Abstract
Pectin is a complex structural heteropolysaccharide that require numerous pectinolytic enzymes for its complete degradation. Polygalacturonase from mesophilic or thermophilic origin are being widely used in fruit and vegetable processing in the recent decades to degrade pectic substances. Recently cold active pectinases are finding added advantages over meso and thermophilic counterparts, to use in industrial scale particularly in food processing industry. They facilitate in conservation of several properties of foods so that the end product retains its naturality and also generates economic benefits. In the present study, Pseudoalteromonas haloplanktis, a well reported marine psychrophile is taken as a model organism for cold active polygalacturonase and is evaluated in comparision to the routinely used mesophilic and thermophilic enzymes by insicio approach. Polygalacturonase sequences from industrially important microbial sources were subjected to MEME and Pfam wherein motifs and domains involved in the conservation were analyzed. Dendrogram revealed sequence level similarity and motifs showed uniform distribution of conserved regions that are involved in important functions. It was also observed through clustalW analysis that the amount of arginine content of psychrophiles is less when compared with thermophiles. Finally, all the modeled enzyme structures were subjected to docking studies using Autodock 4.2 with the substrate polygalacturonic acid and binding energies were found to be −5.73, −6.22 and −7.27 KCals/mole for meso, thermo and psychrophiles respectively which indicates the efficiency of psychrophilic enzymes when compared with its counterparts giving scope for further experimentation to find their better usage in various food industry applications.
Electronic supplementary material
The online version of this article (doi:10.1007/s13197-014-1654-6) contains supplementary material, which is available to authorized users.
Keywords: Pectin, Polygalacturonase, Pseudoalteromonas haloplanktis, Cold active enzymes, Psychrophile
Introduction
Pectin is the most significant and abundant polysaccharide present in the middle lamella and primary cell-wall of all the higher plants. It is made up of 1–4 α-D galacturonic acid units (Alphons et al. 2009; Shefali et al. 2008). Pectin acts as a cementing substance that helps in binding of adjacent cells. Pectin when subjected to enzymatic deesterification forms a polymer known to be pectic acid or polygalacturonic acid (PGA) (Palivanelu 2006). Pectinases or polygalacturonases (PGases) are the enzymes that function as depolymerizers of pectin by cleavage of glycosidic bonds. Commercial use of pectinases started in 1930 in various applications of food industries such as clarification of juice, mashing treatments and enhancing the yield and color of the products (Kertesz 1930; Chawanit et al. 2007; Jose et al. 2008). Most of the enzymes including polygalacturonases that are used commercially for these applications are derived from mesophiles or thermophiles. But in food processing industries, functioning of enzymes at low temperatures is always preferred (20 °C or below) due to certain economic and environmental advantages, such as, energy saving, retention of labile and volatile flavor compounds, prevention of contamination and elimination of any residual enzyme activity, which is inactivation of enzyme when temperature is raised (Adapa et al. 2014). The unique features like high specific activity even at low temperatures and their property of rapid inactivation at moderate to higher temperatures make the psychrophilic genetic resources offer numerous economic and ecological advantages. Low temperature enzymes can reduce the cost of processes at the places where cooling and heating treatments are required. So, enzymes of psychrophilic origin became hot topics, particularly in food processing as they help in the reduction of process cost by skipping heating treatments and also help in retaining the quality of food material. Due to the low temperature food processing, problems of contamination and spoilage can also be minimized (Truong et al. 2001; Pulicherla et al. 2011). As the enzymes from psychrophiles were found to be beneficial in various applications, advances in structural and functional details about psychrophilic enzymes leads to better understanding for using them industrially (Rekha et al. 2013). The psychrophilic organism considered in the present study is Pseudoalteromonas haloplanktis which is a well reported Antarctic marine organism degrading pectin (Gomes and Steiner 2004; Carmen et al. 2012).
An attempt was made to investigate the protein sequences of polygalacturonases from all the three categories of microorgansims such as mesophiles, psychrophiles and thermophiles to characterize their conservation at the amino acid level. Secondary structure analysis was done for various sequences of polygalacturonases that can give information about the functions, homology and structure of the enzyme. MEME identifies structural and functionally key parts of a protein. Motifs in the block diagram help in knowing the building of proteins and also shed light on the evolutionary relationships among proteins. Also, protein binding sites and some of the regulatory elements in the groups of upstream regions from co-regulated genes were identified. From the Pfam database, evolutionarily conserved protein families and annotations about the functions of those families can be known. Further, modeling and docking studies have been carried out to understand the interactions of the enzyme with the substrate which in turn gives information about the stability and activity of the psychrophilic enzyme in comparison with its counterparts.
Materials and methods
Retrieval of polygalacturonase sequences and multiple sequence alignment
Enzyme sequences for the present work were taken from three different habitats like mesophilic, thermophilic and psychrophilic. Fourteen polygalacturonase sequences from bacteria and 18 sequences from fungi that are having huge industrial importance were chosen from National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov) in FASTA format. Only the full length sequences of polygalacturonases were considered for in silico analysis. All the sequences were subjected to multiple sequence alignment using ClustalW to study the residues involved in conservation and also to know about sequence similarity of enzymes from three different catogeries.
Analysis of secondary structures
MEME (Multiple EM for Motif EliCitation) is a well- known tool which allows to discover motifs in protein or DNA sequences. Input parameters such as width of the motif and maximum number of motifs were specified. This tool is helpful to perform motif-motif database search, motif-sequence database search and discovery of motifs etc. The start and end point of amino acid sequences were represented by block diagrams. Location of motifs or sites obtained in the MEME describes the conserved regions that are related to the structural and functional properties of the enzymes in the process of evolution.
The database used in the present study for the analysis of domains is Pfam (Finn et al. 2014) that defines the property of similarity at the sequence level. The sequences were submitted in the form of accession numbers. All the sequences were examined for the domain organization in polygalacturonases.
Phylogenetic analysis and construction of dendrogram
Polygalacturonase sequences from both bacteria and fungi were aligned by using MEGA software version 5.1 (Tamura et al. 2011). Phylogenetic trees were constructed with the help of maximum likelihood method to ensure reliability and stability of Phylogenetic relationships among different strains. Also, the divergence was cross checked by using other methods like maximum parsimony method, UPGMA method and also minimum evolution method using MEGA (Shi et al. 2007).
Acquiring crystal structures
Most prominent sources of polygalacturonases namely Erwinia carotovora and Themotoga maritima were considered for the present work. The crystal structures of mesophile (Erwinia carotovora) and thermophile (Thermotoga maritima) that were deposited in Protein Data Bank (PDB) were retrieved. Structures of less than 2A0 resolution with an R- value of zero which are deposited through X-ray diffraction method rather than NMR were preferred. Psychrophile ‘Pseudoalteromonas haloplanktis’ was selected for the present work and as the X- ray crystallized structure is not available in PDB, the FASTA sequence deposited in NCBI (Accession number: WP_ 004334884.1) was used for homology modeling studies.
Molecular modeling studies
Polygalacturonase protein sequence from psychrophilic organism Pseudoalteromonas haloplanktis was collected from NCBI and searched against PDB using BLAST-P (Russell 2000) to identify the potential templates for carrying out molecular modeling studies. Templates are selected based on the highest similarity and good identity in consideration with E- value. MODELLER 9v10 program was used for homology or comparative modeling of proteins 3D model (Eswar 2008) and was submitted to PROCHECK program of SAVS (Structural Analysis and Verification Server) (Shen and Sali 2006; Laskowski 1996) to determine the stereo chemical quality of the modeled structures based on Ramachandran plot.
Analysis of substrate binding site
Information about the active site was retrieved from PDB database and also determined by the help of multiple dockings between the protein and substrate thereby selecting the residues of best confirmation. Blind docking is carried out by the help of Autodock tool wherein gridbox is used to locate binding site where ligand is observed to bind multiple times. In addition to details from blind docking, residues were also considered based on Q-site finder which uses the interaction energy between protein and van derwaals radius so that energetically favourable regions of the protein were identified. These regions are clustered based on clusters and spatial distribution by the help of sum of interaction energies related to each cluster (Laurie and Jackson 2005).
Molecular docking studies
Preparation of proteins
Structures of mesophilic (1BHE) and thermophilic (3JUR) polygalacturonase enzymes were taken from PDB and are used for docking studies. Hetero atoms in the form of water molecules, cocrystallized molecules and bound ligands were removed. Bonding in HET atoms was corrected and charges were adjusted. The modeled psychrophilic structure was taken and energy minimization or geometry optimization is carried out by using the hyperchem software to optimize the molecular structure until the gradients of potential energy on atoms become negligible. For carrying out docking studies, only half of the tetrameric molecule (1BHE and 3JUR) was considered so as to skip any detrimental conditions during the substrate binding (Adam et al. 2008).
Preparation of ligand
The 2D and 3D structures of polygalacturonic acid was retrieved from chemspider database and are represented in Fig. 1a and b respectively. The structure was downloaded in MOL format and then it was converted to PDB by the help of OPEN BABEL 2.2.3 (O’Boyle et al. 2011). Hydrogen’s were added to the structure using Pymol software and then it was subjected to energy minimization by using hyperchem to avoid stereochemical changes. The obtained energy minimized structures were then further used for docking studies.
Fig. 1.
a: 2D structure of PGA. b: 3D structure of PGA
Docking studies
Graphical User Interface program “AutoDock 4.2” was used to prepare, run, and analyze the protein-ligand docking simulations. Firstly, Kollman united atom charges and hydrogens were added to the receptor PDB file for the preparation of protein. Then ligand was prepared by adding gasteiger charges and also torsions were noticed. Docking software requires pre calculated grid maps for all the atom types present in the flexible molecules being docked and also automatically stores the potential energy generated by the interactions with rigid macromolecules. Grid box has been set around the region of interest in the rigid part of protein. Autogrid uses the algorithm to get the grid results and autodock by Lamarckian Genetic Algorithm (LGA) was chosen for obtaining best conformers. A maximum of 100 generations was considered where the population size was set to 150 with a maximum number of energy evaluations 500,000. As there is no prior knowledge of active site details related to Pseudoalteromonas haloplanktis the structure was submitted to Q-site finder to know the details about the binding pocket. Information about binding pocket residues obtained by Q-site finder was given wherein the grid box is covered for those residues. A grid parameter file (gpf) was generated and the information is passed on to the docking parameter file (dpf) where flexible docking is carried out for that specific area of the protein to optimize the results (Huey 2007; Morris and Lim-Wilby 2008).
Results
Retrieval of polygalacturonase sequences and multiple sequence alignment
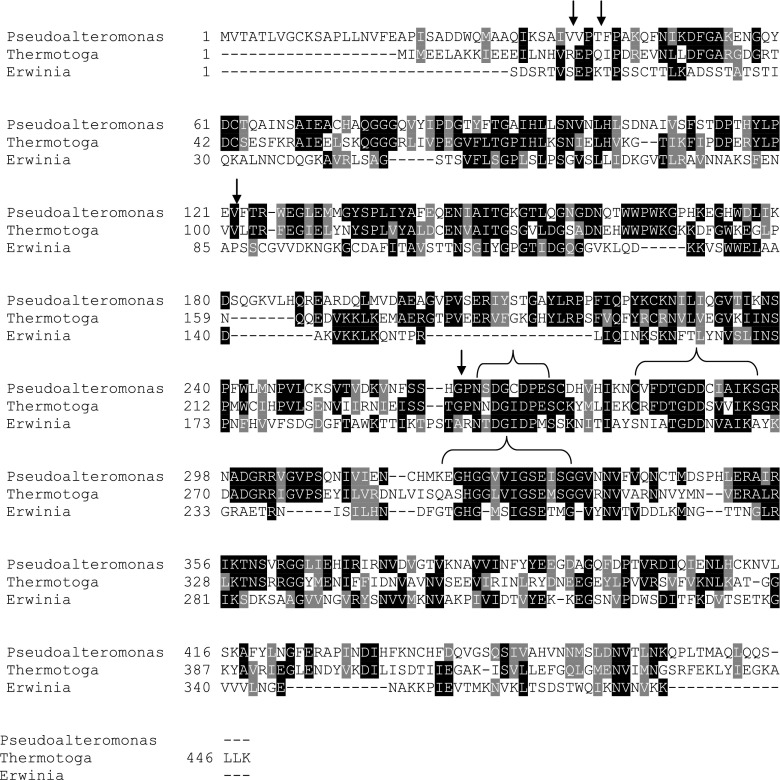
Totally 32 polygalacturonases sequences sourced from bacteria and fungi that are well reported and utmost producers of enzyme were retrieved from NCBI. The details of organisms, their accession numbers and the details about the length of amino acids were specified (Table 1). The selected sources were having mixture of mesophiles, thermophiles and psychrophiles and chosen based on their potentiality in major applications at industrial level as provided in the literature (Adapa et al. 2014). ClustalW result clearly shows that most of the amino acids are highly conserved and some of the significant aminoacids are found to be involved in the formation of the active site (Fig. 2).
Table 1.
Polygalacturonases from different source organisms
| Sl.No. | Name of the organism | Accession number | Length (Amino acids) |
|---|---|---|---|
| Bacteria | |||
| 1. | Pseudomonas syringae | EFW82829.1 | 538 |
| 3. | Enterobacteriaceae | EFV41697.1 | 444 |
| 4. | Xanthomonas translucens | ELQ07192.1 | 558 |
| 5. | Hafnia alvei | EHM46199.1 | 444 |
| 6. | Faecalibacterium prausnitzii | EFQ06666.1 | 510 |
| 7. | Rhizobium leguminosarum | CAK12397.1 | 454 |
| 8. | Lachnospiraceae | EHO52007.1 | 526 |
| 9. | Clostridium hathewayi | EFD00955.1 | 483 |
| 10. | Oribacterium | EFE91614.1 | 526 |
| 11. | Bacteroides clarus | EGF50128.1 | 452 |
| 12. | Enterococcus faecium | EJY53347.1 | 436 |
| 13. | Erwinia pyrifoliae | CAY73958.1 | 397 |
| 14. | Capnocytophaga | EGJ54358.1 | 483 |
| Fungi | |||
| 1. | Rhizoctonia solani | ELU43744.1 | 324 |
| 2. | Magnaporthe oryzae Y34 | ELQ41997.1 | 364 |
| 3. | Colletotrichum gloeosporioides | ELA35572.1 | 373 |
| 4. | Thanatephorus cucumeris | AEK97544.1 | 308 |
| 5. | Penicillium digitatum | EKV09364.1 | 697 |
| 6. | Cryptococcus neoformans | AFR97960.1 | 461 |
| 7. | Fomitiporia mediterranea | EJD03049.1 | 368 |
| 8. | Alternaria tenuissima | AFM35588.1 | 148 |
| 9. | Stereum hirsutum | EIM80998.1 | 362 |
| 10. | Galactomyces citri-aurantii | AFH77948.1 | 367 |
| 11. | Rhizopus delemar | EIE80026.1 | 383 |
| 12. | Colletotrichum higginsianum | CCF33898.1 | 330 |
| 13. | Verticillium dahliae | EGY14086.1 | 469 |
| 14. | Chaetomium thermophilum | EGS18138.1 | 490 |
| 15. | Aspergillus sojae | BAK22527.1 | 363 |
| 16. | Leucoagaricus gongylophorus | ADV30326.1 | 361 |
| 17. | Cryptococcus gattii | ADV21054.1 | 474 |
| 18. | Penicillium griseoroseum | AAC83692.1 | 376 |
Fig. 2.
Multiple sequence alignment of polygalacturonase sequences from meso, thermo and psychrophiles. Arrows represent the most important residues of the active site of the enzyme
Analysis of secondary structures
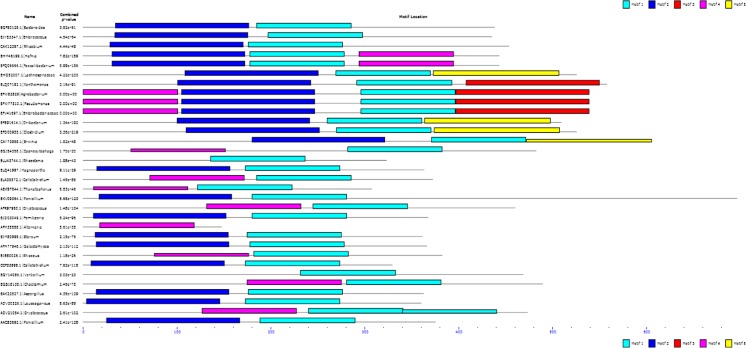
MEME finds five motifs and each of them were present in most of the input sequences (Bailey and Elkan 1994) (Fig. 3). The width and number of occurrences in each of the five motifs were chosen in order to minimize the E-value of the motif. The motif width in the range of 100–200 was specified. Totally, five motifs labeled as 1–5 were considered in the selected sequences. The distribution of the motifs in all the sequences is clearly mentioned in Table 2. Also, other details such as width of motif, sequence information and possible matches were also represented in Table 3. This clearly shows that all most all the organisms contain similar motif showing conservation which signifies motifs role in structural and catalytic attributes.
Fig. 3.
Combined block diagram of motifs in polygalacturonase sequences from various sources
Table 2.
Distribution of motifs among 32 polygalacturonase proteins sequences from different source organisms
| Sl.No. | Name of the organism | Motif 1 | Motif 2 | Motif 3 | Motif 4 | Motif 5 |
|---|---|---|---|---|---|---|
| 1 | Pseudomonas syringae | + | + | + | + | − |
| 2 | Enterobacteriaceae | + | + | + | + | − |
| 3 | Xanthomonas translucens | + | + | + | − | − |
| 4 | Hafnia alvei | + | + | − | + | − |
| 5 | Faecalibacterium prausnitzii | + | + | − | + | − |
| 6 | Rhizobium leguminosarum | + | + | − | − | − |
| 7 | Lachnospiraceae | + | + | − | − | + |
| 8 | Clostridium hathewayi | + | + | − | − | + |
| 9 | Oribacterium | + | + | − | − | + |
| 10 | Bacteroides clarus | + | + | − | − | − |
| 11 | Enterococcus faecium | + | + | − | − | − |
| 12 | Erwinia pyrifoliae | + | + | − | − | + |
| 13 | Capnocytophaga | + | − | − | + | − |
| 14 | Rhizoctonia solani | + | − | − | − | − |
| 15 | Magnaporthe oryzae Y34 | + | + | − | − | − |
| 16 | Colletotrichum gloeosporioides | + | − | − | + | − |
| 17 | Thanatephorus cucumeris | + | − | − | + | − |
| 18 | Penicillium digitatum | + | + | − | − | − |
| 19 | Cryptococcus neoformans | + | − | − | + | − |
| 20 | Fomitiporia mediterranea | + | + | − | − | − |
| 21 | Alternaria tenuissima | − | − | − | + | − |
| 22 | Stereum hirsutum | + | + | − | − | − |
| 23 | Galactomyces citri-aurantii | + | + | − | − | − |
| 24 | Rhizopus delemar | + | − | − | + | − |
| 25 | Colletotrichum higginsianum | + | + | − | − | − |
| 26 | Verticillium dahliae | + | − | − | − | − |
| 27 | Chaetomium thermophilum | + | − | − | + | − |
| 28 | Aspergillus sojae | + | + | − | − | − |
| 29 | Leucoagaricus gongylophorus | + | + | − | − | − |
| 30 | Cryptococcus gattii | + | − | − | + | − |
| 31 | Penicillium griseoroseum | + | + | − | − | − |
| 32 | Agrobacterium | + | + | + | + | − |
Table 3.
Motif information with sequence logo and regular expression of polygalacturonase enzyme

Phylogenetic analysis
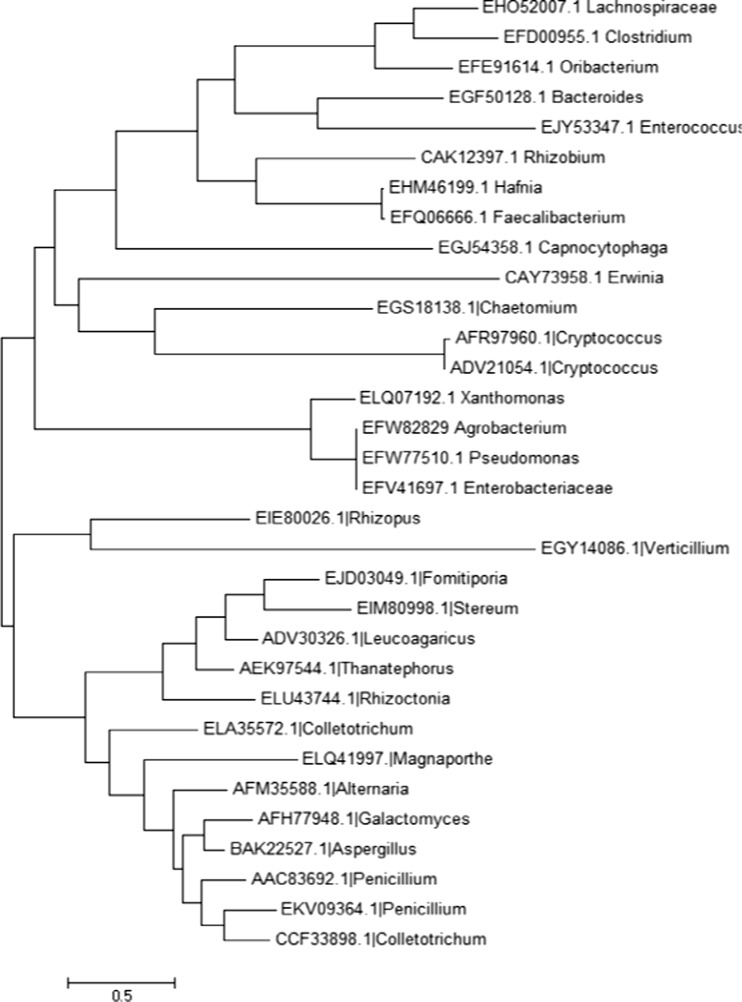
The selected sequences (protein) sourced from both bacteria and fungi were aligned with the ClustalW program. By using the method of maximum likelihood, dendrogram was constructed using the software MEGA where bacteria and fungi appear in two different clusters showing sequence level similarity. Multiple accessions related to fungi namely Rhizopus delemar and Verticillium dahlia showed different clusters showing sequence level similarity. The organisms Magnaporthe, Alternaria, Colletotrichum, Galactomyces, Aspergillus and Pencillium showed different clusters among fungi which are similar at the sequence level. Also, Fomitiporia, Stereum, Leucoagaricus, Thanatephorus and Rhizoctonia which are similar showed different cluster from others. In bacteria, Xanthomonas, Agrobacterium, Pseudomonas and Enterobacteriaceae showed a different group of cluster (Fig. 4). Dendrograms were also constructed by using other methods such as minimum evolution, UPGMA and maximum parsimony which showed an almost similar pattern representing sequence level similarity of polygalacturonases from various sources (Supplementary figures 1, 2, 3).
Fig. 4.
Phylogenetic tree of polygalacturonase sequences of different organisms constructed by maximum likelihood method
Molecular modeling studies
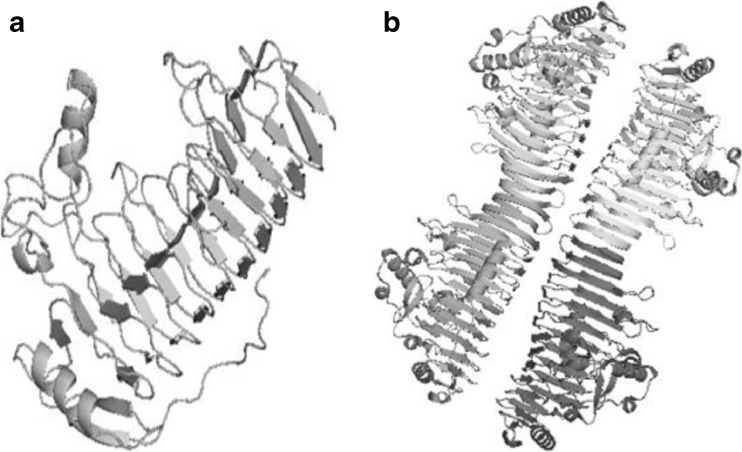
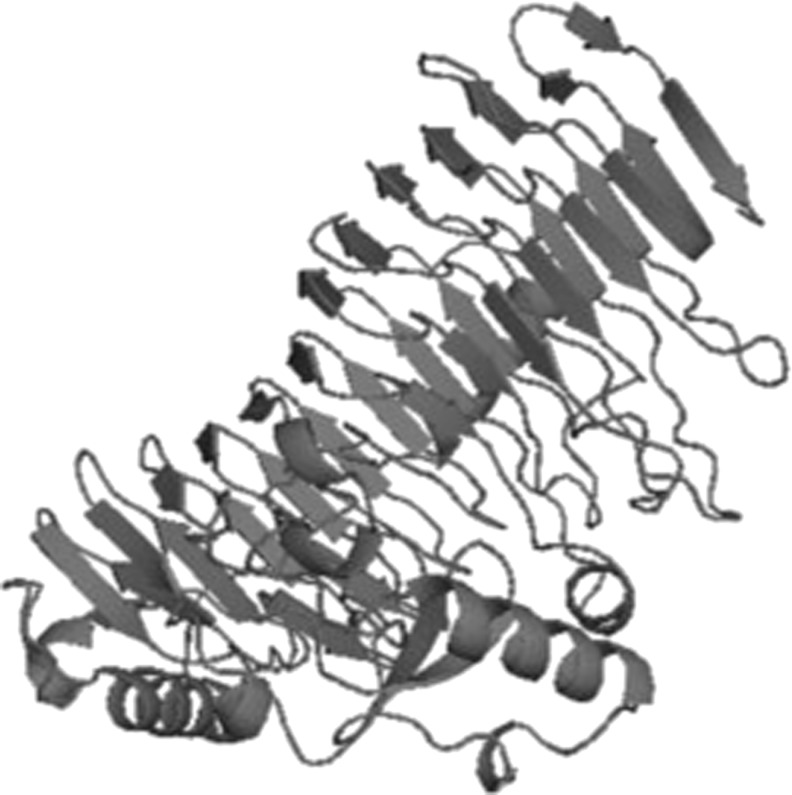
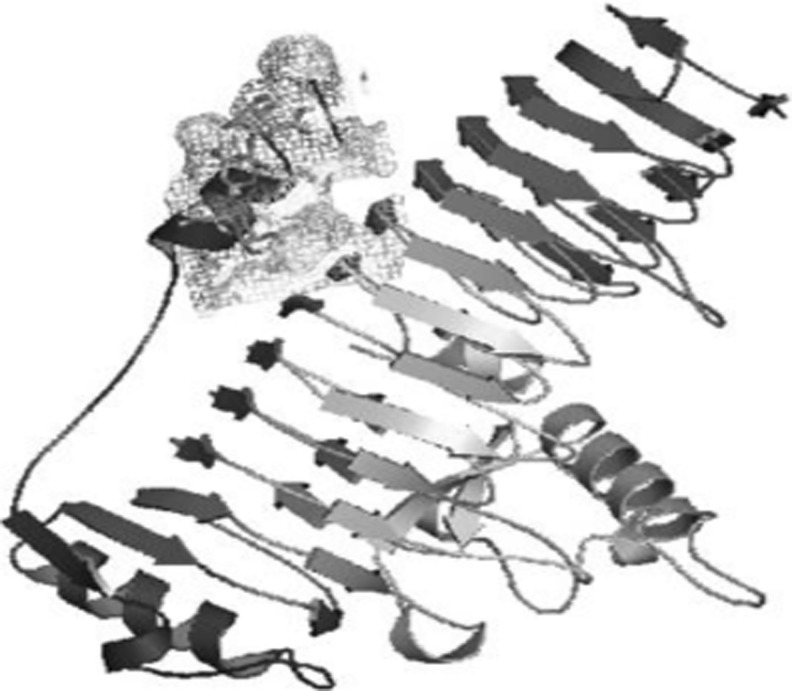
Protein sequence related to mesophile i.e., Erwinia carotovora and thermophile Thermotoga maritima were obtained from PDB bearing IDs 1BHE and 3JUR respectively (Fig. 5). Template structure for modeling protein polygalacturonase from Pseudoalteromonas haloplanktis was searched using BLAST and the highest hit was found to be with 3JUR. By the help of this, a valid 3D model was generated by using Modeler 9v10 (Fig. 6). The model with best DOPE score of −51978.67578 and GA 341 score of 1 was selected. Then finally the model was subjected to further validation studies like PROCHECK and found 81 % of favored region, 13.9 % allowed region and 2.2 % disallowed region.
Fig. 5.
Structure of Polygalacturonase enzyme from a Erwinia carotovora b Thermotoga maritima
Fig. 6.
Modeled Polygalacturonase enzyme from Pseudoalteromonas haloplanktis
Validation of functional sites
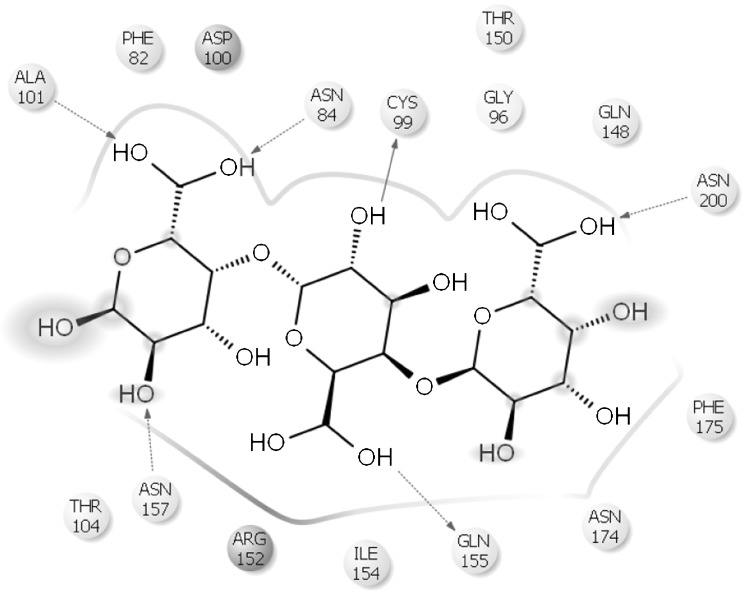
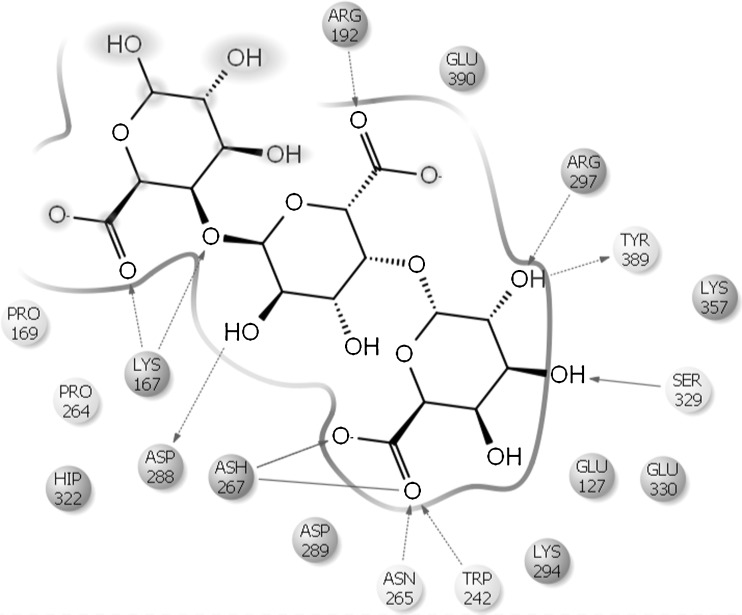
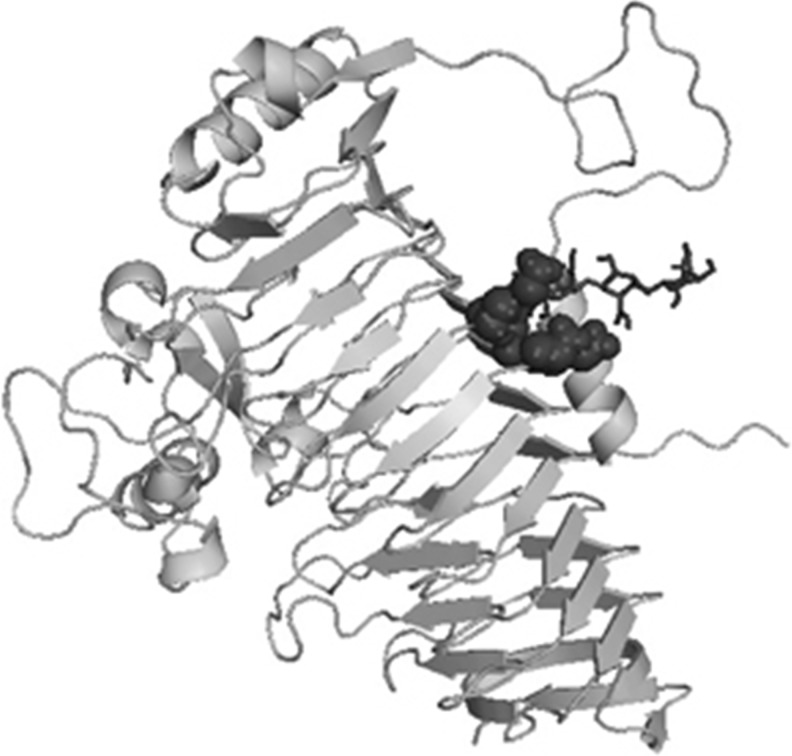
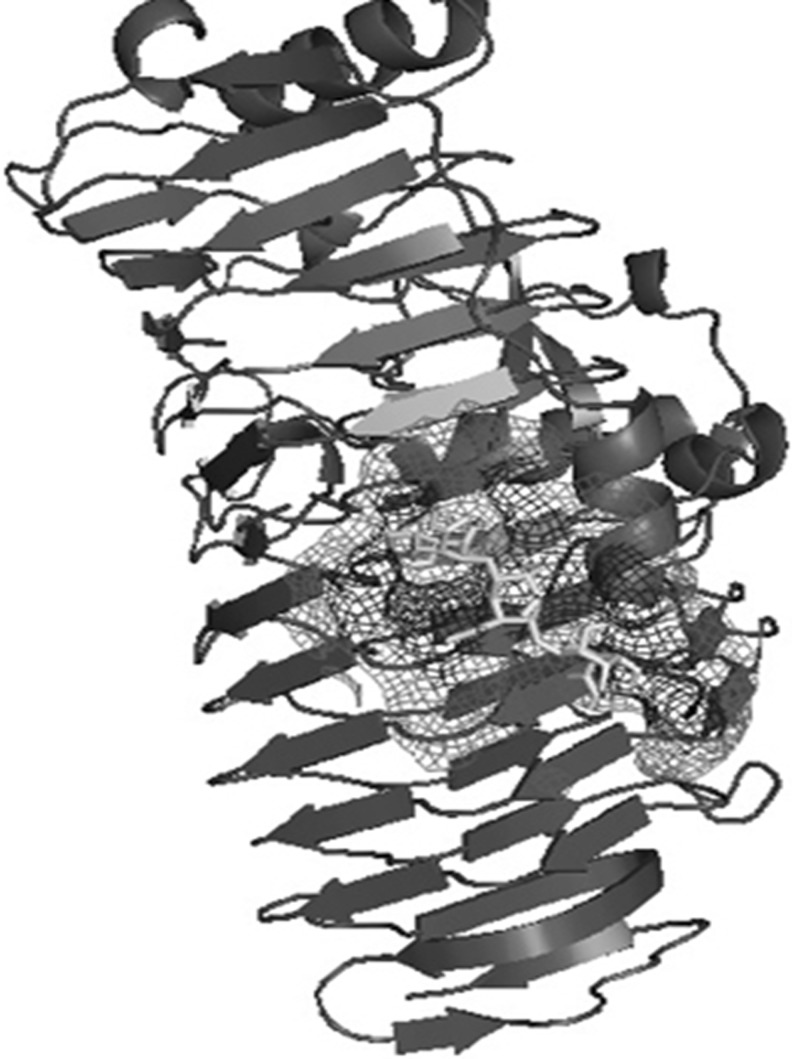
Modelled cold active PGase from P.haloplanktis and the enzymes from Erwinia carotovorum and Thermotoga maritima were further verified for their active site residues by using the blind docking studies. Also, the residues were cross checked with the structures present in PDB associated with a ligand and also by Q- site finder which is found to be same. Active site residues for mesophile, thermophile and psychrophile were tabulated in Table 4 and also represented in Figs. 7, 8 and 9.
Table 4.
Active site residues of polygalacturonase enzyme from all the source organisms
| S.NO | Source | Active site residues |
|---|---|---|
| 1. | Erwinia carotovorum | ALA101,ASN84,CYS99, GLN148, ASN200, THR104,ASN157,GLN155,ASN174,PHE175 |
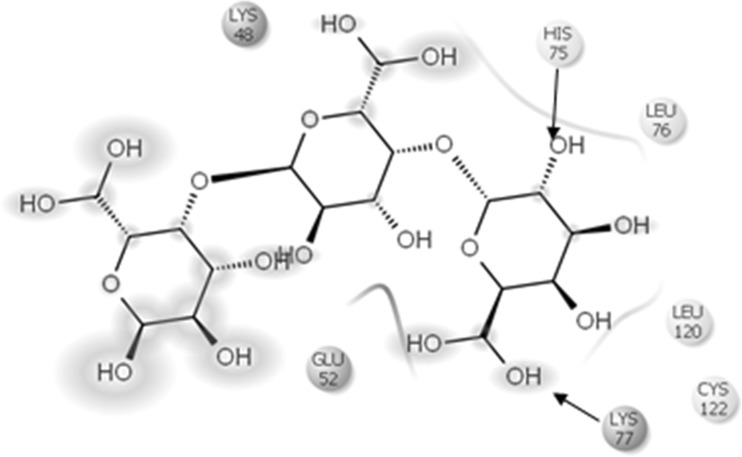
| 2. | Thermotoga maritima | LYS48,HIS75,LEU76,LEU120,CYS122,LYS77,ASP239, LYS329,ILE241,ASP260 |
| 3. | Pseudoalteromonas haloplanktis | ARG192, GLU390, ARG297,TYR389,SER329,TRP242,ASN265,ASP288,LYS167,PRO169 |
Fig. 7.
Polygalacturonase from Erwinia carotovorum docked with PGA
Fig. 8.
Polygalacturonase from Pseudoalteromonas haloplanktis docked with PGA
Fig. 9.
Polygalacturonase from Thermotoga maritima docked with PGA
Molecular docking studies
Polygalacturonase enzyme from mesophile, thermophile and psychrophile were used for further docking studies. Substrate PGA was energy minimized by hyperchem and the energy was found to be −6355.7948 KCals/mole. Protein structures were docked with PGA and their binding energies were calculated (Figs. 10, 11 and 12). Differences in binding affinities were noticed where psychrophilic enzyme has shown to be most favorable. Binding energies for all the three substrates were found to be −5.73, −6.22 and −7.27 KCals/mole for meso, thermo and psychrophiles respectively.
Fig. 10.
Enzyme Polygalacturonic acid docked with PGA (in mesh) from Erwinia carotovora
Fig. 11.
Enzyme Polygalacturonic acid docked with PGA (in spheres) from Pseudoalteromonas haloplanktis
Fig. 12.
Enzyme Polygalacturonic acid docked with PGA (in mesh) from Thermatogo maritima
Discussion
Pectins are the substances present in middle lamella and primary cell wall of higher plants (Shefali et al. 2008). Pectinases are the enzymes that are capable of degrading the pectin and are in use for decades in food and wine making industries (Ribeiro et al. 2010). PGases are a class of pectinases present in fruits and are known to be depolymerizing enzymes (Voragen et al. 2003). Even though the use of PGase is in huge demand in many industries, fruit industry stands first with promising applications such as clarification of juice, in the process of vinification, yield and color enhancement and in the mashing of fruits (Chawanit et al. 2007). Most of the industrial enzymes used were from mesophilic and thermophilic origin out of which thermophilic enzymes are employed due to their property of thermal stability. But it is important to maintain low temperatures (10–12 °C) during many food processes as this enhances the shelf life, aroma, taste and flavor of the product (Molina et al. 2007). Cold active enzymes are very significant in several food industries for processing of food-stuffs as foods should be treated under mild conditions to avoid taste and spoilage of food material (Margesin and Schinner 1994; Russell 1998; Gerday et al. 2000; Feller 2013). Cold active enzymes show very high catalytic activity at low or moderate temperatures and are thermolabile. Psychrozymes have increased flexibility that leads to extreme catalytic activity whereas mesophiles and thermophiles contain rigid protein structures to withstand high temperatures (Hochachka and Somero 1984; Somero 2004; Methe et al. 2005; Adapa et al. 2014; Pulicherla et al. 2011; Rekha et al. 2013). Other most significant and general feature of cold active microorganisms is the modifications in the primary sequences in order to withstand the lower temperatures. Also it was reported that psychrophiles contain greater number of flexible regions when compare to meso and thermophiles which enhances the chances of substrate accessibility. Also, additions and deletions of some of the amino acids in the loop regions nearer to catalytic site also helps to enhance accessibility to substrate accommodation (Marx 2004).
Polygalacturonase from mesophiles, thermophiles and psychrophiles were selected from various sources using bioinformatic tools to study about similarity at the sequence level. ClustalW clearly showed that most of the amino acids were found to be conserved in between meso, thermo and psychrophiles. Amino acids such as aspragine, proline, glycine, aspartic acid were found to be highly conserved (Pickersgill et al. 1998). Arginine content plays a major role in thermal adaptation. Cold active enzymes have a reduced arginine content and decreased number of salt bridges when compared to thermophiles. In the present study also, it has been observed that psychrophilic polygalacturonase Peudoalteromonas haloplanktis has arginine content less than thermophilic organism.
Further analysis of enzymes for their motifs and domain conservation was carried out to understand the possible functions related to the structure building, residues involved in the formation of the active site that helps to know about the stability of the enzyme. This eventually becomes a target for genetic engineering studies and also for further exploring them for large scale industrial production. The domain analysis revealed the information about the conservation which might confer the structural flexibility of the enzyme that influences its catalytic function. Modeled structure of psychrophile Pseudoalteromonas haloplanktis obtained a very good favorable region that has been verified by Ramachandran plot. Even the template structure (3JUR) was also subjected to evaluation which also showed a favorable region of 85 % which is more or less similar to the modeled protein. Docking studies confirmed that cold active enzymes showed a very good affinity and are energetically favourable towards the substrate when compared with mesophilic and themophilic enzymes. Hydrogen bonds are formed between the substrate and the residues of the active site. Presence of multiple H-bonds between active site of the protein and the ligand polygalacturonic acid is significant enough for strong bonding interactions (Patil et al. 2010).
In Erwinia, substrate ligand interactions are observed to be due to the residues ASN84, CYS99, ALA101, GLN155, ASN157 and ASN200 whereas in the case of Thermotoga, most two prominent amino acids between the substrate and the active site include LYS77 and HIS75. In the case of Pseudoalteromonas, similar role is played by LYS167, ARG192, TRP242, ASN265, ARG297 and SER329. The reasonable low binding energy of psychrophiles clearly indicates their higher efficiency when compared with its counterparts. The favorable binding energy of psychrophilic enzyme was found to be encouraging for further research in the perspective of application of cold active enzymes at industrial level particularly in food processing.
Food processing industries prefer low temperature treatments rather than processing at high temperatures as they have the advantages such as retaining nutritional value, avoid spoilage and retain taste which are reported to be very common problems in the area of the food industry (Nakagawa et al. 2004). So one has to explore this area in a greater detail to address the questions related to the efficiency of enzyme at low temperatures, rate of reactions and their economic feasibility with that of normal mesophilic and thermophilic enzymes (Feller and Gerday 2003). Based on our present in silico work, it has been confirmed that cold active enzymes may have much more advantages than the others which makes it worthwhile to evaluate. Further research in this area helps to use the cold adapted enzymes tremendously at industrial level.
Conclusion
Enzymes from psychrophiles were found to be most valuable components in various food processing operations. Their unique properties like high specificity and catalytic activity at low temperatures make them very attractive. In the present work, in silico characterization of polygalacturonases sourced from mesophiles, thermophiles and psychrophiles was done. All the structures of polygalacturonases from three different habitats were used for further docking studies by using PGA as substrate which could become a priliminary step for further in vitro experiments. The cold adapted organism selected here is a well reported marine psychrophile Pseudoalteromonas haloplanktis which has proved to be equally efficient when compared with others and will become a value addition to the industry particularly with respect to fruit and vegetable processing. Future studies with respect to cold active polygalacturonases will help to explore them more at industrial level.
Electronic supplementary material
(DOC 64 kb)
(DOC 66 kb)
(DOC 64 kb)
Contributor Information
L. N. Ramya, Email: nagramya@gmail.com
K. K. Pulicherla, Phone: 91-09488292034, Email: kkpulicherla@gmail.com
References
- Adam J, Kifz Z, Prokop M, Wimmerova M, Koca J. In silico mutagenesis and docking studies of Pseudomonas aeroginosa PA-IIL lectin predicting binding modes and energies. J Chem Inf Model. 2008;48:2234–2242. doi: 10.1021/ci8002107. [DOI] [PubMed] [Google Scholar]
- Adapa V, Ramya LN, Pulicherla KK, Sambasiva Rao KRS (2014) Cold active pectinases: advancing the food industry to the next generation. Appl Biochem Biotechnol 172(5):2324–2337. doi:10.1007/s12010-013-0685-1 [DOI] [PubMed]
- Alphons GJ, Voragen Æ, Gerd-Jan CÆ, René P, Verhoef Æ, Henk A, Schols REVIEW ARTICLE: pectin, a versatile polysaccharide present in plant cell walls. Struct Chem. 2009;20:263–275. doi: 10.1007/s11224-009-9442-z. [DOI] [Google Scholar]
- Bailey TL, Elkan C (1994) “Fitting a mixture model by expectation maximization to discover motifs in biopolymers,” in Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology, pp. 28–36, AAAI Press [PubMed]
- Carmen S, Garcia-Fraga B, Lopez-Seijas J, da Silva AF, Tomas GV (2012) Food industrial processes- methods and equipment. Agricultural and Biotechnological Sciences. ISBN 978-953-307-905-9
- Chawanit S, Surang S, Lerluck C, Vittaya P, Pilanee V, Prisnar S. Screening of pectinase producing bacteria and their efficiency in biopulping of paper mulberry bark. Sci Asia. 2007;33:131–135. doi: 10.2306/scienceasia1513-1874.2007.33.131. [DOI] [Google Scholar]
- Eswar N (2008) Methods Mol Biol 426:145. [PMID: 18542861] [DOI] [PubMed]
- Feller G. (2013) Psychrophilic enzymes: from folding to function and Biotechnology. Sci 13:512840. doi:10.1155/2013/512840 [DOI] [PMC free article] [PubMed]
- Feller G, Gerday C. Psychrophilic enzymes: hot topics in cold adaptation. Nat Rev Microbiol. 2003;1:200–208. doi: 10.1038/nrmicro773. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. Nucleic acids research. Database Issue. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerday C, Aittaleb M, Bentahir M, Chessa JP, Claverie P, Collins T, D’Amico S, Dumont J, Garsoux G, Georlette D, Hoyouz A, Lonhienne T, Meuwis MA, Feller G. Cold- adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol. 2000;18:103–107. doi: 10.1016/S0167-7799(99)01413-4. [DOI] [PubMed] [Google Scholar]
- Gomes J, Steiner W (2004) The biocatalytic potential of extremophiles and extremozymes. Food Technol Biotechnol 42(4):223–235
- Hochachka PW, Somero GN, editors. Biochemical adaptations. Princeton: Princeton University Press; 1984. pp. 355–449. [Google Scholar]
- Huey R (2007) J Comput Chem 28:1145. [PMID: 17274016] [DOI] [PubMed]
- Jose M, Rodriguez N, Natividad O, Manuel P, Maria DB. Pectin hydrolysis in a free enzyme membrane reactor: an approach to the wine and juice clarification. Food Chem. 2008;107(112):119. [Google Scholar]
- Kertesz Z (1930) A new method of enzymatic clarification of unfermented apple juice. US patent no. 1.932.833, New York State Agricultural Experimentation Station (Geneva) Bull. No. 689
- Laskowski RA (1996) J Biomol NMR 8:477. [PMID: 9008363] [DOI] [PubMed]
- Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- Margesin R, Schinner F. Properties of cold-adapted micro-organisms and their potential role in biotechnology. J Biotechnol. 1994;33:1–14. doi: 10.1016/0168-1656(94)90093-0. [DOI] [Google Scholar]
- Marx JC. A perspective on cold enzymes: current knowledge and frequently asked questions. Cell Mol Biol. 2004;50(5):643–655. [PubMed] [Google Scholar]
- Methe BA, Nelson KE, Deming JW, Momen B, Melamud E, Zhang XJ, Moult J, Madupu R, Nelson WC, Dodson RJ, Brinkac LM, Daugherty SC, Durkin AS, DeBoy RT, Kolonay JF, Sullivan SA, Zhou LW, Davidsen TM, Wu M, Huston AL, Lewis M, Weaver B, Weidmann JF, Khouri H, Utterback TR, Feldblyum TV, Fraser CM. The psychrophilic lifestyle as revealed by the genome sequence of Colwelliapsychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci U S A. 2005;102:10913–10918. doi: 10.1073/pnas.0504766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina AM, Swiegers JH, Varela C, Pretorius IS, Agosin E. Influence of wine fermentation temperature on the synthesis of yeast-derived volatile aroma compounds. Appl Microbiol Biotechnol. 2007;77:675–687. doi: 10.1007/s00253-007-1194-3. [DOI] [PubMed] [Google Scholar]
- Morris GM, Lim-Wilby M (2008) Methods Mol Biol 443:365. [PMID: 18446297] [DOI] [PubMed]
- Nakagawa T, Nagaoka T, Taniguchi S, Miyaji T, Tomizuka N. Isolation and characterization of psychrophilic yeasts producing cold-adapted pectinolytic enzymes. Lett Appl Microbiol. 2004;38:383–387. doi: 10.1111/j.1472-765X.2004.01503.x. [DOI] [PubMed] [Google Scholar]
- O’Boyle NM, Banck M, Craig AJ, Morley C, Vandermeersch T, Hutchison GR. Open babel: an open chemical toolbox. J Chem Inf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palivanelu P. Polygalacturonases: active site analyses and mechanism of action. Indian J Biotechnol. 2006;5:148–162. [Google Scholar]
- Patil R, Das S, Stanley A. Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS ONE. 2010;5(8):1–10. doi: 10.1371/journal.pone.0012029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill R, Smith D, Worboys K, Jenkins J. Crystal structure of polygalacturonase from 434 Erwinia carotovora ssp. carotovora. J Biol Chem. 1998;273:24660–24664. doi: 10.1074/jbc.273.38.24660. [DOI] [PubMed] [Google Scholar]
- Pulicherla KK, Ghosh M, Kumar PS, Sambasiva Rao KRS. Psychrozymes- the next generation industrial enzymes. J Marine Sci Res Dev. 2011;1:102. doi: 10.4172/2155-9910.1000102. [DOI] [Google Scholar]
- Rekha VPB, Mrinmoy Ghosh, Vijayanand Adapa, Sung-Jong Oh, Pulicherla KK, and Sambasiva Rao KRS (2013) Optimization of polygalacturonase production from a newly isolated Thalassospira frigidphilosprofundus to use in pectin hydrolysis: statistical approach. BioMed Research International. Volume 2013, ID 750187. doi: 10.1155/2013/750187 [DOI] [PMC free article] [PubMed]
- Ribeiro DS, Henrique SMB, Oliveira LS, Macedo GA, Fleuri LF. Enzymes in juice processing: a review. Int J Food Sci Technol. 2010;45:635–641. doi: 10.1111/j.1365-2621.2010.02177.x. [DOI] [Google Scholar]
- Russell NJ. Molecular adaptations in psychrophilic bacteria: potential for biotechnological applications. Adv Biochem Eng Biotechnol. 1998;61:1–21. doi: 10.1007/BFb0102287. [DOI] [PubMed] [Google Scholar]
- Russell NJ (2000) Extremophiles 4:83. [PMID: 10805562] [DOI] [PubMed]
- Shefali G, Mukesh K, Krishna Kant S, Lavanya MN, Ramesh Chander K. Production and recovery of an alkaline exo-polygalacturonase from Bacillus subtilis RCK under solid-state fermentation using statistical approach. Bioresour Technol. 2008;99:937–945. doi: 10.1016/j.biortech.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Shen MY, Sali A (2006) Protein Sci 15:2507. [PMID: 17075131] [DOI] [PMC free article] [PubMed]
- Shi GY, Jie TY, Bing TH, Hua WK, Wei CK. Chin J Agric Biotechnol. 2007;4:33–38. doi: 10.1017/S1479236207001234. [DOI] [Google Scholar]
- Somero GN. Adaptation of enzymes to temperature: searching for basic “strategies”. Comp Biochem Physiol B Biochem Mol Biol. 2004;139:321–333. doi: 10.1016/j.cbpc.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong LV, Tuyen H, Helmke E, Binh LT, Schweder T. Cloning of two pectate lyase genes from the marine Antarctic bacterium Pseudoalteromonas haloplanktis strain ANT/505 and characterization of the enzymes. Extremophiles. 2001;5(1):35–44. doi: 10.1007/s007920000170. [DOI] [PubMed] [Google Scholar]
- Voragen F, Schols H and Visser R Eds. (2003) Advances in pectin and pectinase research. Annals of Botany 94, 479–480
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 64 kb)
(DOC 66 kb)
(DOC 64 kb)