Abstract
Large data sets are becoming more common in fMRI and, with the advent of faster pulse sequences, memory efficient strategies for data reduction via principal component analysis (PCA) turn out to be extremely useful, especially for widely used approaches like group independent component analysis (ICA). In this commentary, we discuss results and limitations from a recent paper on the topic and attempt to provide a more complete perspective on available approaches as well as discussing various issues to consider related to large group PCA for group ICA. We also provide an analysis of computation time, memory use, and number of dataloads for a variety of approaches under multiple scenarios of small and extremely large data sets.
Keywords: independent component analysis, principal component analysis, RAM, memory
Introduction
A recent paper1, presented some solutions for dealing with large group functional brain imaging studies that require data reduction with principal component analysis (PCA), most notably, as in group independent component analysis (GICA), which has become widely used in the analysis of functional magnetic resonance imaging (fMRI) data2. This is an important topic, especially given the rapid increase in data sharing3,4 as well as in the size of data sets being collected5. Current emphasis on projects such as the human connectome project (plus ongoing and future disease-related extensions) as well as the NIH focus on “big data” further motivate the need for approaches that can easily scale to extremely large numbers of subjects.
In the following sections, we found that there are a few points in the paper1 that need clarification, ranging from subtle (perhaps unintentional) mischaracterization of standard approaches to shallow representation of the functionality of existing tools, notably the group ICA of fMRI toolbox (GIFT; http://mialab.mrn.org/software/gift). For example, the paper1 claims improved accuracy over existing approaches and also claims that available PCA implementations are not scalable in memory (RAM) to large numbers of subject. However both of these points are slightly misleading. We present these points and others in four sections starting first with a discussion of a comparison of accuracy, then memory (RAM) use, followed by a Pareto front optimality analysis comparing ten different PCA strategies, and finally attempt to present the available methods in the context of their initial introduction to fMRI data. Finally, we present our conclusions and summarize the results.
Accuracy of Results
We start with the issue of accuracy. In the paper1 there is an extensive amount of text and multiple figures devoted to comparing the accuracy of some existing and proposed PCA solutions. However, as we discuss, the performance comparisons and most figures in the paper are fairly unnecessary, and the highlighted differences with respect to GIFT due to the PCA decomposition are not accurate. The key issue is that the comparison is between subject-level whitening + PCA versus PCA directly on preprocessed data, although the pretext in the paper1 is a comparison of different PCA approaches only. As it turns out, the ‘errors’ discussed therein are merely related to the degree to which the variance associated with the scaling of subject-level data is preserved during the whitening step. If one instead compares PCA approaches without subject-level whitening (easily done in GIFT), then one finds that all of the PCA approaches discussed are comparable in accuracy (see Table 1). This issue appears to stem from an implicit assumption that the optimal/ideal approach to group PCA revolves around evaluating it directly from raw (preprocessed) data, instead of initially whitening the subject-level data (i.e., not incorporating the eigenvalues in the matrix of subject-specific spatial eigenmaps).
Table 1.
Summary of various group ICA approaches (in chronological order): The computation times were obtained from resting state fMRI data9 containing s = 1600 subjects, v = 66745 in-brain voxels, t = 100 time points and extracting n = 75 group principal components. Here we opted not to whiten the subject-level data prior to group PCA in order to match the assumption in the original paper1. Analyses were run on an 80-core Linux Centos OS release 6.4 with 512 GB and on an 8-core Windows Desktop with 4GB RAM. For Windows desktop, we report the computational times of only the PCA analyses that could be fit on 4GB RAM. For EM PCA2, MPOWIT and SMIG, the maximum number of iterations was set to 100 (on Linux server) and 40 (on Windows desktop) to limit the maximum number of dataloads per subject in each iteration. To compute the estimation error, we use L2-norm of the difference between the eigenvalues obtained from PCA methods and those from the temporal concatenation approach. Only errors greater than 1×10−6 are reported, and for iterative PCA methods like MPOWIT, EM PCA2, and SMIG we also report the number of iterations required to converge. Here the ranking is based on RAM use only. We also report the explained variance (EV) for each method using temporal concatenation as the reference.
| Software (Name) | Date introduced | Average rank (based on least memory use only) |
Compute time (min) (80-core Linux, 512 GB RAM; loading time irrelevant) |
Compute time (min) (8-core Windows Desktop 4GB RAM; loading time dominates) |
|---|---|---|---|---|
| A. GIFT (EVD) | 200118 | 9 | 60.15, EV = 100% | * |
| Original GIFT group ICA approach. | ||||
| B. GIFT (3-step PCA, STP) | 200412,23 | 3-5 (depending on blocksize) |
27.96, error=0.05 EV – 99.9% ~2min loading data |
67.97, error=0.05 EV = 99.9% ~39min loading data |
| A 3-step PCA method implemented early
in the GIFT toolbox and similar to the MIGP approach. Subsampled Time PCA
(STP)14 is a
more recent generalization which avoids whitening in the intermediate group PCA step during the group PCA space update. To compute the memory required, we selected a value of 10 for the number of subjects in each group. The top 500 components were retained in each intermediate group PCA. | ||||
| C. MELODIC (Temporal Concat.) | 200924 | 9 | 60.15 | * |
| Original MELODIC group ICA approach after adoption of temporal concatenation as the default. | ||||
| D. GIFT (EVD Full Storage) | 2009 (GroupICAT v2.0c) | 7 | 87.78, EV = 100% | * |
| Covariance is computed using two data-sets at a time (Time × Time) or one data-set at a time (voxels × voxels). | ||||
| E. GIFT (EVD Packed Storage) | 2009 (GroupICAT v2.0c) | 6 | 915.32, EV = 100% | * |
| Only lower triangular portion of covariance matrix is stored. Covariance is computed in the same way as GIFT (Full Storage). | ||||
| F. GIFT (EM PCA1) | 2010 (GroupICAT v2.0d)8,9 | 8 | 152, iter=496 EV = 100% |
* |
| Expectation maximization assuming all data is in memory. | ||||
| G. GIFT (EM PCA2) | 2010 (GroupICAT v2.0d)8,9 | 1 | 312.18, error=0.09 EV = 100% |
2305, error=6.58 EV = 100% |
| Expectation maximization by loading one data-set at a time. | ||||
| H. GIFT (MPOWIT) | 201314,15 | 3 | 33.19, iter=7 EV = 100% |
278.89, iter=7 EV = 100% |
| Multi power iteration method (MPOWIT)
is an extension of subspace iteration. Typically we select a block multiplier which is
5 times the number of components to be extracted from the data to speed up the convergence of desired eigenvectors. In Figure 1, we show the memory required by MPOWIT when one dataset is loaded at a time. | ||||
| I. MELODIC (SMIG) | 20141 | 1 | 106.9, iter=32 EV = 100% |
2310, iter=32 EV = 100% |
| MELODIC subspace iteration. Memory required will be the same as EM PCA2. | ||||
| J. MELODIC (MIGP) | 20141 | 4 | 48.48, error=1.87 EV = 100% ~2min loading data |
47.51, error=1.58 EV = 100% ~39min loading data |
| MELODIC variation of 3-step PCA method
in GIFT. Memory required is slightly higher compared to EM PCA2 and
SMIG. Here we used m = 2t – 1. | ||||
Ultimately whether this assumption is accurate depends on the problem at hand. Specifically, it comes down to how noise components should be handled and to what end principal components (PCs) will be used. For example, strong subject-specific noise components, highly explanatory of the variance across subjects, may be over-emphasized if their eigenvalues are carried forward to the group PCA estimation. This may also be the case if very weak noise components are whitened before proceeding to group PCA estimation, though the weakest components are typically discarded before whitening. If the ultimate goal is a group ICA estimation, then group PCs (i.e., the eigenmaps) are far less meaningful than the group ICs (consider an orthogonal transformation to the PC space which would produce an apparent “error” according to the paper1 but would still lead to the same end ICA result). In addition, an ICA-oriented simulation (ideally one which fully controlled implicit linear and higher-order dependencies between sources) would be more meaningful than the source generation approach described in the paper1, which has been previously criticized in 6,7. Ultimately more research on this topic is needed. Of note, GIFT supports both approaches, using the expectation maximization (EM) PCA algorithm8,9 and we report those estimates below (Figure 1).
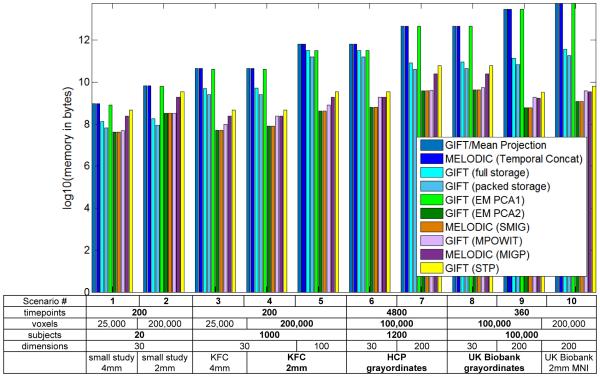
Figure 1.
Memory use of various approaches implemented in MELODIC and GIFT.
On another point related to accuracy, the paper1 proposed a “3-step” MIGP (MELODIC's Incremental Group-PCA). However MIGP can be seen to be a slight variation of the familiar “3-step” PCA available in the GIFT toolbox since 200410-12. This is most evident from the second paragraph of the Conclusions section where the parallelization scheme described therein essentially outlines the steps in the original 3-step PCA. Unlike 3- step PCA, however, MIGP “propagates” the singular values of the current group-level estimate to the following iteration; 3-step PCA does not propagate the singular values of each group forward and, thus, yields group PCs that are different from those obtained by concatenating the entire data13. In other words, retaining the singular values of the current group PCs is key to approximate the group PCs from full concatenation; the singular values of the subject-level data, on the other hand, only relate to the “raw” versus whitened subject- level data debate mentioned above, not the approximation to full concatenation. A generalization of 3-step PCA (and MIGP) called subsampled time PCA (STP)14,15 addresses the issue of preserving group-level singular values throughout and further improves flexibility between accuracy and execution speed. The accuracy of STP in our experiments was fairly comparable with other PCA methods and higher than MIGP (see Table 1).
Use of Memory (RAM)
Another key aspect is an emphasis on RAM efficient implementations of PCA. The paper1 makes the claim that current solutions to this problem do not exist. However, this is not accurate as the GIFT software has implemented multiple memory efficient approaches since 2004 (see Table 1 for more details). The EM PCA implementation (which is as ‘good’ in minimizing RAM as the best solution presented in the paper1) was announced on the GIFT listserv in early 2010, is included in the release notes (http://mialab.mrn.org/software/gift/version_history.html) and manual, and was also discussed in Allen et al.8,9.
Furthermore, perhaps due to a lack of familiarity with GIFT, the memory estimates in Fig. 1 of the paper1 for GIFT (either m = n or m = 2n) are incorrect since the GIFT toolbox implements the same “mathematically equivalent” approach mentioned for the full temporal concatenation (see Table 1). Specifically, instead of estimating the voxels × voxels covariance matrix of temporal correlations for each subject and then averaging over subjects, one can estimate the (subjects × timepoints) × (subjects × timepoints) covariance matrix of spatial correlations (note the GIFT toolbox covariance matrix is always computed along the smallest dimensions of the data by default). Of note, both GIFT and other tools contain many options and, while the default settings typically provide good guidance for the user, there are a number of available options that, depending on the problem, we anticipate would be explored by any user.
In order to provide a more complete and accurate picture of the current state of memory for the two software tools, we have made significant corrections to the original Fig. 1 in the paper1. We have included four approaches (GIFT/Mean Projection, SMIG, MIGP, Temporal concatenation) from the original Fig. 11, two covariance computation strategies (full storage and packed storage), and three approaches that have been in GIFT for years (EM PCA1, EM PCA2 and 3-step PCA/STP) as well as the recently developed Multi power iteration (MPOWIT). Of note, Figure 1 includes the original GIFT approach (introduced in 2001) as well as the original MELODIC group ICA approach using temporal concatenation (introduced in 2009; tensor decomposition was used prior to that16).
Erhardt et al.17 provides extensive comparisons of multiple approaches for group ICA, including various group PCA approaches, and back-reconstruction methods (e.g. PCA-based, spatio-temporal (dual) regression). One of the important take-homes from 17 is that the subject-level PCA dimensionality should be higher than the group-level PCA dimensionality. This has been the GIFT default since 2010, despite the claim in the paper1 that “typically” m =n.
Optimality Analysis
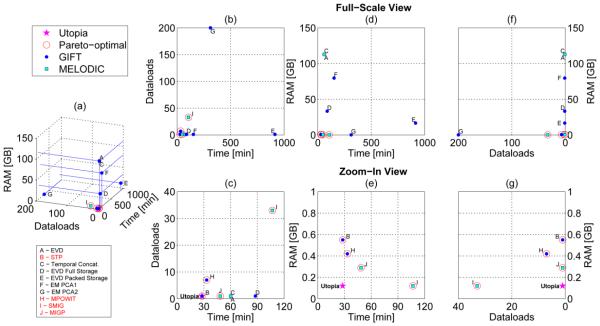
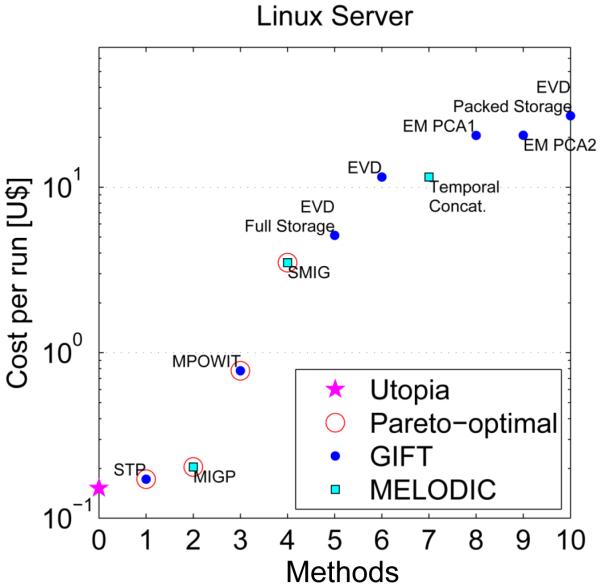
Here we study the optimality of the 10 PCA approaches presented in Table 1 (see the experimental setting description therein). We consider three different response measurements in our analysis: computation time in minutes, RAM memory used in GB, and number of dataloads. Since none of the three criteria alone suffices to establish preference of one method over the other, we considered all three simultaneously and determined the set of Pareto-optimal methods based on the set of non-dominated points in the three-dimensional space of response measurements (Figure 2, panel (a)). The non-dominated points are those that cannot be outperformed simultaneously in all criteria by any other point. These are called Pareto-optimal and are indicated with a red circle in Figures 2-3. The Pareto-optimal collection effectively outlines the trade-offs between each optimal method. Methods outside of the so-called Pareto-front are non-optimal since at least one method in the Pareto-front outperforms them in all criteria. In practice, a method from the Pareto-front should be selected as a result of constraints from the actual problem at hand. For didactical purposes, we then assigned the following fictitious costs in U$ for each response measurement: U$0.10 per GB of RAM per hour, U$0.10 per data-transfer from HD to RAM per 1600 subjects (i.e., per dataload in Figure 2), and U$0.10 per hour waiting for results to be obtained. The resulting fictitious costs are presented in Figure 3.
Figure 2.
Optimality analysis highlighting the set of Pareto-optimal methods on a Linux server. The utopia point combines the best performance measurements across all Pareto- optimal points.
Figure 3.
Fictitious cost analysis on a Linux server. Values obtained using fictitious costs in U$ for each response measurement: U$0.10 per GB of RAM per hour, U$0.10 per data- transfer from HD to RAM per 1600 subjects (i.e., per dataload in Figures 2), and U$0.10 per hour waiting for results to be obtained.
Figure 2 shows that STP, MPOWIT, SMIG and MIGP are Pareto-optimal. Panel (e) indicates that STP and MPOWIT can significantly improve execution time with respect to SMIG and MIGP at the cost of a small increase in RAM use. Note that accuracy (in terms of error with respect to the eigenvalues of full concatenation) was not included as a criterion in the optimality analysis. This was because all methods (except for MIGP and STP) attained very low L2-norm error (< 1×10−6). The errors reported in Table 1 suggest STP makes a better approximation to the full concatenation than MIGP. Figure 3 suggests that STP and MIGP are both very cheap as compared to other techniques. Given the lower accuracy of STP and MIGP with respect to full concatenation, one recommendation is to use these approaches to initialize the more accurate iterative approaches (MPOWIT and SMIG), which should result in faster convergence. Note that these comparisons are meant to be descriptive and helpful, but it should be kept in mind that, though we tried to minimize other factors by using single-user workstations, for measures such as computation time there are many contributing factors and our results are not comprehensive in this regard. In addition, for the beginning user the number of PCA options can be a bit hard to sift through. In this case, we would recommend one of the Pareto-optimal approaches (e.g. STP, MIGP, MPOWIT, or SMIG) which can all handle large data sets and converge reasonably quickly.
Clarity and (Selective) History
In this section we respond to a few claims in the paper1 which are either incomplete or not accurate. First, the statement “Current approaches…cannot be run using the computational facilities available to most researchers” is not accurate as we explained in the previous sections. In the paper1 the sentence after this claim transitions to a discussion of the original group ICA paper from our group18 and claims “There can be a significant loss of accuracy…”, but fails to mention that this is not true if following the recommendations in 17 or using the default settings in the GIFT toolbox. The paper1 also claims that “the amount of memory required is proportional to the number of subjects analyzed”, which is also untrue as described in the previous section. We have already addressed the “typically m=n” claim, and regarding “important information may be lost unless m is relatively large (which in general is not the case when using this approach)” is also not true (see 17,19,20).
In addition, as a more minor point, the paper1 refers (just prior to Equation 8) to the “power method” in the section on small memory iterative group PCA (SMIG). However, to clarify, power iteration methods estimate in deflationary mode (i.e., one component at a time) and, in contrast, the proposed approach estimates all components in parallel (i.e., symmetric mode) and, thus, it is more accurate to call the proposed approach a subspace iteration approach. Subspace iteration has been previously proposed21,22, but was not cited in the paper1. In contrast to subspace iteration, SMIG uses a normalization step that stems from the particular optimization problem proposed in the paper1 instead of the typical QR factorization. Normalization is beneficial in subspace iteration to control the size of the eigenvalues of the covariance matrix powers and avoid ill-conditioned situations in the final SVD. The normalization in SMIG, however, does not allow a check for convergence, which is why the parameter “a” needs to be selected in advance. As an alternative, we have proposed in other work14,15 a different normalization scheme, the MPOWIT algorithm, which enables us to efficiently check for convergence and stop iterating, thus limiting the number of dataloads, rather than just iterating a fixed amount of times.
Conclusions
In summary, we have attempted to provide some corrections and commentary on the important issue of memory efficient approaches for large group PCA that are needed for the widely used group ICA approach.
Highlights.
Group ICA of fMRI on very large data sets is becoming more common
GIFT (since 2009) and MELODIC (since 2014) enable analysis of thousands of subjects
We compare ten available approaches including a Pareto optimal analysis
We provide new analyses and comments on “Group-PCA for very large fMRI datasets”
Acknowledgements
The work was in part funded by NIH via a COBRE grant P20GM103472 and grants R01EB005846 and 1R01EB006841.
Appendix
Group ICA on fMRI data is typically performed by doing subject-level PCA before stacking data-sets temporally across subjects18. We assume that 100% variance is retained in subject-level PCA for demonstration purposes. Let Zi be the original data of subject i (having zero mean) and is of dimensions voxels by time points. PCA reduced data Yi (Equation 3) is computed by performing eigen value decomposition on the covariance matrix Ci using the equations shown below (Equations 1 and 2) where ν is the number of voxels:
| (1) |
| (2) |
| (3) |
Whitening normalizes the variances of components using the inverse square root of eigen values matrix Λi (Equation 3). Covariance matrix of Yi is unitary or in other words eigen values of all components are 1. Therefore, group PCA space obtained by stacking whitened data across subjects Y is not comparable to group PCA space extracted from original data Z. However, if whitening is not used and only the eigen vectors Fi are used in projection (i.e., Yi = ZiFi), eigen values information of each subject are propagated into the group PCA. We used a subset of 100 pre-processed fMRI subjects9 to compare the group PCA on original Z data and group PCA on stacked subject-level PCA with no whitening in the subject-level PCA. Table 1 shows the explained variance by each PCA method using the temporal concatenation on original data Z as the ground truth. It is evident that all PCA methods capture at least 99% explained variance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Smith SM, Hyvarinen A, Varoquaux G, Miller KL, Beckmann CF. Group-PCA for very large fMRI datasets. Neuroimage. 2014 Nov 1;101:738–749. doi: 10.1016/j.neuroimage.2014.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Calhoun VD, Adalı T. Multi-subject Independent Component Analysis of fMRI: A Decade of Intrinsic Networks, Default Mode, and Neurodiagnostic Discovery. IEEE Reviews in Biomedical Engineering. 2012;5:60–73. doi: 10.1109/RBME.2012.2211076. PMC23231989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mennes M, Biswal BB, Castellanos FX, Milham MP. Making data sharing work: the FCP/INDI experience. Neuroimage. 2013 Nov 15;82:683–691. doi: 10.1016/j.neuroimage.2012.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wood D, King M, Landis D, Courtney W, Wang R, Kelly R, Turner J, Calhoun VD. Harnessing modern web application technology to create intuitive and efficient data visualization and sharing tools. Frontiers in Neuroinformatics. 2014;8 doi: 10.3389/fninf.2014.00071. PMC Journal - In Process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Ugurbil K. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med. 2010 May;63:1144–1153. 2906244. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Calhoun VD, Potluru V, Phlypo R, Silva R, Pearlmutter B, Caprihan A, Plis SM, Adalı T. Independent component analysis for brain fMRI does indeed select for maximal independence. PLoS ONE. 2013;8:PMC3757003. doi: 10.1371/journal.pone.0073309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Silva RF, Plis SM, Adali T, Calhoun VD. A statistically motivated framework for simulation of stochastic data fusion models applied to multimodal neuroimaging. Neuroimage. 2014 Nov 15;102:92–117. doi: 10.1016/j.neuroimage.2014.04.035. Pt 1. PMC Journal - In Process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Allen EA, Erhardt EB, Damaraju E, Gruner W, Segall JM, Silva RF, Havlicek M, Rachakonda S, Fries J, Kalyanam R, Michael AM, Caprihan A, Turner JA, Eichele T, Adelsheim S, Bryan A, Bustillo J, Clark VP, Feldstein-Ewing S, Filbey FM, Ford C, Hutchison K, Jung RE, Kiehl KA, Kodituwakku P, Komesu Y, Mayer AR, Pearlson GD, Sadek J, Stevens M, Teuscher U, Thoma RJ, Calhoun VD. A baseline for the multivariate comparison of resting state networks. Biennial Conference on Resting State / Brain Connectivity Milwaukee; WI. 2010. P. J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Allen EA, Erhardt EB, Damaraju E, Gruner W, Segall JM, Silva RF, Havlicek M, Rachakonda S, Fries J, Kalyanam R, Michael AM, Caprihan A, Turner JA, Eichele T, Adelsheim S, Bryan AD, Bustillo J, Clark VP, Feldstein Ewing SW, Filbey F, Ford CC, Hutchison K, Jung RE, Kiehl KA, Kodituwakku P, Komesu YM, Mayer AR, Pearlson GD, Phillips JP, Sadek JR, Stevens M, Teuscher U, Thoma RJ, Calhoun VD. A baseline for the multivariate comparison of resting-state networks. Front Syst Neurosci. 2011;5:2, 3051178. doi: 10.3389/fnsys.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Juarez M, White T, Pearlson GD, Bustillo JR, Lauriello J, Ho BC, Bockholt HJ, Clark VP, Gollub R, Magnotta V, Machado G, Calhoun VD. Functional connectivity differences in first episode and chronic schizophrenia patients during an auditory sensorimotor task revealed by independent component analysis of a large multisite study. Proc. HBM; San Francisco, CA. 2009. [Google Scholar]
- [11].Abbott C, Juarez M, White T, Gollub RL, Pearlson GD, Bustillo JR, Lauriello J, Ho BC, Bockholt HJ, Clark VP, Magnotta V, Calhoun VD. Antipsychotic Dose and Diminished Neural Modulation: A Multi-Site fMRI Study. Progress in Neuro-Psychopharmacology & Biological Psychiatry. 2011;35:473–482. doi: 10.1016/j.pnpbp.2010.12.001. PMC Pending #255577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Machado G, Juarez M, Clark VP, Gollub RL, Magnotta V, White T, Calhoun VD. Probing Schizophrenia With A Sensorimotor Task: Large-Scale (N=273) Independent Component Analysis Of First Episode And Chronic Schizophrenia Patients. Proc. Society for Neuroscience; San Diego, CA. 2007. [Google Scholar]
- [13].Zhang H, Zuo XN, Ma SY, Zang YF, Milham MP, Zhu CZ. Subject order-independent group ICA (SOI-GICA) for functional MRI data analysis. Neuroimage. 2010 Jul 15;51:1414–1424. doi: 10.1016/j.neuroimage.2010.03.039. [DOI] [PubMed] [Google Scholar]
- [14].Rachakonda S, Silva R, Liu J, Adalı T, Calhoun VD. Memory Efficient PCA Approaches For Large Group ICA. submitted. [Google Scholar]
- [15].Rachakonda S, Calhoun VD. Efficient Data Reduction in Group ICA Of fMRI Data. Proc. HBM; Seattle, WA. 2013. [Google Scholar]
- [16].Beckmann CF, Smith SM. Tensorial extensions of independent component analysis for multisubject FMRI analysis. NeuroImage. 2005;25:294–311. doi: 10.1016/j.neuroimage.2004.10.043. [DOI] [PubMed] [Google Scholar]
- [17].Erhardt EB, Rachakonda S, Bedrick EJ, Allen EA, Adali T, Calhoun VD. Comparison of multi-subject ICA methods for analysis of fMRI data. Hum Brain Mapp. 2011 Dec;32:2075–2095. 3117074. doi: 10.1002/hbm.21170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Calhoun VD, Adalı T, Pearlson GD, Pekar JJ. A Method for Making Group Inferences from Functional MRI Data Using Independent Component Analysis. Human Brain Mapping. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Erhardt E, Allen E, Wei Y, Eichele T, Calhoun VD. SimTB, a simulation toolbox for fMRI data under a model of spatiotemporal separability. NeuroImage. 2012;59:4160–4167. PMC3690331. doi: 10.1016/j.neuroimage.2011.11.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Allen EA, Erhardt E, Wei Y, Eichele T, Calhoun VD. Capturing inter-subject variability with group independent component analysis of fMRI data: a simulation study. NeuroImage. 2012;59:4141–4159. doi: 10.1016/j.neuroimage.2011.10.010. PMC Pending #327594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tutishaurser H. Simultaneous Iteration Method for Symmetric Matrices. Mathematik. 1970;16:205–223. [Google Scholar]
- [22].Saad Y. Numerical Methods for Large Eigenvalue Problems. Halstead Press; 1992. [Google Scholar]
- [23].Egolf E, Kiehl KA, Calhoun VD. Group ICA of fMRI Toolbox (GIFT) Proc.HBM. 2004 [Google Scholar]
- [24].Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Matthews PM, Beckmann CF, Mackay CE. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci U S A. 2009 Apr 28;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]