Abstract
The Hippo pathway is a potent regulator of cellular proliferation, differentiation, and tissue homeostasis. Here we review the regulatory mechanisms of the Hippo pathway and discuss the function of Yes-associated protein (YAP)/transcriptional coactivator with a PDZ-binding domain (TAZ), the prime mediators of the Hippo pathway, in stem cell biology and tissue regeneration. We highlight their activities in both the nucleus and the cytoplasm and discuss their role as a signaling nexus and integrator of several other prominent signaling pathways such as the Wnt, G protein-coupled receptor (GPCR), epidermal growth factor (EGF), BMP/transforming growth factor beta (TGFβ), and Notch pathways.
Keywords: extracellular matrix, stem cells, regeneration, primary cilia, Wnt, Notch
The Hippo pathway: signal integrator of cell proliferation and differentiation
Coordinated regulation of stem cell proliferation and differentiation is essential to maintain tissue mass and homeostasis. The Hippo pathway was initially identified through genetic mosaic screens for suppressors of tissue overgrowth in Drosophila melanogaster [1,2]. Later studies identified that the general components and functions of the pathway are highly conserved in mammals [3]. Salient properties of the Hippo pathway are in regulating cell proliferation, survival, and differentiation – all of which are particularly important processes in tissue development and regeneration. Deregulation of this pathway is furthermore implicated in cancer development [4–6].
Downstream effectors of the Hippo kinase cascade are the paralogs and transcriptional coactivators YAP and TAZ (also known as WWTR1) [3]. The Hippo kinase cascade is the major regulator of YAP/TAZ by phosphorylating and thereby inhibiting YAP/TAZ nuclear activities. Recent studies have expanded the complexity of YAP/TAZ considerably, identifying new upstream regulatory components and revealing its integration with other signaling pathways such as the GPCR [7–13], Wnt [14–17], TGFβ [18–20], EGF [21–24], and Notch [25–28]. In addition to expanding the number of inputs that modulate YAP/TAZ, the downstream mechanisms of YAP/TAZ have also grown in complexity. Besides transcriptional regulation, new physiological functions mediated by either nuclear or cytoplasmic localization of YAP/TAZ have been revealed and are providing insights into how YAP/TAZ integrates and modulates the cellular microenvironment.
Drawing on studies in both Drosophila and mammals, we highlight recent findings and provide a current overview of the molecular regulation of the Hippo network in mammals, with a particular focus on the key downstream effectors YAP and TAZ. We highlight the role of YAP/TAZ as a nexus and integrator for multiple prominent pathways and signaling organelles that play key roles in the control of cell fate and tissue regeneration.
At the core of the Hippo pathway: phosphorylation-dependent regulation
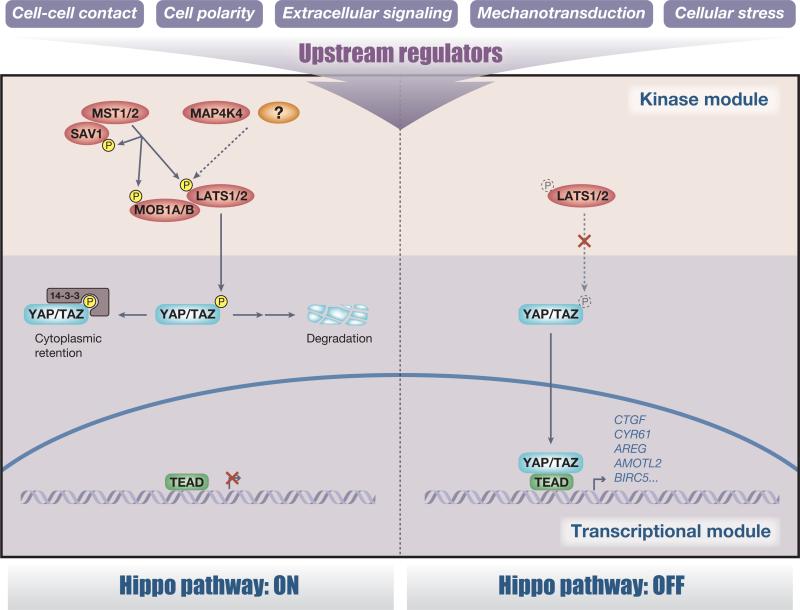
The Hippo pathway comprises a large network of proteins with the central components of a regulatory kinase module and a transcriptional module (Figure 1). Many components of the Hippo pathway are highly conserved from Drosophila to mammals, including mammalian STE20-like protein kinase 1/2 (MST1/2) (Hippo homolog), Salvador family WW domain-containing protein 1 (SAV1) (Salvador homolog), large tumor suppressor 1/2 (LATS1/2) (Warts homolog), MOB kinase activator 1A/B (MOB1A/B) (Mats homolog), MAP4K4 (Misshapen homolog), YAP and TAZ (Yorkie homolog), and TEA domain family members 1–4 (TEAD1–4) (Scalloped homolog). Although the core components of the Hippo pathway are extensively conserved, a divergent role of the upstream regulators through evolution has become apparent (Box 1).
Figure 1.
The core of the Hippo pathway. The core components of the Hippo pathway comprise a regulatory serine–threonine kinase module and a transcriptional module. In mammals, this kinase module comprises two groups of kinases: mammalian STE20-like protein kinase 1 (MST1) (also known as STK4) and MST2 (also known as STK3) and large tumor suppressor 1 (LATS1) and LATS2 in combination with their activating adaptor proteins Salvador family WW domain-containing protein 1 (SAV1) and MOB kinase activator 1A/B (MOB1A/B), respectively. MAP4K4 has recently been identified as an alternative activator of LATS independent of MST. The transcriptional module comprises the transcriptional coactivators Yes-associated protein (YAP) and transcriptional coactivator with a PDZ-binding domain (TAZ) and several transcription factors, of which the most prominently studied are TEA domain family members 1–4 (TEAD1–4). In a classical view, when the upstream kinase module is activated, MST1/2 phosphorylates SAV1, MOB1A/B, and LATS1/2 to activate the LATS1/2 kinases, which in turn directly phosphorylate YAP and TAZ on multiple serine residues leading to cytoplasmic retention of YAP/TAZ via a 14-3-3 interaction. Furthermore, the phosphorylation of YAP/TAZ by LATS1/2 primes YAP/TAZ for proteasomal degradation. The requirement for MST1/2 to phosphorylate LATS1/2 is cell-type dependent, suggesting the existence of additional kinases (drawn as a question mark) that regulate the activating phosphorylation of LATS1/2. By contrast, when the kinase module is inactivated, hypophosphorylated YAP/TAZ translocate into the nucleus and induce target gene expression.
The Hippo pathway responds to various upstream stimuli from the cellular microenvironment including mechanical signals, cellular stress, extracellular stimuli, polarity, and adhesion cues that are integrated through upstream regulators. These upstream regulators promote the activation of the Hippo kinase cascade by activating the mammalian Ser/Thr kinases LATS1/2, which phosphorylate and promote either the cytoplasmic retention of YAP and TAZ via a 14-3-3 interaction or the degradation of YAP/TAZ, ultimately preventing TEAD-mediated transcription [1,3]. Activation of the upstream kinase module thus concludes in inhibition of the transcriptional output module. One of the key questions concerning the Hippo pathway is the molecular mechanism of LATS activation. In a classical view, MST1/2-mediated phosphorylation of LATS1/2 at the hydrophobic motif is necessary for LATS kinase activation [29]. However, the requirement of MST1/2 for LATS1/2 phosphorylation is context- and cell type-dependent. Loss of MST1/2 in mouse embryonic fibroblasts does not significantly affect LATS1/2 phosphorylation and activation [30]. Furthermore, the forced expression of plasma membrane-targeting Warts (the Drosophila homolog of LATS1/2) partially rescues the eye disc phenotype of Hippo (the Drosophila homolog of MST1/2) mutant flies, suggesting the existence of additional kinases that regulate the activity-dependent phosphorylation of LATS1/2 or Warts [31]. Indeed, an alternative pathway activating Warts was identified [32]. In the Drosophila adult midgut, the protein kinase Misshapen activates Warts to negatively regulate the activity of Yorkie and its target gene Upd3 (a JAK–STAT pathway ligand) in differentiating enteroblasts, thereby restricting intestinal stem cell division. Notably, depletion of Hippo in enteroblasts had little effect on midgut proliferation. Moreover, an increase in midgut proliferation resulting from Misshapen knockdown in enteroblasts was not rescued by Hippo overexpression, suggesting that Hippo and Misshapen are not redundant in this cell type [32]. Similarly, the mammalian Misshapen homolog MAP4K4 activates LATS2, suggesting Misshapen/MAP4K4 is an evolutionarily conserved MST1/2-independent activator of LATS1/2, although it is not entirely clear whether Misshapen/MAP4K4 directly phosphorylates and activates LATS [32]. Taken together, these observations suggest a cell type-dependent requirement of Hippo (MST1/2) and Misshapen (MAP4K4) for Warts (LATS1/2) activation and Yki (YAP/TAZ) inhibition. We speculate that these direct mediators of LATS activation (and thereby YAP and TAZ inhibition) could be regulated and fine-tuned on cellular stimulation, depending on the cellular ratios and differential regulation of these two LATS-activating kinases. Elucidating this context dependency and the spatiotemporal hierarchy in the dynamics of LATS activation will reveal further insights into the Hippo pathway.
Besides phosphorylation, recent studies have also revealed additional levels of regulation via protein stability of the Hippo kinase/transcription modules [33–37] and by scaffolding the components of the Hippo pathway [38]. Some of these regulatory mechanisms are highlighted in Box 2. This complexity adds an extra layer of divergent regulation of the Hippo pathway.
Upstream components of the Hippo pathway
Spatial regulation of LATS activation
Epithelial cells adhere mutually through cell–cell junctions such as tight, adherens, and basolateral, dividing the plasma membrane into an apical domain and a basolateral domain and thereby establishing an apical–basal polarity. Studies conducted in both D. melanogaster and mammals have revealed that many components of cell–cell junction- and cell polarity-related protein complexes regulate the Hippo pathway. Moreover, these regulators of YAP and TAZ are especially important in tissue organogenesis and in preventing cancer progression [39–41].
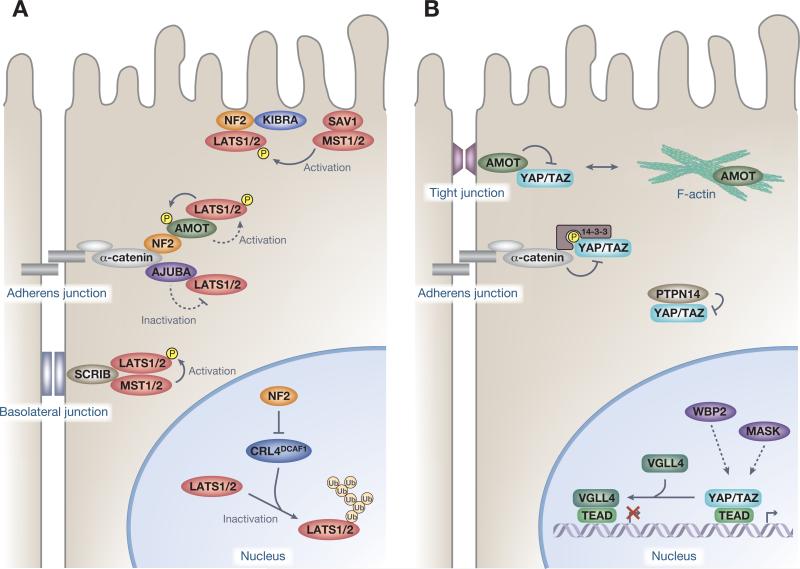
While YAP and TAZ are found in the cytosol and the tight junctions in dense cells, these transcriptional coactivators translocate to the nucleus and activate TEAD-mediated transcription in sparse cells [42,43]. This regulation is mediated primarily via homophilic cell–cell contacts that activate the kinase module [42,44]. In addition, the basolateral junction proteins, including mammalian Scribble (SCRIB) facilitate the activation of the core kinases MST and LATS, thereby inhibiting the nuclear activities of YAP/TAZ in mammary epithelial cells [45] (Figure 2A). Loss of cell polarity via induction of the epithelial–mesenchymal transition (EMT) causes SCRIB delocalization and TAZ activation, which confers self-renewal and tumor-initiation capacities on breast cancer stem cells [45]. Furthermore, the apical membrane-associated FERM-domain protein neurofibromatosis 2 (NF2) and the WW and C2 domain-containing protein kidney and brain expressed protein (KIBRA) (also known as WWC1) form a complex to activate the core kinase cassette [46–48]. NF2 is a potent tumor suppressor, mutations of which are associated with neurofibromatosis type II [5], and is a potent activator of the core Hippo kinase module [1]. Recent studies have revealed the complexity and multilevel regulation of LATS kinases by NF2 (Figure 2A). NF2 interacts with numerous proteins that are localized at the plasma membrane [31], including α-catenin and Angiomotin (AMOT) at adherens junctions [49], but also localizes in the nucleus [50]. At the plasma membrane of cultured cells, NF2 was shown to recruit LATS kinases and coordinate their activating phosphorylation by the MST–SAV1 kinase complex [31], highlighting the plasma membrane as a critical subcellular compartment for Hippo signal transduction. At adherens junctions in the inner cells of preimplantation embryos, NF2 directly interacts with α-catenin and AMOT to recruit LATS kinases, which in turn activates the kinase module of the Hippo pathway and thereby regulates lineage specification in preimplantation embryos [49]. Interestingly, adherens junctions-localized LATS kinases phosphorylate AMOT to further stabilize the AMOT–LATS interaction [49], establishing a potential positive feedback of Hippo pathway activation. By contrast, however, when LATS kinases are recruited to adherens junctions by the LIM domain-containing protein AJUBA in response to cytoskeletal tension, LATS-mediated phosphorylation of YAP and TAZ is impaired [51,52]. Therefore, it is not only LATS localization but also other unknown factors that affect the activation state of LATS. Moreover, another report showed that NF2 inhibits the E3 ubiquitin ligase CRL4DCAF1-mediated ubiquitylation of LATS1/2 in the nucleus [50]. In NF2-mutant tumors, CRL4DCAF1-mediated ubiquitylation of LATS1 results in its proteasomal degradation, while that of LATS2 leads to loss of its kinase activity potentially as a result of conformational change [50]. Together, these observations suggest that NF2 activates LATS kinases at multiple levels, yet the precise mechanism remains unclear.
Figure 2.
Regulators of Yes-associated protein (YAP)/transcriptional coactivator with a PDZ-binding domain (TAZ) activities. (A) Spatial organization and regulation of the kinase module. Neurofibromatosis 2 (NF2) (also known as Merlin) activates large tumor suppressor (LATS) kinases at multiple levels. NF2 recruits LATS at the plasma membrane to coordinate the activating phosphorylation by the mammalian STE20-like protein kinase (MST)–Salvador family WW domain-containing protein 1 (SAV1) kinase complex. NF2 directly interacts with α-catenin and Angiomotin (AMOT) to recruit LATS kinases at the adherens junction, which in turn activate LATS. NF2 inhibits the E3 ubiquitin ligase CRL4DCAF1-mediated ubiquitylation of LATS1/2 in the nucleus to inhibit proteasomal degradation of LATS1 and ubiquitylation-induced conformational change of LATS2. Scribble (SCRIB) recruits the core kinases MST and LATS to facilitate LATS activation at the basolateral junction. LATS recruitment by AJUBA at adherens junctions inhibits LATS kinase activity. (B) Physical regulation of the transcription module. AMOT, protein tyrosine phosphatase 14 (PTPN14), and α-catenin inhibit the activity of the transcription module by complexing YAP and TAZ, thus sequestering and preventing their nuclear access. AMOT interacts with and thereby causes cytoplasmic sequestration of YAP and TAZ by recruiting them to tight junctions. PTPN14 also causes cytoplasmic localization of YAP and decreased nuclear YAP activity. The adherens junctions protein α-catenin also regulates YAP by sequestering YAP–14-3-3 protein complexes in the cytoplasm. WW domain-binding protein 2 (WBP2) and multiple ankyrin repeats single KH domain-containing protein (MASK) interact with nuclear YAP and TAZ, which enhances YAP and TAZ transcriptional coactivator properties, but the precise mechanism remains unknown. Vestigial-like family member 4 (VGLL4) represses target gene expression by competing with YAP–TEA domain family (TEAD) interactions.
Physical regulation of YAP and TAZ
In addition to regulating YAP and TAZ through phosphorylation by LATS, other upstream branches of the Hippo pathway directly inhibit the activity of the transcription module by complexing YAP/TAZ, thus sequestering and preventing their nuclear access (Figure 2B). These branches include AMOT [53–56], protein tyrosine phosphatase 14 (PTPN14) [57,58], and α-catenin [59]. The AMOT–YAP/TAZ interaction is mediated via PPxY motifs of AMOT and WW domains of YAP and TAZ and thus does not require YAP and TAZ phosphorylation by the kinase module [53–55]. This regulatory interaction between PPxY motifs and WW domains is widespread throughout the Hippo pathway [60]. PTPN14 also utilizes its PPxY motifs to directly interact with WW domains of YAP, causing cytoplasmic localization of YAP and decreased YAP activity [57,58]. The adherens junction protein α-catenin also regulates YAP by sequestering YAP–14-3-3 protein complexes in the cytoplasm [59]. However, unlike AMOT, the kinase module that mediates YAP phosphorylation is required for the interaction between α-catenin and YAP, as 14-3-3 binds to phosphorylated YAP only [3]. Notably, α-catenin also provides a scaffold for the LATS activation/inactivation process (Figure 2A).
AMOT is a filamentous actin (F-actin)-binding protein that predominantly localizes at tight junctions and F-actin competes with YAP for binding to AMOT [61]. Interestingly, recent studies revealed that LATS1/2 phosphorylate AMOT within a conserved F-actin-binding domain [49,61–63], which inhibits AMOT binding to F-actin thereby potentiating its inhibitory effects on YAP and TAZ activity [49,61,62]. LATS kinases can therefore inhibit nuclear YAP and TAZ activity both directly and indirectly. Most studies support an inhibitory role of AMOT on YAP/TAZ [35,49,53–55,61,62], but a recent report indicated that AMOT binds to YAP to support YAP–TEAD mediated transcription in the nucleus and thereby stimulate YAP-mediated hepatic tumorigenesis in NF2-deficient mice [64].
Additional regulatory mechanisms of YAP/TAZ activity, divergent from its main regulation via sequestration, have also become apparent (Figure 2B). WW domain-binding protein 2 (WBP2) interacts with YAP and TAZ in a WW domain- and PPxY motif-dependent manner, enhancing YAP and TAZ transcriptional coactivator properties [65,66]. Suppression of WBP2 expression prevents the overgrowth phenotype of Warts mutant flies and inhibits TAZ-driven transformation of MCF10A mammary epithelial cells [65,66]. Multiple ankyrin repeats single KH domain-containing protein (MASK) has also been shown to potentiate YAP activity through direct binding [67,68]. Recent data suggest that vestigial-like family member 4 (VGLL4) is a natural inhibitor of YAP–TEAD interaction that represses target gene expression [69,70]. The Tondu domains of VGLL4 interact with TEAD transcription factors (TFs) and may compete for common binding sites with YAP and TAZ [69,70]. The physiological/pathological roles of these regulations are primarily characterized in YAP/TAZ-induced tumorigenesis. VGLL4 is downregulated in lung [70] and gastric [71] cancers and high MASK1 expression is associated with poor prognosis for breast cancer patients [67].
Nuclear functions of YAP and TAZ
The most widely studied role of YAP and TAZ is as transcriptional coactivators. YAP and TAZ do not contain any DNA-binding domains and thus bind to and elicit their transcription activation via cognate TFs (Figure 3A). Accumulating evidence shows that the TEAD family of TFs are major mediators of YAP and TAZ nuclear activity and therefore gene output [40,43,70,72,73] (Figure 1). Nonetheless, YAP and TAZ do utilize other TFs, and some contextual transcriptional responses are therefore mediated by additional TFs such as SMADs, p63, RUNX, and PAX [3,74], which diversifies the transcriptional output of the Hippo pathway.
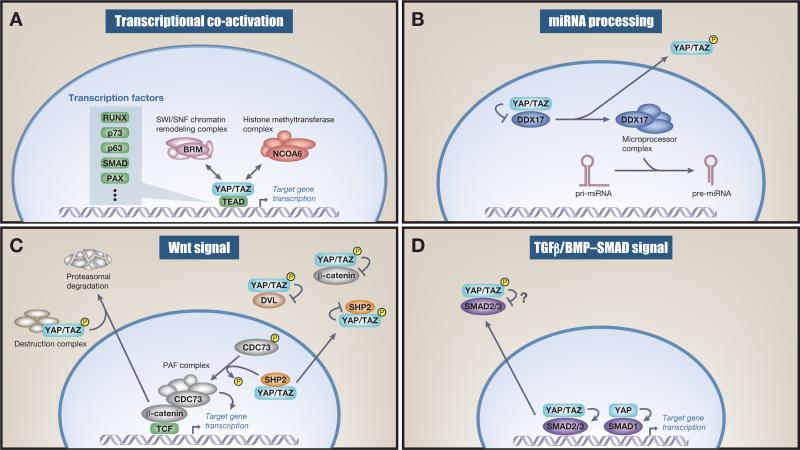
Figure 3.
Function of nuclear and cytoplasmic Yes-associated protein (YAP) and transcriptional coactivator with a PDZ-binding domain (TAZ). (A) Transcriptional coactivation. Nuclear YAP and TAZ bind to transcription factors (TFs) and recruit the NCOA6 methyltransferase complex as well as the BRM-containing SWI/SNF chromatin-remodeling complex, which mediates TF target-specific gene transcription. (B) miRNA processing. Nuclear YAP inhibits DDX17, a regulatory component of the microprocessor complex. This nuclear inhibition decreases global miRNA on YAP stimulation. Of note, some specific miRNAs are upregulated on YAP nuclear activation. (C) Wnt signaling. Cytoplasmic YAP and TAZ sequester β-catenin and in some instances recruit β-TrCP to the destruction complex. This leads to nuclear β-catenin downregulation and therefore dampens Wnt signaling. Cytoplasmic YAP and TAZ also negatively regulate Wnt signaling by sequestering β-catenin and disheveled (DVL). YAP and TAZ dictate nuclear and cytoplasmic localization of SHP2 and therefore its function. This feeds into the Wnt signaling pathway because nuclear SHP2 dephosphorylates parafibromin (CDC73), which leads to increased β-catenin transcriptional activity. (D) Transforming growth factor beta (TGFβ)/BMP–SMAD signal. Cytoplasmic YAP and TAZ sequester SMAD2/3 in the cytoplasm and therefore inhibit SMAD2/3-mediated signaling, although these mechanisms seem not to be universal and somewhat controversial. Nuclear YAP activates SMAD1-mediated transcription.
A functional explanation of how YAP and TAZ mechanistically regulate TEAD transcriptional activity has been revealed [75–78]. TAZ (and YAP) interacts with components of the SWI/SNF chromatin-remodeling complex [77,78] and YAP (and TAZ) recruits a methyl transferase complex into proximity with the TF, which leads to increased histone (H3K4) methylation and transcription of target genes [75,76]. These complex formations are functionally mediated via interactions between WW domains of YAP/TAZ and PPxY motifs of components of chromatin-remodeling/methyltransferase complexes [75–78] (Figure 3A). It will be interesting to learn whether the same mechanism holds true for other YAP/TAZ-utilized TFs. In some instances YAP/TAZ also operate as transcriptional corepressors for additional target genes [43,79,80]. Intriguingly this nuclear function also requires TEAD TFs [43,80].
In addition, nuclear YAP has also been found to regulate miRNA processing. Nuclear YAP/TAZ bind and sequester DEAD box helicase 17 (DDX17) (also known as p72) to repress its association with Microprocessor, a complex that regulates miRNA processing (Figure 3B). Therefore, the activation of YAP/TAZ in the nucleus is also a general mechanism of impaired global miRNA biogenesis [81]. Interestingly, nuclear YAP/TAZ conversely stimulate the biogenesis of certain miRNAs, such as miR-16, -21, -23a, -29, -107, and -152, by increasing Dicer via the Let-7 family of miRNAs [82,83]. YAP and TAZ can thus function as both positive and negative mediators of miRNA processing, depending on their targets.
YAP/TAZ as a signaling network
The Hippo pathway does not appear to have a unique extracellular ‘Hippo pathway-specific’ receptor/ligand interaction that exclusively regulates this pathway. Instead, it is integrated with a network of other prominent signaling pathways that impinge and rely on the Hippo pathway for their comprehensive functions. Moreover, YAP/TAZ serve as a cellular manifold and directly mediate transcription of several of these receptors and ligands that when expressed activate YAP/TAZ signaling. As the activity of constitutive positive feedback loops ultimately would lead to futile cycles and to pathological conditions, these must be tightly regulated, but how unrestrained YAP and TAZ activity is sensed and contained at the cellular level is currently not well understood.
Besides their widely recognized role in cancer [4], the physiological importance of YAP/TAZ has also been established in regeneration, development, and stem cell biology. We focus on a few tissues and biological contexts where the Hippo pathway has been most extensively studied and discuss these findings within the framework of other signaling pathways and cellular signaling organelles.
YAP/TAZ and Wnt signaling in the intestine
YAP/TAZ, the Hippo Pathway, and Wnt signaling are profoundly integrated [84]. In the absence of a Wnt signal, β-catenin remains phosphorylated, resulting in its degradation and the transcriptional inhibition of Wnt target genes [85]. The cytoplasmic retention of YAP/TAZ via its phosphorylation results in the sequestration of β-catenin in the cytoplasm [86] and in some instances the recruitment of β-TrCP to this complex, leading to the degradation of β-catenin [14,87] (Figure 3C). In addition, cytoplasmic YAP/TAZ interact with disheveled (DVL) to inhibit Wnt target gene transcription [15,16]. The tyrosine phosphatase SHP2 (encoded by the PTPN11 gene), like YAP and TAZ [42], shuttles between the nucleus and the cytoplasm where its nuclear localization promotes β-catenin-dependent transcription [88,89]. SHP2 is excluded from the nucleus in high-density cells by its interaction with phosphorylated YAP and TAZ [89], suggesting that cytoplasmic YAP/TAZ functionally terminate Wnt signaling through at least three separate mechanisms [15,16,87]. By contrast, nuclear YAP/TAZ promotes nuclear β-catenin, which mediates T cell factor (TCF)-dependent transcription (Figure 3C). Nuclear YAP/TAZ also facilitate nuclear translocation of SHP2, which in turn dephosphorylates CDC73 (also known as parafibromin) to stimulate CDC73–β-catenin binding and ultimately activate Wnt target gene transcription [88–90], providing additional functional links between Wnt and Hippo signaling (Figure 3C). A negative regulator of both β-catenin and YAP was also recently identified. Protein kinase C zeta (PKCζ) was identified to phosphorylate and thereby inhibit both β-catenin and YAP, highlighting the atypical PKCs as mediators of intestinal homeostasis and regeneration by modulating both YAP and β-catenin activity [91]. A general picture therefore emerges whereby nuclear YAP/TAZ potentiates Wnt signaling whereas cytoplasmic YAP/TAZ, due to an activated Hippo pathway, inhibits Wnt signaling [15,16,92]. In addition, YAP has also been reported to be a β-catenin target gene [17] and thus, in this instance, active Wnt signaling increases YAP abundance and hence transcriptional activity, suggesting that Wnt target genes can also feed back on the Hippo pathway and potentiate its signaling. The close reciprocal and complex relationship between the Hippo pathway and Wnt has been well studied in the intestine [16,93]. Indeed, YAP/TAZ expression is predominantly localized to the bottom of crypts where Lgr5+ intestinal stem cells can be found [16,27,85,87]. While YAP/TAZ are not essential for intestinal homeostasis [87,94], they are required under stress conditions where intestinal proliferation is necessary for survival, such as in the regeneration of damaged tissue after dextran sodium sulfate treatment [93]. In addition, nuclear YAP/TAZ are important mediators of adenomatous polyposis coli (APC)-mediated hyperplasia due to a combination of both increased nuclear β-catenin and YAP/TAZ signaling [87,93], a phenotype that can be mimicked by genetically inducing intestinal YAP/TAZ nuclear hyperactivity [27,93,95,96]. The utilization of an in vivo intestine-specific gene transfer technique revealed a dual role for YAP/TAZ in renewal of the intestinal epithelium. In this process YAP/TAZ use two distinct TFs [96]: TEAD [43,72,73] for stem and progenitor cell proliferation and Krüppel-like factor 4 (Klf4) for differentiation into goblet cells [96].
While these findings suggest a proliferative role of YAP in intestinal homeostasis, other reports observed an inhibitory effect of YAP in intestinal regeneration and Wnt-driven hyperplasia [16,86] (Figure 3C). Moreover, known target genes of YAP/TAZ–TEAD include BMP4 and DKK1 [97,98], which are negative regulators of Wnt signaling and crypt formation [85,97,99] (Figure 4). Future studies should examine how other pathways that are known to be differentially induced on challenge in the intestine [100], such as inflammation [101], integrate these responses via Lgr5, Wnt, and the Hippo pathway.
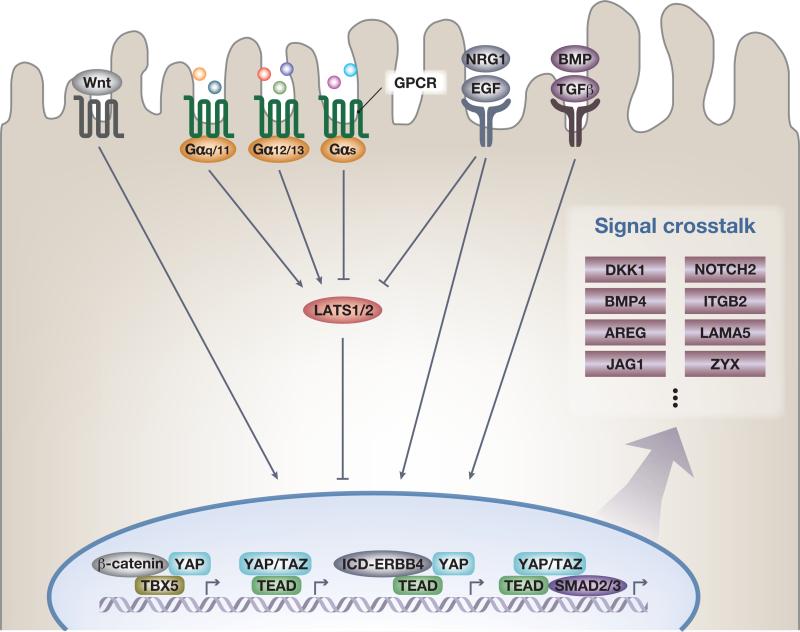
Figure 4.
Signal crosstalk with Yes-associated protein (YAP) and transcriptional coactivator with a PDZ-binding domain (TAZ). Wnt signaling activates YAP and TAZ by inhibiting the formation of the β-catenin destruction complex that in some instances functions as a cytoplasmic sink and causes degradation of YAP and TAZ. Nuclear YAP can form a complex with β-catenin and the transcription factor TBX5 to promote antiapoptotic gene expression, including BCL2L1 and BIRC5. G protein-coupled receptor (GPCR) signaling is relayed via small G proteins. Activation of Gα12/13 and Gαq/11 induces YAP and TAZ nuclear translocation and activity, whereas active Gαs represses YAP and TAZ nuclear activity, which causes either activation or inhibition of large tumor suppressor (LATS) kinase activity. The family of epidermal growth factor (EGF) ligands activates YAP and TAZ in both a LATS-dependent and -independent manner. EGF receptor (EGFR) stimulation inhibits the kinase module by dissociating phosphoinositide-dependent kinase-1 (PDK1) from the Hippo pathway components [LATS, mammalian STE20-like protein kinase 1 (MST), and Salvador family WW domain-containing protein 1 (SAV1)], preventing activation of LATS by MST. EGFR–Ras–MAPK signaling also enhances the AJUBA family protein Wilms’ tumor protein 1-interacting protein (WTIP) binding to LATS, which inhibits LATS activity toward YAP. In addition, a cytoplasmic intracellular domain of the ERRB4 receptor (ICD-ERBB4) is cleaved and released on neuregulin 1 (NRG1) stimulation, which in turn binds to YAP and promotes target gene expression. On transforming growth factor beta (TGFβ) signal activation, nuclear YAP and TAZ form a complex with SMAD2, SMAD3, and TEA domain family (TEAD) transcription factors, coordinating a protumorigenic transcriptional program. YAP and TAZ also directly promote transcription of several receptors and ligands to mediate endocrine and paracrine signal crosstalk and potential feedback loops. Notable examples are the Wnt signal inhibitor DKK1, ligands of the BMP/TGFβ family BMP4, the EGF-like growth factor AREG, NOTCH2, the Notch ligand Jagged1 (JAG1), integrin/extracellular matrix (ECM) signaling components such as β2-integrin (ITGB2) and the α5 subunit of laminin 511 (LAMA5), and the focal adhesion component zyxin (ZYX).
TGFβ and BMP signaling
The TGFβ superfamily is a group of secreted morphogens that are potent regulators of stem cell pluripotency and development [102,103]. TGFβ binds to its cognate serine–threonine kinase receptor to induce phosphorylation and activation of SMAD TFs [102]. The Hippo pathway is integrated with TGFβ and BMP signaling on multiple levels [18–20,79,97,104] (Figures 3D and 4).
In human embryonic stem cells, impairment of the nuclear translocation of TAZ results in the sequestration of a complex comprising TAZ, SMAD2, and SMAD3 in the cytosol [19] (Figure 3D). However, this cytoplasmic function is not universal, as conflicting data have recently been published in a range of cell lines [105,106]. On activation, nuclear YAP/TAZ form a complex with SMAD2/3 to bind TEAD TFs [18] or OCT4, which mediates pluripotency [79]. On loss of YAP/TAZ, these stem cells lose pluripotency potential and differentiate [19,79,107]. In addition, nuclear YAP supports SMAD1 target gene expression downstream of BMP [108]. YAP plays a role in regulating both embryonic and adult lung progenitor cell differentiation by directly specifying the epithelial cell lineage [109,110]. Yap deficiency leads to epithelial progenitors that are unable to properly respond to local TGFβ-induced cues [110], resulting in the differentiation of progenitors into epithelial airway cells [109]. Moreover, increased nuclear YAP activity leads to dedifferentiation of secretory cells into basal-like cells, which is directly mediated via a YAP–p63 interaction [109]. However, it remains to be determined whether YAP/TAZ-mediated, TGFβ-dependent transcription is regulated exclusively via SMADs [104] or whether it may also require TEADs [18,79] or context-dependent partnering with diverse TFs as has been recently observed. It is a task for the future to discover exactly what is regulated via SMADs, TEADs, and potentially other TFs and how these diverse complexes are regulated.
YAP/TAZ and ligands of the BMP/TGFβ family can also create a positive feedback loop that controls the Hippo pathway [20,97,111–113]. TAZ induces BMP4 transcription and promotes cell migration revealing a TAZ/TEAD/BMP signaling axis in mammary epithelial cells [97]. In addition, integrin-linked kinase, an integrin-associated actin and tubulin cytoskeleton-interacting effector that mediates TGFβ signaling, was found to be a negative regulator of the Hippo kinase cascade in cancer cells by inhibiting MYPT1, which leads to inhibition of NF2 and nuclear accumulation of YAP/TAZ [21].
GPCR signaling
A prominent role for GPCRs and their cognate ligands as regulators of Hippo signaling has been established [7–10]. GPCRs are coupled to small heterotrimeric G proteins and YAP/TAZ nuclear activities are either activated or inhibited depending on the G proteins coupled to the active receptors. Notable ligands such as the bioactive lipids lysophosphatidic acid (LPA) and sphingosine 1-phosphate (S1P) work through GPCRs [7,9] via Gα12/13 and are potent inhibitors of the Hippo kinase module and thereby activators of nuclear YAP/TAZ (Figure 4). By contrast, ligands such as epinephrine work via GαS-coupled GPCRs to inhibit YAP/TAZ through PKA-mediated activation of LATS kinases [114,115]. PKA is also implicated as a widespread activator of LATS on other physiological stimuli [114]. GPCR-driven pathologies can be mediated via dysregulated YAP/TAZ [7,11–13,116] and examples of FDA-approved drugs targeting GPCRs modifying YAP/TAZ activity are known [10,117]. Several notable GPCR ligands play crucial roles in intestinal regeneration [118], development [119], and stem cell pluripotency [120,121] – biological processes where YAP and TAZ are known to be key mediators [6,74,107]. Importantly, S1P–YAP signaling was shown to be important for cardiac precursors in zebrafish [119], but whether these effects by LPA, S1P, and other GPCR ligands in general are mediated via activation of YAP/TAZ transcriptional activity in vivo in nonpathological conditions is currently not well understood. It is worthwhile to expand these correlative relationships with loss-of-function experiments in mammalian animal models.
EGF family ligands regulate YAP and TAZ
YAP and TAZ are activated by the EGF family of ligands (Figure 4). Studies in mammalian systems have so far focused on cell culture systems, where YAP/TAZ are facilitators of EGF-mediated cell migration and transcription [22–24]. The regulation of YAP/TAZ via EGF-like molecules is twofold. EGF inhibits the kinase module via phosphoinositide-dependent kinase-1 and AJUBA proteins, which leads to impairment of SAV1's role in the kinase complex [22,23]. In addition, a cytoplasmic intracellular domain (ICD) of ERBB4 is cleaved and released on neuregulin 1 (NRG1) stimulation that in turn binds to and facilitates nuclear translocation of YAP. This effect is independent of LATS activity [24,122,123] (Figure 4). Moreover, activation of YAP/TAZ results in the transcription of the EGF-like growth factor AREG [21], providing crosstalk between Hippo and EGF signaling [21–23]. The biological role of the EGF-like molecules in the regulation of YAP/TAZ is currently not well understood in vivo. As they are prudent modulators of cancer etiology [124] and stem cell biology [103], it will be interesting to delineate precisely what is mediated via the Hippo pathway.
Notch signaling
A direct link between Notch signaling and YAP has become apparent in several systems, such as the developing embryo [28,41,56,125–128], neuronal stem cells [25], mature hepatocytes [26], and hepatocyte-derived stem cells [129]. The ligand Jagged1 (JAG1) [26] and the receptor NOTCH2, both known to be critical determinants of biliary cell fate [130,131], are direct YAP–TEAD target genes [26,27,126,129] (Figure 4). Inducing YAP activity in vivo can reprogram hepatocytes via NOTCH activation into progenitor cells [27,129].
Notch signaling is also a well-known regulator of satellite cells [132]. In skeletal muscle, YAP maintains satellite cells undifferentiated via TEAD, partly by repressing the pro-differentiating gene transcription program mediated by MYOD1 and MEF2 [111,133,134]. However, it remains to be elucidated whether YAP mediates this effect in satellite cells directly via NOTCH.
Hippo and NOTCH signaling play an important role during mouse embryogenesis from the morula to the blastocyst stage when the trophectoderm (TE), which gives rise to the placenta, and the inner cell mass (ICM), which gives rise to the embryo proper, lineages are specified. NOTCH–RBPJ and YAP–TEAD4 combinatorially bind to distinct promoter regions that directly regulate the transcription of the TE lineage determining TF CDX2 [28,41,56,125–128].
Reciprocal interaction with the microenvironment, cellular tension, and the cytoskeleton
Cell shape, cell–cell interactions, mechanotransduction, and matrix adhesiveness and stiffness are all prudent mediators of cellular state, identity, and niche [135–139]. The forces elicited by these interactions are dynamically regulated and are especially important during processes where the cell experiences drastic changes, such as during development, cell differentiation, and regeneration [74,135–138] – all processes where the Hippo pathway has been shown to be an important mediator [3,74,84,137,140]. RHO activity and the actin cytoskeletal organization are robust regulators of YAP/TAZ activity [52,139,141–145]. YAP/TAZ localizes to the nucleus and is transcriptionally active in cells grown on high-rigidity substrate, under static or cyclic high tension, or in cells lacking cell–cell contact [139–142,146,147]. The regulation of YAP/TAZ by these biological cues has been shown to impact lineage commitment [148], of which directing mesenchymal stem cell (MSC) fate is best described. High tension induced by cell spreading or by culturing on a hard surface promotes the nuclear localization of YAP/TAZ and drives MSCs toward the osteoblast lineage. By contrast, growth on a soft surface retains YAP/TAZ in the cytoplasm and promotes adipogenesis, a phenotype that can be recapitulated by depletion of TAZ [142,147,149]. YAP/TAZ also induce the expression of a range of cellular tension regulators such as zyxin (a component of focal adhesions) and the matricellular protein CYR61, as well as integrins and their ligands [43,113,136,146,150–152]. YAP/TAZ thereby serve as nuclear relays [135] of mechanical signals exerted by extracellular matrix (ECM) rigidity, tension, and cell shape.
These same integrin/ECM signaling proteins can also influence YAP/TAZ regulation [20,43,146,151,152]. For example, in skeletal stem cells the membrane-anchored metalloproteinase MT1-MMP was found to activate the β1-integrin/RHO GTPase signaling cascade, which triggered the nuclear accumulation of YAP/TAZ and promoted skeletal stem cell lineage commitment [150].
The Hippo pathway and signaling organelles
Cellular organelles that serve as signaling hubs are important integrators of YAP/TAZ activity. The Hippo pathway both controls and is regulated by the primary cilia [94,153–155], a microtubule-based solitary sensory signaling organelle comprising a special part of the plasma membrane in most eukaryotic quiescent cells [156]. The primary cilia respond to mechanical signals [157] and dynamically localize and regulate a range of receptors mediated by YAP/TAZ, such as GPCRs, sonic hedgehog (shh), the TGFβ family, receptor tyrosine kinases, and Wnt receptors [7,10,14,17,22,23,158]. This regulation of receptors is predominantly mediated through the cilia transition zone, the cellular gatekeeper for receptors destined for cilial localization [156]. Members of the nephrocystin (NPHP) family, established primary cilia-associated proteins [159], induce TAZ (and YAP) activity. NPHP4 binds to LATS and negatively regulates LATS activity, whereas NPHP3 and NPHP9 bind directly to TAZ and potentially compete with 14-3-3 binding to relieve cytoplasmic retention [160,161]. Therefore, members of the NPHP family increase transcriptional activity of YAP and TAZ [153]. Furthermore, TAZ binds the primary cilia-associated protein GLIS family zinc finger 3 (Glis3), facilitating Glis3-mediated transcription [154]. In addition, SHH, the prototypical primary cilia signaling ligand [162], activates YAP transcription, abundance, and activity [158]. SHH binds to the inhibitory transmembrane protein Patched and thereby relieves the GPCR-like molecule Smoothened from its inhibition [163], an effect that could be mediated via GPCR signaling. The upstream Hippo kinase module likewise integrates with the primary cilia, as MST and SAV1 (Figure 1) inhibit the disassembly of primary cilia by MST-mediated phosphorylation of AURKA, an important regulator of the primary cilia disassembly complex, and by association with the NPHP transition-zone complex, thereby inducing an increase in steady-state primary cilial abundance [153]. A range of diseases caused by dysfunctional primary cilia are termed ciliopathies [156,164] and, intriguingly, TAZ knockout animals develop polycystic kidneys [94] and express lower levels of multiple genes important for primary cilial morphogenesis; thus, these mice have malfunctioning cilia and exhibit a phenotype reminiscent of known human ciliopathies [94,164]. By contrast, TAZ depletion in tissue culture cells such as RPE1 cells induce ciliogenesis [155]. It is worth noting that the primary cilia are turned over at each cell cycle and that YAP/TAZ causes cell cycle progression; however, since the primary cilia are present in quiescent cells only, it needs to be fully determined which of these effects are causative or correlative.
The endolysosomal system is also a mediator of the Hippo pathway [165–167]. Through a genetic screen for conditional growth suppressor mutants, Myopic [the Drosophila homolog of the endosomal regulator His-domain protein tyrosine phosphatase (HD-PT) also known as PTPN23] was identified as a regulator of Yorkie endosomal localization [168]. A series of Hippo interactome studies also link endosomal compartments to the Hippo pathway [165–167]. Yorkie (the fly YAP/TAZ homolog, see Box 1) is recruited onto endolysosomes via Leash, a member of the arrestin family, which leads to degradation and inhibition of Yorkie nuclear activity [165]. Overexpression of Warts induces Yorkie-containing vesicular structures, suggesting that phosphorylation of Yorkie facilitates its vesicular localization [165]. Although it remains unclear whether similar regulation of YAP and TAZ occurs in mammalian systems, a role for the autophagosome/lysosome system in YAP protein stability in mice has been observed [169]. YAP accumulates in the kidney of tuberous sclerosis complex 1 (TSC1)-deficient mice due to impaired autophagy and thereby promotes abnormal cell proliferation and polycystosis, reminiscent of human perivascular epithelioid cell tumors from TSC patients. How lysosomal regulation of YAP can be reconciled with previous reports [170] that indicate proteasomal degradation of YAP is currently unclear. However, these findings suggest an intriguing new mode of regulation, as this organelle structure has been shown to not only downregulate surface receptors but also to dynamically respond to intra- and extracellular stimuli [171,172] and serve as an integrator for diverse cellular signaling events [124,172].
With the realization that multiple signaling pathways converge and integrate the Hippo pathway, it will be important to further delineate the interactions and dynamic spatiotemporal reciprocal regulation of the Hippo pathway to and from cellular signaling hubs such as the primary cilia and the endolysosomal system to fully understand the contextual signaling mediated by YAP and TAZ and the complete Hippo pathway.
Concluding remarks
The Hippo pathway extends beyond a simple kinase cascade targeting the transcription module and several proteins have been discovered to either completely or partially bypass the core kinase module to regulate YAP and TAZ. Studies in both Drosophila and mammals have broadened the Hippo pathway network, which now extends to components of cell–cell junction protein complexes, additional kinases, cytoskeletal regulators, cellular organelles, and, most recently, cellular stress responses (Box 2). Numerous pathways converge on multiple levels with YAP/TAZ and it is being revealed that YAP/TAZ serve as potent manifolds by partnering with seemingly distant signaling molecules for contextual cellular decision making.
Some of the initial in vivo Hippo pathway studies in flies [173] and mammals [27,174] highlighted the importance of this pathway in organ growth control as well as its therapeutic potential in regeneration and in tumorigenesis. These studies laid the foundation for the immense interest in and research into this pathway. Many important questions remain to be answered (Box 3). Recent findings showing that increasing YAP activity in various organs can reprogram fully differentiated cells into an undifferentiated state in vivo [77,109,129] clearly indicate a powerful therapeutic capacity in tissue regeneration and repair. Inhibiting the upstream Hippo kinase module should be a prime target in regenerative medicine [100] and research in cellular plasticity should be explored in the future. A clear challenge for this therapeutic strategy will be to fine-tune the activation state of YAP/TAZ, recollecting the interdependence between cancer and regeneration [4,6,100].
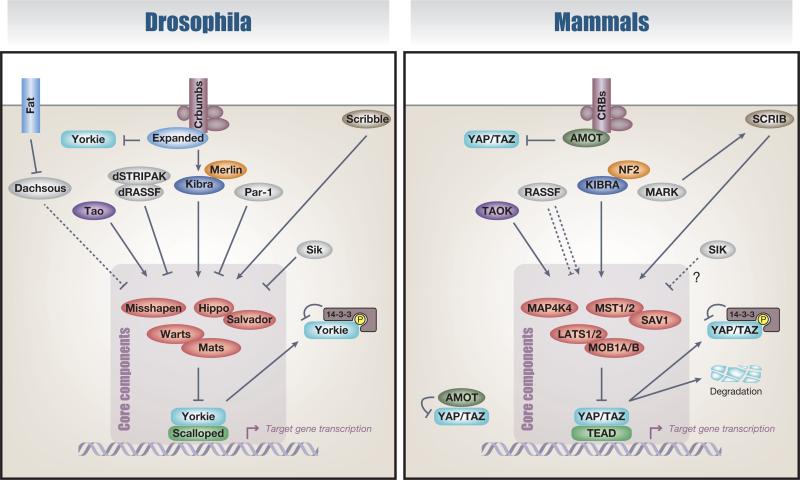
Box 1. Evolutionary divergence of the Hippo pathway.
The general structure of the Hippo pathway was drafted by studies conducted in Drosophila melanogaster (reviewed in [1]) and a wealth of studies in both flies and mammals since then has considerably expanded the realization of the Hippo pathway network as an evolutionarily conserved growth- and cell fate-controlling integrator. The core architecture of the Hippo pathway is highly conserved in evolution and homologous components are found in most major animal phyla, predating the origin of the Metazoa [175,176].
Multiple upstream branches of the Hippo pathway also regulate the core components. In D. melanogaster, Tao [thousand and one amino acid protein kinases (TAOKs) in mammals] phosphorylates Hippo (the Drosophila homolog of MST1/2) to activate its kinase activity [177,178], Par-1 [MAP/microtubule affinity-regulating kinases (MARKs) in mammals] phosphorylates Hippo to restrict its activity [179], Sik [salt-inducible kinases (SIKs) in mammals] phosphorylates Salvador (the Drosophila homolog of SAV1) to inhibit its association with the core kinase module [180], and RAS association domain-containing family protein (RASSF) competes with Salvador for Hippo and recruits a Drosophila PP2A phosphatase complex (dSTRIPAK) to dephosphorylate and inactivate Hippo [181,182]. However, except for TAOK [175,176], the functions of these upstream regulators do not appear to be conserved in mammals: MARK acts downstream of liver kinase B1 (LKB1) to activate the core kinase module through SCRIB [183]; the phosphorylation site in Salvador by Sik is not conserved in human SAV1 [180]; and multiple RASSF isoforms in mammals have different roles in the mammalian Hippo pathway [184,185]. Also, while YAP and TAZ are targeted for degradation in mammals, the phosphodegron responsible for this degradation is not conserved in Yorkie [170]. A plausible explanation for this divergence is an evolutionary shift at the base of the arthropod lineage [186]. In this transition, fly Fat gained novel domains that are required for restricting Dachsous to regulate the levels of Warts (the Drosophila homolog of LATS1/2) [186]. Unlike fly Fat, mouse Fat4 does not appear to regulate the Hippo pathway in murine liver [186]. Nevertheless, an additional mechanism linking mouse Fat4 to Yap is possible because a genetic interaction between Fat4 and Yap in mouse embryonic neuroepithelium has been reported [187]. In mammals, CRBs (crumbs family members) polarity complex forms a complex with AMOT [104], which binds and sequesters YAP and TAZ through its PPxY motif [53–55]. AMOT is likely to have been present in the last common ancestor of all bilaterian animals but was lost during arthropod evolution [186]. By contrast, the fly Crumbs-binding protein Expanded [188–190] gained a novel C-terminal domain that contains a PPxY motif and is thus able to interact with Yorkie [186,191,192]. The Crumbs complex thereby exchanges AMOT for Expanded at the base of the arthropod lineage. Expanded also regulates the Hippo pathway by forming a complex with Merlin (NF2 in mammals) and Kibra in flies [48,193]. Therefore, although groundbreaking discoveries continue to be pioneered in the fly [32,144], fundamental differences exist in the regulatory mechanisms of core Hippo components between flies and mammals.
Figure I. Evolutionary divergence of the Hippo pathway.
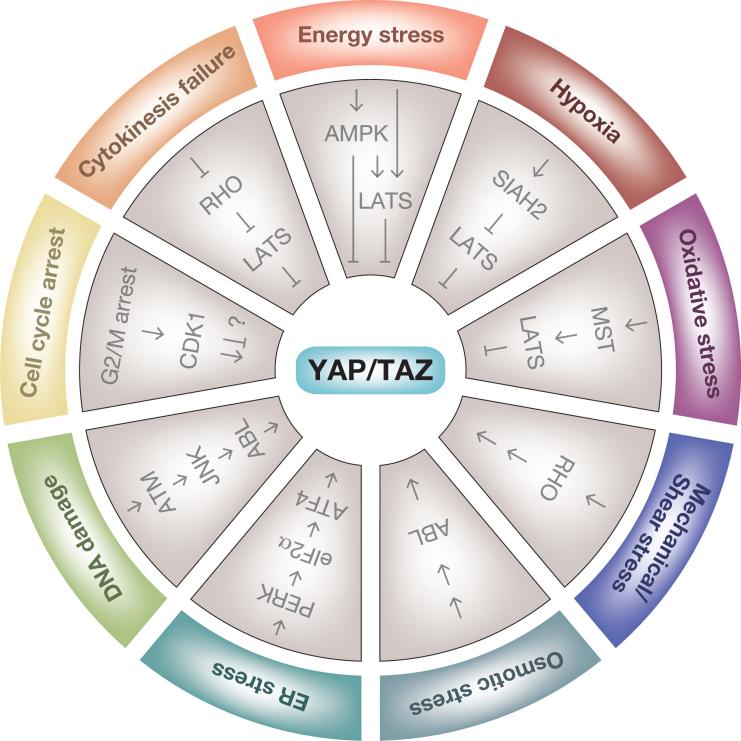
Box 2. YAP and TAZ regulation in response to cellular stress.
Cells are constantly subjected to external and internal stresses that endanger their integrity. YAP and TAZ are regulated in response to a range of these cellular stresses. Cytokinesis failure and extra centrosomes alter small G protein signaling and thereby activate LATS2 kinase. LATS2 in turn inhibits YAP and TAZ and stabilizes p53, thus arresting cells at G1 phase to prevent tumorigenesis [143]. Treatment of cancer cells with antitubulin drugs exerts a mitotic block, which activates cyclin-dependent kinase 1 (CDK1) [194]. CDK1 directly phosphorylates and inhibits YAP function to induce apoptosis. However, the role of CDK1-mediated YAP phosphorylation is controversial as mitotic phosphorylation of YAP by CDK1 promotes mitotic defects and potentiates oncogenic functions of YAP [195]. DNA damage activates the tyrosine kinase ABL through ATM–JNK signaling, which in turn phosphorylates YAP to stimulate proapoptotic functions of YAP in complex with p73 [196]. Endoplasmic reticulum (ER) stress has also recently been implicated in YAP regulation. The ER stress transducer PKR-like ER kinase (PERK) phosphorylates eukaryotic initiation factor 2α (eIF2α) during the adaptive stage of the unfolded protein response (UPR), which suppresses general protein synthesis and specifically induces the translation of ATF4, which regulates YAP transcription [197]. ER stress thus induces YAP expression through the PERK–eIF2α–ATF4 axis to prevent cell death during the adaptive stage of the UPR, while prolonged ER stress activates Hippo signaling to inhibit YAP and promote apoptosis [197].
YAP and TAZ likewise respond to external stresses evoked by these cellular microenvironments. Hyperosmotic stress induces tyrosine phosphorylation of TAZ by the ABL kinase, which facilitates the interaction between TAZ and nuclear factor of activated T cells 5 (NFAT5) to inhibit NFAT5 function in osmoregulatory transcription [198]. YAP/TAZ are activated by interstitial flow-driven shear stress and promote osteogenic differentiation of mesenchymal stem cells [147,199]. Rho GTPase appears to be involved in this regulation, although the precise mechanism remains unknown [148]. Oxidative stress evoked by ischemia and reperfusion in the mouse heart activates the Hippo pathway to antagonize a functional YAP–FOXO1 complex, leading to enhanced oxidative stress-induced cell death [200]. By contrast, hypoxia deactivates the Hippo pathway by destabilizing LATS2 through SIAH2 ubiquitin ligase-induced degradation [33]. Energy stress, such as inhibition of glucose metabolism and ATP production, induces AMP-activated protein kinase (AMPK)-mediated phosphorylation of Angiomotin-like 1 (AMOTL1) to stabilize and increase AMOTL1, which in turn stimulates LATS [35]. Furthermore, LATS also senses and is activated by glucose starvation in an AMPK-independent manner [201]. In addition, AMPK inhibits YAP by directly phosphorylating it at least two distinct sites [201,202]; therefore, energy stress inhibits YAP nuclear activity by several mechanisms [35,201,202].
Figure I. Yes-associated protein (YAP) and transcriptional coactivator with a PDZ-binding domain (TAZ) regulation in response to cellular stress.
Box 3. Outstanding questions.
How is LATS activated in different contexts?
How does the temporal and spatial organization and regulation of the Hippo pathway occur?
Does the Hippo pathway regulate additional effectors besides YAP and TAZ?
How do YAP and TAZ differ and how are their nuclear/cytoplasmic functions regulated?
What underlies the context-dependent physiological roles and activities of YAP and TAZ?
How can the Hippo pathway be therapeutically utilized in regeneration and cancer?
YAP and TAZ are major downstream effectors of the Hippo pathway.
YAP and TAZ function as signaling manifolds in the nucleus and the cytoplasm.
The Hippo pathway interacts on multiple levels with many other signaling pathways and organelles.
The Hippo network is a potent regulator of tissue regeneration and stem cell biology.
Acknowledgments
The authors apologize to colleagues whose work could not be cited owing to space limitations. The authors thank all members of the Guan laboratory for insightful discussion and critical comments. Work in the Guan laboratory was supported by a National Institutes of Health (NIH) grant (CA132809 and EYO226116) to K-L.G. C.G.H. is supported by a Postdoctoral Fellowship from the Danish Council for Independent Research/Natural Sciences. T.M. is supported by the Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowships for Research Abroad.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pan D. The hippo signaling pathway in development and cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh H, Irvine KD. Yorkie: the final destination of Hippo signaling. Trends Cell Biol. 2010;20:410–417. doi: 10.1016/j.tcb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varelas X. The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development. 2014;141:1614–1626. doi: 10.1242/dev.102376. [DOI] [PubMed] [Google Scholar]
- 4.Moroishi T, et al. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer. 2015;15:73–79. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvey KF, et al. The Hippo pathway and human cancer. Nat. Rev. Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 6.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev. Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu FX, et al. Regulation of the Hippo–YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mo JS, et al. Regulation of the Hippo–YAP pathway by protease-activated receptors (PARs). Genes Dev. 2012;26:2138–2143. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller E, et al. Identification of serum-derived sphingosine-1-phosphate as a small molecule regulator of YAP. Chem. Biol. 2012;19:955–962. doi: 10.1016/j.chembiol.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Bao Y, et al. A cell-based assay to screen stimulators of the Hippo pathway reveals the inhibitory effect of dobutamine on the YAP-dependent gene transcription. J. Biochem. 2011;150:199–208. doi: 10.1093/jb/mvr063. [DOI] [PubMed] [Google Scholar]
- 11.Liu G, et al. Kaposi sarcoma-associated herpesvirus promotes tumorigenesis by modulating the Hippo pathway. Oncogene. 2014 doi: 10.1038/onc.2014.281. Published online September 8, 2014. http://dx.doi.org/10.1038/onc.2014.281. [DOI] [PMC free article] [PubMed]
- 12.Yu FX, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell. 2014;25:822–830. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng X, et al. Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell. 2014;25:831–845. doi: 10.1016/j.ccr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Azzolin L, et al. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–1456. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 15.Varelas X, et al. The Hippo pathway regulates Wnt/β-catenin signaling. Dev. Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Barry ER, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konsavage WM, Jr, et al. Wnt/β-catenin signaling regulates Yes-associated protein (YAP) gene expression in colorectal carcinoma cells. J. Biol. Chem. 2012;287:11730–11739. doi: 10.1074/jbc.M111.327767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiemer SE, et al. The transcriptional regulators TAZ and YAP direct transforming growth factor beta-induced tumorigenic phenotypes in breast cancer cells. J. Biol. Chem. 2014;289:13461–13474. doi: 10.1074/jbc.M113.529115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varelas X, et al. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 20.Serrano I, et al. Inactivation of the Hippo tumour suppressor pathway by integrin-linked kinase. Nat. Commun. 2013;4:2976. doi: 10.1038/ncomms3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, et al. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat. Cell Biol. 2009;11:1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan R, et al. Regulation of Hippo pathway by mitogenic growth factors via phosphoinositide 3-kinase and phosphoinositide-dependent kinase-1. Proc. Natl Acad. Sci. U. S. A. 2013;110:2569–2574. doi: 10.1073/pnas.1216462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy BV, Irvine KD. Regulation of Hippo signaling by EGFR–MAPK signaling through Ajuba family proteins. Dev. Cell. 2013;24:459–471. doi: 10.1016/j.devcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haskins JW, et al. Neuregulin 1-activated ERBB4 interacts with YAP to induce Hippo pathway target genes and promote cell migration. Sci. Signal. 2014;7:ra116. doi: 10.1126/scisignal.2005770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, et al. Genome-wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and Hippo pathway effectors by Notch1. Stem Cells. 2012;30:741–752. doi: 10.1002/stem.1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tschaharganeh DF, et al. Yes-associated protein up-regulates Jagged-1 and activates the Notch pathway in human hepatocellular carcinoma. Gastroenterology. 2013;144:1530–1542. doi: 10.1053/j.gastro.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 28.Rayon T, et al. Notch and Hippo converge on cdx2 to specify the trophectoderm lineage in the mouse blastocyst. Dev. Cell. 2014;30:410–422. doi: 10.1016/j.devcel.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hergovich A. Regulation and functions of mammalian LATS/NDR kinases: looking beyond canonical Hippo signalling. Cell Biosci. 2013;3:32. doi: 10.1186/2045-3701-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou D, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin F, et al. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Q, et al. The conserved Misshapen–Warts–Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Dev. Cell. 2014;31:291–304. doi: 10.1016/j.devcel.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma B, et al. Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat. Cell Biol. 2015;17:95–103. doi: 10.1038/ncb3073. [DOI] [PubMed] [Google Scholar]
- 34.Lignitto L, et al. Proteolysis of MOB1 by the ubiquitin ligase praja2 attenuates Hippo signalling and supports glioblastoma growth. Nat. Commun. 2013;4:1822. doi: 10.1038/ncomms2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeRan M, et al. Energy stress regulates Hippo–YAP signaling involving AMPK-mediated regulation of Angiomotin-like 1 protein. Cell Rep. 2014;9:495–503. doi: 10.1016/j.celrep.2014.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salah Z, et al. Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Res. 2011;71:2010–2020. doi: 10.1158/0008-5472.CAN-10-3516. [DOI] [PubMed] [Google Scholar]
- 37.Bae SJ, et al. NEDD4 controls intestinal stem cell homeostasis by regulating the Hippo signalling pathway. Nat. Commun. 2015;6:6314. doi: 10.1038/ncomms7314. [DOI] [PubMed] [Google Scholar]
- 38.Heidary Arash E, et al. Arhgef7 promotes activation of the Hippo pathway core kinase Lats. EMBO J. 2014;33:2997–3011. doi: 10.15252/embj.201490230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearson HB, et al. SCRIB expression is deregulated in human prostate cancer, and its deficiency in mice promotes prostate neoplasia. J. Clin. Invest. 2011;121:4257–4267. doi: 10.1172/JCI58509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu-Chittenden Y, et al. Genetic and pharmacological disruption of the TEAD–YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cockburn K, et al. The Hippo pathway member Nf2 is required for inner cell mass specification. Curr. Biol. 2013;23:1195–1201. doi: 10.1016/j.cub.2013.05.044. [DOI] [PubMed] [Google Scholar]
- 42.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim NG, et al. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl Acad. Sci. U. S. A. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cordenonsi M, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 46.Baumgartner R, et al. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 47.Genevet A, et al. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu J, et al. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirate Y, et al. Polarity-dependent distribution of Angiomotin localizes Hippo signaling in preimplantation embryos. Curr. Biol. 2013;23:1181–1194. doi: 10.1016/j.cub.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li W, et al. Merlin/NF2 loss-driven tumorigenesis linked to CRL4DCAF1-mediated inhibition of the hippo pathway kinases Lats1 and 2 in the nucleus. Cancer Cell. 2014;26:48–60. doi: 10.1016/j.ccr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das Thakur M, et al. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. 2010;20:657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rauskolb C, et al. Cytoskeletal tension inhibits Hippo signaling through an Ajuba–Warts complex. Cell. 2014;158:143–156. doi: 10.1016/j.cell.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao B, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan SW, et al. Hippo pathway-independent restriction of TAZ and YAP by Angiomotin. J. Biol. Chem. 2011;286:7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang W, et al. Angiomotin-like proteins associate with and negatively regulate YAP1. J. Biol. Chem. 2011;286:4364–4370. doi: 10.1074/jbc.C110.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leung CY, Zernicka-Goetz M. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat. Commun. 2013;4:2251. doi: 10.1038/ncomms3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X, et al. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene. 2013;32:1266–1273. doi: 10.1038/onc.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W, et al. PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 2012;26:1959–1971. doi: 10.1101/gad.192955.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlegelmilch K, et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem. Sci. 2010;35:627–633. doi: 10.1016/j.tibs.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 61.Mana-Capelli S, et al. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol. Biol. Cell. 2014;25:1676–1685. doi: 10.1091/mbc.E13-11-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adler JJ, et al. Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc. Natl Acad. Sci. U. S. A. 2013;110:17368–17373. doi: 10.1073/pnas.1308236110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dai X, et al. Phosphorylation of Angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J. Biol. Chem. 2013;288:34041–34051. doi: 10.1074/jbc.M113.518019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yi C, et al. The p130 isoform of Angiomotin is required for Yap-mediated hepatic epithelial cell proliferation and tumorigenesis. Sci. Signal. 2013;6:ra77. doi: 10.1126/scisignal.2004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chan SW, et al. WW domain-mediated interaction with Wbp2 is important for the oncogenic property of TAZ. Oncogene. 2011;30:600–610. doi: 10.1038/onc.2010.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang X, et al. Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador–Warts–Hippo pathway. Cell Death Differ. 2011;18:1346–1355. doi: 10.1038/cdd.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sansores-Garcia L, et al. Mask is required for the activity of the Hippo pathway effector Yki/YAP. Curr. Biol. 2013;23:229–235. doi: 10.1016/j.cub.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sidor CM, et al. Mask proteins are cofactors of Yorkie/YAP in the Hippo pathway. Curr. Biol. 2013;23:223–228. doi: 10.1016/j.cub.2012.11.061. [DOI] [PubMed] [Google Scholar]
- 69.Koontz LM, et al. The Hippo effector Yorkie controls normal tissue growth by antagonizing Scalloped-mediated default repression. Dev. Cell. 2013;25:388–401. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang W, et al. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP–TEAD transcriptional complex. Cell Res. 2014;24:331–343. doi: 10.1038/cr.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiao S, et al. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:166–180. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 72.Vassilev A, et al. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 74.Mo JS, et al. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15:642–656. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oh H, et al. Yorkie promotes transcription by recruiting a histone methyltransferase complex. Cell Rep. 2014;8:449–459. doi: 10.1016/j.celrep.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qing Y, et al. The Hippo effector Yorkie activates transcription by interacting with a histone methyltransferase complex through Ncoa6. Elife. 2014 doi: 10.7554/eLife.02564. Published online July 15, 2014. http://dx.doi.org/10.7554/eLife.02564. [DOI] [PMC free article] [PubMed]
- 77.Skibinski A, et al. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell Rep. 2014;6:1059–1072. doi: 10.1016/j.celrep.2014.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oh H, et al. Genome-wide association of Yorkie with chromatin and chromatin-remodeling complexes. Cell Rep. 2013;3:309–318. doi: 10.1016/j.celrep.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beyer TA, et al. Switch enhancers interpret TGF-β and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 2013;5:1611–1624. doi: 10.1016/j.celrep.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 80.Kim M, et al. Transcriptional co-repressor function of the Hippo pathway transducers YAP and TAZ. Cell Rep. 2015;11:270–282. doi: 10.1016/j.celrep.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 81.Mori M, et al. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chaulk SG, et al. The Hippo pathway effectors TAZ/YAP regulate Dicer expression and microRNA biogenesis through Let-7. J. Biol. Chem. 2014;289:1886–1891. doi: 10.1074/jbc.C113.529362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tumaneng K, et al. YAP mediates crosstalk between the Hippo and PI3K–TOR pathways by suppressing PTEN via miR-29. Nat. Cell Biol. 2012;14:1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Piccolo S, et al. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 85.Clevers H, et al. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 86.Imajo M, et al. A molecular mechanism that links Hippo signalling to the inhibition of Wnt/β-catenin signalling. EMBO J. 2012;31:1109–1122. doi: 10.1038/emboj.2011.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Azzolin L, et al. YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 88.Takahashi A, et al. SHP2 tyrosine phosphatase converts parafibromin/Cdc73 from a tumor suppressor to an oncogenic driver. Mol. Cell. 2011;43:45–56. doi: 10.1016/j.molcel.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tsutsumi R, et al. YAP and TAZ, Hippo signaling targets, act as a rheostat for nuclear SHP2 function. Dev. Cell. 2013;26:658–665. doi: 10.1016/j.devcel.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 90.Mosimann C, et al. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with β-catenin/Armadillo. Cell. 2006;125:327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 91.Llado V, et al. Repression of intestinal stem cell function and tumorigenesis through direct phosphorylation of β-catenin and Yap by PKCζ. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.007. Published online February 4, 2015. http://dx.doi.org/10.1016/j.celrep.2015.01.007. [DOI] [PMC free article] [PubMed]
- 92.Heallen T, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cai J, et al. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010;24:2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hossain Z, et al. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc. Natl Acad. Sci. U. S. A. 2007;104:1631–1636. doi: 10.1073/pnas.0605266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou D, et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc. Natl Acad. Sci. U. S. A. 2011;108:E1312–E1320. doi: 10.1073/pnas.1110428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Imajo M, et al. Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat. Cell Biol. 2015;17:7–19. doi: 10.1038/ncb3084. [DOI] [PubMed] [Google Scholar]
- 97.Lai D, Yang X. BMP4 is a novel transcriptional target and mediator of mammary cell migration downstream of the Hippo pathway component TAZ. Cell. Signal. 2013;25:1720–1728. doi: 10.1016/j.cellsig.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 98.Seo E, et al. SOX2 regulates YAP1 to maintain stemness and determine cell fate in the osteo–adipo lineage. Cell Rep. 2013;3:2075–2087. doi: 10.1016/j.celrep.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haramis AP, et al. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303:1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- 100.Forbes SJ, Rosenthal N. Preparing the ground for tissue regeneration: from mechanism to therapy. Nat. Med. 2014;20:857–869. doi: 10.1038/nm.3653. [DOI] [PubMed] [Google Scholar]
- 101.Taniguchi K, et al. A gp130–Src–YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Massague J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schuldiner M, et al. Effects of eight growth factors on the differentiation of cells derived from human embryonic stem cells. Proc. Natl Acad. Sci. U. S. A. 2000;97:11307–11312. doi: 10.1073/pnas.97.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Varelas X, et al. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β–SMAD pathway. Dev. Cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 105.Nallet-Staub F, et al. Cell density sensing alters TGF-β signaling in a cell-type-specific manner, independent from Hippo pathway activation. Dev. Cell. 2015;32:640–651. doi: 10.1016/j.devcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Narimatsu M, et al. Distinct polarity cues direct Taz/Yap and TGFβ receptor localization to differentially control TGFβ-induced Smad signaling. Dev. Cell. 2015;32:652–656. doi: 10.1016/j.devcel.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 107.Lian I, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Alarcon C, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-β pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao R, et al. Yap tunes airway epithelial size and architecture by regulating the identity, maintenance, and self-renewal of stem cells. Dev. Cell. 2014;30:151–165. doi: 10.1016/j.devcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mahoney JE, et al. The Hippo pathway effector Yap controls patterning and differentiation of airway epithelial progenitors. Dev. Cell. 2014;30:137–150. doi: 10.1016/j.devcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Judson RN, et al. The Hippo pathway member Yap plays a key role in influencing fate decisions in muscle satellite cells. J. Cell Sci. 2012;125:6009–6019. doi: 10.1242/jcs.109546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee MJ, et al. YAP and TAZ regulate skin wound healing. J. Invest. Dermatol. 2014;134:518–525. doi: 10.1038/jid.2013.339. [DOI] [PubMed] [Google Scholar]
- 113.Diepenbruck M, et al. Tead2 expression levels control the subcellular distribution of Yap and Taz, zyxin expression and epithelial–mesenchymal transition. J. Cell Sci. 2014;127:1523–1536. doi: 10.1242/jcs.139865. [DOI] [PubMed] [Google Scholar]
- 114.Kim M, et al. cAMP/PKA signalling reinforces the LATS–YAP pathway to fully suppress YAP in response to actin cytoskeletal changes. EMBO J. 2013;32:1543–1555. doi: 10.1038/emboj.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yu FX, et al. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 2013;27:1223–1232. doi: 10.1101/gad.219402.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Plouffe SW, et al. Disease implications of the Hippo/YAP pathway. Trends Mol. Med. 2015;21:212–222. doi: 10.1016/j.molmed.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Park HW, Guan KL. Regulation of the Hippo pathway and implications for anticancer drug development. Trends Pharmacol. Sci. 2013;34:581–589. doi: 10.1016/j.tips.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yun CC, Kumar A. Diverse roles of LPA signaling in the intestinal epithelium. Exp. Cell Res. 2014;333:201–207. doi: 10.1016/j.yexcr.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fukui H, et al. S1P–Yap1 signaling regulates endoderm formation required for cardiac precursor cell migration in zebrafish. Dev. Cell. 2014;31:128–136. doi: 10.1016/j.devcel.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 120.Avery K, et al. Sphingosine-1-phosphate mediates transcriptional regulation of key targets associated with survival, proliferation, and pluripotency in human embryonic stem cells. Stem Cells Dev. 2008;17:1195–1205. doi: 10.1089/scd.2008.0063. [DOI] [PubMed] [Google Scholar]
- 121.Nagata Y, et al. Entry of muscle satellite cells into the cell cycle requires sphingolipid signaling. J. Cell Biol. 2006;174:245–253. doi: 10.1083/jcb.200605028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Komuro A, et al. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 123.Omerovic J, et al. Ligand-regulated association of ErbB-4 to the transcriptional co-activator YAP65 controls transcription at the nuclear level. Exp. Cell Res. 2004;294:469–479. doi: 10.1016/j.yexcr.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 124.Tomas A, et al. EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol. 2014;24:26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nishioka N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 126.Home P, et al. Altered subcellular localization of transcription factor TEAD4 regulates first mammalian cell lineage commitment. Proc. Natl Acad. Sci. U. S. A. 2012;109:7362–7367. doi: 10.1073/pnas.1201595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yagi R, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 128.Strumpf D, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- 129.Yimlamai D, et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tchorz JS, et al. Notch2 signaling promotes biliary epithelial cell fate specification and tubulogenesis during bile duct development in mice. Hepatology. 2009;50:871–879. doi: 10.1002/hep.23048. [DOI] [PubMed] [Google Scholar]
- 131.Boulter L, et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat. Med. 2012;18:572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bjornson CR, et al. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 2012;30:232–242. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tremblay AM, et al. The Hippo transducer YAP1 transforms activated satellite cells and is a potent effector of embryonal rhabdomyosarcoma formation. Cancer Cell. 2014;26:273–287. doi: 10.1016/j.ccr.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 134.Watt KI, et al. Yap is a novel regulator of C2C12 myogenesis. Biochem. Biophys. Res. Commun. 2010;393:619–624. doi: 10.1016/j.bbrc.2010.02.034. [DOI] [PubMed] [Google Scholar]
- 135.Wang N, et al. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 136.Seguin L, et al. Integrins and cancer: regulators of cancer stemness, metastasis, and drug resistance. Trends Cell Biol. 2015:25–234–240. doi: 10.1016/j.tcb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gaspar P, Tapon N. Sensing the local environment: actin architecture and Hippo signalling. Curr. Opin. Cell Biol. 2014;31:74–83. doi: 10.1016/j.ceb.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 138.Engler AJ, et al. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 139.Zhao B, et al. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cui Y,HF, et al. Cyclic stretching of soft substrates induces spreading and growth. Nat. Commun. 2015;6:6333. doi: 10.1038/ncomms7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wada K, et al. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 142.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 143.Ganem NJ, et al. Cytokinesis failure triggers Hippo tumor suppressor pathway activation. Cell. 2014;158:833–848. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Fletcher GC, et al. The Spectrin cytoskeleton regulates the Hippo signalling pathway. EMBO J. 2015;34:940–954. doi: 10.15252/embj.201489642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Deng H, et al. Spectrin regulates Hippo signaling by modulating cortical actomyosin activity. Elife. 2015 doi: 10.7554/eLife.06567. Published online March 31, 2015. http://dx.doi.org/10.7554/eLife.06567. [DOI] [PMC free article] [PubMed]
- 146.Chang C, et al. A laminin 511 matrix is regulated by TAZ and functions as the ligand for the α6Bβ1 integrin to sustain breast cancer stem cells. Genes Dev. 2015;29:1–6. doi: 10.1101/gad.253682.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kim KM, et al. Shear stress induced by an interstitial level of slow flow increases the osteogenic differentiation of mesenchymal stem cells through TAZ activation. PLoS One. 2014;9:e92427. doi: 10.1371/journal.pone.0092427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Halder G, et al. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 149.Hong JH, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 150.Parsons JT, et al. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 2010;11:633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wu DD, et al. Matricellular protein Cyr61 bridges lysophosphatidic acid and integrin pathways leading to cell migration. J. Biol. Chem. 2014;289:5774–5783. doi: 10.1074/jbc.M113.533042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tang Y, et al. MT1-MMP-dependent control of skeletal stem cell commitment via a β1-integrin/YAP/TAZ signaling axis. Dev. Cell. 2013;25:402–416. doi: 10.1016/j.devcel.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kim M, et al. The MST1/2–SAV1 complex of the Hippo pathway promotes ciliogenesis. Nat. Commun. 2014;5:5370. doi: 10.1038/ncomms6370. [DOI] [PubMed] [Google Scholar]
- 154.Kang HS, et al. Glis3 is associated with primary cilia and Wwtr1/TAZ and implicated in polycystic kidney disease. Mol. Cell. Biol. 2009;29:2556–2569. doi: 10.1128/MCB.01620-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kim J, et al. Actin remodelling factors control ciliogenesis by regulating YAP/TAZ activity and vesicle trafficking. Nat. Commun. 2015;6:6781. doi: 10.1038/ncomms7781. [DOI] [PubMed] [Google Scholar]
- 156.Czarnecki PG, Shah JV. The ciliary transition zone: from morphology and molecules to medicine. Trends Cell Biol. 2012;22:201–210. doi: 10.1016/j.tcb.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Battle C, et al. Intracellular and extracellular forces drive primary cilia movement. Proc. Natl Acad. Sci. U. S. A. 2015;112:1410–1415. doi: 10.1073/pnas.1421845112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fernandez LA, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sang L, et al. Mapping the NPHP–JBTS–MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Habbig S, et al. The ciliopathy disease protein NPHP9 promotes nuclear delivery and activation of the oncogenic transcriptional regulator TAZ. Hum. Mol. Genet. 2012;21:5528–5538. doi: 10.1093/hmg/dds408. [DOI] [PubMed] [Google Scholar]
- 161.Frank V, et al. Mutations in NEK8 link multiple organ dysplasia with altered Hippo signalling and increased c-MYC expression. Hum. Mol. Genet. 2013;22:2177–2185. doi: 10.1093/hmg/ddt070. [DOI] [PubMed] [Google Scholar]
- 162.Briscoe J, Therond PP. The mechanisms of hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013;14:416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 163.Sharpe HJ, et al. Regulation of the oncoprotein Smoothened by small molecules. Nat. Chem. Biol. 2015;11:246–255. doi: 10.1038/nchembio.1776. [DOI] [PubMed] [Google Scholar]
- 164.Hildebrandt F, et al. Ciliopathies. N. Engl. J. Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kwon Y, et al. The Hippo signaling pathway interactome. Science. 2013;342:737–740. doi: 10.1126/science.1243971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang W, et al. Defining the protein–protein interaction network of the human Hippo pathway. Mol. Cell. Proteomics. 2014;13:119–131. doi: 10.1074/mcp.M113.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Couzens AL, et al. Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase–phosphatase interactions. Sci. Signal. 2013;6:rs15. doi: 10.1126/scisignal.2004712. [DOI] [PubMed] [Google Scholar]
- 168.Gilbert MM, et al. A screen for conditional growth suppressor genes identifies the Drosophila homolog of HD-PTP as a regulator of the oncoprotein Yorkie. Dev. Cell. 2011;20:700–712. doi: 10.1016/j.devcel.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liang N, et al. Regulation of YAP by mTOR and autophagy reveals a therapeutic target of tuberous sclerosis complex. J. Exp. Med. 2014;211:2249–2263. doi: 10.1084/jem.20140341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhao B, et al. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCFβ-TRCP. Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J. Clin. Invest. 2015;125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jewell JL, et al. Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 2013;14:133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Justice RW, et al. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 174.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hilman D, Gat U. The evolutionary history of YAP and the Hippo/YAP pathway. Mol. Biol. Evol. 2011;28:2403–2417. doi: 10.1093/molbev/msr065. [DOI] [PubMed] [Google Scholar]
- 176.Sebe-Pedros A, et al. Premetazoan origin of the Hippo signaling pathway. Cell Rep. 2012;1:13–20. doi: 10.1016/j.celrep.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Boggiano JC, et al. Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo–Salvador–Warts tumor suppressor pathway. Dev. Cell. 2011;21:888–895. doi: 10.1016/j.devcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Poon CL, et al. The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador–Warts–Hippo pathway. Dev. Cell. 2011;21:896–906. doi: 10.1016/j.devcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 179.Huang HL, et al. Par-1 regulates tissue growth by influencing Hippo phosphorylation status and Hippo–Salvador association. PLoS Biol. 2013;11:e1001620. doi: 10.1371/journal.pbio.1001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wehr MC, et al. Salt-inducible kinases regulate growth through the Hippo signalling pathway in Drosophila. Nat. Cell Biol. 2013;15:61–71. doi: 10.1038/ncb2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Polesello C, et al. The Drosophila RASSF homolog antagonizes the Hippo pathway. Curr. Biol. 2006;16:2459–2465. doi: 10.1016/j.cub.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ribeiro PS, et al. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell. 2010;39:521–534. doi: 10.1016/j.molcel.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 183.Mohseni M, et al. A genetic screen identifies an LKB1–MARK signalling axis controlling the Hippo–YAP pathway. Nat. Cell Biol. 2014;16:108–117. doi: 10.1038/ncb2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Praskova M, et al. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem. J. 2004;381:453–462. doi: 10.1042/BJ20040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ikeda M, et al. Hippo pathway-dependent and -independent roles of RASSF6. Sci. Signal. 2009;2:ra59. doi: 10.1126/scisignal.2000300. [DOI] [PubMed] [Google Scholar]
- 186.Bossuyt W, et al. An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene. 2014;33:1218–1228. doi: 10.1038/onc.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cappello S, et al. Mutations in genes encoding the cadherin receptor–ligand pair DCHS1 and FAT4 disrupt cerebral cortical development. Nat. Genet. 2013;45:1300–1308. doi: 10.1038/ng.2765. [DOI] [PubMed] [Google Scholar]
- 188.Chen CL, et al. The apical–basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl Acad. Sci. U. S. A. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ling C, et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl Acad. Sci. U. S. A. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Robinson BS, et al. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr. Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Badouel C, et al. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 192.Oh H, et al. Phosphorylation-independent repression of Yorkie in Fat–Hippo signaling. Dev. Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hamaratoglu F, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 194.Zhao Y, et al. YAP-induced resistance of cancer cells to antitubulin drugs is modulated by a Hippo-independent pathway. Cancer Res. 2014;74:4493–4503. doi: 10.1158/0008-5472.CAN-13-2712. [DOI] [PubMed] [Google Scholar]
- 195.Yang S, et al. CDK1 phosphorylation of YAP promotes mitotic defects and cell motility and is essential for neoplastic transformation. Cancer Res. 2013;73:6722–6733. doi: 10.1158/0008-5472.CAN-13-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Basu S, et al. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 197.Wu H, et al. Integration of Hippo signalling and the unfolded protein response to restrain liver overgrowth and tumorigenesis. Nat. Commun. 2015;6:6239. doi: 10.1038/ncomms7239. [DOI] [PubMed] [Google Scholar]
- 198.Jang EJ, et al. TAZ suppresses NFAT5 activity through tyrosine phosphorylation. Mol. Cell. Biol. 2012;32:4925–4932. doi: 10.1128/MCB.00392-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zhong W, et al. Mesenchymal stem cell and chondrocyte fates in a multishear microdevice are regulated by Yes-associated protein. Stem Cells Dev. 2013;22:2083–2093. doi: 10.1089/scd.2012.0685. [DOI] [PubMed] [Google Scholar]
- 200.Shao D, et al. A functional interaction between Hippo–YAP signalling and FoxO1 mediates the oxidative stress response. Nat. Commun. 2014;5:3315. doi: 10.1038/ncomms4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang W, et al. AMPK modulates Hippo pathway activity to regulate energy homeostasis. Nat. Cell Biol. 2015;17:490–499. doi: 10.1038/ncb3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mo JS, et al. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat. Cell Biol. 2015;17:500–510. doi: 10.1038/ncb3111. [DOI] [PMC free article] [PubMed] [Google Scholar]