and Bos taurus (Table 5). Amongst these matched peptides, 04 peptides, viz. IPAVFK, VLVLDTDYK, TPEVDDEALEK and ALPMHIR have been reported to be ACE inhibitory. From these 04 peptides, VLVLDTDYK and ALPMHIR have also been reported to possess antimicrobial and antioxidant property, respectively (Table 2). Peptide PTPEGDLEILLQK though was returned in in silico prediction and was found in actual samples too, the biological activity of it remains unexplored. Peptide EVDDEALEK was neither identified by in silico method nor does it find any reporting in literature but was found in the hydrolysate samples. The remaining 02 peptides from 07 matched, VYVEELK is documented against antioxidant and SLAMAASDISLLDAQSAPLR and IDALNENK against ACE inhibitory and immunomodulatory properties, respectively (Table 2).
The results from PeptideCutter were found to be parallel, when related to those from ‘ENZYME(S) ACTIVITY’ of BIOPEP analysis, throughout. From both, PeptideCutter and BIOPEP, the peptide ALCSEK, as from α la sequence of Bubalus bubalis, was the only active fragment documented in the database with its biological activity being antibacterial. The BIOPEP analysis returned 04 peptides with biological activity when ß lg sequence of Bubalus bubalis and Bos taurus was tested for action of trypsin, individually. Since the peptides were same, the obtained peptides have been presented in only one table i.e. Table 6. Amongst these, ALPMHIR and GLDIQK were documented as ACE inhibitory and IPAVFK and VLVLDTDYK as antibacterial in the database. However, peptide GLDIQK as returned by BIOPEP, was not identified in the hydrolysate samples. Moreover, peptides ALPMHIR and VLDTDYK are reported, respectively, as antioxidative and ACE inhibitory in the literature (see Table 2 for references), but the same is not reflected in the BIOPEP database.
Table 6.
[In-silico] - List of active fragments and activity from ß lg sequence of Bubalus bubalis
| No. | Sequence | Location | Name | Activity |
|---|---|---|---|---|
| 1 | ALPMHIR | {160–166} | beta-lactokinin (fr. of ß-lg: 142–148) | ACE inhibitory |
| 2 | GLDIQK | {27–32} | ACE inhibitor (b-lg 9–14) | ACE inhibitory |
| 3 | IPAVFK | {96–101} | f (78–83) of bovine b-lactoglobulin | Antibacterial |
| 4 | VLVLDTDYK | {110–118} | f (92–100) of bovine b-lactoglobulin | Antibacterial |
Upon analyzing the sequences with the tool ‘PROFILES OF POTENTIAL BIOLOGICAL ACTIVITY’ from BIOPEP, α la sequence returned 37 probable peptides with ACE inhibition property and 06 with antioxidative. With the same tool, ß lg sequence of Bubalus bubalis and Bos taurus, returned 66 and 64 ACE inhibitors respectively and 14 antioxidative peptides. Only 02 peptides, and that too only ACE inhibitory, i.e. ALPMHIR and VLDTDYK, corresponded with the resulting peptides from the optimized hydrolysate. Peptide VLDTDYK was though predicted as an ACE inhibitor from ß lg, in the BIOPEP analysis, the generation of peptide was not predicted in the in silico proteolysis, either by PeptideCutter or ENZYME(S) ACTION tool. However, the peptide was identified in the optimized hydrolysate. Antioxidant activity of ALPMHIR was not found in the library though the peptide is known for this property as per works of Mullally et al. (1997).
Apart peptides discussed so far, the peptides identified from the optimized hydrolysate, also included IIVTQTMK, SLAMAASDISLLDAQSAPLR, DAQSAPLR, IPAVF and PAVFVF reported with ACE inhibition activity (Table 2), AASDISLLDAQSAPLR bactericidal and ALKALPMHIR with antioxidant activity. These peptides were not identified in either of the databases, be it in silico proteolysis by PeptideCutter; with the consideration of degree of hydrolysis; or without, as in case of ‘ENZYME(S) ACTION’. These peptides remained unidentified even during in silico release of the sequences as well as were not found as biologically active peptides in the databases. These peptides, along with their respective biological activity(ies) have been reported by various researchers at various instances, in the literature, but the same is not reflected in the BIOPEP database when ‘PROFILES OF POTENTIAL BIOLOGICAL ACTIVITY’ tool is applied on the sequences of α la or ß lg.
Conclusion
Computer aided simulation or in silico experimentations are a potential way for prediction and validation. However, there were some gaps that got addressed with the current study. Although, with PeptideCutter software, proteolysis prediction was done by workers like Pripp (2005) and Otte et al. (2007), the latter also maintained that comparison of experimental and theoretical results remain unavoidable for better dependability. Thus, a more updated or frequently updated database is imperatively required for better prediction and/or validation. However, this is only possible when the researchers submit their findings to these databases in the form of peptides and their properties. Since, there are many different web portals handling and offering access to different databases with different tools, the heterogeneity makes it difficult for the researcher to select a specific database for submission. An integration of the existing databases, with the consent of the scientific communities or creation of a dedicated Bioactive Peptide Bank; attempting to handle all the bioactive peptides from all the known food sources i.e. animal and plant kingdom; is likely to address this problem. This will not only benefit the further researchers in pre-analysis and promote prediction research but also provide detailed information, resulting in more refined and precise outcomes. Secondly, introduction of more flexibility within the tools, that are currently not available for performing simulated proteolysis, could assist in achieving more precise predictions. Such flexibilities may include factors that directly affect the enzymatic proteolysis such as pH, time, temperature and enzyme substrate ratio, in particular, and thus the generation of bioactive peptides of different molecular weight and length, and form important variables for predicting proteolytic effect of enzyme on substrate. Availability of such variations currently lack for in silico predictions. Such tools with sufficient flexibility, be available for predictive and bioactive property targeted generation of peptides. The tools are to be chiefly those that directly form the manipulative independent variables for effective generation of these bioactive peptides, as pointed out. However, an interpretation of the discussed in silico results may also be an indication towards relative non generation of antihypertensive or antioxidant peptides at lower level of hydrolysis. It is essential to use the word ‘relative’ owing to the comparison of the results from the simulation; that resulted in generation of more general bioactive motifs; versus actual experimentation that was optimized with focus on generation of bioactive peptides with specific bioactivity. An effort towards more integrated and updated databases will not only benefit the scientific community but also be a reference centre for correctness of research.
Electronic supplementary material
(DOCX 196 kb)
Acknowledgments
Thankful acknowledgement to the Director, National Dairy Research Institute for the providing economic assistance in the form of Senior Research Fellowship (Post Graduate Studies) constituted by Indian Council of Agricultural Research, Pusa, New Delhi and other infrastructural amenities for conducting the presented research work.
Abbreviations
- AA
Amino acid
- ACE
Angiotensin I converting enzyme
- DPPH
2 diphenyl 1 picryl hydrazyl
- LCMS/MS
Liquid chromatography–mass spectrometry
- QSAR
Quantity-structure-activity-relationship
- WPC
Whey protein concentrate
References
- Adler-Nissen J. In enzymatic hydrolysis of food proteins. London: Elsevier Applied Science Publishers; 1986. pp. 32–35. [Google Scholar]
- Ames BN, Shigenaga MK, Hagen TM. In: Oxidants, antioxidants, and the degenerative diseases of aging. USA: Proceedings of National Academy of Sciences; 1993. pp. 7915–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YW, Alli I (2012) In silico assessment: suggested homology of chickpea (Cicer arietinum L.) legumin and prediction of ACE inhibitory peptides from chickpea proteins using BLAST and BIOPEP analyses. Food Res Int. doi:10.1016/j.foodres.2012.07.006
- Cheung H-S, Wang F-L, Ondetti MA, Sabo EF, Cushman DW. Binding of peptide substrates and inhibitors of angiotensinconverting enzyme. J Biol Chem. 1980;255:401–407. [PubMed] [Google Scholar]
- Chobert JM, El-zahar K, Sitohy M, Dalgalarrondo M, Metro F, Choiset Y, Haertle T. Angiotensin I converting enzyme (ACE) inhibitory activity of tryptic peptides of ovine beta lactoglobulin and of milk yoghurts obtained by using different starters. Lait. 2005;85:141–152. doi: 10.1051/lait:2005005. [DOI] [Google Scholar]
- Cushman DW, Cheung HS. Spectrophotometric assay and properties of angiotensin converting enzyme of Rabbit lung. Biochem Pharmacol. 1971;20(7):1637–1648. doi: 10.1016/0006-2952(71)90292-9. [DOI] [PubMed] [Google Scholar]
- Dalsgaard TK, Heegaard CW, Larsen LB. Plasmin digestion of photooxidized milk proteins. J Dairy Sci. 2008;91:2175–2183. doi: 10.3168/jds.2007-0843. [DOI] [PubMed] [Google Scholar]
- Dziuba J, Niklewicz M, Iwaniak A, Darewicz M, Minkiewicz P. Bioinformatic aided prediction for release possibilities of bioactive peptides from plant proteins. Acta Aliment. 2004;33(3):227–235. doi: 10.1556/AAlim.33.2004.3.3. [DOI] [Google Scholar]
- Dziuba M, Dziuba B, Iwaniak A. Milk proteins as precursors of bioactive peptides. ACTA Sci Pol Technol Aliment. 2009;8(1):71–90. doi: 10.17306/j.afs.2014.1.1. [DOI] [PubMed] [Google Scholar]
- Hernandez-ledesma B, Miralles B, Amigo L, Ramos M, Recio I. Identification of antioxidant and ACE inhibitory peptides in fermented milk. J Sci Food Agric. 2005;85(6):1041–1048. doi: 10.1002/jsfa.2063. [DOI] [Google Scholar]
- Kamau SM, Lu RR. The effect of enzymes and hydrolysis conditions on degree of hydrolysis and DPPH radical scavenging activity of whey protein hysrolysates. Curr Res Dairy Sci. 2010;1994–5434:1–8. [Google Scholar]
- Konard B, Dabrowska A, Szołtysik M, Pokora M, Zambrowicz A, Chrzanowska J. The evaluation of dipeptidyl peptidase (DPP)-IV, a-glucosidase and angiotensin converting enzyme (ACE) inhibitory activities of whey proteins hydrolyzed with serine protease isolated from Asian pumpkin (Cucurbita ficifolia) Int J Pept Res Ther. 2014 doi: 10.1007/s10989-014-9413-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G-H, Le G-W, Shi Y-H, Shrestha S. Angiotensin I-converting enzyme inhibitory peptides derived from food proteins and their physiological and pharmacological effects. Nutr Res. 2004;24:469–486. doi: 10.1016/S0271-5317(04)00058-2. [DOI] [Google Scholar]
- Lopez-Fandino R, Otte J, Van-Camp J. Physiological, chemical and technological aspects of milk protein derived peptides with antihypertensive and ACE-inhibitory activity. Int Dairy J. 2006 [Google Scholar]
- Madureira AR, Tavares T, Gomes AMP, Pintado ME, Malcata FX. Invited review: physiological properties of bioactive peptides obtained from whey proteins. J Dairy Sci. 2010;93:437–455. doi: 10.3168/jds.2009-2566. [DOI] [PubMed] [Google Scholar]
- Mullally MM, Meisel H, FitzGerald RJ. Identification of a novel angiotensin I converting enzyme inhibitory peptide corresponding to a tryptic fragment of bovine ß lactoglobulin. FEBS Lett. 1997;402:99–101. doi: 10.1016/S0014-5793(96)01503-7. [DOI] [PubMed] [Google Scholar]
- Otte J, Shalabya SM, Zakora M, Pripp AH, El-Shabrawyb SA. Angiotensin-converting enzyme inhibitory activity of milk protein hydrolysates: Effect of substrate, enzyme and time of hydrolysis. Int Dairy J. 2007;17:488–503. doi: 10.1016/j.idairyj.2006.05.011. [DOI] [Google Scholar]
- Pellegrini A, Dettling C, Thomas U, Hunziker P. Isolation and characterization of four bactericidal domains in the bovine ß lactoglobulin. Biochim Biophys Acta. 2001;1526:131–140. doi: 10.1016/S0304-4165(01)00116-7. [DOI] [PubMed] [Google Scholar]
- Pellegrini A, Thomas U, Bramaz N, Hunziker P, Fellenberg RV. Isolation and identification of three bactericidal domains in the bovine α lactalbumin molecule. Acta Biochim Biophys Sin. 1999;1426(3):439–448. doi: 10.1016/S0304-4165(98)00165-2. [DOI] [PubMed] [Google Scholar]
- Pihlanto-leppala A, Koskinen P, Piilola K, Tupasela T, Korhonen H. Angiotensin I converting enzyme inhibitory properties of whey protein digests: Concentration and characterization of active peptides. J Dairy Res. 2000;67:53–64. doi: 10.1017/S0022029999003982. [DOI] [PubMed] [Google Scholar]
- Pripp AH. Initial proteolysis of milk proteins and its effect on formation of ACE inhibitory peptides during gastrointestinal proteolysis: a bioinformatic, in silico, approach. Eur Food Res Technol. 2005;221(5):712–716. doi: 10.1007/s00217-005-0083-1. [DOI] [Google Scholar]
- Pripp AH, Sorensen R, Stepaniak L, Sorhaug T. Relationship between proteolysis and angiotensin-I-converting enzyme inhibition in different cheeses. LWT Food Sci Technol. 2006;36:677–683. doi: 10.1016/j.lwt.2005.03.018. [DOI] [Google Scholar]
- Rodriguez-carrio J, Fernandez A, Riera FA, Suarez A. Immunomodulatory activities of whey ß lactoglobulin tryptic digested fractions. Int Dairy J. 2014;34(1):65–73. doi: 10.1016/j.idairyj.2013.07.004. [DOI] [Google Scholar]
- Roufik S, Gauthier SF, Dufour E, Turgeon SL. Interactions between bovine β Lactoglobulin A and Various bioactive peptides as studied by front face fluorescence spectroscopy. J Agric Food Chem. 2006;54(14):4962–4969. doi: 10.1021/jf060506m. [DOI] [PubMed] [Google Scholar]
- Shimada K, Fujikawa K, Yahara K, Nakamura T. Antioxidative properties of xanthane on the autoxidation of soyabean oil in cyclodextrin emulsion. J Agric Food Chem. 1992;40(6):945–948. doi: 10.1021/jf00018a005. [DOI] [Google Scholar]
- Vermeirssen V, Bent-van-der A, Camp-van J, Amerongen-van A, Verstraete W. A quantitative in silico analysis calculates the angiotensin I converting enzyme (ACE) inhibitory activity in pea and whey protein digests. Biochimie. 2004;86:231–239. doi: 10.1016/j.biochi.2004.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 196 kb)