Abstract
Purpose
To evaluate the prognostic impact of cell-of-origin (COO) subgroups, assigned using the recently described gene expression–based Lymph2Cx assay in comparison with International Prognostic Index (IPI) score and MYC/BCL2 coexpression status (dual expressers).
Patients and Methods
Reproducibility of COO assignment using the Lymph2Cx assay was tested employing repeated sampling within tumor biopsies and changes in reagent lots. The assay was then applied to pretreatment formalin-fixed paraffin-embedded tissue (FFPET) biopsies from 344 patients with de novo diffuse large B-cell lymphoma (DLBCL) uniformly treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) at the British Columbia Cancer Agency. MYC and BCL2 protein expression was assessed using immunohistochemistry on tissue microarrays.
Results
The Lymph2Cx assay provided concordant COO calls in 96% of 49 repeatedly sampled tumor biopsies and in 100% of 83 FFPET biopsies tested across reagent lots. Critically, no frank misclassification (activated B-cell–like DLBCL to germinal center B-cell–like DLBCL or vice versa) was observed. Patients with activated B-cell–like DLBCL had significantly inferior outcomes compared with patients with germinal center B-cell–like DLBCL (log-rank P < .001 for time to progression, progression-free survival, disease-specific survival, and overall survival). In pairwise multivariable analyses, COO was associated with outcomes independent of IPI score and MYC/BCL2 immunohistochemistry. The prognostic significance of COO was particularly evident in patients with intermediate IPI scores and the non–MYC-positive/BCL2-positive subgroup (log-rank P < .001 for time to progression).
Conclusion
Assignment of DLBCL COO by the Lymph2Cx assay using FFPET biopsies identifies patient groups with significantly different outcomes after R-CHOP, independent of IPI score and MYC/BCL2 dual expression.
INTRODUCTION
Diffuse large B-cell lymphoma (DLBCL) is the most frequent non-Hodgkin lymphoma subtype and represents a morphologically, biologically, and clinically heterogeneous group of malignant diseases.1 More than a decade ago, comparison of gene expression profiling (GEP) of DLBCLs with profiling of normal B cells at different stages of development provided classification into two distinct subtypes: germinal center B-cell–like (GCB) and activated B-cell–like (ABC) subtypes.2,3 This cell-of-origin (COO) classification not only defined subgroups with distinct biology and pathogenesis4 but also identified groups of patients with different outcomes after treatment.5,6
The initial requirement for fresh frozen biopsies and microarray technology has proven to be an insurmountable obstacle to implementation of COO molecular subtyping in routine clinical practice. To overcome these barriers, several immunohistochemistry (IHC) –based algorithms have been proposed.7–9 However, these are limited by their binary nature (not identifying 10% to 15% of biopsies unclassified by GEP) as well as significant interlaboratory and interobserver variability.10 These factors have contributed to the highly discordant literature regarding the prognostic significance of COO subtypes as determined by IHC.11,12 With evidence emerging that novel therapeutic agents have selective activity in ABC and GCB subtypes,13–16 an accurate and reproducible assay for determining COO is imperative to support clinical trials and ultimately identify patients who will benefit from these agents.
Recent improvements in technology have provided the opportunity to use formalin-fixed paraffin-embedded tissue (FFPET) biopsies for reliable GEP.17 We recently reported the feasibility of applying a digital gene expression–based test to FFPET samples for COO assignment.18 The Lymph2Cx assay was shown to be a highly accurate test, with excellent concordance of COO assignment between laboratories.
Over the last 2 years, the assessment of MYC and BCL2 protein expression has emerged as a prognostic biomarker for outcome of patients diagnosed with DLBCL.19–22 In one analysis, it was proposed that the prognostic power of COO was entirely related to more frequent inclusion of MYC/BCL2 dual expressers in the ABC subtype.21 Herein, we demonstrate the consistency and reproducibility of COO assignment using the Lymph2Cx assay and apply the assay to a large patient cohort, uniformly treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP), to investigate the relationship between COO, MYC/BCL2 dual expression, and International Prognostic Index23 (IPI) score with respect to defining prognosis in patients with DLBCL.
PATIENTS AND METHODS
Patient Population
Pretreatment FFPET tumor biopsies of patients diagnosed with de novo DLBCL according to the 2008 WHO classification,1 as determined through standardized review by expert hematopathologists (A.M., P.F., G.W.S., and R.D.G.), were used in this study. Patients were included if they were age ≥ 16 years, were treated with R-CHOP with curative intent at the British Columbia Cancer Agency, and had a matched source of frozen biopsy material to facilitate future genetic analyses. Patients were excluded if they had primary mediastinal large B-cell lymphoma, primary or secondary CNS involvement at diagnosis, previous diagnosis of an indolent lymphoproliferative disorder, positive HIV serology, or either secondary malignancy or major medical comorbidity precluding treatment with curative intent. No selection was based on tumor content of the biopsy.
Patients with advanced-stage disease, defined as Ann Arbor stage III or IV or Ann Arbor stage I or II with either B symptoms or bulk disease (≥ 10 cm) or disease that could not be encompassed within a single involved-field radiation port, were intended to receive six to eight cycles of R-CHOP. All other patients, defined as having limited-stage disease, were eligible to receive three cycles of R-CHOP and involved-field radiation therapy or four cycles of R-CHOP without radiation therapy, if fluorodeoxyglucose positron emission tomography scan was negative after three cycles of R-CHOP. The study was approved by the University of British Columbia–British Columbia Cancer Agency Research Ethics Board.
GEP
Digital GEP was performed on the NanoString platform17 (NanoString Technologies, Seattle, WA) using 200 ng RNA extracted from FFPET biopsies with the Qiagen AllPrep DNA/RNA FFPE kit (Qiagen, Hilden, Germany). The Lymph2Cx 20-gene gene expression–based assay for DLBCL COO was applied as described by Scott et al18 (Data Supplement). COO was not assigned in FFPET biopsies where tumor content was < 10% (Data Supplement lists experiments supporting use of this tumor content threshold). The reagent lots of NanoString code set had identical assignment of the reporter probe fluorescent tag to the target gene.
Tissue Microarray and IHC
All patient cases underwent a centralized review to determine tumor content (reported as percentage of total cell number) and select the area of the paraffin block for tissue microarray (TMA) construction. Duplicate 0.6-mm cores were assembled on three TMAs. After TMA construction, 4-μm slides were cut, and IHC staining was performed on a Benchmark XT platform (Ventana Medical Systems, Tucson, AZ) using the antibodies listed in Data Supplement Table 1. Protein expression was independently determined and recorded as the percentage of positive tumor cells in 10% increments by two hematopathologists (A.M. and P.F.), allowing the application of different thresholds to define positivity of these stains. Unless otherwise stated, the cutoff points established by Johnson et al19 were used for MYC (≥ 40% positive tumor cells) and BCL2 (≥ 50% positive tumor cells). Patient cases with discordant results were evaluated by three pathologists (A.M., P.F., and R.D.G.) at a multiheaded microscope to reach consensus.
Statistical Analysis
Baseline patient characteristics were compared between groups using χ2 and t tests. Time to progression (TTP; event: progression/relapse or death resulting from lymphoma or acute treatment toxicity), progression-free survival (PFS; event: progression/relapse or death resulting from any cause), disease-specific survival (DSS; event: death resulting from lymphoma or acute treatment toxicity), and overall survival (OS; event: death resulting from any cause) were measured from the time of initial pathologic diagnosis. TTP, PFS, DSS, and OS were estimated using the Kaplan-Meier method. Differences in outcome between groups were assessed using the log-rank test. Cox proportional hazard models were used to test the prognostic utility of the groupings when used either alone or in combination with other prognostic factors. Data were analyzed using GraphPad Prism (GraphPad Prism [version 6.0] for Mac; GraphPad Software, La Jolla, CA) and SPSS software (SPSS [version 14.0] for Windows; SPSS, Chicago, IL). Two-sided P < .05 was considered significant.
RESULTS
Lymph2Cx Assay Technical Validation
Previously, the ability of the Lymph2Cx assay to produce concordant results across two laboratories was demonstrated using nonadjacent scrolls of FFPET biopsies that were independently extracted and assayed.18 Here we extended these findings, with a new lot of NanoString (NanoString Technologies) code set, by exploring the effect of using either nonadjacent scrolls or different FFPET blocks from the same diagnostic biopsy on the Lymph2Cx score and subtype assignment. In total, RNA from nonadjacent scrolls from 39 patient cases and different blocks from 10 patient cases were assayed (results shown in Data Supplement Fig 1A). The mean difference in the linear predictor score (LPS) between the samples was 95 points (standard deviation, 87 points). For comparison, the LPS would need to change by 526 points for the subtype to change from GCB to ABC or vice versa. Consistent with this, repeated sampling resulted in concordant COO assignment in 96% of biopsies, with one biopsy shifting from ABC to the unclassified group and the other from GCB to unclassified.
To determine whether the Lymph2Cx assay provided stable subtyping between different code set reagent lots, the assay was run on RNA samples from 83 patient cases of DLBCL using reagents from Scott et al18 and a second reagent lot. Without calibration, the bias in the LPS across the 83 patient cases between the code set lots was 52 points (Data Supplement Fig 1B). When reference oligonucleotides were used to correct for hybridization differences between the lots, this bias was reduced to 26 points (Data Supplement Fig 1C). The concordance of COO calls across the two reagent lots was 100% (Data Supplement Fig 1D). Therefore, the Lymph2Cx assay assigned COO consistently across reagent lots and on repeated sampling of biopsies. Critically, the low variability observed between LPSs translated into no patient cases shifting between the definitive COO subtypes (ie, ABC to GCB or vice versa).
COO Is a Prognostic Biomarker in DLBCL
COO was determined in a cohort of 344 patients uniformly treated with R-CHOP. The median follow-up of living patients was 6.5 years (range, 0.75 to 13.2 years). Comparison of the baseline characteristics of the study group with the British Columbia Cancer Registry–based population of patients with de novo DLBCL treated with R-CHOP at the British Columbia Cancer Agency, over the same time period, showed the cohort was broadly representative of the general population with DLBCL (Table 1). The significantly lower proportion of patients with ≥ two extranodal sites (P < .001) in the study cohort may reflect the exclusion of core needle biopsies and fine-needle aspirates from this study. The outcomes in the study cohort were not significantly different from the registry-based population (Data Supplement Fig 2).
Table 1.
Demographic and Clinical Characteristics of Study Cohort
| Characteristic | Study Cohort (N = 344) | Population-Based Registry (n = 1,194) | P |
|---|---|---|---|
| Age, years | .09 | ||
| Median (range) | 64 (16-92) | 66 (16-93) | |
| > 60, No. (%) | 205 (60) | 771 (65) | |
| Sex, No. (%) | .49 | ||
| Male | 214 (62) | 718 (60) | |
| Female | 130 (38) | 476 (40) | |
| Stage, No. (%) | .11 | ||
| I/II | 165 (49) | 514 (44) | |
| III/IV | 176 (51) | 666 (56) | |
| Missing | 3 | 14 | |
| LDH, No. (%) | .34 | ||
| Normal | 152 (48) | 571 (51) | |
| > ULN | 164 (52) | 545 (49) | |
| Missing | 38 | 78 | |
| ECOG PS, No. (%) | .11 | ||
| 0-1 | 230 (67) | 736 (63) | |
| 2-4 | 111 (33) | 437 (37) | |
| Missing | 3 | 21 | |
| Extranodal sites, No. (%) | < .001 | ||
| 0-1 | 291 (85) | 880 (75) | |
| ≥ 2 | 50 (15) | 297 (25) | |
| Missing | 3 | 17 | |
| IPI score, No. (%) | .29 | ||
| Low (0-1) | 116 (35) | 349 (31) | |
| Intermediate (2-3) | 150 (46) | 511 (46) | |
| High (4-5) | 63 (19) | 250 (23) | |
| Not calculable | 15 | 84 |
NOTE. Bold font indicates significance at P < .05.
Abbreviations: ECOG PS, Eastern Cooperate Oncology Group performance status; IPI: International Prognostic Index; LDH, lactate dehydrogenase; ULN, upper level of normal.
The Lymph2Cx assay was used to assign COO in 99% (339 of 344) of the patient biopsies, with five biopsies being excluded on the basis of tumor content < 10% (Data Supplement Fig 3). The assay assigned a COO for 335 (99%) of these 339 patient cases, with the GEP of four patient cases not passing the previously established quality control criteria.18 Of the entire DLBCL cohort, 32% (108 of 335) were ABC subtype, 56% (189 of 335) were GCB subtype, and 11% (38 of 335) were unclassified.
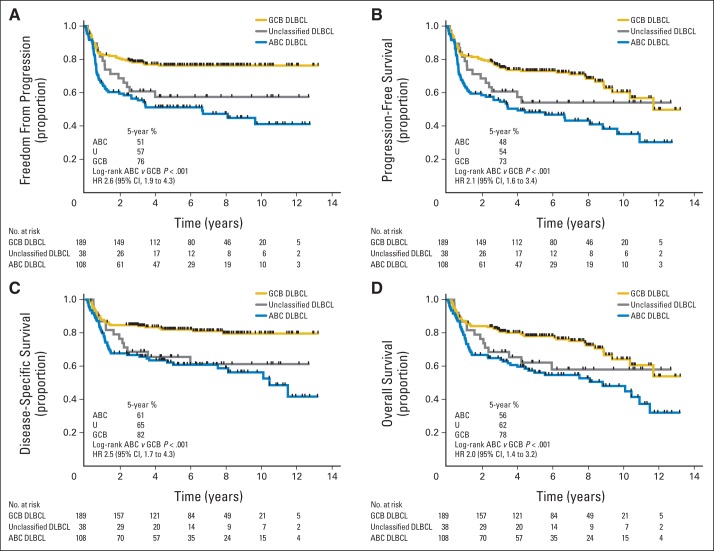
The ABC group experienced significantly inferior outcomes compared with the GCB group (log-rank P < .001 for TTP, PFS, DSS, and OS; Fig 1). Interestingly, no patients with GCB DLBCL experienced relapse after 50 months, despite 110 patients in that group having follow-up beyond that time. Comparison of clinical characteristics between patients with ABC and GCB DLBCL (Table 2) showed that the proportion of patients with stage III to IV disease was greater in the ABC group, despite similar proportions of limited- and advanced-stage disease, as previously defined. Given the different treatments administered in limited- versus advanced-stage disease, we examined the effect of COO on outcome in these two groups separately, again observing significantly inferior outcomes in the ABC subtype (Data Supplement Figs 4 and 5).
Fig 1.
Outcomes in patients with diffuse large B-cell lymphoma (DLBCL) after treatment with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone according to cell of origin. Curves shown for (A) time to progression, (B) progression-free survival, (C) disease-specific survival, and (D) overall survival. ABC, activated B-cell–like DLBCL; GCB, germinal center B-cell–like DLBCL; HR, hazard ratio; U, unclassified DLBCL.
Table 2.
Demographic, Clinical, and Pathologic Characteristics by DLBCL COO Subtype
| Characteristic | ABC DLBCL (n = 108) | GCB DLBCL (n = 189) | Unclassified DLBCL (n = 38) | P (ABC v GCB) |
|---|---|---|---|---|
| Age, years | .30 | |||
| Median (range) | 66.5 (16-86) | 62 (16-92) | 60.5 (20-87) | |
| Sex, No. (%) | .31 | |||
| Male | 71 (66) | 113 (60) | 25 (66) | |
| Female | 37 (34) | 76 (40) | 13 (34) | |
| B symptoms, No. (%) | .61 | |||
| Absent | 66 (62) | 122 (65) | 22 (58) | |
| Present | 40 (38) | 65 (35) | 16 (42) | |
| Missing | 2 | 2 | 0 | |
| Bulk (> 10 cm), No. (%) | .54 | |||
| Absent | 82 (77) | 135 (74) | 28 (74) | |
| Present | 24 (23) | 47 (26) | 10 (26) | |
| Missing | 2 | 7 | 0 | |
| Disease stage, No. (%) | .61 | |||
| Limited | 32 (30) | 61 (33) | 10 (26) | |
| Advanced | 75 (70) | 125 (67) | 28 (74) | |
| Missing | 1 | 3 | 0 | |
| IPI factors | ||||
| Age, years, No. (%) | .36 | |||
| ≤ 60 | 39 (36) | 79 (42) | 19 (50) | |
| > 60 | 69 (64) | 110 (58) | 19 (50) | |
| Stage, No. (%) | < .001 | |||
| I/II | 40 (37) | 108 (58) | 15 (39) | |
| III/IV | 67 (63) | 79 (42) | 23 (61) | |
| Missing | 1 | 2 | 0 | |
| LDH, No. (%) | .24 | |||
| Normal | 44 (44) | 88 (51) | 16 (44) | |
| > ULN | 56 (56) | 83 (49) | 20 (56) | |
| Missing | 8 | 18 | 2 | |
| ECOG PS, No. (%) | .17 | |||
| 0-1 | 69 (64) | 135 (72) | 24 (63) | |
| 2-4 | 38 (36) | 52 (28) | 14 (37) | |
| Missing | 1 | 2 | 0 | |
| Extranodal sites, No. (%) | .25 | |||
| 0-1 | 90 (84) | 166 (89) | 31 (82) | |
| ≥ 2 | 17 (16) | 21 (11) | 7 (18) | |
| Missing | 1 | 2 | 0 | |
| IPI score, No. (%) | .04 | |||
| Low (0-1) | 31 (30) | 73 (41) | 10 (27) | |
| Intermediate (2-3) | 46 (45) | 82 (46) | 21 (57) | |
| High (4-5) | 26 (25) | 25 (14) | 6 (16) | |
| Not calculable | 5 | 9 | 1 | |
| FISH, No. (%) | ||||
| BCL2 | < .001 | |||
| Normal | 91 (94) | 93 (54) | 28 (82) | |
| Breakapart | 6 (6) | 78 (46) | 6 (18) | |
| Fail | 11 | 18 | 4 | |
| MYC | .07 | |||
| Normal | 84 (90) | 132 (82) | 31 (97) | |
| Breakapart | 9 (10) | 29 (18) | 1 (3) | |
| Fail | 15 | 28 | 6 | |
| BCL6 | .005 | |||
| Normal | 71 (71) | 153 (85) | 19 (56) | |
| Breakapart | 29 (29) | 27 (15) | 15 (44) | |
| Fail | 8 | 9 | 4 | |
| MYC/BCL2 double hit* | < .001 | |||
| Absent | 100 (100) | 144 (86) | 35 (100) | |
| Present | 0 (0) | 23 (14) | 0 (0) | |
| Fail | 8 | 22 | 3 | |
| IHC, No. (%) | ||||
| BCL2 | < .001 | |||
| Negative (< 50%) | 16 (15) | 78 (42) | 14 (37) | |
| Positive (≥ 50%) | 90 (85) | 106 (58) | 24 (63) | |
| Fail | 2 | 5 | 0 | |
| MYC | < .001 | |||
| Negative (< 40%) | 42 (39) | 126 (68) | 24 (63) | |
| Positive (≥ 40%) | 66 (61) | 58 (32) | 14 (37) | |
| Fail | 0 | 5 | 0 | |
| MYC/BCL2 dual expressers† | < .001 | |||
| Absent | 51 (47) | 148 (80) | 28 (74) | |
| Present | 57 (53) | 36 (20) | 10 (26) | |
| Fail | 0 | 5 | 0 |
NOTE. Bold font indicates significance at P < .05.
Abbreviations: ABC, activated B cell–like; COO, cell of origin; DLBCL, diffuse large B-cell lymphoma; ECOG PS, Eastern Cooperate Oncology Group performance status; FISH, fluorescent in situ hybridization; GCB, germinal center B cell–like; IPI, International Prognostic Index; LDH, lactate dehydrogenase; ULN, upper level of normal.
Breakapart of MYC and BCL2 loci.
Tumors that express MYC and BCL2 proteins.
Relationship Between COO, IPI Score, and MYC/BCL2 IHC
There was a greater proportion of patients with high IPI score in the ABC subgroup (Table 2). IPI score defined groups with significantly different outcomes after R-CHOP (Data Supplement Fig 6), raising the possibility that the observed prognostic power of COO may be adequately represented by IPI score alone. However, pairwise multivariable analyses including COO and IPI score demonstrated that the prognostic impact of COO is independent of IPI score (Table 3). The prognostic value added by COO was particularly evident when examining the outcomes in patients with intermediate IPI scores (ABC v GCB: 5-year TTP, 53% v 74%; log-rank P = .003; Data Supplement Figs 6E and 6F).
Table 3.
Multivariable Analyses Including COO and Other Prognostic Factors
| Variable | TTP |
PFS |
DSS |
OS |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| Pairwise multivariable analysis 1 | ||||||||||||
| COO: ABC v GCB | 2.2 | 1.5 to 3.4 | < .001 | 1.9 | 1.3 to 2.7 | .001 | 2.0 | 1.3 to 3.2 | .003 | 1.7 | 1.1 to 2.5 | .01 |
| IPI score:* | < .001 | < .001 | < .001 | < .001 | ||||||||
| Intermediate v low | 2.1 | 1.2 to 3.6 | .009 | 2.3 | 1.4 to 3.7 | .001 | 1.9 | 1.0 to 3.7 | .05 | 2.1 | 1.2 to 3.7 | .006 |
| High v low | 4.8 | 2.7 to 8.6 | < .001 | 4.3 | 2.5 to 7.5 | < .001 | 6.2 | 3.2 to 11.9 | < .001 | 5.3 | 3.0 to 9.5 | < .001 |
| Pairwise multivariable analysis 2 | ||||||||||||
| COO: ABC v GCB | 2.2 | 1.4 to 3.4 | < .001 | 1.9 | 1.3 to 2.8 | .001 | 2.0 | 1.0 to 3.1 | .004 | 1.8 | 1.2 to 2.4 | .008 |
| IHC: MYC positive/BCL2 positive v non–MYC positive/BCL2 positive | 1.6 | 1.0 to 2.4 | .04 | 1.4 | 0.9 to 2.0 | .12 | 1.9 | 1.2 to 3.0 | .009 | 1.5 | 1.0 to 2.3 | .05 |
| Multivariable analysis | ||||||||||||
| COO: ABC v GCB | 1.9 | 1.2 to 3.1 | .005 | 1.7 | 1.2 to 2.6 | .008 | 1.7 | 1.0 to 2.8 | .05 | 1.5 | 1.0 to 2.3 | .06 |
| IHC: MYC positive/BCL2 positive v non–MYC positive/BCL2 positive | 1.4 | 0.9 to 2.2 | .15 | 0.8 | 0.6 to 1.9 | .31 | 1.6 | 1.0 to 2.7 | .07 | 1.3 | 0.8 to 2.1 | .25 |
| IPI score:* | < .001 | < .001 | < .001 | < .001 | ||||||||
| Intermediate v low | 2.1 | 1.2 to 3.6 | .01 | 2.2 | 1.4 to 3.7 | .001 | 1.9 | 1.0 to 3.7 | .05 | 2.1 | 1.2 to 3.6 | .007 |
| High v low | 4.4 | 2.4 to 8.0 | < .001 | 4.1 | 2.3 to 7.1 | < .001 | 5.5 | 2.8 to 10.8 | < .001 | 4.9 | 2.7 to 8.9 | < .001 |
NOTE. Bold font indicates significance at P < .05.
Abbreviations: ABC, activated B cell–like; COO, cell of origin; DSS, disease-specific survival; GCB, germinal center B cell–like; HR, hazard ratio; IHC, immunohistochemistry; IPI, International Prognostic Index; OS, overall survival; PFS, progression-free survival; TTP, time to progression;
Low (0-1), intermediate (2-3), and high (4-5).
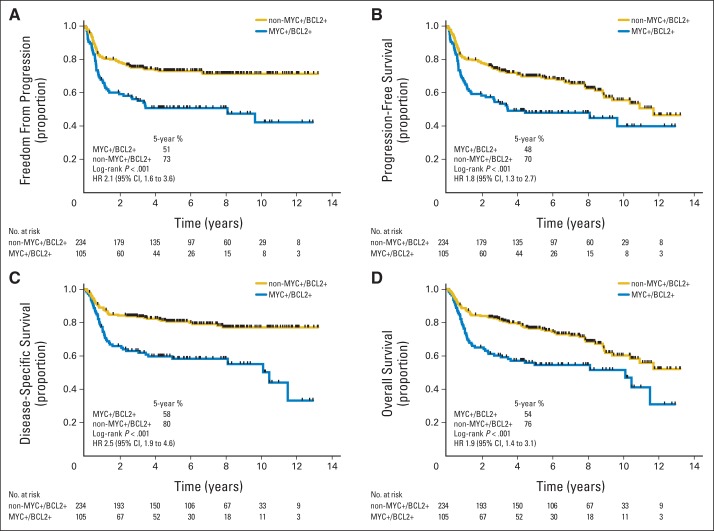
Dual positivity for MYC and BCL2 proteins (MYC positive/BCL2 positive) was seen in 31% (105 of 339) of the tumors by IHC (Data Supplement Table 2). Patients with MYC-positive/BCL2-positive tumors had significantly inferior outcomes compared with patients with tumors that did not display both MYC and BCL2 positivity (non–MYC positive/BCL2 positive; log-rank P < .001 for TTP, PFS, DSS, and OS; Fig 2). The concordant results when a different antibody was used to detect BCL2 (ie, E17 clone) or a different threshold, as proposed in Hu et al,21 was applied to define BCL2 positivity are shown in Data Supplement Table 3 and Data Supplement Figures 7 and 8. The patient cohort in this study included 88 patient cases from our prior description of the prognostic significance of MYC and BCL2 IHC.19 When outcomes were examined with these patient cases excluded, the associations between MYC/BCL2 IHC and outcomes in this independent cohort were still evident (Data Supplement Table 4; Data Supplement Fig 9).
Fig 2.
Outcomes in patients with diffuse large B-cell lymphoma after treatment with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone according to groups defined by immunohistochemistry for MYC and BCL2. Curves shown for (A) time to progression, (B) progression-free survival, (C) disease-specific survival, and (D) overall survival. Positivity for MYC was defined as ≥ 40% of tumor cells stained and for BCL2 as ≥ 50% of tumor cells stained. HR, hazard ratio.
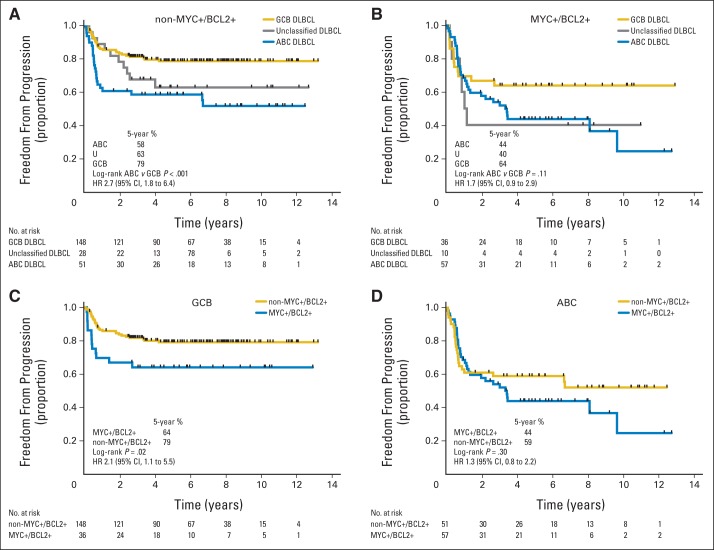
COO and MYC/BCL2 IHC identified high-risk groups that were similar in size (32% v 31%) and had comparable outcomes (5-year TTP, 51% v 51%). Consistent with previous reports,19–21 the proportion of ABC tumors that were MYC positive/BCL2 positive was significantly higher than the proportion of GCB tumors (Table 2). In pairwise multivariable analyses including COO and MYC/BCL2 IHC as variables, COO remained significant, demonstrating that COO had prognostic power beyond that conferred merely by enrichment of MYC-positive/BCL2-positive patient cases of the ABC subtype (Table 3). To explore this further, outcomes were examined within the groups defined by MYC/BCL2 IHC. In the non–MYC-positive/BCL2-positive group, patients with ABC DLBCL had significantly inferior outcomes compared with those with GCB DLBCL (Fig 3A; Data Supplement Fig 10). However, COO did not provide statistically significant risk stratification within the MYC-positive/BCL2-positive group (Fig 3B; Data Supplement Fig 11). Meanwhile, MYC/BCL2 IHC identified groups with different outcomes in the GCB subtype but not in the ABC group (Figs 3C and 3D; Data Supplement Figs 12 and 13). Thus, COO and MYC/BCL2 IHC both contributed to context-specific risk stratification. Finally, when COO, IPI and MYC/BCL2 IHC were included in multivariable analyses, COO remained significantly associated with TTP and PFS (Table 3). These results were also observed when the alternative threshold for BCL2, as proposed by Hu et al,21 was used (Data Supplement Table 3; Data Supplement Figs 14 to 17) and when different groupings of IPI scores were employed (Data Supplement Tables 5 and 6).
Fig 3.
Time to progression for patients with diffuse large B-cell lymphoma (DLBCL) after treatment with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone according to cell of origin and immunohistochemistry for MYC and BCL2. Curves shown for patients with (A) non–MYC-positive/BCL2-positive status, (B) MYC-positive/BCL2-positive status, (C) germinal center B-cell–like (GCB) subtype, and (D) activated B-cell–like (ABC) subtype. HR, hazard ratio; U, unclassified DLBCL.
DISCUSSION
Recently, the Lymphoma/Leukemia Molecular Profiling Project developed a fully validated new digital gene expression–based assay (Lymph2Cx) for COO determination in FFPET biopsies.18 This assay was trained against the so-called gold standard of COO assignment using GEP on fresh frozen tissue, locked down, and then tested in an independent cohort. Separating DLBCL into COO groups identifies tumors with distinct biologies4 and clinical behaviors5,6 and, with targeted agents currently in clinical trials, represents a logical next step toward precision medicine for this disease. Our study was undertaken to determine the prognostic impact of COO assigned using the Lymph2Cx assay in a large cohort of patients treated with R-CHOP.
Previously, we showed that Lymph2Cx assigned COO with excellent concordance between independent laboratories.18 The results here extend these findings, demonstrating the reproducibility and consistency of COO assigned using Lymph2Cx with regard to lot-to-lot reagent performance and sampling within and between FFPET biopsy blocks. In this study, we applied the Lymph2Cx assay to biopsies where the tumor content was ≥ 10%. In silico dilution experiments of high–tumor content biopsies with a range of reactive lymph node RNA samples (Data Supplement) demonstrated that even at a simulated 10% tumor content, the misclassification rate (ABC to GCB or vice versa) was still lower than that reported for IHC-based algorithms by Gutiérrez-García et al.11 Further testing using dilution with other tissues may be useful to confirm 10% as an appropriate tumor content threshold for a clinical assay. If confirmed, this would make the Lymph2Cx assay broadly applicable to almost all FFPET blocks from patients with DLBCL.
Application of the Lymph2Cx assay to a large number of FFPET biopsies from patients with de novo DLBCL uniformly treated with R-CHOP stratified these patients into groups with significantly different outcomes. The size of the study cohort allowed exploration of the prognostic value of COO in comparison with other prognostic tools. Although the IPI remains the most powerful tool for risk stratification, COO assignment provides additional prognostic information, particularly evident in the intermediate IPI score group.
Recently, we19 and others20 described an IHC-based prognostic tool using antibodies to detect MYC and BCL2 protein expression. Patients with tumors that expressed both MYC and BCL2 (dual expressers) had significantly inferior outcomes in comparison with patients with tumors that were non–MYC positive/BCL2 positive, using defined thresholds.19–21,24 However, optimal thresholds for positivity for BCL2 and MYC are still being defined, with each study using different values.19–22,24 Previously, we reported that the proportion of dual expressers was higher in ABC/non-GCB compared with GCB DLBCL, classified using a combination of GEP and IHC,19 a finding that is confirmed in this study.
Hu et al21 also observed this greater frequency of MYC and BCL2 dual expressers in the ABC group in a large cohort of patients treated with R-CHOP, where COO was assigned by IHC-based algorithms or GEP using microarrays on RNA from FFPET material.21 By showing that outcome differences between COO subtypes disappeared when stratified into the two IHC-defined MYC/BCL2 groups, their data suggested that the prognostic significance of COO was entirely attributable to enrichment of dual expressers within the ABC subtype. In contrast, our results present a more complex picture, with COO and MYC/BCL2 IHC providing independent prognostic value. Within the non–MYC-positive/BCL2-positive group, COO stratified patients into groups with significantly different outcomes when treated with R-CHOP. However, the effect of COO on outcome was not statistically significant in MYC/BCL2 dual expressers. On examining the effect of MYC/BCL2 IHC in the individual COO subtypes, dual expressers exhibited inferior outcomes in GCB DLBCL; however, this effect was not evident in the ABC subgroup. Thus, these assays provide complementary prognostic information. These results were also observed when the definitions of MYC and BCL2 IHC positivity from Hu et al21 were used. Furthermore, although there is established literature supporting the value of COO as a predictive biomarker,13–16 the potential for MYC/BCL2 IHC to select patients for targeted therapies is an area of active research. Although the contradictory findings between these two large retrospective studies may reflect the differences in baseline clinical characteristics of the study cohorts related to the methods used to assemble them, it is also plausible that the findings are the result of the difference in methodology used to assign COO. The method employed in that study relied on GEP using RNA derived from FFPET, with the unclassified patient cases being designated either GCB or ABC using IHC, thus producing a binary COO classification. It is unclear how accurately those assignments reflect the gold standard of GEP from fresh frozen tissue.
In conclusion, Lymph2Cx is an accurate assay for DLBCL COO assignment in widely available FFPET biopsies. In contrast to results obtained with IHC,10–12 the consistency of this assay provides the opportunity to bring uniformity to how these GEP-defined subtypes are assigned and subsequently studied. In this first, to our knowledge, large cohort of R-CHOP–treated patients where COO was determined using this assay, Lymph2Cx separated patients into groups with significantly different outcomes, independent of IPI score and MYC/BCL2 IHC. We anticipate that over the next few years, with the emergence of agents with selective activity in ABC or GCB DLBCL,13–15 the determination of COO will become part of the foundation for optimal patient care.
Footnotes
See accompanying editorial on page 2835
Supported by the Terry Fox Foundation and by the British Columbia Cancer Foundation (D.W.S.); A.M. holds fellowship awarded by the Mildred Scheel Cancer Foundation.
Authors' disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Disclosures provided by the authors are available with this article at www.jco.org.
AUTHOR CONTRIBUTIONS
Conception and design: David W. Scott, Anja Mottok, Daisuke Ennishi, Joseph M. Connors, Randy D. Gascoyne
Provision of study materials or patients: Kerry J. Savage, Laurie H. Sehn, Lisa M. Rimsza, Randy D. Gascoyne
Collection and assembly of data: David W. Scott, Anja Mottok, Daisuke Ennishi, Pedro Farinha, Susana Ben-Neriah, Garrett S. Barry, Pau Abrisqueta, Merrill Boyle, Barbara Meissner, Adele Telenius, Kerry J. Savage, Graham W. Slack, Joseph M. Connors, Lisa M. Rimsza, Randy D. Gascoyne
Data analysis and interpretation: David W. Scott, Anja Mottok, Daisuke Ennishi, George W. Wright, Susana Ben-Neriah, Robert Kridel, Garrett S. Barry, Christoffer Hother, Laurie H. Sehn, Christian Steidl, Louis M. Staudt, Joseph M. Connors, Randy D. Gascoyne
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Prognostic Significance of Diffuse Large B-Cell Lymphoma Cell of Origin Determined by Digital Gene Expression in Formalin-Fixed Paraffin-Embedded Tissue Biopsies
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
David W. Scott
Honoraria: Celgene
Consulting or Advisory Role: Celgene
Patents, Royalties, Other Intellectual Property: Named inventor on pending patent describing gene expression profiling in prognostication in classical Hodgkin lymphoma; as member of Lymphoma/Leukemia Molecular Profiling Project, potentially named inventor on pending patent on use of gene expression profiling to assign cell-of-origin in diffuse large B-cell lymphoma
Anja Mottok
No relationship to disclose
Daisuke Ennishi
No relationship to disclose
George W. Wright
Patents, Royalties, Other Intellectual Property: Inventor on submitted patent for determining cell of origin, subtype, and prognosis for lymphoma samples via gene expression
Pedro Farinha
No relationship to disclose
Susana Ben-Neriah
No relationship to disclose
Robert Kridel
No relationship to disclose
Garrett S. Barry
Patents, Royalties, Other Intellectual Property: British Columbia Cancer Agency (Inst)
Christoffer Hother
No relationship to disclose
Pau Abrisqueta
No relationship to disclose
Merrill Boyle
No relationship to disclose
Barbara Meissner
No relationship to disclose
Adele Telenius
No relationship to disclose
Kerry J. Savage
Honoraria: Seattle Genetics, Celgene, Bristol-Myers Squibb
Consulting or Advisory Role: Seattle Genetics, Bristol-Myers Squibb
Research Funding: Roche (Inst)
Laurie H. Sehn
Honoraria: Roche/Genentech, Amgen, Gilead Sciences, Lundbeck, Janssen Pharmaceuticals, Celgene, Seattle Genetics, Pfizer
Consulting or Advisory Role: Roche/Genentech, Amgen, Gilead Sciences, Lundbeck, Janssen Pharmaceuticals, Celgene, Seattle Genetics, Pfizer
Graham W. Slack
Honoraria: Janssen Pharmaceuticals, Seattle Genetics
Consulting or Advisory Role: Seattle Genetics, Celgene, Seattle Genetics (Inst), Celgene (Inst)
Christian Steidl
Consulting or Advisory Role: Affimed Therapeutics
Patents, Royalties, Other Intellectual Property: British Columbia Cancer Agency (Inst)
Louis M. Staudt
Research Funding: Pharmacyclics (Inst), Celgene (Inst)
Patents, Royalties, Other Intellectual Property: National Institutes of Health
Joseph M. Connors
Research Funding: Seattle Genetics, Takeda Pharmaceutical, Roche
Lisa M. Rimsza
Honoraria: Ventana Medical Systems, Celgene
Consulting or Advisory Role: Ventana Medical Systems
Research Funding: Merck
Patents, Royalties, Other Intellectual Property: University of Arizona (Inst)
Randy D. Gascoyne
Honoraria: Roche, Genentech, Celgene, Janssen, Seattle Genetics
Consulting or Advisory Role: Roche, Genentech, Celgene, Janssen, Seattle Genetics, NanoString Technologies
Speakers' Bureau: Seattle Genetics
Patents, Royalties, Other Intellectual Property: As member of Lymphoma/Leukemia Molecular Profiling Project, potentially named inventor on patent licensed to NanoString Technologies (Seattle, WA)
REFERENCES
- 1.Swerdlow SH, Campo E, Harris NL, et al. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues (ed 4) Lyon, France: IARC Press; 2008. [Google Scholar]
- 2.Alizadeh AA, Elsen MB, Davis ER, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 3.Wright G, Tan B, Rosenwald A, et al. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 6.Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 8.Choi WW, Weisenburger DD, Greiner TC, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15:5494–5502. doi: 10.1158/1078-0432.CCR-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer PN, Fu K, Greiner TC, et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol. 2010;29:200–207. doi: 10.1200/JCO.2010.30.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong D, Rosenwald A, Chhanabhai M, et al. Immunohistochemical prognostic markers in diffuse large B-cell lymphoma: Validation of tissue microarray as a prerequisite for broad clinical applications—A study from the Lunenburg Lymphoma Biomarker Consortium. J Clin Oncol. 2007;25:805–812. doi: 10.1200/JCO.2006.09.4490. [DOI] [PubMed] [Google Scholar]
- 11.Gutiérrez-García G, Cardesa-Salzmann T, Climent F, et al. Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood. 2011;117:4836–4843. doi: 10.1182/blood-2010-12-322362. [DOI] [PubMed] [Google Scholar]
- 12.Read JA, Koff JL, Nastoupil LJ, et al. Evaluating cell-of-origin subtype methods for predicting diffuse large B-cell lymphoma survival: A meta-analysis of gene expression profiling and immunohistochemistry algorithms. Clin Lymphoma Myeloma Leuk. 2014;14:460–467. doi: 10.1016/j.clml.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunleavy K, Pittaluga S, Czuczman MS, et al. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood. 2009;113:6069–6076. doi: 10.1182/blood-2009-01-199679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nowakowski GS, LaPlant B, Macon WR, et al. Lenalidomide combined with R-CHOP overcomes negative prognostic impact of non-germinal center B-cell phenotype in newly diagnosed diffuse large B-cell lymphoma: A phase II study. J Clin Oncol. 2015;33:251–257. doi: 10.1200/JCO.2014.55.5714. [DOI] [PubMed] [Google Scholar]
- 15.Roschewski M, Staudt LM, Wilson WH. Diffuse large B-cell lymphoma-treatment approaches in the molecular era. Nat Rev Clin Oncol. 2014;11:12–23. doi: 10.1038/nrclinonc.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molina TJ, Canioni D, Copie-Bergman C, et al. Young patients with non-germinal center B-cell-like diffuse large B-cell lymphoma benefit from intensified chemotherapy with ACVBP plus rituximab compared with CHOP plus rituximab: Analysis of data from the Groupe d'Etudes des Lymphomes de l'Adulte/Lymphoma Study Association phase III trial LNH 03-2B. J Clin Oncol. 2014;32:3996–4003. doi: 10.1200/JCO.2013.54.9493. [DOI] [PubMed] [Google Scholar]
- 17.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 18.Scott DW, Wright GW, Williams PM, et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123:1214–1217. doi: 10.1182/blood-2013-11-536433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson NA, Slack GW, Savage KJ, et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3452–3459. doi: 10.1200/JCO.2011.41.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green TM, Young KH, Visco C, et al. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3460–3467. doi: 10.1200/JCO.2011.41.4342. [DOI] [PubMed] [Google Scholar]
- 21.Hu S, Xu-Monette ZY, Tzankov A, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: A report from the International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121:4021–4031. doi: 10.1182/blood-2012-10-460063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horn H, Ziepert M, Becher C, et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood. 2013;121:2253–2263. doi: 10.1182/blood-2012-06-435842. [DOI] [PubMed] [Google Scholar]
- 23.A predictive model for aggressive non-Hodgkin's lymphoma. The International Non-Hodgkin's Lymphoma Prognostic Factors Project. N Engl J Med. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 24.Perry AM, Alvarado-Bernal Y, Laurini JA, et al. MYC and BCL2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with rituximab. Br J Haematol. 2014;165:382–391. doi: 10.1111/bjh.12763. [DOI] [PubMed] [Google Scholar]