Abstract
In this contribution, recent developments in the design of biocatalysts are reviewed with particular emphasis in the de novo strategy. Studies based on three different reactions, Kemp elimination, Diels-Alder and retro-aldolase, are used to illustrate different success achieved during the last years. Finally, a section is devoted to the particular case of designed metalloenzymes. As a general conclusion, the interplay between new and more sophisticated engineering protocols and computational methods, based on molecular dynamics simulations with Quantum Mechanics/Molecular Mechanics potentials and fully flexible models, seems to constitute the bed rock for present and future successful design strategies.
1. Introduction
Enzymes are biological catalysts that speed up chemical reactions making them compatible with life. Often, these catalysts show important advantages with respect to non-natural catalysts such as their chemo-, regio- and stereoselectivity and the ability to work under mild conditions of temperature and presure. Although the knowledge about the origin of enzymatic efficiency to catalyse chemical reactions is still not complete, there have been numerous studies that have provided a solid understanding about some of the key factors in biocatalysis.1,2,3,4
Many reactions employed in the chemical and biochemical industry need of efficient catalysts to improve their rate, their selectivity and their environmental friendship. Obviously enzymes are excellent candidates to fulfil these requirements but, in order to extend their applicability to different purposes under different conditions, they must be modified using protein engineering techniques. First, enzymes need to be stable enough in different conditions of pH, temperature, solvent composition and substrate concentration. In addition, many of the chemical reactions carried out in the industry do not have a natural counterpart and then new enzymes with new functions should be developed. During the last years, different protein designs have been proposed to catalyse new chemical reactions. These designs can be classified, according to the strategy used in their development, into those based on directed evolution, rational design, or a combination of them named as semi-rational design.5
Directed evolution6 applies to strategies based in natural evolution to tailor the properties of molecules instead of organisms.7 Random mutations or recombination can be done to evolve proteins in the laboratory identifying the successful variants by screening or by selection. New proteins with new desired functions can be obtained after some mutations or recombining protein fragments. The advantage of this strategy is that no structural information is needed and that distant regions of the sequence space can be explored. However, in most cases changes introduced far from the active site provoke minor effects in the kinetic properties of the protein. Another limitation of this strategy is that a minimum threshold of activity is required to start the cycles of mutations and screening.
Although in the next future we should be able to design a particular amino acid sequence that will fold into a particular structure with the desired function, rational design refers, in these days, to the introduction of direct mutations of selected residues on specific positions of an already existing protein.8 Mutations of few residues can lead to important changes in the active site while the global structure of the protein remains essentially unaltered. These mutations are driven by the analysis of the data obtained from different sources, ranging from X-ray to simulations. In particular, when computational analysis of the enzymatic activity is used to guide the mutations, one usually refers to this strategy as computational design.9
These two strategies can be combined into an efficient procedure for the design of new proteins with new functions. In the semi-rational approach a new function is obtained from rational design and the activity of the resulting protein is improved by means of several directed evolution cycles.
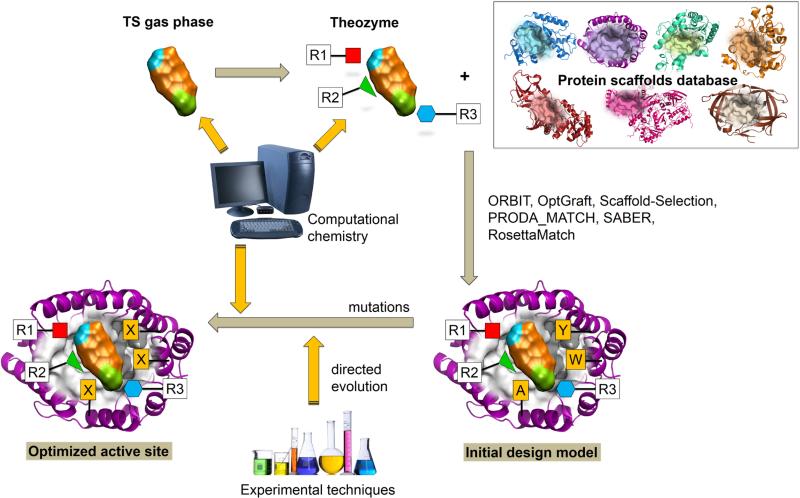
The strategies for the computational design of new enzymes can start from different protein scaffolds. These can be i) immune-globulins, proteins that were already used to produce catalytic antibodies (CAs) more than thirty years ago;10 ii) a promiscuous protein showing catalytic properties for a secondary reaction that is redesigned to improve or to change this activity; and iii) a protein without specific catalytic properties that is used as a scaffold to support a design from scratch, or de novo design, of a new activity.9,11 The first group of proteins, CAs, are obtained using as hapten a Transition State Analogue (TSA), a stable molecule resembling the Transition State (TS) structure of the reaction. While showing certain catalytic activity, their efficiency is low compared to that of natural enzymes, which can be attributed to either the nature of immune-globulins, to the lack of adequacy of the TSA to the real TS of the target reaction, or to the fact that these proteins bind the products so tightly that is the major flaw in their ability to rival enzymes for catalytic efficiency. The use of these proteins in the design process seems to decline in the last years. Instead, other protein structures seem more adequate for the process of designing new biocatalysts. The active sites of natural enzymes are an obvious starting point for this process. These active sites, composed by few residues that promote a particular chemical reaction, can be redesigned for a new function after few mutations.12 This redesign process can take advantage from the different promiscuous behaviours that can appear in a natural enzyme. Enzymes promiscuity with respect to different substrates, different catalytic activities or different reaction conditions provides the raw material for the redesign of new properties.13 The promiscuous activity in natural enzymes, which could be already known or discovered by chance, can be also induced by rational design or directed evolution. Finally, de novo design is based in the knowledge of the chemical reaction to be catalysed and, in particular, in the TS and the physicochemical principles governing its stabilization (see Figure 1). This information is obtained from computational simulations. Mayo and co-workers pioneered the field of the de novo design of enzymes14 converting the E. coli protein thioredoxin into a primitive esterase. In particular, a histidine nucleophile was introduced into an engineered surface active site reaching a 180-fold rate acceleration. The process made use of the ORBIT program to identify enzyme-like active sites within a pre-existing protein scaffold.15 This program explores simultaneously the conformational and sequence space of the designed biocatalysts.16 The actual boom in de novo design starts with three significant works about three different reaction for which no natural enzymes are known: the Kemp elimination,17 the Diels-Alder reaction18 and the retro-aldolase reaction.19 In all the three cases, which will be described in the following sections, the design process employed the Rosetta software developed at University of Washington. A previous step for the design process is the knowledge of the reaction TS and the design of a minimalist active site to stabilize its charge distribution. This active site, often known as a ‘theozyme’,20 consists of disembodied amino acids placed at adequate positions around the substrate to promote the reaction. At this stage, quantum mechanical methods are needed to properly localize the TS in this reduced model, as far as changes happen at the electronic level. In the next step the minimal active site model must be placed into an existing protein structure, what is done using the RosettaMatch module.21 This module tests different protein scaffolds grafting the ‘theozyme’ at each of the possible active-site positions. After trying different rotamers, a match is obtained when the substrate can be positioned without clashing to the protein backbone and key interactions observed in the ‘theozyme’ are conserved. The third step of the process is the design of the resulting candidates to optimize the surrounding residues for catalysis.22,23 This step consists in several cycles of sequence design and protein optimization. For this purpose protein residues are classified as designable or repackable, according to their distance to the ligand. The optimization or packing procedure is based in a simple repulsive potential. The last step consists in the ranking or scoring of the designed sequences.24,25 The hundreds or thousands of resulting designs must be evaluated before production. With this purpose each of the resulting designs is scored using different criteria that include ligand affinity and protein stability. Only those better placed in the final ranking will be expressed and tried experimentally.
Figure 1.
Schematic representation of the de novo design of new enzymes
Different improvements have been incorporated to this strategy during the last years. Usually, programs for protein design consider fixed the backbone of the protein during the design and optimization steps26 although, for instance, Rosetta program was recently modified to consider flexible backbones.27 The optimization in the sequence space can be expanded considering also non-canonical amino acids.28,29,30 Algorithm improvements have been also achieved taking advantages of player strategies and social evolution.31 Obviously, although Rosetta is probably the best-known suite of programs for enzyme design, there exist alternatives. For instance, the matching of the ideal ‘theozyme’ into an existing protein scaffold can be also carried out with OptGraft,32 Scaffold-Selection,33 and PRODA_MATCH.34,35 A different strategy is employed by the SABER program.36 Instead of placing the ideal active site into selected proteins, this program searches for proteins already presenting the catalytic residues in the adequate relative positions, thus requiring the introduction of a lower number of mutations.
In spite of all these improvements the de novo design strategy still suffers from limitations, as demonstrated by the fact that the rate of successful designs is quite low and, in addition, the best ones display reaction rate enhancements far from those of natural enzymes. As pointed out by Baker,37 a design can fail at different levels: i) the proposed ‘theozyme’ can be not a perfect model for the real TS of the enzymatic reaction, ii) the active site designed into a given protein scaffold can be distorted with respect to the ideal geometry, and iii) the effect of the long range electrostatic interactions and/or protein dynamics can be incompatible with catalysis. In addition, a more complete design strategy should not consider exclusively the TS stabilization but the whole reaction process and, in particular, the differential stabilization of the TS with respect to the reactants. In other words, the enzyme must be designed considering not only the preorganization of the active site around the TS but also the reorganization that occurs during the chemical process and that can impose an energy penalty during the barrier climbing process. In a review published in 2008, we already distinguished between “structure-based strategies” and “reaction analysis-based strategies”.7 Enzyme designs based in a ‘theozyme’ would fall in the first category while the limitations pointed out in current strategies could be overcome with methods that make use of the analysis of reaction profiles. Obviously the computational cost of this second strategy, which is based in the use of Molecular Dynamics (MD) with hybrid Quantum Mechanics/Molecular Mechanics (QM/MM) potentials, is much higher and then it has to be used in combination with the first one.
Many excellent reviews about the design of new biocatalysts have been recently published.5,7,9,11,38,39,40,41,42,43,44,45 This review will focus mainly on the advances obtained in the field of the de novo design during the last years, emphasizing the new achievements of the different computational strategies employed in the design process. The three aforementioned reactions, Kemp, Diels-Alder and retro-aldolase, will be used to illustrate these points. An additional section will be devoted to the particularities of the design of metal-containing enzymes.
2. Kemp elimination
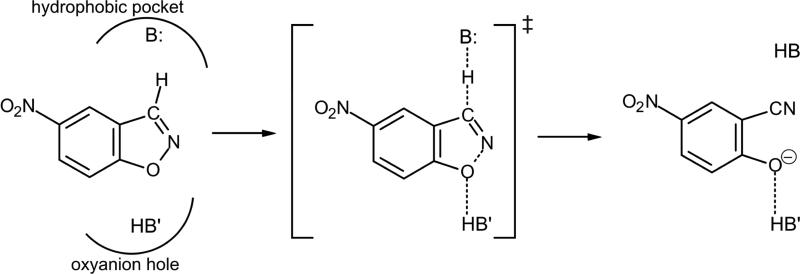
The Kemp elimination, that consists in the conversion of benzisoxazoles into salicylonitriles, is a well-studied reaction that takes place by means of an E2 elimination mechanism (see Scheme 1). The reaction implies a proton transfer from a carbon atom of the substrate to a base, thus creating an anion species with the concomitant opening of the ring.46,47,48 The reaction appear to be very sensitive to the base strength, having tested hydroxide ion,46,47 amines46,47 or the conjugated base of acids.48 Since no naturally occurring enzyme has been identified to catalyse the reaction, and together with the fact that it is not a very complex reaction, it has been used as a benchmark of different protocols to design new enzymes.
Scheme 1.
Representation of the base-catalyzed Kemp elimination
2.1. Antibodies and other catalysts
The first designed biocatalysts of Kemp elimination (34E4 and 35F10), with rate acceleration greater than 108, were catalytic antibodies generated in response to a positively charged hapten mimicking the TS geometry.49 Other cationic haptens have been used to get catalytic antibodies that catalyze this reaction, such as 4B2 and 6C2,50 or the 13G551, being the latter, together with the previously mentioned 34E4, the ones showing the highest catalytic activity. Following a similar experimental protocol, an antibody with an active site consisting of a hydrophobic pocket with a conserved lysine residue was generated against a reactive diketone hapten. This antibody, 38C2, showed activity for a variety of reactions that proceed via an enamine intermediate and, interestingly, also present promiscuous activity for the base-catalysed Kemp elimination.52
The use of X-ray diffraction has allowed obtaining details, at atomic level, of the structures of the originally designed catalytic antibodies, as well as variants generated by mutation of key residues. Analysis of these structures together with kinetic studies, has contributed to the elucidation of the key features responsible of the catalytic activity of these new enzymes. Thus, the active site of 34E4 shows conformational plasticity that can be responsible of a reduced efficiency since only one of the possible orientations of the substrate is reactive. An optimal microenvironment must be present in the hydrophobic pocket to impose the proper positioning of the substrate to interact with the conjugated base, usually Glu or Asp, and to modulate the pKa of the base, since water molecules could stabilize their non-protonated form. It has been also observed that buried water molecules found in the active site can stabilize the negatively charged developed in the phenoxide oxygen atom of the substrate. Finally, π stacking interactions between a Trp residue and the highly polarizable TS has been also proposed as the origin of catalysis in 34E4.53,54,55 In the case of 13G5 antibody, solved X-ray structures suggest that Asp and Glu would act as a base to abstract the proton from the substrate and to orient a water molecule to stabilize the leaving group, respectively.56 Kemp elimination is dramatically sensitive to the medium, as illustrated by the fact that the reaction with acetate is over 107 folds slower in water than in acetonitrile, as a consequence of the stabilization of carboxylate anions in aqueous environments.57 Theoretical studies based on quantum chemical calculations on model systems in gas phase have been used to propose the most appropriate base and acid moieties to predict catalytic functionalities and to design haptens to produce improved catalytic antibodies.58
Two independent investigations showed that familiar “off-the-shelf” proteins, serum albumins, catalyse the same Kemp elimination at similar rates than the previous catalytic antibodies. In this case, the base was a lysine side-chain amino group located in a hydrophobic pocket.59,60,61 It has been demonstrated that these albumin based catalysts, that uses a lysine as a base, are less sensitive to the environment. Based on quantum mechanical optimized geometries on gas phase and in solution represented by continuum models, this different environmental dependent behaviour has been rationalized.62
A different scaffold employed to design a catalyst for the Kemp elimination has been the polyethyleneimine, highly branched polymers with primary, secondary and tertiary amines linked by ethylene units that can be modified by alkylation or acylation of their amine groups. These enzyme-like catalysts were named as synzymes.63 More recently, a new Kemp bio-catalysts has been designed by a single mutation in a small regulatory protein calmodulin (CaM). Native CaM, upon binding Ca(II), opens the hydrophobic pockets in each of its domains which, when introducing a dehydrated carboxylate, allows its accommodation to productively interact with the Michaelis complex and stabilize the TS of the reaction.64,65 Additionally, this new enzyme serves as a calcium ion sensor. Engineering of model protein cavities to catalyse the Kemp elimination has been also obtained by a single mutation of Met to His in the artificial L99A cavity in T4 Lysozyme.66 The presence of the new catalytic base, His, that abstract the proton from the substrate in a hydrophobic buried cavity is enough to catalyse the process, demonstrating that protein model cavities may provide templates for studying protein design in simplified environments.
These studies demonstrate the possibility of designing new enzymes by a reduced number of mutations in the native protein scaffold. Nevertheless, the catalytic efficiency of these new biocatalysts is still far from the most efficient enzymes.
2.2. de novo design of new enzymes
The first example of de novo protein design for Kemp elimination was published in 2008 by Houk, Tawfik, Baker and co-workers using the strategy described in the introduction.17 The significance of this new protocol based on cutting-edge computational methods was emphasized by Nanda, who stressed the importance of combining experimental and computational tools.67 In their work, different non-immunoglobulin proteins were used as starting scaffolds selected by means of Rosetta software.24,25,26 In particular, from previously designed theozymes computed by means of quantum mechanical calculations in reduced models, two active-site motifs were considered differing in the choice of catalytic base; one utilized a carboxylate group for benzisoxazole deprotonation, whereas the other exploited a histidine side chain backed up by an aspartate or glutamate to regulate its pKa and tautomeric state. In addition, a hydrogen-bond donor was provided to stabilize the developing negative charge on the phenolic oxygen atom, and π –stacking interactions were included for TS stabilization. Fifty-nine designs, each containing 10 to 15 mutations, were experimentally characterized and eight of them resulted to be active. The X-ray diffraction analysis of one of the newly produced enzymes, KE07, was solved and demonstrated to superimpose well with the computational model. Interestingly, although the best computationally designed enzyme was more than one order of magnitude less efficient than comparable antibody catalysts, their activity was significantly improved by directed evolution.17 KE07 was used as a template for further incorporation of up to 8 mutations providing a 106-fold rate acceleration, but with a marked decrease in the overall stability.68 In a different study using the KE70 as template, new computationally guided optimizations where carried out including backbone flexibility. This, combined with further directed evolution render a 400-fold increased activity.69 The highest active new catalyst originally obtained by Tawfik and co-workers, KE59, presented the inconvenience of being the least stable. The strategy to improve the performance of this template was to carry out a number of fold-stabilizing mutations prior to directed evolution cycles. Sixteen rounds of mutation and selection led to more than 2,000-fold increase in catalytic efficiency.70
Jorgensen and co-workers studied free energy surfaces to explore the reaction mechanism of four of the designed Kemp elimination catalysts17 by means of free-energy perturbation (FEP) methods within hybrid QM/MM potentials based on Monte Carlo (MC) simulations.71 Some approximations were considered in the simulations, like keeping the protein backbone fixed, the use of a semiempirical Hamiltonian (PM3) to describe the QM region, or keeping the interatomic distance between the proton donor and acceptor atoms as constant. According to their results, the catalytic mechanisms of Kemp elimination catalysed by KE07, KE10(V131N), KE15 were found to be concerted with proton transfer more advanced in the TS than the breaking of the isoxazolyl N-O bond, while a step-wise mechanism preceded by the proton transfer was found for the KE16. The trend predicted from the computed free energy barriers, by comparison with the experimental rate constants, was not encouraging.71 The application of this strategy to the study of the mechanism of KE59, KE70 and several mutants rendered similar conclusions.72 The study of the effect of the mutation Glu50Asp in the 34E4 catalytic antibody, showed an increase in the free energy barrier of 2.4 kcal·mol−1,73 in good agreement with experimental observation. Again, the same methodology allowed justifying the promiscuity of the 4B2 antibody in catalysing the allylic isomerization and the Kemp elimination reactions.74
Different approaches based on density functional theory (DFT) methods, ranging from reduced models in gas phase to those including the full protein and solvent water molecules in ONIOM QM/MM MD simulations, were carried out by Houk and coworkers to rationalize the experimental data of several designed catalysts of Kemp elimination.75 Computed activation barriers were in qualitative agreement with experiments, allowing distinguishing the inactive from the active catalysts but with a weak correlation with the energy barriers deduced from the experimentally determined rate constants.
Warshel and coworkers45,76,77 have recently pointed out that the design of a new catalyst must be focused not only in placing the reacting fragments in proper places around the substrate but in optimizing the full environment preorganization. This preorganization accounts for the required electrostatic properties of the enzyme to lower the activation free energy of the reaction, a property that depends not only on residues of the active site. Using an Empirical Valence Bond (EVB) approach, these authors studied several Kemp eliminases (catalytic antibodies, albumines and de novo designs) remarking the difficulties of designing a correctly preorganized environment adapted for the small changes in the electronic distribution observed during the formation of the TS. A larger preorganization of the environment is closely related to a smaller reorganization of the protein needed to attain the TS of the reaction. This reorganization implies an energy penalty that should be minimized to improve the catalysts design, an aspect that was emphasized also by Labas et al.78
A different approach, based on studies of failed designs of new enzymes, has been applied for the development of more efficient catalysts. In particular, Mayo and co-workers used an iterative approach starting from an inactive protein scaffold, HG-1, that was designed to convert the xylan binding pocket of a thermostable xylanase into a Kemp eliminase.79 Based on MD simulations and analysis of structures obtained from X-ray crystallography, the authors found that the inactivity could be due to bonded waters and high flexibility of residues in the active site. These findings guided the design of a more embedded binding pocket, which moved the active site deeper into the protein and resulted in an active Kemp eliminase, HG-2. MD simulations of this last structure let to an additional mutation that provided a better packing around the substrate by reducing the active site conformational heterogeneity observed in HG-2. The new designed protein, HG-3, exhibited activity comparable to the best Rosetta design.79 This new catalyst was proved to be particularly amenable to direct evolution based on multiple rounds of mutagenesis and screening. The turnover number for proton transfer of the best variant obtained in the Hilvert laboratory after 17 rounds of mutagenesis and screening, HG3.17, was comparable to the efficiency of many natural enzymes.80 The three factors that were suggested to be decisive in the improvement of the Kemp eliminase activity from HG-3 to HG3.17 were: 1) the extraordinary high shape complementarity between the binding pocket of the protein and the substrate (while two possible conformers of substrate were able to bind the pocket of HG3, only one orientation was detected in HG3.17); 2) the ligand alignament with Asp127, the catalytic base, was optimized by evolution resulting in an unusual short hydrogen bond; and 3) a new catalytic group, Gln50, stabilized the development of a negative charge in the TS.80,81
Recently, hybrid QM/MM MD simulations for the Kemp elimination in this new HG3.17 enzyme have provided a deeper insight into the origin of its catalytic efficiency.82 In this case, we have demonstrated the presence of different conformations with significant different reactivity. The larger reactivity has been demonstrated to be related with a better electrostatic preorganization of the environment that creates a more favourable electrostatic potential for the reaction to proceed. In HG3.17, efforts to improve the catalytic properties must be focused in possible mutations increasing the preorganization and decreasing the reorganization around the oxyanion hole. Mutations should be considered not only in the first shell of residues but in further shells since protein electrostatics is a long range property.
3. Diels Alder
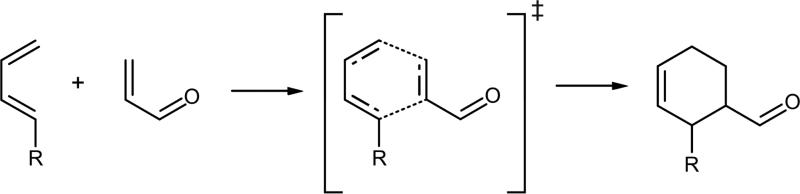
Reactions involving the forming of two carbon-carbon bonds, such as the Diels-Alder reaction (see Scheme 2), are very powerful tools in organic synthesis and they have been identified in several metabolites. Nevertheless, the number of enzymes catalysing this cycloadition reaction, if any, is very low.83,84 In fact, a recent work has claimed for the first in vitro characterization of an enzyme committed to the catalysis of a [4+2] cycloaddition reaction.85
Scheme 2.
Representation of the Diels Alder cycloaddition reaction
High regioselectivity (orto and para isomers are favoured with respect to the meta), diastereoselectivity (endo conformers are favoured with respect to the exo) and enantioselectivity (the reaction creates up to four new stereocenters) are probably key features that are searched when designing new enzymes catalysing the addition of a diene with a dienophile.9,11 The enhanced reaction rate measured in new designed Diels-Alderases has been proposed to be due to a lowering of the entropy of activation by binding the two substrate molecules, also known as “entropy trap”.86 However, a reduction in the activation enthalpy has been also proposed as the origin of catalysis.87
3.1 catalytic antibodies
The first designed catalytic antibody (1E9) showing activity as a Diels-Alderase was reported by Hilvert and co-workers.88 Braisted and Schultz, one year later, designed another catalytic antibody (39A11) with a different hapten.89 Series of site-directed mutagenesis carried out by Romesberg and Schultz allowed obtaining a mutant that displays an increase of one order of magnitude in its catalytic activity.90 Three-dimensional crystal structures of these antibodies complexed with their respective TSA have suggested an almost perfect shape complementarity between protein cavity and the substrates in the TS conformation.91,92 A deep analysis of 39A11 structure and its mutant revealed the presence of not only a favourable packing but hydrogen bond interactions that could be responsible of the control of the relative geometries of the bound substrates and the electronic distribution in the dienophile.92 A theoretical study on the germline catalytic antibody and its matured form, 39A11, based on hybrid QM/MM MD simulations, suggested a complex indirect effect through coupled movements of the backbone of the protein and the substrate in the germline, after mutation.93 These results stress the importance of backbone flexibility in docking studies and in the design of new enzymes.
A combination of experimental studies and quantum mechanical gas phase calculations, carried out by Gouverneur et al., was focused on the stereoselectivity of newly generated catalytic antibodies. In particular, the cycloaddition between trans-1-N-acylamino-1,3-butadiene and N,N-dimethylacrylamide was used to explore the control of the endo/exo and absolute configuration of the eight possible reaction products.94 These substrates were in fact the ones used in the de novo design of new Diels-Alderase, as shown in next section. A new catalytic antibody, 13G5, was made to catalyse the disfavoured exo Diels-Alder transformation, yielding a single regioisomer in high enantiomeric excess.95 This surprising absolute enantioselectivity of 13G5 was explained based on gas phase quantum mechanical calculations followed by docking and MD simulations.96
3.2. de novo design of new enzymes
The first de novo design of an enzyme catalysing the bimolecular Diels-Alder cycloaddition reaction, with high stereoselectivity and substrate specificity was performed by Siegel et al.18 After studying the normal-electron-demand reaction by quantum mechanics in gas phase, frontier molecular orbital analysis dictated that narrowing the energy gap between the highest occupied molecular orbital (HOMO) of the diene with the lowest unoccupied molecular orbital (LUMO) of the dienophile should reduce the activation energy and thus it would increase the rate of the reaction. This was achieved by positioning a hydrogen bond acceptor to interact with the former (raising the energy of the HOMO) and a hydrogen bond donor to interact with the later (thus lowering the energy of the LUMO). These electrostatic requirements were added to efforts to improve the binding of the substrates in optimal position in the active site of the designed enzyme. The design process starts from generating three dimensional models of minimal active sites (theozymes) that, in this case, consist in a glutamine or an asparagine to accept hydrogen bond from the N-H moiety of the diene carbamate, and a serine, threonine or tyrosine to donate a hydrogen bond to the carbonyl oxygen of the dienophile amide moiety. Afterwards, RosettaMatch21 program was used to search stable protein scaffolds that allow the theozyme to be placed without making substantial steric clashes with the protein backbone. Then, RosettaDesign24,25 was used to optimize the set of putative stable proteins. And finally, a reduced number of designs were selected for experimental validation proving that only two of them (DA_20 and DA_42) had catalytic activity. After rounds of directed evolution, further improvement of catalytic activity of DA_20_10 (100-fold over the original design) was obtained, with high stereoselectivity that favoured formation of only one of the stereoisomers (3R,4S). De novo design can be improved if backbone flexibility is considered. In this sense, the search for adequate conformations can be enhanced with creative strategies such as the one used by the Foldit multiplayer online game.97 In this game, players compete and collaborate solving structure refinements problems where backbone flexibility is considered. This strategy was applied to remodel the backbone of DA_20_10 Diels-Alderase to enable additional interactions with the substrates, resulting in a design (CE6) with a modest increase of the enzymatic activity of about 20-fold in kcat.98 Rounds of directed evolution were used by Hilvert and coworkers to obtain a Diels-Alderase (CE20) 6-times more efficient than CE6, reshaping the active site pocket for a better substrate preorganization and TS stabilization.99
A different semirational combinatorial design protocol has been used by Brinck and co-workers to design new Diels-Alderases with significant rate accelerations.100,101,102 The computational protocol employs molecular docking, MD and quantum chemical calculations at DFT level to find reasonable candidate protein and substrate combinations for which protein-substrate interactions were optimized. The estimation of the free energy of activation is based on two steps; the free energy difference going from the bound state to conformations named as the “near attack-conformation” (NAC),103 and the energy required to go from NAC to the TS. The first term is computed from the MD simulations while the second term is derived from the DFT calculations. In particular, efforts were focused in finding and optimizing a protein structure for a suitable oxyanion hole that would stabilize the LUMO of the dienophile,101 or in developing a route based on in situ acid-base catalysed diene generation.102
Other strategies to obtain new Diels-Alderases based on the use of metalloenzymes will be presented in section 5.
4. Retro-Aldol reaction
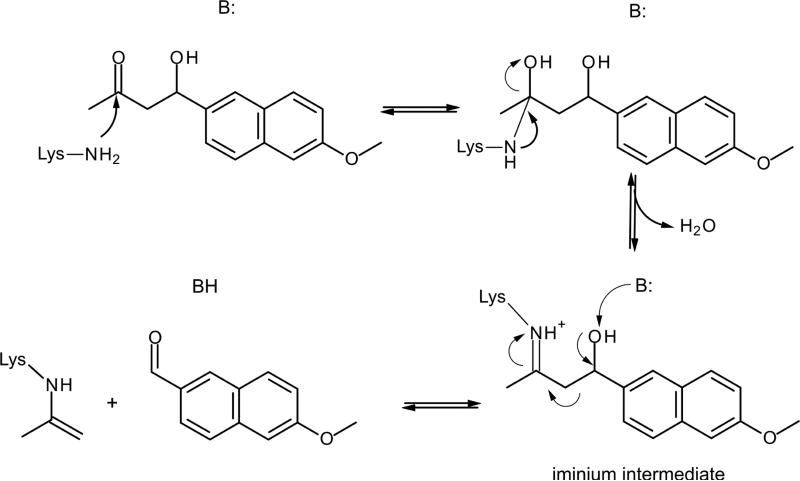
The Retro-Aldol reaction, in contrast to previous presented reactions, consists in a multi-step process that results in a C-C bond breaking. The reaction mechanism involves a nucleophilic lysine and the formation of an iminium intermediate (see Scheme 3).
Scheme 3.
Representation of the Retro-Aldol reaction mechanism
To account for the multistep reaction mechanisms, the designed new enzyme has to present an active site simultaneously compatible with different TSs and intermediates. Thus, there must be a compromise between structural reorganization during the catalytic process and the preorganization of the active site for such multistep enzyme catalysed reaction. The enzyme must present some motions to reach some degree of active site reorganization to stabilize each TS. Hilvert, Houk and co-workers have stated that these motions may be divided into intrastep and interstep reorganization motions.104
4.1 catalytic antibodies
The first catalytic antibodies that showed activity for the Aldol reaction, 38C2 and 33F12,105 were generated to catalyse the carbon-carbon bond formation. These antibodies showed to also catalyse the Retro-Aldol reaction.106,107 Later, new aldolase catalytic antibodies were designed with special attention to the enantio- and diastereoselectivity.108 The mechanism for antibody catalysis of this reaction mimics that used by natural class I aldolase enzymes that utilize the ε-amino group of an active site lysine residue to form a Shiff base with the substrate (see Scheme 3). X-ray crystallographic studies on 33F12 confirmed the presence of a lysine residue, deeply buried in a hydrophobic pocket at the base of the binding site, thereby accounting for its perturbed pKa.109 Further structural analysis by X-ray diffraction studies gave a direct observation of the enamine intermediate.110 Characterization of the Aldol reaction mechanism and the proposal of a TSA was made by Arnó and Domingo based on quantum mechanical calculations within reduced models.111
4.2. de novo design of new enzymes.
The first de novo protein design for the Retro-Aldol reaction was published in 2008 by Baker and co-workers.19 In this case, four different theozymes were prepared all presenting a lysine as a Schiff base. The four active site motifs optimized with quantum mechanical methods differ in the general acid/base for deprotonation of the β alcohol: Lys/Asp dyad, Tyr, His/Asp dyad or a water molecule. The first proposal was analogous to that with D-2-deoxyribose-5-phosphate aldolase, while the other three motifs were inspired on the active site of catalytic antibodies, as described above. Using Rosetta program24,25 for different active site geometries of the four different motifs that fitted into the binding pockets of 71 protein scaffolds, a total of 72 designs were selected for experimental characterization. The backbone geometries and side-chain orientation of two apo structures solved by X-ray diffraction studies showed an excellent agreement with those of the designed enzymes. 32 of them showed detectable retro-aldolase activity, although the rate enhancements were between two and three orders of magnitude lower than the observed in the catalytic antibodies.107,108
A computational study of these de novo designed enzymes, based on MD simulations, showed that the protein distorted the designed geometry of the substrate-enzyme intermediate thus suggesting that the substrate and the catalytic residues were poorly packed in the models. These results emphasize that the assumption of the rigid backbone during the computational enzyme designs is one important limitation. Further studies on one of the most active new enzymes, corresponding to the last motif, revealed that the catalysis was largely due to hydrophobic substrate binding interactions and, to a lesser extent, to the shifting of the pKa of the catalytic lysine.112 Structural analysis of covalently bound mechanism based inhibitor showed to be similar but not identical to the design models.113 Thus, while the substrate appears to be well oriented in the binding pocket through hydrophobic interactions with the naphthyl moiety, as predicted in the model, the relative orientation with the catalytic lysine is different. The bound water network appears to be more extensive, and the bound substrate analog exhibits more conformational heterogeneity than typical native enzyme-inhibitor complexes.
In an attempt to improve the efficiency of the new designed enzymes, Hilvert, Baker and co-workers optimized the protein via directed evolution,114,115 from the previously computationally retro-aldolases that were designed from scratch.19 The specific activity was boosted more than 4,400-fold by random mutagenesis and screening, affording catalytic efficiencies approaching those of natural enzymes. Surprisingly, dramatic changes were detected in the active site by comparison with the original structure. In particular, the reactive catalytic lysine and the ordered water molecule were unexpectedly abandoned and another lysine residue was found at another binding pocket created during the optimization process.115 The drawback of the original computational design was the assumption of a rigid protein scaffold during the simulations.115 More recently, Hilvert and co-workers have designed a new Retro-Aldolase by directed evolution from the previously computationally designed RA110.116 The original activity, that accelerated substrate cleavage by a factor of 12000 over background, was increased more than 1000-folds. The X-ray structure of the new catalyst with an inhibitor showed, again, substantial deviations from the designed model. Finally, Baker and co-workers have investigated an alternative active site design in which the catalytic water molecule was replaced by the side chain of a glutamic acid. After site-directed evolution, the rate enhanced of the new enzyme, RA110.4-6, was still below the best values obtained for catalytic antibodies and the previous example.
5. Artificial Metalloenzymes
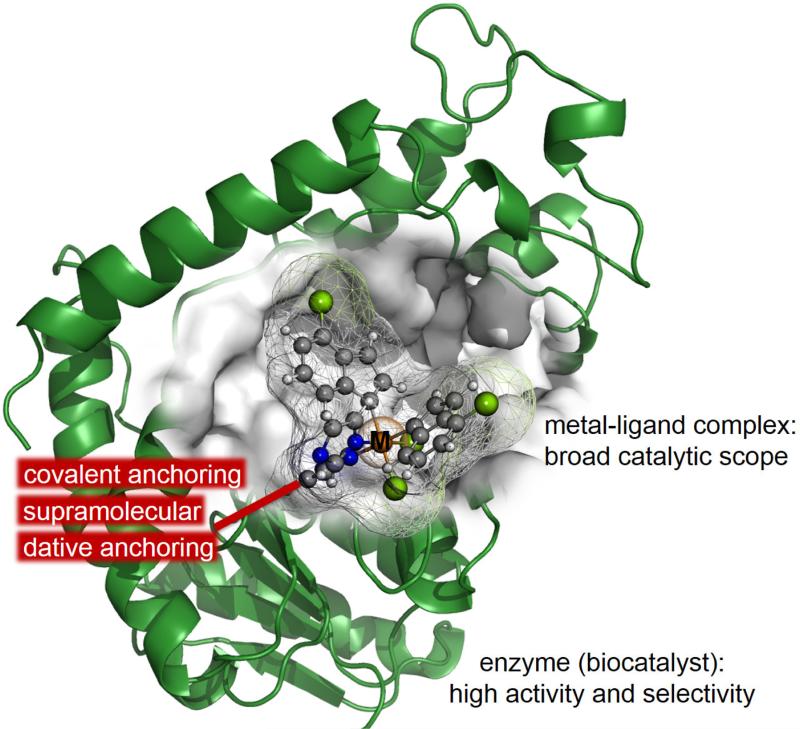
Metalloproteins, which represents almost half of the enzymes in nature, catalyse some of the most difficult biological transformations by complex reaction mechanisms.117,118 This is the origin of the extensive work devoted to the development of strategies for the design of artificial metalloenzymes as stressed in recent reviews.119,120,121,122,123,124,125,126 One of the main targets in this field is to merge the attractive properties of homogeneous catalysis and biocatalysis (see Figure 2). It is envisioned that artificial metalloenzymes can combine the best of both worlds; the broad catalytic scope, a hallmark of homogeneous catalysis, and the high activity and selectivity under mild conditions that characterize enzymatic catalysis. The main structural characteristic of metalloenzymes is the presence of a metal-ligand complex that is anchored to the biomolecule host and provides the first coordination sphere whereas the host provides the second one. Two different anchoring strategies can be used; noncovalent, which includes anchoring through supramolecular and dative interactions, and covalent anchoring, usually through a cysteine residue.117,118,119,120 Nevertheless, non-native functionalities has also been expanded through the incorporation into the protein scaffold of metal-containing cofactors not found in nature.118 In nature, proteins incorporate metal cofactors such as haem by means of non-covalent and covalent interactions.118 The de novo design of an artificial metalloenzyme is also related with the modelling of the protein scaffold from first principles. This can imply the redesign of a critical loop to improve the key side-chain interactions established with the substrate.127 Other efforts have been focused in the engineering of metal-binding sites containing cysteine residues in an existing three-helix boundle,128 or the generation of combinatorial libraries of de novo proteins (named “artificial superfamilies”) containing, for instance, four-helix bundles and presenting catalytic activity for a variety of functions.129 It is then not surprising that the border between the de novo protein design and the redesign based on existing proteins is not well defined.126,130
Figure 2.
Schematic representation of an artificial metalloenzyme, where a metal ligand complex is posed in the cavity of an enzyme by means of different possible anchoring strategies.
As exposed above, due to the large number of designed metalloenzymes, only selected examples will be highlighted in this review. In particular, we will first focus on metalloenzymes with catalytic activity in the Diels-Alder reaction. A section devoted to the design of proteins with zinc hydrolytic centers will be followed by examples of heme no-heme artificial enzymes, and we will finish with some examples of technologies based on the supramolecular anchoring approach.
5.1 Diels-Alder
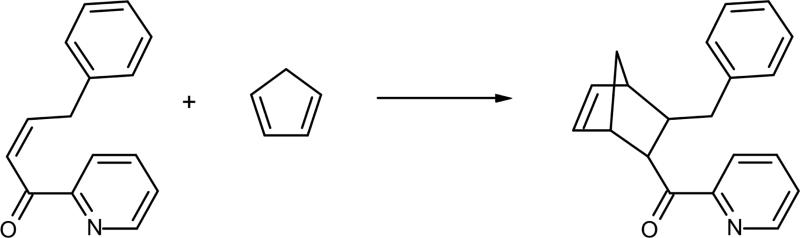
A metalloenzyme that catalyses the Diels-Alder cycloadition of azachalcone (see Scheme 4) have been created by designing a Cu(II)-specific binding site with coordinating amino acids at geometrically appropriate positions in a thermoestable host protein (tHisF).131,132 The catalytic activity has been assumed to be due to the five-membered chelate structure made by the lone electron pairs at the carbonyl O atom and the pyridine N atom, activating the substrate by lowering the energy of the LUMO. Authors observed notable enantioselectivity, enhanced endo-selectivity, and higher reaction rate. The same asymmetric Diels-Alder reaction has been used to design an artificial metalloenzyme from a different protein scaffold, a sterol carrier protein type 2 like domain (SCP-2L), obtaining a promising enantioselectivity of 25% ee and good endo selectivity133
Scheme 4.
Representation of the Diels-Alder cycloadition between azachalcone and cyclopentadiene
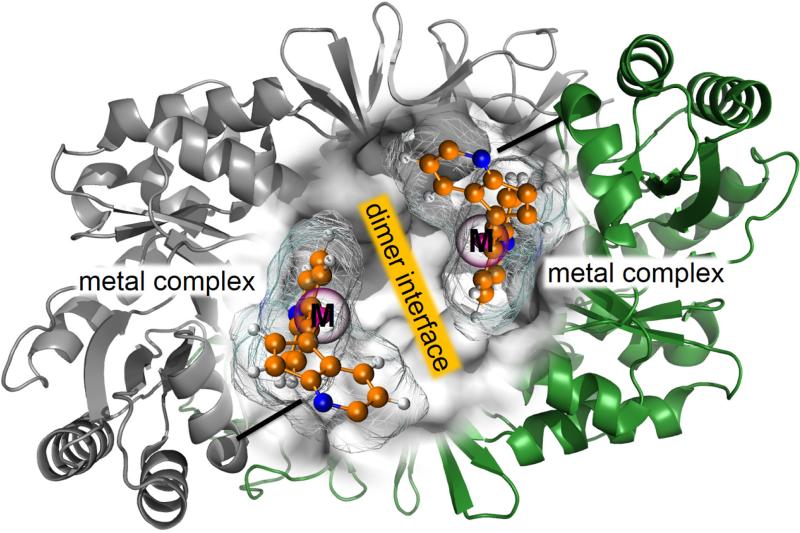
Roelfes group, using a different strategy, has also studied the Diels-Alder cycloadition between azachalcone and cyclopentadiene as a benchmark reaction to design new metaloenzymes.134 First, an active site is generated on the dimer interface of the Lactococcal multidrug resistance Regulator (LmrR) (see Figure 3). A copper(II) phenantroline complex was covalent anchored in the hydrophobic pocket of the protein using a cysteine conjugation strategy. The resulting artificial metaloenzyme showed excellent enantioselectivity (up to 97%) and diastereoselectivity (endo/exo 95:5).134
Figure 3.
Schematic representation of of proposed artificial metalloenzymes based on Cu(II) complexes grafted on the dimer interface of a protein.
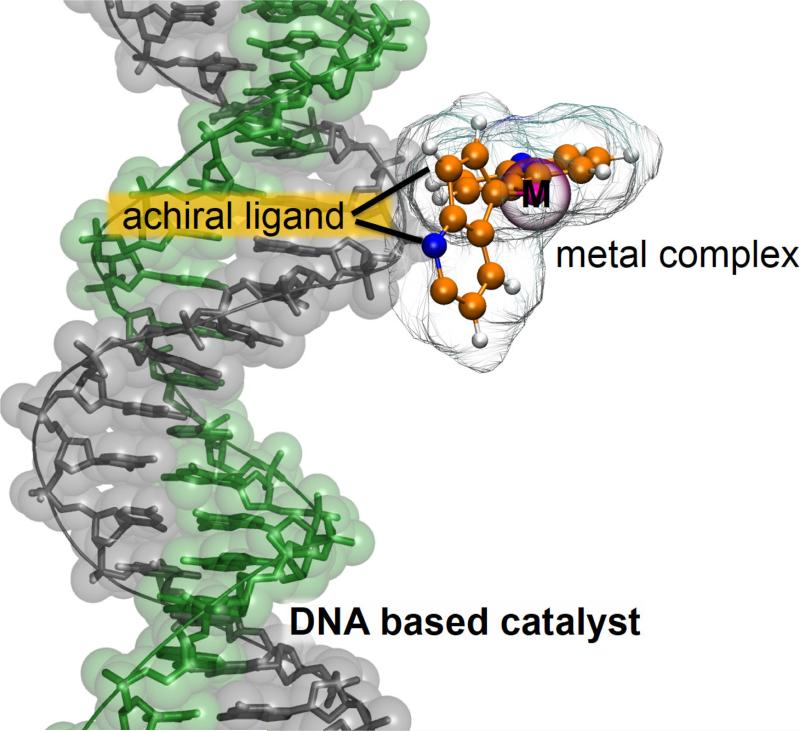
Similar to proteins, DNA can offer a defined chiral second coordination sphere for the construction of an artificial metalloenzyme, as introduced by Roelfes and Feringa for the asymmetric Diels-Alder catalysis.135,136,137 A DNA-based catalyst was designed using a copper complex of a non-chiral ligand that can bind to DNA (see Figure 4). The active Cu2+ center was brought into proximity of the DNA double helix, allowing transfer of chirality from DNA to the product of Cu2+-catalyzed Diels-Alder reaction. Products were obtained with very high diastereoselectivities and enantioselectivities (83-98%).135,136 As explained, the catalytic ensemble comprises an intercalator and a copper-chelating moiety. In the first generation, these two moieties were linked by a spacer, whereas in the second generation both sites are integrated into a single bipyridine moiety and thus the reactive copper center is brought into closer contact with the chiral environment. These findings underline the fact that the design of the ligand is an important factor in the development of this kind of artificial catalysts.117 The second generation of catalysts are also capable of enhancing the rate of the reaction up to two orders of magnitude.137
Figure 4.
Schematic representation of of proposed artificial metalloenzymes based on modular assembly of a copper complex of a non-chiral ligand to a DNA scaffold.
5.2 Zinc Hydrolytic centers
In the preceding section we have presented two different strategies for the design of Diels-Alder metalloenzymes, based on the complexation of a Cu(II) metal ion, or a metal complex. In this section we focus in the case metalloenzyme designs based in the complexation of just metal ions such as Zn(II) or Fe (II).
As stated in a recent review by Alexandrova and co-workers,138 the selection of the catalytic metal in the process of metalloenzymes evolution can be due to factors other than optimizing catalysis alone, such as metal availability or toxicity. Enzymes evolved under these restrictions are good enough for vital purposes but they do not necessarily reach the best catalytic performance. Consequently, these limitation provides an excellent opportunity to improve the catalytic properties of these enzymes in the laboratory substituting the metal for another one. In this sense, computational predictions based on DFT calculations have shown that β-lactamases could be improved via replacement of zinc by nickel.139 It must be taken into account that one of the main limitations in the computational study of metalloenzymes is the description of metal ions and its coordination shell. Typically, classical force fields and semiempirical methods, and also some DFT functional or ab initio treatments, can lead to erroneous descriptions of the coordination index of the metal and in particular its change during the reaction process.
Active site redesign changing metal ions, has been found to be a promising strategy to improve not only the primary catalytic reaction but also secondary processes in promiscuous enzymes. As an example, Leitgeb and Nidetzky enhanced the secondary hydrolase activity of a promiscuous oxygenase β-Diketone-cleaving enzyme by changing Fe2+ to Zn2+ in the active site.140
Another strategy to design metalloproteins was reported by the Baker group as an extension of their de novo enzyme design methodology to exploit the reactivity of metal ions in existing enzyme active sites.141 In particular, the method was based in the redesign of existing suitable native crystallized proteins containing mononuclear zinc centers with at least one free coordination site. TS models of organophosphate hydrolase reaction were modelled into the zinc sites and additional hydrogen-bond interactions were identified using their RosettaMatch program. After maturation using directed evolution, a 107-fold increase in the catalytic activity was obtained. Warshel and co-workers have suggested that designs on mononuclear zinc metalloenzyme can be improved by optimizing the preorganization of the environment, which is the key factor in enzyme catalysis.142
Recently, Pecoraro and co-workers have designed efficient hydrolytic zinc sites to create an artificial hydrolase, with catalytic activity that compares well with natural carbonic anhydrases.143,144,145 X-ray crystallography showed that the designed protein contained two different metal ions; Zn(II), which is important for catalysis, and a Hg(II) ion, which provides structural stability. In these hydrolytic zinc metalloenzymes, it was demonstrated the influence of location of the active site formed by the metal and three histidine residues in a three-stranded coiled-coil oligopeptide.
Kuhlman and co-workers serendipitously discovered that a computationally designed Zn(II) site at the interface of a homodimer (MID1-Zn) could effectively catalyse carboxyester and phosphoester hydrolysis.146,147 A Rosetta-based approach was used for the rational design of a protein monomer to form a zinc-mediated, symmetric homodimer. The computationally designed zinc-mediated protein showed a good overall agreement with the X-ray structure,146 and suggested that the clefts formed by the protein-protein interactions were well suited for creating enzyme active sites.147
5.3 heme no-heme artificial enzymes
This section deals with catalysis by bimetallic centers, where one of the metals forms part of a heme complex and the other is directly complexed by enzymatic residues. Example of key reactions that are catalysed by heme no-heme artificial enzymes are the reduction of NO to NO2 and the reduction of O2 to H2O. Nitric oxide reductase (NOR), is a metalloenzyme that participate in the nitrification pathway and is critical for life. A first example of the success of designing a NOR artificial enzyme is the conversion from a myoglobin by introducing a non-heme FeB site in the distal heme pocket. One glutamate residue and two histidines were required in this FeB active site for both iron binding and NOR activity.148,149 The introduction of a second glutamate residue in a myoglobin protein scaffold, that appears to interact with a water molecule forming a hydrogen bonding network, was demonstrated to increase 100-fold the NOR activity. It was suggested the importance of this hydrogen bonding network in facilitating the proton delivery for NOR reactivity.150 Replacing the Fe metal in the no-heme center by other cations shows that Cu dependent NOR activity was significantly higher than that of Fe in the same metal binding site.151 Molecular dynamics simulations on a mutant with an additional distal histidine (L29H) and a distal tyrosine (F43Y) in the heme pocket, to mimic the active site of cytochrome cd1 nitrite reductase from Ps. aeruginosa, revealed that the new protein contains the necessary structural features of the later. This strategy provides rationality for protein design and can guide further protein engineering.152
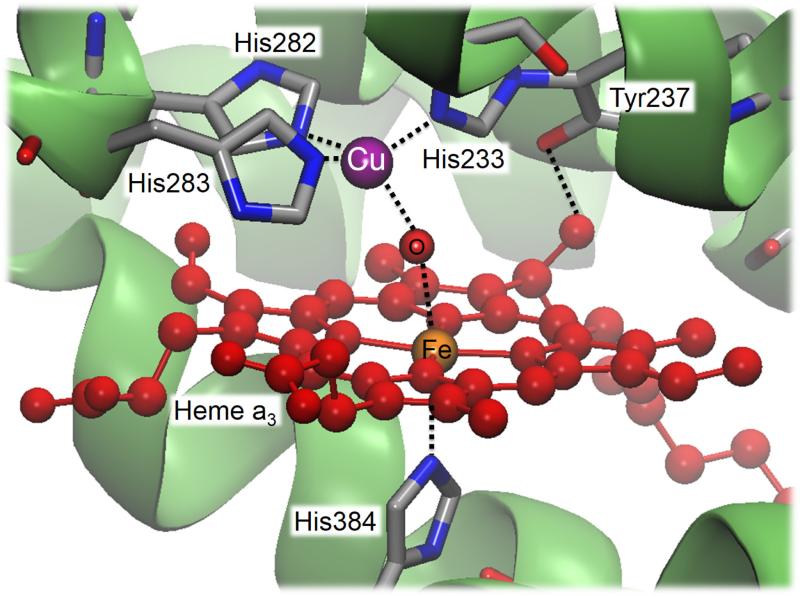
In a similar way, myoglobins have been also engineering into a heme copper oxidase (HCO) that performs the reduction of oxygen to water, by introducing a histidine residue near the heme site to create a CuB site. Thus, the corresponding active site consists of a heterobinuclear center containing a heme Fe and a histidine ligated Cu (CuB) as well as a post-translationally installed crosslink between a tyrosine residue and a close lying histidine (see Figure 5). The designed functional metalloenzyme showed a HCO activity with over one thousand turnovers.153,154 Genetic incorporation of an unnatural aminoacid, which mimics the cross-linked Tyr-His ligand, allowed stressing the effect of Tyr and His with and without cross-linking (obtained after a selected mutation that avoided its formation). The new designed model achieved higher activity for O2-reduction, with an eightfold increase in selectivity and a threefold increase in catalytic turnover.154
Figure 5.
Crystal structure of the binuclear center of cytochrome ba3 oxidase (PDB code 1XME).
5.4 Supramolecular Anchoring Approach
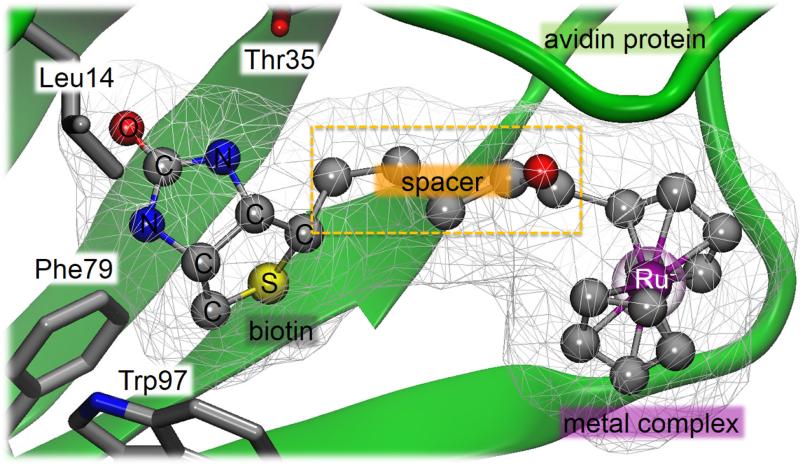
The concept of introducing a homogeneous catalyst within a protein environment to create artificial metalloenzymes was first proposed by Wilson and Whitesides in the early 1970s.155 Examples of biomolecular scaffolds capable of embedding a catalytically active transition metal complex include avidin, streptavidin, bovine serum albumin and myoglobin.134 The extraordinary affinity of the molecule biotin for either avidin or streptavidin makes this case as one of the most employed systems. Moreover, these protein scaffolds present a binding pocket that is large enough to bind the organometallic catalyst and still leave space for the substrate.117,134 Biotin is covalently linked to a catalyst precursor, a metal complex, ensuring that the metal moiety is quantitatively incorporated within the host protein upon stoichiometric addition of (strep)avidin (see Figure 6).156 Another important advantage of these hybrid supramolecular catalysts is that, since the biotynilated catalyst precursor and the host protein are developed separately of one another, they can be optimized separately through synthetic and molecular biology approaches.126
Figure 6.
Schematic representation of artificial metalloenzymes based on the biotinavidin technology.
Computational simulations of protein-ligand docking and experimental studies based on electron paramagnetic resonance spectroscopy, chemical modifications and mutagenesis studies, suggest that a vanadyl ion is located within the biotin-binding pocket and interacts only via the second coordination sphere contacts with strep(avidin).157 Many reactions have been catalysed following this synthetic strategy.156,158 Interestingly, a theoretical study based on MD simulations has shown significant flexibility of the biotinylated ligand in artificial metalloenzymes based on streptavidin.159 The creation of a bifunctional artificial metalloenzyme with a reduced flexibility was generated by introducing a base (an aspartate or a glutamate residue) into the streptavidin in concert with a docked biotinylated rhodium(III) complex to enable asymmetric carbon-hydrogen activation.160 A similar strategy has been used to improve the catalytic performance in the transfer hydrogenation of imines by an in silico design of an artificial metalloenzyme. The designed structure, confirmed by X-ray crystallography, contained a suitable positioned histidine residue that firmly anchor, via a dative bond, a biotinylated rhodium piano stool complex within streptavidin.161
An integrated computational approach, combining protein-ligand docking, quantum mechanical, and QM/MM calculations, has been used to provide a deep insight into the mechanistic behaviour of the first artificial metalloenzyme generated by supramolecular anchoring approach able to reduce enantioselectivily cyclic imines.162 The identification of the TS geometries leading to (R)- and (S)-reduction products in the presence of the full hybrid system allowed the authors to identify the distinctive pattern of interactions between the substrate and the binding site, responsible of the observed results.
6 Future perspectives
The design of new biocatalysts is an ambitious target that requires a detailed knowledge of the reaction to be catalyzed, and how natural enzymes achieve their catalytic efficiency in mild environmental conditions. In this regard, the combination of experimental and computational techniques has been revealed as a very reasonable strategy. As mentioned by Hilvert and co-workers, precision is essential for development of new biocatalysts based on the combination of de novo computational design with laboratory evolution techniques.80 New and more sophisticated engineering protocols are required to approach to the rate constants usually observed in native enzyme catalysed processes.
The success in the design of new enzymes with new functions has potential applications in different chemical and biochemical industries. This review has been focused on the advances obtained in the field of the de novo design during the last years using examples of increasing complexity. Kemp, Diels-Alder and retro-aldol, three of the most popular benchmarks reactions in the field designing new enzymes have been used to illustrate recent achievements. The first one has been chosen due to its simplicity and the fact that no natural enzyme catalyses this reaction. Diels-Alder is a bimolecular reaction that can produce many different isomers. The control of the products specificity is a feature that, in fact, can be more important than enhancing its rate constant. Finally, the design of new biocatalysts for the retro-aldol reaction represents an additional difficulty since the reaction takes place in several chemical steps. Thus, the new enzyme must properly represent the features of several TSs and, at least, one intermediate. Conformational changes of the protein to form the Michaelis complex (preorganization) has to take place along the reaction to stabilize different intermediates of the different steps. This requires a coupling between the reaction progress and the conformational changes of the protein to get a proper preorganization for each chemical step. Finally, a section has been devoted to the particularities of the design of metal-containing enzymes, a promising field due to the combination of the best features of homogeneous catalysis and enzyme catalysis; the broad catalytic scope and the high activity and selectivity under mild conditions, respectively. Nevertheless, since one or more metal ions, or metal complexes, are involved in the process, the difficulties of designing this kind of biocatalysts dramatically increase.
From the pioneer work of Mayo and co-workers in the de novo design of an esterase,14 several groups have focused the attention into the design of catalysts for the three mentioned reactions by means of similar strategies, still with a limited success. Different improvements have been incorporated during the past decade, such as partially considering the flexibility of the backbone of the template protein or the use of non-canonical amino acids. Nevertheless, the rate enhancement reached by these new biocatalysts is usually far from those of natural enzymes. As mentioned by Baker,37 and stressed in the introduction section of this review, the de novo based design strategies can fail due to an imperfect theozyme, which does not represent the real TS of the reaction, a distortion of the designed active site into a given protein scaffold, or due to the effect of the long range electrostatic interactions and/or protein dynamics that can be incompatible with catalysis. Nevertheless, long range electrostatic interactions, their knowledge and simulation, can be used as a guide to the design of the new enzyme, instead of considering them as a limitation. Mutations far from the active site can be proposed to improve this kind of interactions, and the flexibility of the protein must be considered in any step of the design protocol. It must be kept in mind that while efforts have been focused exclusively in the chemical step of the reaction to be catalyzed, it will be interesting to extend these methodologies to analyze the influence of mutations on steps like substrate binding or product release. This could improve the substrate affinity and/or the catalytic turnover, respectively. Increasing the preorganization of the active site would likely improve the catalytic efficiency. In addition, a more complete design strategy should consider not only the TS stabilization but the differential stabilization of the TS with respect to the reactants. Thus, further improvements on the de novo design of new enzymes will be probably based in better localization and characterization of the TS and reactants by means of high level computational methods. The use of flexible molecular models treated by QM/MM multiscale methods can be the bedrock of future successful studies. A proper sampling of the protein conformational landscape, performed by Molecular Dynamics simulations, could give also information related with the capability of a designed enzyme to catalyze a multistep-process. In these more complex targets, the active site of the enzyme has to be prepared not only to stabilize the TS but to accommodate and stabilize the system for different steps. In this refinement, the improved packing should be considered as not only a mechanical embedding but also an electrostatic one. The role of not only residues belonging to the first shell of the active site should be taken into account.
In the next future we should be able to design a particular amino acid sequence that will fold into a particular structure with the desired function from scratch. This is the final goal in rational design of proteins.
Highlights.
- The design of new biocatalysts requires a detailed knowledge of the reaction to be catalysed.
- The combination of experimental and computational techniques is a very reasonable strategy.
- New enzymes have been designed by combination of de novo techniques with laboratory evolution.
- Kemp, Diels-Alder and retro-aldol reactions have been used to illustrate recent achievements.
- Flexible molecular models treated by QM/MM multiscale methods can be used in future studies.
Acknowledgements
This work was supported by the Spanish Ministerio de Economía y Competitividad for project CTQ2012-36253-C03, Universitat Jaume I (project P1·1B2014-26), Generalitat Valenciana (PROMETEOII/2014/022 and ACOMP/2014/277 projects), Polish National Center for Science (NCN) (grant 2011/02/A/ ST4/00246, 2012–2017), the Polish Ministry of Science and Higher Education (“Iuventus Plus” program project no. 0478/IP3/2015/73, 2015-2016) and the USA National Institute of Health (ref. NIH R01 GM065368). Authors acknowledge computational resources from the Servei d'Informàtica of Universitat de València on the ‘Tirant’ supercomputer and the Servei d'Informatica of Universitat Jaume I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olsson MHM. Chem. Rev. 2006;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 2.Benkovic SJ, Hammes-Schiffer S. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Viloca M, Gao J, Karplus M, Truhlar DG. Science. 2004;303:186–195. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- 4.Martí S, Roca M, Andrés J, Moliner V, Silla E, Tuñón I, Bertrán J. Chem. Soc. Rev. 2004;33:98–107. doi: 10.1039/b301875j. [DOI] [PubMed] [Google Scholar]
- 5.Steiner K, Schwab H. Comput. Struc. Biotech. J. 2012;2:e2012209010. doi: 10.5936/csbj.201209010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carbone MN, Arnold FH. Curr. Opin. Struct. Biol. 2007;17:454–459. doi: 10.1016/j.sbi.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Martí S, Andrés J, Moliner V, Silla E, Tuñón I, Bertran J. Chem. Soc. Rev. 2008;37:2634–2643. doi: 10.1039/b710705f. [DOI] [PubMed] [Google Scholar]
- 8.Morley KL, Kazlauskas RJ. Trends Biotechnol. 2005;23:231–237. doi: 10.1016/j.tibtech.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Kiss G, Celebi-Olcum N, Moretti R, Baker D, Houk KN. Angew. Chem. Int. Ed. 2013;52:5700–5725. doi: 10.1002/anie.201204077. [DOI] [PubMed] [Google Scholar]
- 10.Keinan E, editor. Wiley-VCH; Verlag Weinheim: Catalytic Antibodies. 2005. [Google Scholar]
- 11.D. Hilvert. Annu. Rev. Biochem. 2013;82:447–470. doi: 10.1146/annurev-biochem-072611-101825. [DOI] [PubMed] [Google Scholar]
- 12.Toscano MD, Woycechowsky KJ, Hilvert D. Angew. Chem. Int. Ed. 2007;46:3212–3236. doi: 10.1002/anie.200604205. [DOI] [PubMed] [Google Scholar]
- 13.Humble MS, Berglund P. Eur. J. Org. Chem. 2011:3391–3401. [Google Scholar]
- 14.Bolon DN, Mayo SL. Proc. Natl. Acad. Sci. USA. 2001;98:14274–14279. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahiyat BI, Mayo SL. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 16.Lassila JK, Privett HK, Allen BD, Mayo SL. Proc. Natl. Acad. Sci. USA. 2006;103:16710–16715. doi: 10.1073/pnas.0607691103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Röthlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Nature. 2008;453:190–95. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 18.Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, St. Clair JL, Gallaher JL, Hilvert D, Gelb MH, Stoddard BL, Houk KN, Michael FE, Baker D. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang L, Althoff EA, Clemente FR, Doyle L, Röthlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, III, Hilvert D, Houk KN, Stoddard BL, Baker D. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tantillo DJ, Chen J, Houk KN. Curr. Opin. Chem. Biol. 1988;2:743–750. doi: 10.1016/s1367-5931(98)80112-9. [DOI] [PubMed] [Google Scholar]
- 21.Zanghellini A, Jiang L, Wollacott AM, Cheng G, Meiler J, Althoff EA, Röthlisberger D, Baker D. Protein Science. 2006;15:2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meiler J, Baker D. Proteins: Structure, Function and Bioinformatics. 2006;65:538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 23.Tinberg CE, Khare SD, Dou J, Doyle L, Nelson JW, Schena A, Jankowski W, Kalodimos CG, Johnsson K, Stoddard BL, Baker D. Nature. 2013;501:212–216. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leaver-Fay A, et al. Methods in Enzymology. Vol. 487. Elsevier; 2011. pp. 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richter F, Leaver-Fay A, Khare SD, Bjelic S, Baker D. PLoS ONE. 2011;6:e19230. doi: 10.1371/journal.pone.0019230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandell DJ, Kortemme T. Curr. Opin. Biotech. 2009;20:420–428. doi: 10.1016/j.copbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Huang P-S, Ban Y-EA, Richter F, Andre I, Vernon R, Schief WR, Baker D. PLoS ONE. 2011;6:e24109. doi: 10.1371/journal.pone.0024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renfrew PD, Choi EJ, Bonneau R, Kuhlman B. PLoS ONE. 2012;7:e32637. doi: 10.1371/journal.pone.0032637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mills JH, Khare SD, Bolduc JM, Forouhar F, Mulligan VK, Lew S, Seetharaman J, Tong L, Stoddard BL, Baker D. J. Am. Chem. Soc. 2013;135:13393–13399. doi: 10.1021/ja403503m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drienovská I, Rioz-Martínez A, Draksharapu A, Roelfes G. Chem. Sci. 2015;6:770–776. doi: 10.1039/c4sc01525h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khatib F, Cooper S, Tyka MD, Xu K, Makedon I, Popovic Z, Baker D. Foldit Players. Proc. Natl. Acad. Sci. USA. 2011;108:18949–18953. doi: 10.1073/pnas.1115898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fazelinia H, Cirino PC, Maranas CD. Protein Science. 2008;18:180–195. doi: 10.1002/pro.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malisi C, Kohlbacher O, Höcker B. Proteins. 2009;77:74–83. doi: 10.1002/prot.22418. [DOI] [PubMed] [Google Scholar]
- 34.Lei Y, Luo W, Zhu Y. Protein Science. 2011;20:1566–1575. doi: 10.1002/pro.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang X, Han K, Zhu Y. Protein Science. 2013;22:929–941. doi: 10.1002/pro.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nosrati GR, Houk KN. Protein Science. 2012;21:697–706. doi: 10.1002/pro.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker D. Protein Science. 2010;19:1817–1819. doi: 10.1002/pro.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samish I, MacDermaid CM, Perez-Aguilar JM, Saven JG. Annu. Rev. Phys. Chem. 2011;62:129–149. doi: 10.1146/annurev-physchem-032210-103509. [DOI] [PubMed] [Google Scholar]
- 39.Bornscheuer UT, Huisman GW, Kazlauskas RJ, Lutz S, Moore JC, Robins K. Nature. 2012;485:185–194. doi: 10.1038/nature11117. [DOI] [PubMed] [Google Scholar]
- 40.Barrozo A, Borstnar R, Marloie G, Kamerlin SCL. Int. J. Mol. Sci. 2012;13:12428–12460. doi: 10.3390/ijms131012428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kries H, Blomberg R. D. Hilvert. Curr. Opin. Chem. Biol. 2013;17:221–228. doi: 10.1016/j.cbpa.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 42.Khare SD, Fleishman SJ. FEBS Letters. 2013;587:1147–1154. doi: 10.1016/j.febslet.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 43.Wijma HJ, Janssen DB. FEBS Journal. 2013;280:2948–2960. doi: 10.1111/febs.12324. [DOI] [PubMed] [Google Scholar]
- 44.Damborsky J, Brezovsky J. Curr. Opin Chem. Biol. 2014;19:8–16. doi: 10.1016/j.cbpa.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 45.Frushicheva MP, Mills MJL, Schopf P, Singh MK, Prasad RB, Warshel A. Curr. Opin. Chem. Biol. 2014;21:56–62. doi: 10.1016/j.cbpa.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casey ML, Kemp DS, Paul KG, Cox DD. J. Org. Chem. 1973;38:2294–2301. [Google Scholar]
- 47.Kemp DS, Casey ML. J. Am. Chem. Soc. 1973;95:6670–6680. [Google Scholar]
- 48.Kemp DS. Nature. 1995;373:196–197. doi: 10.1038/373196a0. [DOI] [PubMed] [Google Scholar]
- 49.Thorn SN, Daniels RG, Auditor MTM, Hilvert D. Nature. 1995;373:228–230. doi: 10.1038/373228a0. [DOI] [PubMed] [Google Scholar]
- 50.Genre-Grandpierre A, Tellier C, Loirat M-J, Blanchard D, Hodgson DRW, Hollfelder F, Kirby AJ. Bioorg. Med. Chem. Lett. 1997;7:2497–2502. [Google Scholar]
- 51.Müller R, Debler EW, Steinmann M, Seebeck FP, Wilson IA, Hilvert D. J. Am. Chem. Soc. 2007;129:460–461. doi: 10.1021/ja066578b. [DOI] [PubMed] [Google Scholar]
- 52.James LC, Tawfik DS. Protein Sci. 2001;10:2600–2607. doi: 10.1110/ps.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Debler EW, Ito S, Seebeck FP, Heine A, Hilvert D, Wilson IA. Proc. Nat. Acad. Sci. USA. 2005;102:4984–4989. doi: 10.1073/pnas.0409207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seebeck FP, Hilvert D. J. Am. Chem. Soc. 2005;127:1307–1312. doi: 10.1021/ja044647l. [DOI] [PubMed] [Google Scholar]
- 55.Debler EW, Müller R, Hilvert D, Wilson IA. J. Biol. Chem. 2008;283:16554–16560. doi: 10.1074/jbc.M710256200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Debler EW, Müller R, Hilvert D, Wilson IA. Proc. Nat. Acad. Sci. USA. 2009;106:18539–18544. doi: 10.1073/pnas.0902700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kirby AJ. Acc. Chem. Res. 1997;30:290–296. [Google Scholar]
- 58.Na J, Houk KN, Hilvert D. J. Am. Chem. Soc. 1996;118:6462–6471. [Google Scholar]
- 59.Kikuchi K, Thorn SN, Hilvert D. J. Am. Chem. Soc. 1996;118:8184–8185. [Google Scholar]
- 60.Hollfelder F, Kirby AJ, Tawfik DS. Nature. 1996;383:60–63. doi: 10.1038/383060a0. [DOI] [PubMed] [Google Scholar]
- 61.Hollfelder F, Kirby AJ, Tawfik DS, Kikuchi K, Hilvert D. J. Am. Chem. Soc. 2000;122:1022–1029. [Google Scholar]
- 62.Hu Y, Houk KN, Kikuchi K, Hotta K, Hilvert D. J. Am. Chem. Soc. 2004;126:8197–8205. doi: 10.1021/ja0490727. [DOI] [PubMed] [Google Scholar]
- 63.Hollfelder F, Kirby AJ, Tawfik DS. J. Am. Chem. Soc. 1997;119:9578–9579. [Google Scholar]
- 64.Korendovych IV, Kulp DW, Wu Y, Cheng H, Roder H, DeGrado WF. Proc. Nat. Acad. Sci. USA. 2011;108:6823–6827. doi: 10.1073/pnas.1018191108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moroz OV, Moroz YS, Wu Y, Olsen AB, Cheng H, Mack KL, McLaughlin JM, Raymond EA, Zhezherya K, Roder H, Korendovych IV. Angew. Chem. Int. Ed. 2013;52:6246–6249. doi: 10.1002/anie.201302339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Merski M, Shoichet BK. Proc. Nat. Acad. Sci. USA. 2012;109:16179–16183. doi: 10.1073/pnas.1208076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.V. Nanda. Nat. Chem. Biol. 2008;4:273–275. doi: 10.1038/nchembio0508-273. [DOI] [PubMed] [Google Scholar]
- 68.Khersonsky O, Röthlisberger D, Dym O, Albeck S, Jackson CJ, Baker D, Tawfik DS. J. Mol. Biol. 2010;396:1025–1042. doi: 10.1016/j.jmb.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 69.Khersonsky O, Röthlisberger D, Wollacott AM, Murphy P, Dym O, Albeck S, Kiss G, Houk KN, Baker D, Tawfik DS. J. Mol. Biol. 2011;407:391–412. doi: 10.1016/j.jmb.2011.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khersonsky O, Kiss G, Röthlisberger D, Dym O, Albeck S, Houk KN, Baker D, Tawfik DS. Proc. Nat. Acad. Sci. USA. 2012;109:10358–10363. doi: 10.1073/pnas.1121063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alexandrova AN, Röthlisberger D, Baker D, Jorgensen WL. J. Am. Chem. Soc. 2008;130:15907–15915. doi: 10.1021/ja804040s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sparta M, Alexandrova AN. Mol. Simul. 2011;37:557–571. [Google Scholar]
- 73.Alexandrova AN, Jorgensen WL. J. Phys. Chem. B. 2009;113:497–504. doi: 10.1021/jp8076084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Acevedo O. J. Phys. Chem. B. 2009;113:15372–15381. doi: 10.1021/jp9069114. [DOI] [PubMed] [Google Scholar]
- 75.Kiss G, Röthlisberger D, Baker D, Houk KN. Prot. Sci. 2010;19:1760–1773. doi: 10.1002/pro.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frushicheva MP, Cao J, Chu ZT, Warshel A. Proc. Nat. Acad. Sci. USA. 2010;107:16869–16874. doi: 10.1073/pnas.1010381107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frushicheva MP, Cao J, Warshel A. Biochemistry. 2011;50:3849–3858. doi: 10.1021/bi200063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Labas A, Szabo E, Mones L, Fuxreiter M. Biochim. Biophys. Acta. 2013;1834:908–917. doi: 10.1016/j.bbapap.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 79.Privett HK, Kiss G, Lee TM, Blomberg R, Chica RA, Thomas LM, Hilvert D, Houk KN, Mayo SL. Proc. Nat. Acad. Sci. USA. 2012;109:3790–3795. doi: 10.1073/pnas.1118082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Blomberg R, Kries H, Pinkas DM, Mittl PRE, Grütter MG, Privett HK, Mayo SL, Hilvert D. Nature. 2013;503:418–421. doi: 10.1038/nature12623. [DOI] [PubMed] [Google Scholar]
- 81.Höhne M, Bornscheuer UT. Angew. Chem. Int. Ed. 2014;53:1200–1202. doi: 10.1002/anie.201309591. [DOI] [PubMed] [Google Scholar]
- 82.Swiderek K, Tuñón I, Moliner V, Bertran J. ACS Catalysis. 2014 In Press. [Google Scholar]
- 83.Laschat S. Angew. Chem. Int. Ed. 1996;35:289–291. [Google Scholar]
- 84.Kelly WL. Org. Biomol.Chem. 2008;6:4483–4493. doi: 10.1039/b814552k. [DOI] [PubMed] [Google Scholar]
- 85.Hak Joong Kim Mark W. Ruszczycky, Sei-hyun Choi Yung-nan Liu. Hung-wen Liu. Nature. 2011;473:109–112. doi: 10.1038/nature09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Page MI, Jencks WP. Proc. Natl. Acad. Sci. USA. 1971;68:1678–1683. doi: 10.1073/pnas.68.8.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Golinelli-Pimpaneau B. Curr. Op. Struct. Biol. 2000;10:697–708. doi: 10.1016/s0959-440x(00)00157-3. [DOI] [PubMed] [Google Scholar]
- 88.Hilvert D, Hill KW, Nared KD, Auditor MTM. J. Am. Chem. Soc. 1989;111:9261–9262. [Google Scholar]
- 89.Braisted AC, Schultz PG. J. Am. Chem. Soc. 1990;112:7430–7431. [Google Scholar]
- 90.Romesberg FE, Schultz PG. Bioorg. Med. Chem. Lett. 1999;9:1741–1744. doi: 10.1016/s0960-894x(99)00281-4. [DOI] [PubMed] [Google Scholar]
- 91.Xu J, Deng Q, Chen J, Houk KN, Bartek J, Hilvert D, Wilson IA. Science. 1999;286:2345–2348. doi: 10.1126/science.286.5448.2345. [DOI] [PubMed] [Google Scholar]
- 92.Romesberg FE, Spiller B, Schultz PG, Stevens RC. Science. 1998;279:1929–1933. doi: 10.1126/science.279.5358.1929. [DOI] [PubMed] [Google Scholar]
- 93.Marti S, Andres J, Moliner V, Silla E, Tuñón I, Bertran J. Chem. Eur. J. 2008;14:596–602. doi: 10.1002/chem.200700290. [DOI] [PubMed] [Google Scholar]
- 94.Gouverneur VE, Houk KN, Pascual-Teresa B, Beno B, Janda KD, Lerner RA. Science. 1993;262:204–208. doi: 10.1126/science.8211138. [DOI] [PubMed] [Google Scholar]
- 95.Heine A, Stura EA, Yli-Kauhaluoma JT, Gao C, Deng Q, Beno BR, Houk KN, Janda KD, Wilson IA. Science. 1998;279:1934–1940. doi: 10.1126/science.279.5358.1934. [DOI] [PubMed] [Google Scholar]
- 96.Cannizzaro CE, Ashley JA, Janda KD, Houk KN. J. Am. Chem. Soc. 2003;125:2489–2506. doi: 10.1021/ja020879d. [DOI] [PubMed] [Google Scholar]
- 97.Cooper S, Khatib F, Treuille A, Barbero J, Lee J, Beenen M, Leaver-Fay A, Baker D, Popovic Z, Players F. Nature Letters. 2010;466:756–760. doi: 10.1038/nature09304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Eiben CB, Siegel JB, Bale JB, Cooper S, Khatib F, Shen BW, Players F, Stoddard BL, Popovic Z, Baker D. Nature Biotechnology. 2012;30:190–192. doi: 10.1038/nbt.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Preiswerk N, Beck T, Schulz JD, Milovnik P, Mayer C, Siegel JB, Baker D, Hilvert D. Proc. Natl. Acad. Sci. USA. 2014;111:8013–8018. doi: 10.1073/pnas.1401073111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Linder M, Johansson AJ, Olsson TSG, Liebeschuetz J, Brinck T. J. Chem. Inf. Model. 2011;51:1906–1917. doi: 10.1021/ci200177d. [DOI] [PubMed] [Google Scholar]
- 101.Linder M, Johansson AJ, Olsson TSG, Liebeschuetz J, Brinck T. J. Comput. Aided Mol. Des. 2012;26:1079–1095. doi: 10.1007/s10822-012-9601-y. [DOI] [PubMed] [Google Scholar]
- 102.Linder M, Johansson AJ, Manta B, Olsson P, Brinck T. Chem. Comm. 2012;48:5665–5667. doi: 10.1039/c2cc31502e. [DOI] [PubMed] [Google Scholar]
- 103.Lightstone FC, Bruice TC. J. Am. Chem. Soc. 1996;118:2595–2605. [Google Scholar]
- 104.Smith AJT, Müller R, Toscano MD, Kast P, Hellinga HW, Hilvert D, Houk KN. J. Am. Chem. Soc. 2008;130:15361–15373. doi: 10.1021/ja803213p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wagner J, Lerner RA, Barbas III CF. Science. 1995;270:1797–1800. doi: 10.1126/science.270.5243.1797. [DOI] [PubMed] [Google Scholar]
- 106.Hoffmanm T, Zhong G, List B, Shabat D, Anderson J, Gramatikova S, Lerner RA, Barbas III CF. J. Am. Chem. Soc. 1998;120:2768–2779. [Google Scholar]
- 107.List B, Barbas III CF, Lerner RA. Proc. Natl. Acad. Sci. USA. 1998;95:15351–15355. doi: 10.1073/pnas.95.26.15351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhong G, Lerner RA, Barbas CF., III Angew. Chem. Int. Ed. 1999;38:3738–3741. doi: 10.1002/(sici)1521-3773(19991216)38:24<3738::aid-anie3738>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 109.Barbas CF, III, Heine A, Zhong G, Hoffmann T, Gramatikova S, Björnestedt R, List B, Anderson J, Stura EA, Wilson IA, Lerner RA. Science. 1997;278:2085–2092. doi: 10.1126/science.278.5346.2085. [DOI] [PubMed] [Google Scholar]
- 110.Zhu X, Tanaka F, Lerner RA, Barbas CF, III, Wilson IA. J. Am. Chem. Soc. 2009;131:18206–18207. doi: 10.1021/ja907271a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arnó M, Domingo LR. Org. Biomol. Chem. 2003;1:637–643. doi: 10.1039/b209636f. [DOI] [PubMed] [Google Scholar]
- 112.Lassila JK, Baker D, Herschlag D. Proc. Natl. Acad. Sci. USA. 2010;107:4937–4942. doi: 10.1073/pnas.0913638107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang L, Althoff EA, Bolduc J, Jiang L, Moody J, Lassila JK, Giger L, Hilvert D, Stoddard B, Baker D. J. Mol. Biol. 2012;415:615–625. doi: 10.1016/j.jmb.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Althoff EA, Wang L, Jiang LK, Giger L, Lassila JK, Wang Z, Smith M, Hari S, Kast P, Herschlag D, Hilvert D, Baker D. Prot. Sci. 2012;21:717–726. doi: 10.1002/pro.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Giger L, Caner S, Obexer R, Kast P, Kast P, Baker D, Ban N, Hilvert D. Nat. Chem. Biol. 2013;9:494–498. doi: 10.1038/nchembio.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Obexer R, Studer S, Giger L, Pinkas DM, Grütter MG, Baker D, Hilvert D. ChemCatChem. 2014;6:1043–1050. [Google Scholar]
- 117.Steinreiber J, Ward TR. Coord. Chem. Rev. 2008;252:751–766. [Google Scholar]
- 118.Lu Y, Yeung N, Sieracki N, Marshall NM. Nature. 2009;460:855–862. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Heinisch T, Ward TR. Curr. Op. Chem. Biol. 2010;14:184–199. doi: 10.1016/j.cbpa.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 120.Rosati F, Roelfes G. ChemCatChem. 2010;2:916–927. [Google Scholar]
- 121.Saven JG. Curr. Op. Chem. Biol. 2011;15:452–457. doi: 10.1016/j.cbpa.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ringenberg MR, Ward TR. Chem. Comm. 2011;47:8470–8476. doi: 10.1039/c1cc11592h. [DOI] [PubMed] [Google Scholar]
- 123.Petrik ID, Liu J. Y. Lu. Curr. Op. Chem. Biol. 2014;19:67–75. doi: 10.1016/j.cbpa.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dürrenberger M, Ward TR. Curr. Op. Chem. Biol. 2014;19:99–106. doi: 10.1016/j.cbpa.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 125.Bos J, Roelfes G. Curr. Op. Chem. Biol. 2014;19:135–143. doi: 10.1016/j.cbpa.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 126.Yu F, Cangelosi VM, Zastrow ML, Tegoni M, Plegaria JS, Tebo AG, Mocny CS, Ruckthong L, Qayyum H, Pecoraro VL. Chem. Rev. 2014;114:3495–3578. doi: 10.1021/cr400458x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Murphy PM, Bolduc JM, Gallaher JL, Stoddard BL, Baker D. Proc. Nat. Acad. Sci. USA. 2009;106:9215–9220. doi: 10.1073/pnas.0811070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chakraborty S, Kravitz JY, Thulstrup PW, Hemmingsen L, DeGrado WF, Pecoraro VL. Angew. Chem. Int. Ed. 2011;50:2049–2053. doi: 10.1002/anie.201006413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Patel SC, Bradley LH, Jinadasa SP, Hecht MH. Prot. Sci. 2009;18:1388–1400. doi: 10.1002/pro.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Farinas E, Regan L. Prot. Sci. 1998;7:1939–1946. doi: 10.1002/pro.5560070909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Podtetenieff J, Taglieber A, Bill E, Reijerse EJ, Reetz MT. Angew. Chem. Int. Ed. 2010;49:5151–5155. doi: 10.1002/anie.201002106. [DOI] [PubMed] [Google Scholar]
- 132.Reetz MT. Chem. Rec. 2012;12:391–406. doi: 10.1002/tcr.201100043. [DOI] [PubMed] [Google Scholar]
- 133.Deuss PJ, Popa G, Slawin AMZ, Laan W, Kamer PCJ. Chem Cat Chem. 2013;5:1184–1191. [Google Scholar]
- 134.Bos J, Fusetti F, Driessen AJM, Roelfes G. Angew. Chem. Int. Ed. 2012;51:7472–7475. doi: 10.1002/anie.201202070. [DOI] [PubMed] [Google Scholar]
- 135.Roelfes G, Boersma AJ, Feringa BL. Chem. Comm. 2006:635–637. doi: 10.1039/b516552k. [DOI] [PubMed] [Google Scholar]
- 136.Boersma AJ, Feringa BL. G. Roelfes. Org. Lett. 2007;9:3647–3650. doi: 10.1021/ol7015274. [DOI] [PubMed] [Google Scholar]
- 137.Rosati F, Boersma AJ, Klijn JE, Meetsma A, Feringa BL. G. Roelfes. Chem. Eur. J. 2009;15:9596–9605. doi: 10.1002/chem.200900456. [DOI] [PubMed] [Google Scholar]
- 138.Valdez Crystal E., Smith Quentin A., Nechay Michael R., Alexandrova Anastassia N. Acc. Chem. Res. 2014;47:3110–3117. doi: 10.1021/ar500227u. [DOI] [PubMed] [Google Scholar]
- 139.Valdez Crystal E., Alexandrova Anastassia N. J. Phys. Chem. B. 2012;116:10649–10656. doi: 10.1021/jp302771n. [DOI] [PubMed] [Google Scholar]
- 140.Leitgeb S, Nidetzky B. ChemBioChem. 2010;11:502–505. doi: 10.1002/cbic.200900688. [DOI] [PubMed] [Google Scholar]
- 141.Khare SD, Kipnis Y, Greisen P, Jr, Takeuchi R, Ashani Y, Goldsmith M, Song Y, Gallaher JL, Silman I, Leader H, Sussman JL, Stoddard BL, Tawfik DS, Baker D. Nat. Chem. Biol. 2012;8:294–300. doi: 10.1038/nchembio.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Singh MK, Chu ZT, Warshel A. J. Phys. Chem. B. 2014;118:12146–12152. doi: 10.1021/jp507592g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zastrow ML, Peacock AFA, Stuckey JA, Pecoraro VL. Nat. Chem. 2012;4:118–123. doi: 10.1038/nchem.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zastrow ML, Pecoraro VL. J. Am. Chem. Soc. 2013;135:5895–5903. doi: 10.1021/ja401537t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zastrow ML, Pecoraro VL. Biochemistry. 2013;53:957–978. doi: 10.1021/bi4016617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Der BS, Machius M, Miley MJ, Mills JL, Szyperski T, Kuhlman B. J. Am. Chem. Soc. 2012;134:375–385. doi: 10.1021/ja208015j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Der BS, Edwards DR, Kuhlman B. Biochemistry. 2012;51:3933–3940. doi: 10.1021/bi201881p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yeung N, Lin Y-W, Gao Y-G, Zhao X, Russell BS, Lei L, Miner KD, Robinson H, Lu Y. Nature. 2009;462:1079–1082. doi: 10.1038/nature08620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hino T, Matsumoto Y, Nagano S, Sugimoto H, Fukumori Y, Murata T, Iwata S, Shiro Y. Science. 2010;330:1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 150.Lin Y-W, Yeung N, Gao Y-G, Miner KD, Tian S, Robinson H, Lu Y. Proc. Nat. Acad. Sci. USA. 2010;107:8581–8586. doi: 10.1073/pnas.1000526107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lin Y-W, Yeung N, Gao Y-G, Miner KD, Lei L, Robinson H, Lu Y. J. Am. Chem. Soc. 2010;132:9970–9972. doi: 10.1021/ja103516n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lin Y-W, Nie C-M, Liao L-F. J. Mol. Model. 2012;18:4409–4415. doi: 10.1007/s00894-012-1451-y. [DOI] [PubMed] [Google Scholar]
- 153.Miner KD, Mukherjee A, Gao Y-G, Null EL, Petrik ID, Zhao X, Yeung N, Robinson H, Yu Y. Angew. Chem. Int. Ed. 2012;51:5589–5592. doi: 10.1002/anie.201201981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu X, Yu Y, Hu C, Zhang W, Lu Y, Wang J. Angew. Chem. Int. Ed. 2012;51:4312–4316. doi: 10.1002/anie.201108756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wilson ME, Whitesides GM. J. Am. Chem. Soc. 1978;100:306–307. [Google Scholar]
- 156.Ward TR. Acc. Chem. Res. 2011;44:47–57. doi: 10.1021/ar100099u. [DOI] [PubMed] [Google Scholar]
- 157.Pordea A, Creus M, Panek J, Duboc C, Mathis D, Novic M, Ward TR. J. Am. Chem. Soc. 2008;130:8085–8088. doi: 10.1021/ja8017219. [DOI] [PubMed] [Google Scholar]
- 158.Pordea A, Mathis D, Ward TR. J. Organometallic Chem. 2009;694:930–936. [Google Scholar]
- 159.Panek JJ, Ward TR, Jezierska-Mazzarello A, Novic M. J. Comput. Aided Mol. Des. 2010;24:719–732. doi: 10.1007/s10822-010-9369-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hyster TK, Knörr L, Ward TR, Rovis T. Science. 2012;338:500–503. doi: 10.1126/science.1226132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zimbron JM, Heinisch T, Schmid M, Hamels D, Nogueira ES, Schirmer T, Ward TR. J. Am. Chem. Soc. 2013;135:5384–5388. doi: 10.1021/ja309974s. [DOI] [PubMed] [Google Scholar]
- 162.Muñoz Robles V, Vidossich P, Lledós A, Ward TR, Maréchal J-D. ACS Catal. 2014;4:833–842. [Google Scholar]