Abstract
Type II transmembrane serine proteases (TTSPs) are important in many biological processes. Cell surface expression is critical for TTSP activation and function. To date, the mechanism underlying TTSP cell surface expression is poorly understood. Corin is a TTSP and acts as the pro-atrial natriuretic peptide convertase that is essential for sodium homeostasis and normal blood pressure. In this study, we investigated how cytoplasmic tail sequences may regulate corin expression and activation on the cell surface. By site-directed mutagenesis, we made mouse corin proteins with truncations or point-mutations in the cytoplasmic tail. We expressed the mutants in transfected HEK293 cells and analyzed corin cell surface expression and activation by Western blotting and flow cytometry. We found that corin truncation mutants lacking a Lys-Phe-Gln sequence at residues 71–73 had higher levels of cell surface expression and activation compared with that in wild-type corin. When Lys-71, Phe-72 and Gln-73 residues were mutated together, but not individually, in corin with the full-length cytoplasmic tail, increased levels of cell surface expression and zymogen activation were also observed. These results indicate that residues Lys-71, Phe-72 and Gln-73 serve as a novel retention motif in the intracellular pathway to regulate corin cell surface expression and activation.
Keywords: corin, cytoplasmic tail, intracellular trafficking, natriuretic peptides, transmembrane proteases
1. Introduction
Type II transmembrane serine proteases (TTSPs) are trypsin-like enzymes that participate in diverse biological processes [1–5]. All TTSPs consist of an N-terminal cytoplasmic tail followed by a single-span transmembrane domain and an extracellular region with a C-terminal protease domain. The transmembrane domain serves as an anchor to restrict the protease activity in the desired tissues [1, 6]. The cytoplasmic tails of the TTSPs range from 19 to 161 amino acids in length [1, 6]. To date, their functional significance remains largely unknown.
Corin is a TTSP, consisting of a cytoplasmic tail, a transmembrane domain and an extracellular region with two frizzled, eight LDL receptor, a scavenger receptor and a protease domains [7, 8]. In cardiomyocytes, corin activates the natriuretic peptides that are essential for normal blood volume and pressure [7, 9–11]. Corin deficiency prevents natriuretic peptide processing and causes hypertension in knockout mice [12, 13]. Corin variants have been found in people with hypertension [14–19].
Corin is synthesized in an inactive zymogen form. Upon reaching the cell surface, it is cleaved and activated at a conserved site [20–23]. Previously [24], we found that a DDNN cytoplasmic motif regulated human corin cell surface expression and zymogen activation, indicating that the cytoplasmic tail plays a role in corin intracellular trafficking.
The natriuretic peptides are conserved from fish species to mammals [25]. Corin is also conserved evolutionarily and its homolog exists in the fruit fly Drosophila [26, 27]. Interestingly, the corin cytoplasmic tails in different species differ both in length and sequence (www.ensemble.org). For example, the mouse corin cytoplasmic tail is 67 amino acids longer than the human corin cytoplasmic tail [8]. Moreover, mouse and human corin cytoplasmic tails share little sequence similarities. In particular, mouse corin lacks the DDNN motif that mediates human corin cell surface expression [24].
We hypothesize that different cytoplasmic sequences may be involved in regulating mouse corin cell surface expression. To test this hypothesis, we created mouse corin proteins with altered cytoplasmic lengths and sequences. We expressed these proteins in human embryonic kidney 293 (HEK293) cells and analyzed their cell surface expression and zymogen activation by Western blotting and flow cytometry.
2. Materials and methods
2.1. Expression plasmids
Plasmids encoding mouse corin wild-type (WT) and mutants were constructed by site-directed mutagenesis. All plasmids were pcDNA3.1 (Invitrogen)-based and verified by sequencing. Individual corin mutants are illustrated in Figs. 1–4. Plasmids encoding human WT corin and a splicing variant, hE1a, were described previously [24]. All recombinant corin proteins contained a C-terminal V5 tag for protein detection in Western blotting and flow cytometry.
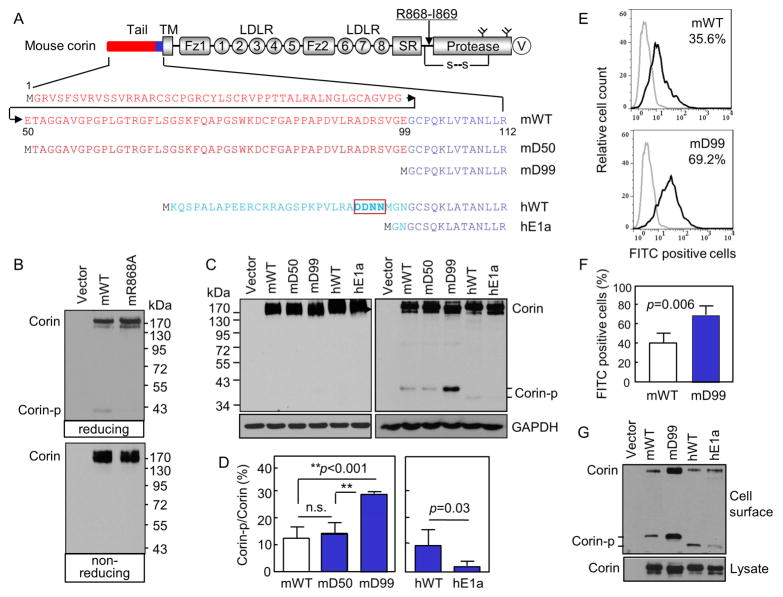
Fig. 1. Corin cytoplasmic tail and activation.
(A) Corin domains and cytoplasmic sequences. An arrow indicates the activation site. A disulfide bond (s-s) links the protease domain and the propeptide. Y-shaped symbols in the protease domain indicate N-glycosylation sites. A V5 tag (V) is at the C-terminus. Cytoplasmic sequences in mouse (m) corin WT and the truncation mutants mD50 and mD99 as well as human (h) corin WT and the hE1a variant are shown. (B, C) Western analysis of corin proteins under reducing (B, top; C, right) and non-reducing (B, bottom; C, left) conditions. (D) Percentages of corin activation (Corin-p vs. Corin bands) were calculated based on densitometry of Western blots represented in C, right panel. n=4 per group. **p<0.001; n.s., not significant. (E, F) Flow cytometric analysis of surface corin in HEK293 cells transfected with WT and mD99 plasmids. Data are mean ± S.D. of corin-positive cells. n=4 per group. (G) Western analysis of cell surface-labeled corin proteins under reducing conditions (top). Corin proteins in cell lysates were controls (bottom).
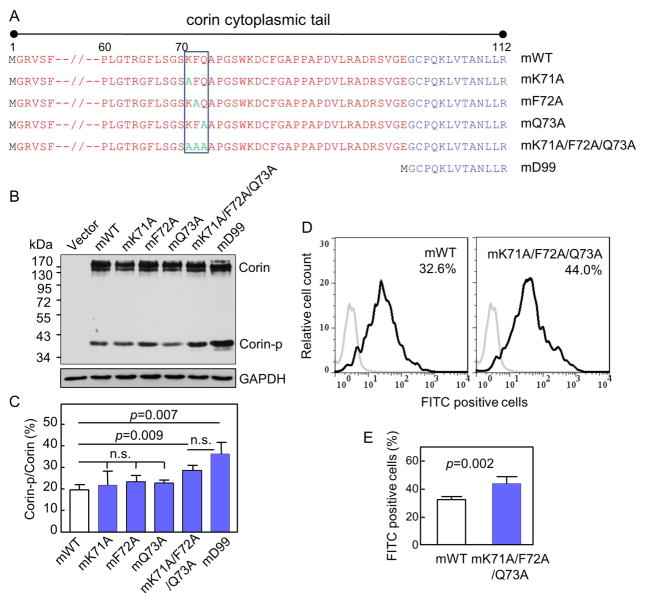
Fig. 4. Analysis of corin mutants with full-length cytoplasmic tails lacking the KFQ motif.
(A) Illustration of corin mutants with full-length cytoplasmic tails. Residues 7–59 are not shown. The KFQ motif is boxed. (B, C) Western analysis of corin mutants in transfected HEK293 cells under reducing conditions. Percentage of corin activation (Corin-p vs. Corin bands) were calculated based on densitometry of Western blots. n=3 per group. (D, E) Flow cytometric analysis of surface corin in HEK293 cells transfected with mWT and mK71A/F72A/Q73A plasmids. Data are mean ± S.D. of corin-positive cells. n=5 per group.
2.2. Cell culture and transfection
HEK293 cells were grown in 6-well plates (Corning) with DMEM and 10% fetal bovine serum (FBS) at 37°C in humidified incubators with 5% CO2 and 95% air. To express corin proteins, the cells were transfected with expression plasmids using TurboFect (Thermo Scientific) or FuGENE HD (Roche Diagnostics) reagents, following manufacturers’ instructions. The cells were cultured in DMEM with 10% FBS at 37°C for 24 h.
2.3. Western blotting
The transfected cells were lysed in a solution containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100 (v/v), 10% glycerol (v/v), and a protease inhibitor mixture (1:100 dilution; Sigma). Lysates were mixed with a loading buffer with (reducing) or without (non-reducing) 5% β-mercaptoethanol and heated at 98°C for 5 min. The proteins were analyzed by SDS-PAGE and Western blotting using an antibody (Invitrogen) against the V5 tag in recombinant corin proteins, as described previously [24].
2.4. Analysis of cell surface proteins
HEK293 cells in 6-well plates were transfected with corin-expressing plasmids and cultured at 37°C for 24 h. Cell surface proteins were labeled with 1 mM biotin-conjugated sulfo-NHS (N-hydroxysulfosuccinimide) (Pierce) at 4°C for 5 min. The reaction was terminated with 100 mM glycine in PBS. After 30 min at 4°C, the cells were lysed. Streptavidin-sepharose beads (Thermo Scientific) were added to the cell lysate and incubated at 4°C for 2 h. The beads were washed with PBS and heated at 98°C in a loading buffer for 1 min. Eluted proteins were analyzed by SDS-PAGE and Western blotting.
2.5. Flow cytometry
Plasmids expressing corin were transfected into cultured HEK293 cells. After 24 h at 37°C, the cells were detached using a 0.02% EDTA solution. Corin proteins on the intact cell surface were analyzed by flow cytometry using an anti-V5 antibody and an FITC-conjugated secondary antibody (Invitrogen) [24]. Life gating was performed using pyridinium iodide (Sigma). Data were acquired on a flow cytometer (FACSCalibur, BD) and analyzed by FlowJo V7.6.5 software (Tree Star).
2.6. Statistical analysis
Data were analyzed using software ImageJ or Prism 5 (GraphPad). Comparisons between two groups were done using Student’s t test. Comparisons among three or more groups were done using ANOVA followed by Bonferroni’s post hoc analysis. P<0.05 was considered to be significant. All data are presented as means ± S.D.
3. Results
3.1. Expression and activation of mouse corin WT and the mD50 and mD99 mutants
Mouse corin is activated at Arg-868 (Fig. 1A). A disulfide bond tethers the protease domain to the propeptide after the cleavage. On Western blots under reducing conditions, the cleaved protease domain fragment (Corin-p) appeared at ~40 kDa (Fig. 1B, top). Under non-reducing conditions, the disulfide bond remained unbroken and the activated corin migrated similarly as corin zymogen at ~190 kDa (Fig. 1B, bottom). In the mR868A mutant lacking the activation site (Fig. 1A), the ~40-kDa band was undetectable (Fig. 1B, top).
The mouse corin cytoplasmic tail consists of 112 amino acids, of which residues 1–99 share little sequence similarity with human corin sequence (Fig. 1A). We made the mouse corin mutants mD50 and mD99 by deleting 49 and 98 residues, respectively (Fig. 1A), and expressed them in HEK293 cells. In Western analysis, mouse corin WT (mWT) and mutants mD50 and mD99 had similar expression levels and migration patterns (Fig. 1C, left). The Western blotting was not sensitive enough to distinguish the corin proteins with different cytoplasmic lengths at ~190–200 kDa. Under reducing conditions, the Corin-p band was detected in the mD99 mutant at a higher level than that in mWT and the mD50 mutant (Fig. 1C, right). Based on densitometry of the Western blots, the Corin-p band in the mD99 mutant represented 29.0 ± 0.3% of the total amount of corin protein, which was higher than that in mWT (12.5 ± 1.2%) and the mD50 mutant (13.9 ± 2.2%) (p values <0.001) (Fig. 1D, left). As controls, human corin WT (hWT) and the hE1a variant were included (Fig. 1A and C). The level of the Corin-p band in hWT was higher than that in the hE1a variant (Fig. 1D, right), as reported previously [24]. On Western blots, the mouse Corin-p band migrated slower than the human Corin-p band, because the mouse fragment contains two N-glycosylation sites (N970 and N1089, Fig. 1A) and hence more N-glycans than that in the human fragment, which contains only one N-glycosylation site [28].
Corin activation occurs on the cell surface but not intracellularly [23, 24]. The Corin-p band thus serves as an indicator for corin cell surface expression and activation. Increased levels of the Corin-p band in the mD99 mutant indicated increased cell surface expression. We confirmed this result by flow cytometry. In mD99 plasmid transfected HEK293 cells, 69.2 ± 4.5% were corin-positive, which was higher than that in mWT plasmid transfected cells (35.6 ± 5.1%) (Figs. 1E and F, p=0.006). We verified these results by Western analysis of biotin-labeled cell surface proteins in the transfected cells. Levels of mD99 protein in the cell surface proteins were higher than that of mWT (Fig. 1G, top), whereas these two proteins had comparable levels in cell lysates (Fig. 1G, bottom). As controls, levels of the Corin-p band in hWT were higher than that in the hE1a variant in the cell surface proteins (Fig. 1G, top), consistent with the results in Fig. 1C and D and our previous reports [24].
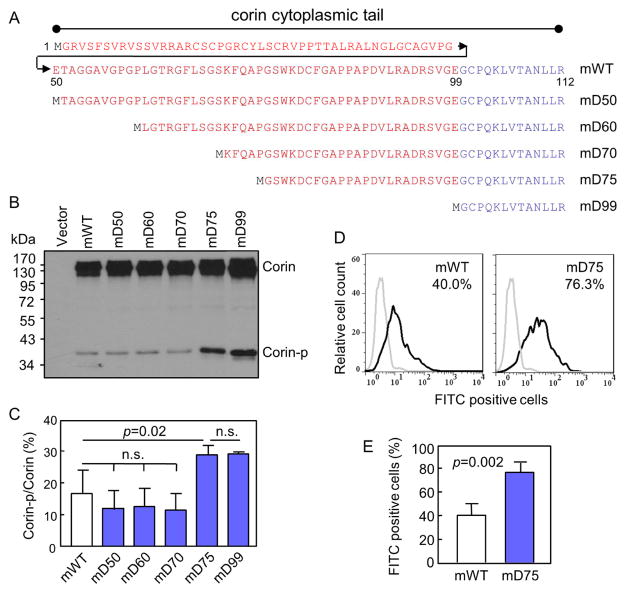
3.2. Analysis of the corin m60, m70 and m75 mutants
Increased levels of cell surface expression and activation in the mD99 mutant indicated that cytoplasmic residues between 50–98 inhibited mouse corin cell surface targeting. To identify such residues, we constructed the mutants mD60, mD70 and mD75 by deleting 59, 69 and 74 cytoplasmic residues, respectively (Fig. 2A). The mutants were expressed in HEK293 cells and analyzed by Western blotting. The results showed that the Corin-p band level in the mD75 mutant was comparable to that in the mD99 mutant but higher than that in mWT and mD60 and mD70 mutants (Fig. 2B). Based on densitometry of the Western blots, percentages of the Corin-p band in the mD75 (28.8 ± 0.02%) and mD99 (29.1 ± 0.3%) mutants were higher than that in mWT (16.6 ± 0.04%) and the mD50 (11.8 ± 2.9%), mD60 (12.5 ± 2.9%) and mD70 (11.5 ± 2.6%) mutants (p values <0.05) (Fig. 2C). The differences between the mD75 and mD99 mutants and among mWT and the mD50, mD60 and mD70 mutants were not significant (Fig. 2C).
Fig. 2. Analysis of corin mutants with truncated cytoplasmic tails.
(A) Illustration of corin mutants with truncated cytoplasmic tails. (B, C) Western analysis of corin mutants in transfected HEK293 cells under reducing conditions. Percentage of corin activation (Corin-p vs. Corin bands) were calculated based on densitometry of Western blots. n=3 per group. (D, E) Flow cytometric analysis of surface corin in HEK293 cells transfected with mWT and mD75 plasmids. Data are mean ± S.D. of corin-positive cells. n=4 per group.
We next compared the cell surface expression of mWT and the mD75 mutant by flow cytometry. In mWT plasmid transfected HEK293 cells, 40.0 ± 5.1% were corin-positive, whereas in mD75 plasmid transfected cells, 76.3 ± 4.4% were corin-positive (Fig. 2D). The difference was highly significant (p=0.002) (Fig. 2E). The results indicated that residues 71–75 had an inhibitory effect on corin cell surface expression.
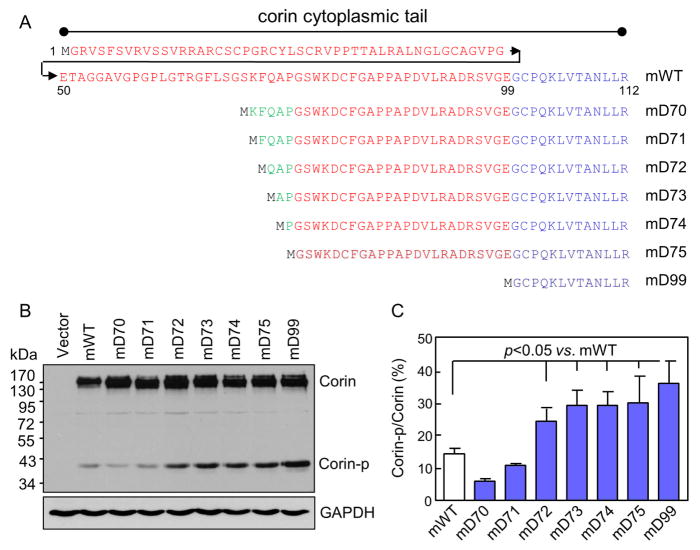
3.3. Analysis of the corin mD71, mD72, mD73 and mD74 mutants
We made four more truncation mutants the mD71, mD72, mD73 and mD74 (Fig. 3A), and expressed them in HEK293 cells. In Western analysis of cell lysates under reducing conditions, levels of the Corin-p band in the mD72, mD73 and mD74 mutants were all higher than that in mWT (Fig. 3B). In contrast, no such increase was observed in the mD70 and mD71 mutants. Based on densitometry of the Western blots, percentages of the Corin-p band were 26.4 ± 2.7, 29.2 ± 2.7, 29.2 ± 2.5 and 30.1 ± 4.7%, respectively, in the mD72, mD73, mD74 and mD75 mutants, which were all higher than that in mWT (16.4 ± 2.1%) (p values <0.05) (Fig. 3C). The results indicated that residues K71, F72 and Q73, individually or together, were responsible for inhibiting corin cell surface expression and activation.
Fig. 3. Analysis of corin mutants with truncated cytoplasmic tails around the KFQ motif.
(A) Illustration of corin mutants with truncated cytoplasmic tails. (B, C) Western analysis of corin truncation mutants in transfected HEK293 cells under reducing conditions. Percentage of corin activation (Corin-p vs. Corin bands) were calculated based on densitometry of Western blots. n=4 per group.
3.4. Analysis of the corin mK71A, mF72A, mQ73A and mK71A/F72A/Q73A mutants
We next made the mK71A, mF72A, mQ73A and mK71A/F72A/Q73A mutants with point mutations in the full-length cytoplasmic tail (Fig. 4A). In Western analysis of lysates from transfected HEK293 cells, levels of the Corin-p band were similar in mWT and the mK71A, mF72A and mQ73A mutants (Fig. 4B and C). In contrast, levels of the Corin-p band in the mK71A/F72A/Q73A mutant were higher than that in mWT (Fig. 4B). Based on densitometry of the Western blots, the percentage of the Corin-p band in the mK71A/F72A/Q73A mutant was 28.7 ± 1.3%, significantly higher than that in mWT (19.6 ± 1.4%; p=0.009) (Fig. 4C). The difference in Corin-p band levels between the mK71A/F72A/Q73A and mD99 mutants was not significant (p>0.05) (Fig. 4B and C). We also examined the cell surface expression of mWT and the mK71A/F72A/Q73A mutant by flow cytometry. In mK71A/F72A/Q73A plasmid transfected cells, 44.0 ± 2.2% cells were corin-positive, which was significantly higher than that in mWT plasmid transfected cells (32.6 ± 1.0%, p=0.002) (Fig. 4D and E).
4. Discussion
Transmembrane proteins represent ~20–30% predicted proteins in eukaryotic genomes [29, 30] and are essential for cell survival and function. Cytoplasmic tails in these proteins play a key role in protein scaffolding, intracellular trafficking, membrane sorting and cell signaling [31–34]. In many transmembrane proteases, the cytoplasmic tails regulate protease translocation, activity, substrate accessibility, and internalization [35–37]. The TTSPs are important in maintaining physiological homeostasis [1, 4]. To date, however, there have been few studies analyzing cytoplasmic domain functions in these proteases.
In this study, we analyzed the role of cytoplasmic sequences in corin cell surface targeting and activation. We expressed mouse corin proteins with truncated cytoplasmic tails in transfected HEK293 cells and found that corin truncation mutants lacking a Lys-Phe-Gln (KFQ) sequence at residues 71–73 had higher levels of cell surface expression and zymogen activation compared with that in mWT (Figs. 1–3). When Lys-71, Phe-72 and Gln-73 residues were mutated together in the full-length corin, the mutant mK71A/F72A/Q73A also exhibited increased levels of cell surface expression and activation (Fig. 4). In contrast, such an increase was not observed in the mutants mK71A, mF72A and mQ73A, in which Lys-71, Phe-72 and Gln-73 residues were mutated individually (Fig. 4). These results indicate that Lys-71, Phe-72 and Gln-73 residues serve as a motif in regulating corin cell surface expression.
Cytoplasmic motifs are known to mediate transmembrane protein translocation and sorting [38, 39]. These motifs are recognized by membrane-associated carrier proteins in the ER and the Golgi for targeting to different subcellular destinations [40–42]. At this time, little is known regarding how corin traffics intracellularly. The KFQ motif we identified in this study has not been reported in other transmembrane proteins. We found that deletion or mutation of this motif enhanced corin cell surface expression, suggesting that the motif may interact with a regulatory element in the ER or the Golgi, thereby reducing corin trafficking to the cell surface. In many transmembrane proteins, ER and Golgi retention signals serve as a mechanism to ensure that proteins are properly folded and glycosylated before being transported to specific subcellular locations [39, 43–45]. Corin proteins are highly N-glycosylated [23]. N-glycans are required for corin targeting to the cell surface [20, 23, 28]. The identification of the KFQ motif should encourage additional studies to understand how this motif regulates corin transport in the intracellular trafficking pathway.
It is interesting to note that the KFQ motif is conserved in mouse and rat corin proteins but not in human corin. Corin proteins in most mammalian species, including monkey, bovine and dog, are similar to human corin containing the DDNN, but not the KFQ, cytoplasmic motif (www.ensemble.org). The different cytoplasmic sequences are likely derived from alternatively spliced corin gene exons [24]. It remains unclear how the exon encoding the KFQ-containing segment was selected preferentially in the rodent corin genes during evolution.
In human corin, the cytoplasmic DDNN motif regulates the cell surface expression [24]. Recently, we identified a human CORIN minor allele encoding a variant with a truncated cytoplasmic tail that lacks the DDNN motif [46]. In transfected cells, the corin variant exhibited poor trafficking in the Golgi and low expression on the cell surface [46]. Individuals carrying this variant CORIN allele have higher levels of unprocessed natriuretic peptides in blood and are more likely to develop hypertension than individuals with the WT allele [46]. These results support the idea that the corin cytoplasmic tail is of functional importance and that genetic mutations altering corin cytoplasmic sequences may impair corin activity and contribute to hypertension. In light of our new findings, further studies to understand the functional significance of the corin cytoplasmic tails in different species may provide new insights into the cellular mechanism that regulates corin expression and activity.
Supplementary Material
Highlights.
Corin is a transmembrane protease that activates the natriuretic peptides
Cell surface expression is important for corin zymogen activation
A novel cytoplasmic motif regulates corin cell surface expression and activation
Acknowledgments
We thank Drs. Meng Liu and Tiantian Zhou for their support. This work was supported in part by grants from the National Institutes of Health (HD064634, HL126697), the National Natural Science Foundation of China (81170247, 81370718), the Ministry of Education of China (213016A) and the Priority Academic Program Development of Jiangsu Higher Education Institutes.
Abbreviations
- FBS
fetal bovine serum
- HEK
human embryonic kidney
- TTSP
type II transmembrane serine protease
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Antalis TM, Bugge TH, Wu Q. Membrane-anchored serine proteases in health and disease. Prog Mol Biol Transl Sci. 2011;99:1–50. doi: 10.1016/B978-0-12-385504-6.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi SY, Bertram S, Glowacka I, Park YW, Pohlmann S. Type II transmembrane serine proteases in cancer and viral infections. Trends Mol Med. 2009;15:303–312. doi: 10.1016/j.molmed.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Hooper JD, Clements JA, Quigley JP, Antalis TM. Type II transmembrane serine proteases. Insights into an emerging class of cell surface proteolytic enzymes. J Biol Chem. 2001;276:857–860. doi: 10.1074/jbc.R000020200. [DOI] [PubMed] [Google Scholar]
- 4.Szabo R, Bugge TH. Membrane-anchored serine proteases in vertebrate cell and developmental biology. Annu Rev Cell Dev Biol. 2011;27:213–235. doi: 10.1146/annurev-cellbio-092910-154247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webb SL, Sanders AJ, Mason MD, Jiang WG. Type II transmembrane serine protease (TTSP) deregulation in cancer. Front Biosci (Landmark Ed) 2011;16:539–552. doi: 10.2741/3704. [DOI] [PubMed] [Google Scholar]
- 6.Bugge TH, Antalis TM, Wu Q. Type II transmembrane serine proteases. J Biol Chem. 2009;284:23177–23181. doi: 10.1074/jbc.R109.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hooper JD, Scarman AL, Clarke BE, Normyle JF, Antalis TM. Localization of the mosaic transmembrane serine protease corin to heart myocytes. Eur J Biochem. 2000;267:6931–6937. doi: 10.1046/j.1432-1033.2000.01806.x. [DOI] [PubMed] [Google Scholar]
- 8.Yan W, Sheng N, Seto M, Morser J, Wu Q. Corin, a mosaic transmembrane serine protease encoded by a novel cDNA from human heart. J Biol Chem. 1999;274:14926–14935. doi: 10.1074/jbc.274.21.14926. [DOI] [PubMed] [Google Scholar]
- 9.Ichiki T, Huntley BK, Heublein DM, Sandberg SM, McKie PM, Martin FL, Jougasaki M, Burnett JC., Jr Corin is present in the normal human heart, kidney, and blood, with pro-B-type natriuretic peptide processing in the circulation. Clin Chem. 2011;57:40–47. doi: 10.1373/clinchem.2010.153908. [DOI] [PubMed] [Google Scholar]
- 10.Wu Q, Xu-Cai YO, Chen S, Wang W. Corin: new insights into the natriuretic peptide system. Kidney Int. 2009;75:142–146. doi: 10.1038/ki.2008.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan W, Wu F, Morser J, Wu Q. Corin, a transmembrane cardiac serine protease, acts as a pro-atrial natriuretic peptide-converting enzyme. Proc Natl Acad Sci USA. 2000;97:8525–8529. doi: 10.1073/pnas.150149097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan JC, Knudson O, Wu F, Morser J, Dole WP, Wu Q. Hypertension in mice lacking the proatrial natriuretic peptide convertase corin. Proc Natl Acad Sci U S A. 2005;102:785–790. doi: 10.1073/pnas.0407234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Shen J, Cui Y, Jiang J, Chen S, Peng J, Wu Q. Impaired sodium excretion and salt-sensitive hypertension in corin-deficient mice. Kidney Int. 2012;82:26–33. doi: 10.1038/ki.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui Y, Wang W, Dong N, Lou J, Srinivasan DK, Cheng W, Huang X, Liu M, Fang C, Peng J, Chen S, Wu S, Liu Z, Dong L, Zhou Y, Wu Q. Role of corin in trophoblast invasion and uterine spiral artery remodelling in pregnancy. Nature. 2012;484:246–250. doi: 10.1038/nature10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong N, Fang C, Jiang Y, Zhou T, Liu M, Zhou J, Shen J, Fukuda K, Qin J, Wu Q. Corin mutation R539C from hypertensive patients impairs zymogen activation and generates an inactive alternative ectodomain fragment. J Biol Chem. 2013;288:7867–7874. doi: 10.1074/jbc.M112.411512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong N, Zhou T, Zhang Y, Liu M, Li H, Huang X, Liu Z, Wu Y, Fukuda K, Qin J, Wu Q. Corin mutations K317E and S472G from preeclamptic patients alter zymogen activation and cell surface targeting. J Biol Chem. 2014;289:17909–17916. doi: 10.1074/jbc.M114.551424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dries DL, Victor RG, Rame JE, Cooper RS, Wu X, Zhu X, Leonard D, Ho SI, Wu Q, Post W, Drazner MH. Corin gene minor allele defined by 2 missense mutations is common in blacks and associated with high blood pressure and hypertension. Circulation. 2005;112:2403–2410. doi: 10.1161/CIRCULATIONAHA.105.568881. [DOI] [PubMed] [Google Scholar]
- 18.Rame JE, Drazner MH, Post W, Peshock R, Lima J, Cooper RS, Dries DL. Corin I555(P568) allele is associated with enhanced cardiac hypertrophic response to increased systemic afterload. Hypertension. 2007;49:857–864. doi: 10.1161/01.HYP.0000258566.95867.9e. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Y, Wu Q. Corin in natriuretic peptide processing and hypertension. Curr Hypertens Rep. 2014;16:415. doi: 10.1007/s11906-013-0415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gladysheva IP, King SM, Houng AK. N-glycosylation modulates the cell-surface expression and catalytic activity of corin. Biochem Biophys Res Commun. 2008;373:130–135. doi: 10.1016/j.bbrc.2008.05.181. [DOI] [PubMed] [Google Scholar]
- 21.Gladysheva IP, Robinson BR, Houng AK, Kovats T, King SM. Corin is co-expressed with pro-ANP and localized on the cardiomyocyte surface in both zymogen and catalytically active forms. J Mol Cell Cardiol. 2008;44:131–142. doi: 10.1016/j.yjmcc.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 22.Knappe S, Wu F, Masikat MR, Morser J, Wu Q. Functional analysis of the transmembrane domain and activation cleavage of human corin: design and characterization of a soluble corin. J Biol Chem. 2003;278:52363–52370. doi: 10.1074/jbc.M309991200. [DOI] [PubMed] [Google Scholar]
- 23.Liao X, Wang W, Chen S, Wu Q. Role of glycosylation in corin zymogen activation. J Biol Chem. 2007;282:27728–27735. doi: 10.1074/jbc.M703687200. [DOI] [PubMed] [Google Scholar]
- 24.Qi X, Jiang J, Zhu M, Wu Q. Human corin isoforms with different cytoplasmic tails that alter cell surface targeting. J Biol Chem. 2011;286:20963–20969. doi: 10.1074/jbc.M110.217570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takei Y, Kawakoshi A, Tsukada T, Yuge S, Ogoshi M, Inoue K, Hyodo S, Bannai H, Miyano S. Contribution of comparative fish studies to general endocrinology: structure and function of some osmoregulatory hormones. J Exp Zool A Comp Exp Biol. 2006;305:787–798. doi: 10.1002/jez.a.309. [DOI] [PubMed] [Google Scholar]
- 26.Irving P, Troxler L, Heuer TS, Belvin M, Kopczynski C, Reichhart JM, Hoffmann JA, Hetru C. A genome-wide analysis of immune responses in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15119–15124. doi: 10.1073/pnas.261573998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan J, Jia H, Ma Z, Ye H, Zhou M, Su L, Liu J, Guo AY. The evolutionary analysis reveals domain fusion of proteins with Frizzled-like CRD domain. Gene. 2014;533:229–239. doi: 10.1016/j.gene.2013.09.083. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Zhou T, Peng J, Xu P, Dong N, Chen S, Wu Q. Distinct roles of N-glycosylation at different sites of corin in cell membrane targeting and ectodomain shedding. J Biol Chem. 2015;290:1654–1663. doi: 10.1074/jbc.M114.606442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleming KG. Riding the wave: structural and energetic principles of helical membrane proteins. Curr Opin Biotechnol. 2000;11:67–71. doi: 10.1016/s0958-1669(99)00056-7. [DOI] [PubMed] [Google Scholar]
- 30.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carson DD. The cytoplasmic tail of MUC1: a very busy place. Sci Signal. 2008;1:pe35. doi: 10.1126/scisignal.127pe35. [DOI] [PubMed] [Google Scholar]
- 32.Hunyady L. Molecular mechanisms of angiotensin II receptor internalization. J Am Soc Nephrol. 1999;10(Suppl 11):S47–56. [PubMed] [Google Scholar]
- 33.Patel BN, Van Vactor DL. Axon guidance: the cytoplasmic tail. Curr Opin Cell Biol. 2002;14:221–229. doi: 10.1016/s0955-0674(02)00308-3. [DOI] [PubMed] [Google Scholar]
- 34.Qin J, Vinogradova O, Plow EF. Integrin bidirectional signaling: a molecular view. PLoS Biol. 2004;2:e169. doi: 10.1371/journal.pbio.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker RP, Urban S. Cytosolic extensions directly regulate a rhomboid protease by modulating substrate gating. Nature. 2015;523:101–105. doi: 10.1038/nature14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajasekaran SA, Anilkumar G, Oshima E, Bowie JU, Liu H, Heston W, Bander NH, Rajasekaran AK. A novel cytoplasmic tail MXXXL motif mediates the internalization of prostate-specific membrane antigen. Mol Biol Cell. 2003;14:4835–4845. doi: 10.1091/mbc.E02-11-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu P, Sun Y, Xu R, Sang Y, Zhao J, Liu G, Cai L, Li C, Zhao S. The interaction between ADAM 22 and 14-3-3zeta: regulation of cell adhesion and spreading. Biochem Biophys Res Commun. 2003;301:991–999. doi: 10.1016/s0006-291x(03)00056-1. [DOI] [PubMed] [Google Scholar]
- 38.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 39.Teasdale RD, Jackson MR. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the golgi apparatus. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- 40.Dancourt J, Barlowe C. Protein sorting receptors in the early secretory pathway. Annu Rev Biochem. 2010;79:777–802. doi: 10.1146/annurev-biochem-061608-091319. [DOI] [PubMed] [Google Scholar]
- 41.Guo Y, Sirkis DW, Schekman R. Protein sorting at the trans-Golgi network. Annu Rev Cell Dev Biol. 2014;30:169–206. doi: 10.1146/annurev-cellbio-100913-013012. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Boulan E, Kreitzer G, Musch A. Organization of vesicular trafficking in epithelia. Nat Rev Mol Cell Biol. 2005;6:233–247. doi: 10.1038/nrm1593. [DOI] [PubMed] [Google Scholar]
- 43.Banfield DK. Mechanisms of protein retention in the Golgi. Cold Spring Harb Perspect Biol. 2011;3:a005264. doi: 10.1101/cshperspect.a005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomord V, Wee E, Faye L. Protein retention and localization in the endoplasmic reticulum and the golgi apparatus. Biochimie. 1999;81:607–618. doi: 10.1016/s0300-9084(99)80118-7. [DOI] [PubMed] [Google Scholar]
- 45.Machamer CE. Golgi retention signals: do membranes hold the key? Trends Cell Biol. 1991;1:141–144. doi: 10.1016/0962-8924(91)90001-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Li H, Zhou J, Wang A, Yang J, Wang C, Liu M, Zhou T, Zhu L, Zhang Y, Dong N, Wu Q. A corin variant identified in hypertensive patients that alters cytoplasmic tail and reduces cell surface expression and activity. Sci Rep. 2014;4:7378. doi: 10.1038/srep07378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.