Abstract
We have used coarse-grained (CG) and united atom (UA) molecular dynamics simulations to explore the mechanisms of protein orientational transition of a model peptide (Aβ42) in a phosphatidylcholine/cholesterol (PC/CHO) lipid bilayer. We started with an inserted state of Aβ42 containing a folded (I) or unfolded (II) K28-A42 lipid insertion domain (LID), which was stabilized by the K28-snorkeling and A42-anchoring to the PC polar groups in the lipid bilayer. After a UA-to-CG transformation and a 1000 ns-CG simulation for enhancing the sampling of protein orientations, we discovered two transitions: I-to-”deep inserted” state with disrupted K28-snorkeling and II-to-”deep surface” state with disrupted A42-anchoring. The new states remained stable after a CG-to-UA transformation and a 200 ns-UA simulation relaxation. Significant changes in the cholesterol-binding domain of Aβ42 and protein-induced membrane disruptions were evident after the transitions. We propose that the conformation of the LID regulates protein orientational transitions in the lipid membrane.
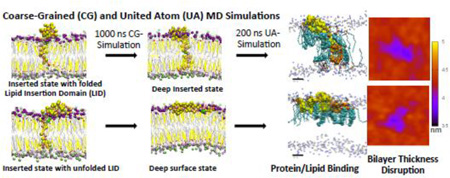
Graphical abstract
I. INTRODUCTION
Membrane protein orientations in lipid membranes play a key role in normal, pathogenic and therapeutic cellular processes such as signal transduction, protein-misfolding disorders, and drug delivery to cells [1–3]. At present, the detailed molecular mechanisms that regulate protein orientational transition and the subsequent protein/lipid interaction in a lipid bilayer that signal various cellular processes remain unclear.
A 42-residue beta-amyloid (Aβ) or Aβ42 represents an ideal membrane-active protein model to study protein orientational transition and protein/lipid interactions [4] in a lipid bilayer. Monomeric Aβ peptides of different lengths are released by the concerted proteolytic cleavage of the amyloid precursor protein by beta and gamma secretases in neurons [5]. Among other shorter alloforms of Aβ, Aβ42 is more neurotoxic and has higher propensity to self-aggregate in aqueous solution or in membrane [6]. The C-terminal (K28-A42) of Aβ42 represents a distinctive lipid insertion domain (LID) containing 13 non-polar amino acid residues bounded, at physiological pH, by a positively charged lysine (K28) and a negatively charged C-terminus (A42) [7]. Interestingly, this LID contains several key elements of protein membrane translocation: (i) a charged K28 that can bind to a lipid target on the upper lipid leaflet, (ii) a charged A42 that can anchor the entire peptide by binding to the lower lipid leaflet, and (iii) a loosely folded transbilayer insertion structure [8–10].
Due to its amphiphilic structure, Aβ42; may exhibit different protein orientational states that strongly depend on the protein conformation and lipid environment [11, 12]. Starting from a “partially inserted” state of Aβ42 as predicted by NMR [7], previous simulation studies [13, 14] have established that the C-terminus (A42) of the protein can descend and anchor to the polar headgroup region of the lower leaflet of the lipid bilayer or undergo a “partially inserted”-to-”fully inserted” orientational state transition within 50 ns of an atomistic simulation. Note that the previous NMR work [7] was performed on a lipid membrane mimic, i.e., a SDS micelle environment, which may not be totally equivalent to the natural lipid bilayer system as used in this work. Protein orientational states, including inserted and surface, of various Aβ peptides have been experimentally verified in various studies [15–17]. Questions remain about the stability of the inserted state and the mechanisms that regulate the transition from the inserted state to other state such as the surface state. In addition, the alterations of the protein/lipid interactions, e.g., protein binding behavior to nearby lipids, membrane disruption and bilayer thickness, due to the transition at atomistic details are unclear.
Using a multi-resolution molecular dynamics (MD) simulation approach, we explored the transition behavior of protein orientational states of Aβ42 on the subnanosecond time scale and at both united atom (UA) and coarse-grained (CG) spatial resolutions in a fully hydrated binary phosphatidycholine/cholesterol (PC/CHO) bilayer. The use of a UA-to-CG mapping of the starting UA structure and the subsequent CG simulation allowed sampling new protein orientational states more effectively than the UA simulation alone [18, 19]. On the other hand, the following reverse CG-to-UA mapping and the subsequent UA simulation [18, 20], which relaxed the reconstructed UA structure, allowed characterizations of the protein conformation and the subsequent lipid/protein interactions in the new protein orientational states with high spatial resolutions.
We started with a UA inserted state of Aβ42 containing a folded (I) or unfolded (II) LID in a phosphatidylcholine/cholesterol (PC/CHO) bilayer that mimics a partially folded or unfolded membrane-active protein embedded in the cholesterol-enriched raft domain of the cell membrane [21]. Our previous UA simulation studies [13, 14] have established the dual-stabilizing, protein/lipid interaction mechanism of Aβ42 via the K28-snorkeling and A42-anchoring to the upper and lower leaflets of the PC/CHO bilayer for both the I and II complexes. The multi-resolution MD simulation approach used here explores new protein orientational state transitions of Aβ42 and to characterize the subsequent protein/lipid interactions in the folded I and unfolded II systems. Our major goal was to understand how the folding of the LID (I vs. II) in a protein influences the protein orientational transition and the subsequent protein/lipid interaction behavior in a well-defined, model protein/membrane system.
Since Aβ is an important amyloidogenic peptide in the brain [22], protein-interaction events with neuronal membranes [11, 23], such as the protein orientational transitions and subsequent protein/interactions of Aβ within the lipid bilayer studied here, are important molecular events for understanding the complex pathogenic amyloid cascade pathways [5, 6, 16, 24–27] that lead to Alzheimer’s. Both computer simulations and complementary experiments such as NMR and single-molecule imaging and spectroscopy are urgently needed to uncover the basic mechanisms governing these events [28]. There have been a number of recent MD simulations of Aβ peptide in or on model membranes with different starting structures [13, 14, 29–33]. Simulations of aggregation and release processes [30, 31, 33] of Aβ are relevant to this work, because they represent the future molecular events that take place after the completion of the predicted protein orientational transitions. In addition, our predicted structures of the new protein orientational states may signal further downstream folding/unfolding, binding and self-aggregation phenomena in the complex membrane-associated amyloid cascade pathways that may contribute to the pathogenesis of Alzheimer’s.
II. MATERIALS AND METHODS
A. Starting Structures of Protein/Lipid/Water/Ion Complexes
1. Protein
We used the human Aβ42 in this study, and its sequence [34] is:
DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIA
The N-terminus and C-terminus are D1 and A42, respectively. Aβ42 has two segments, an N-terminal D1-N27 (non-LID) and a C-terminal K28-A42 (LID), which is in bold. The LID or lipid insertion domain has 13 non-polar residues, G29 to I41, flanked by the polar K28 and A42 residues. The non-LID contains mainly polar residues. At neutral pH, there are six negatively charged residues: E3, D7, E11, E22, D23 and a deprotonated C-terminus or A-42, and three positively charged residues: R5, K16 and K28. Therefore, Aβ42 has a net charge of −3.
2. Lipid
We used two lipid molecules, 1-palmitoyl-2-oleoyl-phosphatidylcholine (PC) and cholesterol (CHO). Here PC has a zwitterionic polar headgroup, a glycerol backbone and two acyl chains; whereas CHO has a small polar group, four bulky ring groups and a hydrocarbon tail. We identified 5 structural groups in PC: NC3, PO4, GLY, sn-1 chain and sn-2 chain. Here, NC3 contains the positively charged trimethylammonium, PO4 the negatively charged phosphate, sn-1 chain the saturated C16:0 acyl chain, and sn-2 chain the mono-unsaturated C18:1 acyl chain with an unsaturated bond at the C9 and C10 positions. For CHO, the three structural groups are ROH, Rings and Chain, representing the polar 3β-OH, four fused rings and the tail chain, respectively. A detailed labeling scheme and the chemical structures of PC and CHO are given in Figs. S1 and S2 in Supporting Information. NC3, PO4, GLY and ROH are all polar lipid groups and are capable of forming hydrogen bonds with the polar residues of Aβ42.
3. Protein/lipid/water/ion complexes
We focused on two starting Aβ42/lipid/water/ion complexes, I (initial) and II (initial). In both complexes Aβ42 was in a fully inserted orientational state. These two starting structures were extracted from the previously published D1 and D3 trajectories at 10 and 200 ns [14]. A detailed description of the UA structures and the procedure to generate the D1 and D3 trajectories are given elsewhere [14]. The unfolding extent of Aβ42, expressed as the number of residues exhibiting non-hydrogen-bonded structures [35] divided by the total number of residues in each segment of the peptide and multiplied by 100, was 51.9% in the non-LID and 26.7% in the LID of the I (initial), and 40.7% in the non-LID and 60% in the LID of the II (initial) complex. Therefore the LID was more unfolded in the II (initial) complex than in the I (initial) complex. Three positive counterions, Na+ were present to ensure a charge neutral system for each starting structure. Water was also present in the two starting structures.
B. MD Simulations of Protein/Lipid/Water/Ion Complexes
1. UA-to-CG mapping method to generate starting CG complexes
A forward mapping procedure, UA-to-CG mapping, was used to transform the starting UA Aβ42/lipid/water/ion complexes to the corresponding CG complexes. Protein and lipid molecules were transformed using an AWK script (atom2cg_v2.1.awk) [36] and an in-house C program, respectively, based on the UA-to-CG mapping scheme of the Martini model [36, 37]. These programs assigned the coordinates of the CG beads to the corresponding centers of mass of the atoms according to the mapping scheme. In general, four heavy atoms were transformed to one CG bead for both protein and lipids.
Direct transformation of the original UA water [38] to CG water from the starting atomic structure was not possible due to the steric conflict among CG water beads after transformation and also the non-uniformly distributed interfacial water at the lipid/water interface. Therefore, all water molecules were removed from the UA starting complexes and replaced by randomized CG water. Similarly, UA sodium ions were removed and replaced by CG sodium ions. In the Martini Model, one CG water represents four water molecules but one CG sodium ion represents one sodium ion [36]. Our starting CG simulation system consisted of 574 PC, 383 CHO, and 16,789 water with a box size of ~ 13.8 × 15.9 × 13.8 nm3.
2. CG MD simulations
CG MD simulations were performed on the initial CG Aβ42/lipid/water/ion complexes. We used the MARTINI 2.1 CG force field for protein [37], ion and water, and MARTINI 2.0 CG force field for lipids [36]. For protein, the CG force field requires the secondary structure of the protein. This secondary structure was determined from the atomic coordinates of each starting complex using the DSSP (Define Secondary Structure of Proteins) algorithm [35]. A leapfrog integrator with a 25 fs time step was used. For the non-bonded interactions, a Leonard-Jones potential with cutoff at 1.2 nm and shifted from 0.9 to 1.2 nm, and a coulomb potential shifted from 0 to 1.2 nm were used. A v-scale thermostat [39] with a coupling time of 1 ps to maintain a fixed temperature of 310K, and a Berendsen barostat [40] with a coupling time of 1 ps and a compressibility of 1×10−4 bar−1 to achieve a semi-isotropic pressure of 1 bar along the x-y and z-directions were employed. To ensure effective phase sampling, four independent simulation replicates, each with a different initial velocity distribution but identical coordinates of the CG beads, were generated for each starting CG structures. A 1000 ns-CG simulation was performed on each replicate under constant number, pressure and temperature (NPT) condition with x-y-z periodic boundaries.
3. CG-to-UA mapping to reconstruct UA complexes from CG complexes
A reverse mapping procedure, CG-to-UA mapping, was performed to reconstruct the high-resolution UA structures from the 1000 ns simulated CG structures. Detailed steps of this reverse mapping procedure have been published by Rzepiela et al. 2010 [18]. Briefly, the first step involved the reconstruction of an initial UA complex from the CG complex by placing the constituent atoms randomly inside each corresponding CG bead within a radius of 3 Å for lipid and protein but 2.1 Å for water. A restrained simulated annealing (RSA) MD simulation was then performed for 100 ps. Here, an external restraining harmonic potential with a force constant of 12,000 kJ mol−1nm−2 was applied to lipid and protein atoms to keep the center of mass of those reconstituted atoms in close proximity to the center of the corresponding CG bead. For water, a smaller harmonic force constant of 400 kJ mol−1nm−2 was used for reconstructing four water molecules from a single CG water bead. Also, the temperature was varied linearly from a high temperature TH to the target temperature of 310K. For our case, the TH for the protein and lipid reconstruction was at 1300K and that for the water and ion at 400K.
Due to the random placement of atoms within the corresponding CG bead, large forces occurred at the beginning of the simulation that caused instability of the simulation. To solve this issue, a time varying force threshold or cap was applied in order to achieve a balance of simulation stability while maintaining efficient phase sampling of the energy-minimized structure throughout the simulation. In this work, the force threshold started from 15,000 kJ mol−1 nm−1 and increased linearly at a rate of 100 kJ mol−1 nm−1 ps−1. The RSA simulation was performed at a constant volume with a 2 fs integration step. After the RSA, the external restraining potential was slowly removed in 10 ps to create the reconstructed UA complex. Our starting UA simulation system consisted of 574 PC, 383 CHO, and 67,099 water with a size of ~ 13.8 × 15.9 × 13.7 nm3.
4. UA simulations of reconstructed UA complexes
To ensure that the reconstructed UA structures are properly relaxed and equilibrated, a 200 ns atomistic simulation was performed on each reconstructed UA structure. Again, to improve phase sampling of different conformations, two independent simulation replicates, each with a different initial velocity distribution but identical coordinates, were generated for each of the reconstructed UA complex. The details of the UA simulations have been published elsewhere [13, 14]. Briefly, molecular dynamics simulations were performed under NPT conditions using Gromacs 4.0 [41–44] with Berger et al. [45] and Holtje et al. [46] lipid parameters and a modified GROMOS87 force field [47, 48]. The effect of force field on behavior specific to this system has been discussed recently in detail [13]. Periodic boundary conditions were applied each direction. A SPC model [38] was used for water. Electrostatic interactions were estimated by Particle-Mesh-Ewald method with the direct space cutoff set to 1.0 nm [49, 50]. Bond lengths were constrained by LINCS [51]. A leapfrog integrator with a 2 fs time step was used to integrate the motion of the systems. Temperature baths of 310 K were coupled to water, lipids and protein separately, using a v-rescale thermostat [39] with a coupling time of 0.05 ps. A Berendsen barostat [40] with a coupling time of 1 ps kept the systems at an isotropic pressure of 1 atm.
C. Data Analysis
1. Protein secondary structures
The DSSP algorithm [35] was used to analyze the secondary structures and their time evolution of Aβ42 peptide in all reconstructed and relaxed UA simulated complexes. In DSSP, a secondary structure was assigned to each amino acid residue of the protein based on the atomic coordinates of the backbone NH and CO dipoles between two amino acid residues using an electrostatic model. According to the repeating pattern and locations of the residues, DSSP assigned 6 hydrogen-bonded structures: 3-turn helix (G), 4-turn helix (H) and 5-turn helix (I), beta-sheet (E), isolated beta-bridge (B) and turn (T), and two non-hydrogen bonded structures: coil (C) and bend (S), at each time frame of all the UA simulations. Four reduced secondary structures, helix (GHI), beta (EB), turn (T) and loop (SC), were used in this study to facilitate the analysis of the hydrogen-bonded (GHI, EB NS T) and non-hydrogen bonded (SC) behavior of the amino acid residues in Aβ42. The Visual Molecular Dynamics (VMD) program [52] was used to generate snapshots of proteins with detailed secondary structures as well as nearby lipids in both CG and UA representations.
2. Residue-specific protein/lipid interaction dynamics
Residue-specific protein/lipid interaction dynamics was analyzed using the minimum distance analysis tool, g_mindist from GROMACS [53]. This tool calculates the minimum distance between any atom of a protein group, e.g., the K28 or A42 side chain of a residue of Aβ42, to any atom of a lipid group, e.g., the structural group PO4 or GLY of PC or ROH of CHO, for each time frame in a simulated trajectory. This minimum distance dynamics analysis was applied to trajectories in both UA and CG representations.
3. Residue-specific hydrogen bonding profile
Residue-specific protein/lipid, protein/protein and protein/solvent hydrogen bonding profile was examined using the hydrogen-bonding analysis tool, g_hbond from GROMACS [53]. This tool identifies the hydrogen bond partners between two groups, e.g., a specific residue of a protein (donor) and a lipid, protein residue or solvent (acceptor). Hydrogen bonding between the two matched pair is assigned when the threshold bonding distance and angle between the donor-acceptor pair atoms are less than or equal to the default values of 3.5 Å and 30°, respectively, from GROMACS [53]. We used this tool to examine the K28 (donor) hydrogen bonding profile with the acceptor from lipid (PC and CHO), protein (residues other than K28) and solvent (water) in the UA complexes. Other than identifying the acceptors, the percentage of time that K28 was engaged in hydrogen bonding with each identified acceptor within the last 50 ns of the 200 ns simulation was also determined for each UA trajectory. In addition, average K28 hydrogen bonding profile was also generated by calculating the average K28 hydrogen bonding time across all repeated replicates of each I or II complex.
4. Annular and non-annular lipid and solvent analysis
To analyze the structures of the molecules near the protein, annular (AL) and non-annular (nAL) lipids (PC and CHO) and solvent (w) surrounding the protein were identified from the simulated trajectories using the molecule selection tool, g_select from GROMACS [53]. This tool identifies the group of lipid or solvent molecules that are within a threshold distance between any atom of the protein and any atom of the surrounding lipid or solvent frame-by-frame in a given trajectory. In this study, AL-PC, nAL-PC, AL-CHO, nAL-CHO, AL-w and nAL-w were identified for each time frame in the UA trajectory with a threshold of 0.5 nm [12]. The selection of AL and nAL molecules was performed for each time frame.
5. Transbilayer density profile
The time-averaged transbilayer density profiles for different lipid structural groups: PO4 of PC, GLY of PC, ROH of CHO, solvent (w) and protein: all atoms or a selected side chain group (e.g., NH3 of K28) were calculated for each time frame, and then averaged over a given length of time for both AL and nAL regions. The time-averaged density for each group is given by
| (1) |
Here, ni(z) is the number of particles for each lipid or protein group within a thin z-slice at a given z-coordinate and A is the cross-sectional or x-y area of the simulation system, N is the total number of time frames for the calculations. In this study, Δz = Zs/100, where Zs was the Z-dimension of the simulation box which depended on the amount of explicit solvent on the top and bottom of the lipid bilayer. The z-axis was parallel to the normal of the PC/CHO lipid bilayer in all trajectories. We used the analysis tool, g_density from GROMACS [53] for the density profile calculation, averaged over the last 50 ns of each 200 ns UA trajectory for each simulation replicate. In addition, average density profile was also generated by calculating the average of all time-averaged density profiles across all repeated replicates of each I or II complex.
6. Order parameter of lipid
A time-averaged orientational order parameter S(n) at a given carbon number position (n) was used to quantify the segmental orientation order of the PC acyl chains with respect to the normal (z-axis) of the bilayer over a given length of time based on the following equation:
| (2) |
Here, θn is the angle between the z-axis and a vector Cn (= rn+1 – rn−1) joining the nearest-neighbor carbons on each side of the carbon atom n, where rn+1 and rn−1 are the position vectors of the n+1 and n−1 carbon positions of the chain. Again, N is the number of time frames for the averaging. The square bracket denotes the average over all the participating lipids in the AL or nAL regions. The order parameter calculation was performed on both sn-1 and sn-2 chains using a molecular order tool, g_order from GROMACS [53] averaged over the last 50 ns of each 200 ns UA trajectory for each simulation replicate. In addition, average order parameters were also generated by calculating the average of the time-averaged order parameters across all repeated replicates of each I or II complex.
7. Bilayer thickness maps
Time-averaged bilayer thickness maps of a lipid bilayer were calculated over a given length of time using a grid-based membrane analysis tool, GridMAT-MD [54], for all our UA simulations. This tool reads the coordinates of all the phosphorus atoms in the lipid bilayer, determines the shortest z-distances between those from the top layer and those from the bottom layer, and generates a 2-dimensional thickness matrix for each time frame. Upon averaging the matrix over a given time range, the time-averaged thickness map was created for a given trajectory using an in-house R script. In addition, we identified the location of the protein by assigning a large “thickness” value e.g., 6 nm, for each identified protein atom on the x-y plane and overlaid them on the thickness map frame-by-frame. The time-averaged thickness and protein location maps were created over the last 50 ns of each 200 ns UA trajectory. The average (avg) and uncertainty thickness maps were also generated from the average and standard error pixel-by-pixel calculations of all repeated replicates of each I or II complex. To identify the thickness near the periodic boundary, a larger, repeated or tiled image along the x- and y- directions, is presented for each thickness map.
3. RESULTS
A. MD Simulations of Protein/Lipid/Water/Ion Complexes
From the two starting UA structures of Aβ42/lipid/water/ion complexes, 8 CG and 16 reconstructed UA simulation replicates were generated using the multi-resolution MD simulations. The 4 computational steps are given below.
1. Creation of starting CG complexes
A UA-to-CG mapping transformed 2 UA starting structures to 2 corresponding starting CG structures (Table 1). The starting UA I (initial) and II (initial) structures of the Aβ42/lipid/water/ion complexes are illustrated in Figures 1A and 1D, respectively. A successful transformation of the protein and lipid atoms into corresponding CG beads that preserved the orientations and conformations of the side chains and backbone of Aβ42 and key structural groups of PC and CHO was evident for each starting structure.
Table 1. Protein orientational states and protein unfolding behavior of Aβ42 in PC/CHO bilayers.
The protein orientational states and unfolding % of Aβ42 in the I and II systems before (I (initial) or II (initial)) and after 1000 ns-CG or after 200 ns-UA MD simulations are shown. For the 1000 ns-CG simulation (I-rep* or II-rep*), the unfolding % was calculated after the CG-to-UA reverse mapping. For the 200 ns-UA simulation, (I-rep*-* or II-rep*-*), the unfolding % was based on the last 50 ns of the simulation with the average and the standard error of the mean given. Average unfolding % across all 4 replicates in CG (I (avg)-CG or II (avg)-CG) or 8 replicates in UA (I (avg)-UA or II (avg)-UA) and the standard error of the mean are also presented. * represents replicate number 0–3.
| System | Simulation conditions |
Protein orientational statea |
Unfolding %c D1-N27 (non-LID) |
Unfolding %c K28-A42 (LID) |
Unfolding %c D1-A42 |
|---|---|---|---|---|---|
| I (initial) | 0 | Inserted | 51.9 | 26.7 | 42.9 |
| I-rep0 | 1000 ns-CG | Deep inserted | 40.7 | 26.7 | 35.7 |
| I-rep0-0 | 200 ns-UA | Deep inserted-Ca | 53.2 ± 1.4 | 47.1 ± 0.3 | 51.0 ± 0.3 |
| I-rep0-1 | 200 ns-UA | Deep inserted-Ca | 32.4 ± 0.8 | 48.4 ± 0.7 | 38.1 ± 0.7 |
| I-rep1 | 1000 ns-CG | Deep inserted | 63.0 | 26.7 | 50.0 |
| I-rep1-0 | 200 ns-UA | Deep inserted | 63.9 ± 0.9 | 55.5 ± 1.5 | 60.9 ± 1.5 |
| I-rep1-1 | 200 ns-UA | Deep inserted | 73.3 ± 1.2 | 61.5 ± 0.9 | 69.1 ± 0.9 |
| I-rep2 | 1000 ns-CG | Deep inserted | 59.3 | 20.0 | 45.2 |
| I-rep2-0 | 200 ns-UA | Deep inserted | 66.8 ± 1.0 | 39.6 ± 0.3 | 57.1 ± 0.3 |
| I-rep2-1 | 200 ns-UA | Deep inserted-Pb | 57.4 ± 0.7 | 28.0 ± 0.5 | 46.9 ± 0.5 |
| I-rep3 | 1000 ns-CG | Deep inserted | 48.1 | 40.0 | 45.2 |
| I-rep3-0 | 200 ns-UA | Deep inserted | 44.4 ± 0.7 | 41.9 ± 0.8 | 43.5 ± 0.8 |
| I-rep3-1 | 200 ns-UA | Deep inserted | 42.3 ± 1.3 | 68.1 ± 1.3 | 51.5 ± 1.3 |
| I (avg)-CG | 1000 ns-CG | Deep inserted | 52.8 ± 5.1 | 28.3 ± 4.2 | 44.0 ± 2.6 |
| I (avg)-UA | 200 ns-UA | Deep inserted | 54.2 ± 4.9 | 48.8 ± 4.5 | 52.3 ± 3.5 |
| II (initial) | 0 | Inserted | 40.7 | 60.0 | 47.6 |
| II-rep0 | 1000 ns-CG | Deep surface | 85.2 | 100 | 90.5 |
| II-rep0-0 | 200 ns-UA | Deep surface | 59.0 ± 0.6 | 99.3 ± 0.5 | 73.4 ± 0.5 |
| II-rep0-1 | 200 ns-UA | Deep surface | 69.5 ± 1.0 | 98.8 ± 0.7 | 80.1 ± 0.9 |
| II-rep1 | 1000 ns-CG | Deep surface | 44.4 | 100 | 64.3 |
| II-rep1-0 | 200 ns-UA | Deep surface | 41.6 ± 0.6 | 63.3 ± 2.3 | 49.3 ± 1.2 |
| II-rep1-1 | 200 ns-UA | Deep surface | 36.9 ± 0.7 | 52.5 ± 1.9 | 42.5 ± 1.1 |
| II-rep2 | 1000 ns-CG | Deep surface | 63.0 | 100 | 76.2 |
| II-rep2-0 | 200 ns-UA | Deep surface | 62.6 ± 0.9 | 89.3 ± 0.9 | 72.1 ± 0.9 |
| II-rep2-1 | 200 ns-UA | Deep surface | 57.9 ± 1.3 | 100.0 ± 0.1 | 72.9 ± 0.8 |
| II-rep3 | 1000 ns-CG | Deep surface | 48.1 | 66.7 | 54.8 |
| II-rep3-0 | 200 ns-UA | Deep surface | 62.9 ± 0.1 | 55.0 ± 0.5 | 60.1 ± 0.3 |
| II-rep3-1 | 200 ns-UA | Deep surface | 53.3 ± 0.4 | 65.7 ± 1.1 | 57.7 ± 0.6 |
| II (avg)-CG | 1000 ns-CG | Deep surface | 60.2 ± 9.2 | 91.7 ± 8.3 | 71.4 ± 8.5 |
| II (avg)-UA | 200 ns-UA | Deep surface | 55.4 ± 3.9 | 78.0 ± 7.4 | 63.5 ± 4.7 |
Protein orientational substate of Aβ42 with K28 hydrogen-bonded with CHO polar group.
Protein orientational substate of Aβ42 with K28 hydrogen-bonded with PC polar group.
Unfolding % was based on the % of residues in the non-hydrogen-bonded SC structures in the LID (27 residues total), the non-LID (15 residues total) or the whole chain (42 residues total).
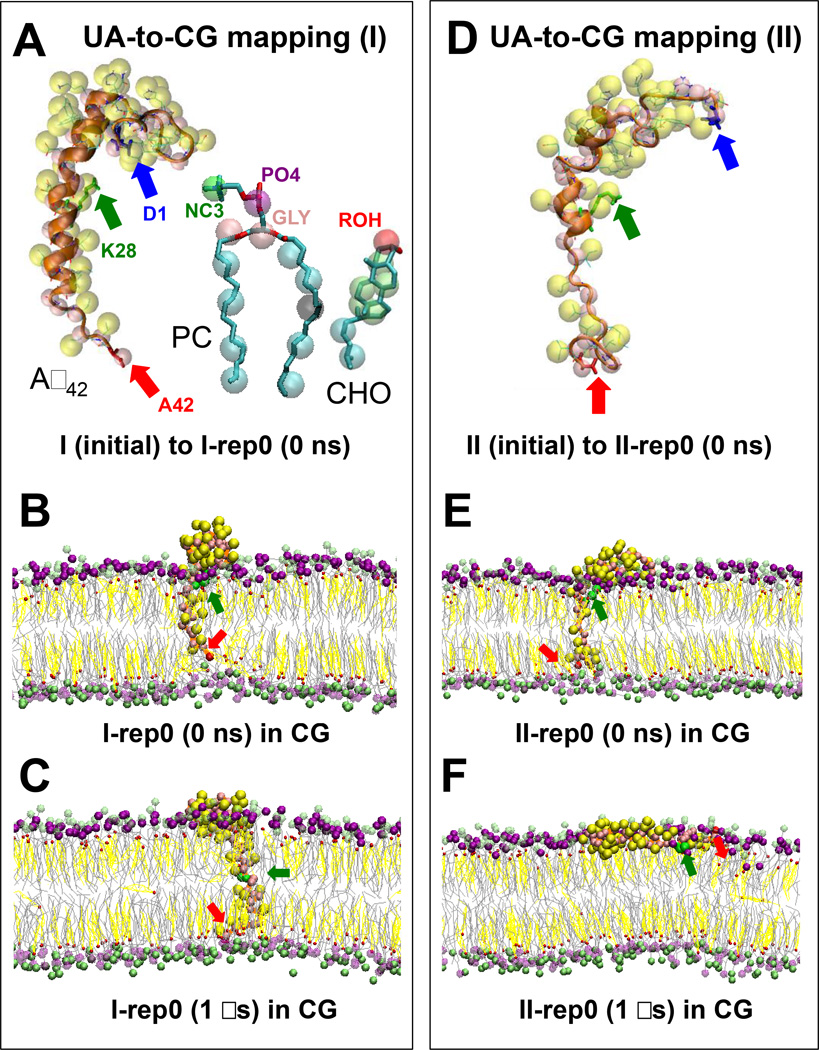
FIG. 1. UA-to-CG mapping and CG simulations of protein/lipid/water/ion complexes.
UA and CG structures of Aβ42 PC and CHO in the I (initial) and II (initial) II (initial) (D) complexes are shown. An orange ribbon (A, D) highlights the secondary structure of the UA protein, whereas yellow and pink spheres represent the side chain and backbone CG beads, respectively, of the CG protein. Blue, green and red arrows mark the residues D1, K28 and A42, respectively, of Aβ42. In CG-PC (A), the polar CG-beads in the headgroup (NC3 in green, PO4 in purple and GLY in pink) and the non-polar beads in the saturated (C16:0) sn-1 chain (4 blue beads) and mono-unsaturated (C18:1) sn-2 chain (four blue and one black beads) are shown. A single black bead represents the C=C of PC. In CG-CHO (A), 1 polar CG bead (ROH in red), 4 green CG beads for the non-polar fused rings and 2 blue beads for the non-polar acyl chain are shown. Representative CG simulation replicates (I-rep0 and II-rep0) before (B, E) and after (C, F) 1 µs CG simulations for the I (B, C) and II (E, F) complexes are shown. The polar NC3 (green) and PO4 (purple) beads of PC and the polar ROH (red) beads of CHO are highlighted, whereas the lines joining the other lipid CG-beads are given by gray (PC) and yellow (CHO) lines. CG water and ions were present in all CG simulations but are not shown.
2. Simulation of CG complexes
A 1000 ns-CG simulation was performed on each of the 4 independent simulation replicates generated from each starting CG complex. As noted in Materials and Methods, each replicate had identical initial coordinates but a different randomly generated initial velocity distribution. Hence, a total of 8 CG replicates were generated: I-rep0, I-rep1, I-rep2 and I-rep3 for the I complex and II-rep0, II-rep1, II-rep2 and II-rep3 for II complex. Table 1 summarizes these 8 CG replicates. For the I complex, the beginning (0 ns) and ending (1000 ns) CG structures of a representative replicate I-rep0 are shown in Figures 1B and 1C, respectively. Similarly, for the II complex, the CG structures of a representative replicate II-rep0 are shown in Figures 1E and 1F, respectively. For clarity, water and ions present in CG simulation are not shown.
3. Creation of reconstructed UA complexes
A CG-to-UA mapping was used to reconstruct the UA structures from the 1000 ns-equilibrated CG structures. These reconstructed UA structures provided the “starting” coordinates for the UA simulation that followed. Successful reconstruction of the UA atoms from the corresponding CG beads for both protein and lipid molecules were observed in all replicates. Figures 2A and 2D illustrate the accuracy of the assignment of the atomic coordinates of protein and lipids and the successful reconstruction of the protein secondary structure from the protein CG beads in representative replicates I-rep0 and II-rep0, respectively.
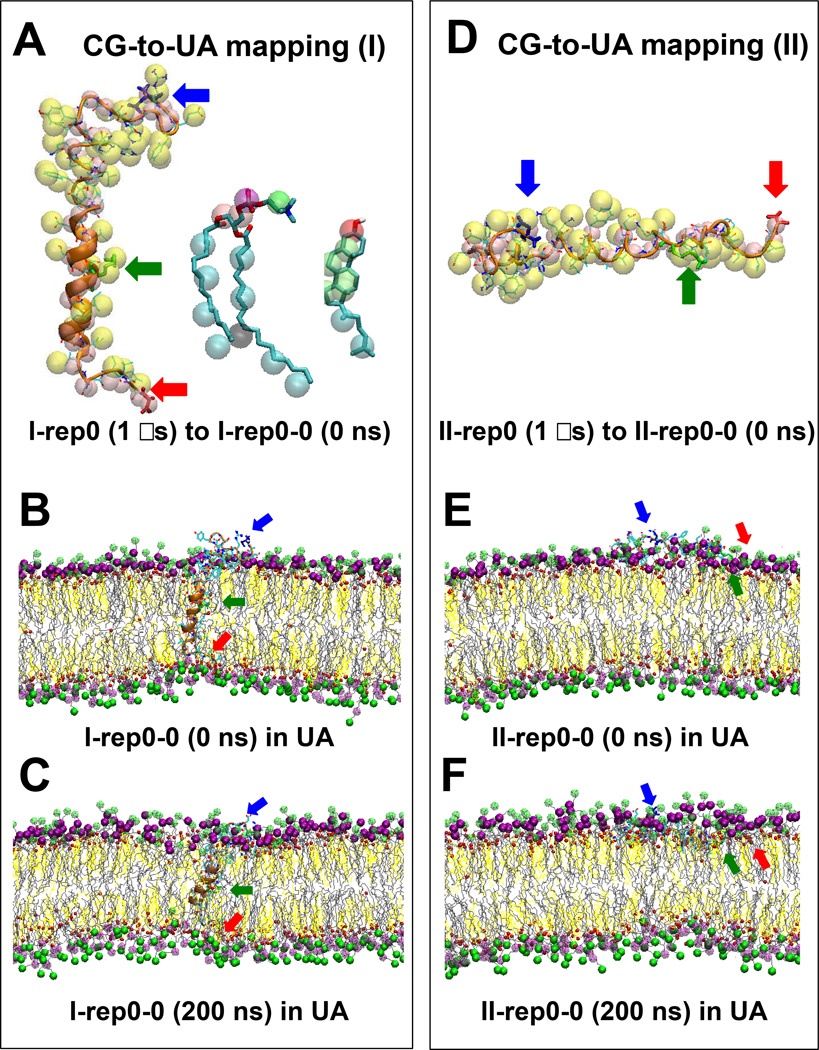
FIG. 2. CG-to-UA mapping and UA simulations of protein/lipid/water/ion complexes.
Overlapping structures of Aβ42, PC and CHO in the representative CG I-rep0 and UA I-rep0-0 replicates (A), and Aβ42 in the representative CG II-rep0 and UA II-rep0-0 replicates (D) using CG-to-UA mapping are shown. Shown are representative UA structures, (I-rep0-0 and II-rep0-0), of two Aβ42/lipid/water/ion complexes before (B, E) and after (C, F) 200 ns UA simulations for the I (B, C) and II (E, F) complexes. Refer to the legends of Figure 1 for details of the atomic group labeling.
4. Relaxation of reconstructed UA complexes
A 200 ns-UA simulation was used to relax each of the 8 reconstructed UA replicates. Here 2 independent replicates were generated for each reconstructed UA replicate. In total, 16 replicates were generated: I-rep0-0, I-rep0–1, I-rep1-0, I-rep1-1, I-rep2-0, I-rep2-1, I-rep3-0 and I-rep3-1 for the I complex and II-rep0-0, II-rep0–1, II-rep1-0, II-rep1-1, II-rep2-0, II-rep2-1, II-rep3-0 and II-rep3-1 for the II complex. Table 1 summarizes these UA simulation replicates. For the I complex, the beginning (0 ns) and ending (1000 ns) UA structures of a representative replicate I-rep0-0 are shown in Figures 2B and 2C, respectively. Similarly, for the II complex, the structures of a representative replicate II-rep0-0 are shown in Figures 2E and 2F, respectively. Again, water and ions present in the UA simulations are not shown for clarity.
B. Exploration of Protein Orientational States
VMD and transbilayer density profile analysis were used to explore protein orientational states from the above CG and UA trajectories.
1. Fully inserted state in starting complexes
By our design rationale, the protein was in a “fully inserted” state in the starting complexes. Figure 3A illustrates the VMD visualization of the D1-N27 (non-LID) and K28-A42 (LID) fragments of Aβ42 of the I (initial) complex before the UA-to-CG mapping. Here the elongated LID segment (orange) of Aβ42 was embedded entirely inside the lipid bilayer with its long helical axis in a transbilayer vertical orientation, parallel to the z-axis or normal to the bilayer plane. The bilayer plane was defined by the polar headgroups of PC. In contrast, most of the globular non-LID segment (yellow) was exposed to water and the polar region of the upper lipid leaflet of the lipid bilayer. Figure 3D shows the average minimum distance from each protein residue to the nearest lipid, CHO or PC, for this complex. Similar protein orientation was found in the II (initial) complex, and also in all 8 CG starting structures after the UA-to-CG mapping.
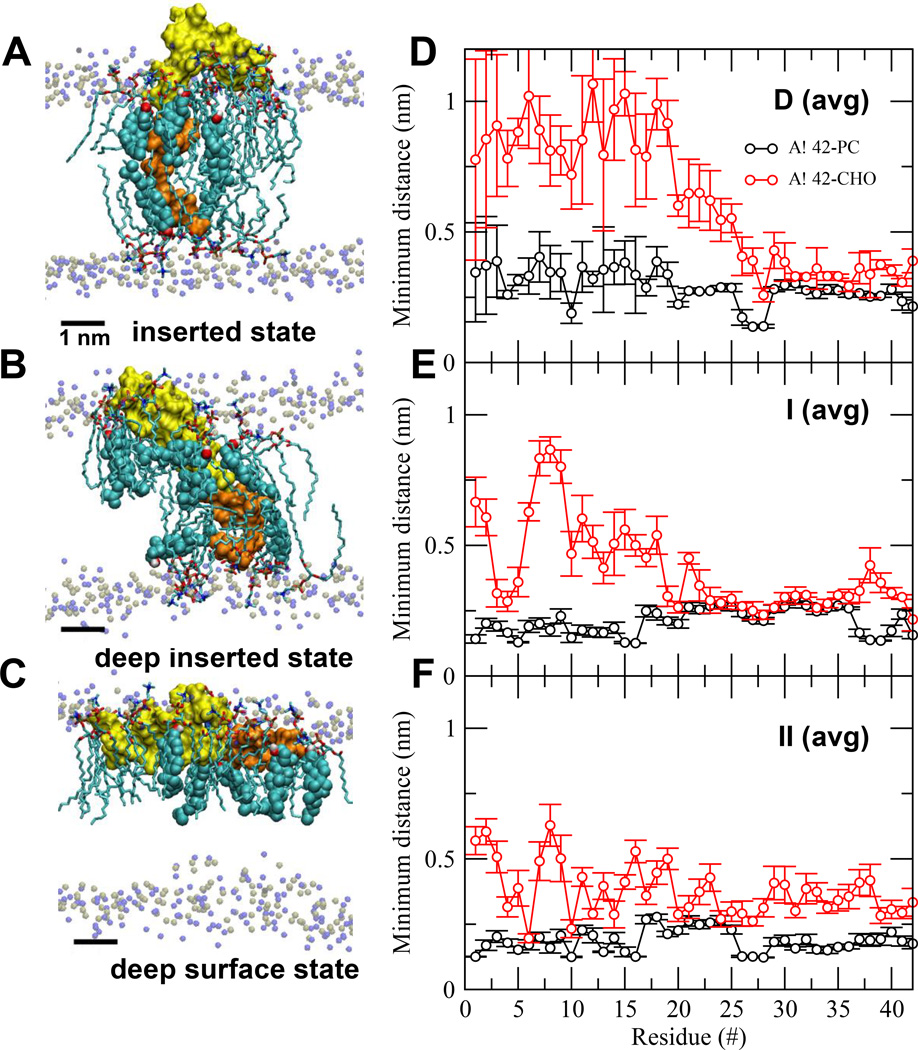
FIG. 3. Protein orientational states and lipid/protein interactions in Aβ42/lipid/water/ion complexes.
UA structures of inserted (A), deep inserted (B) and deep surface (C) states of Aβ42 in Aβ42/lipid/water/ion complexes. D1-N27 or non-LID (yellow) and K28-A42 or LID (orange) segments of the protein are rendered in surface representation. The polar phosphate (silver) and NC3 (blue) of all PC are shown. The annular (AL) lipids, AL-CHO (color spheres) and AL-PC (sticks), are shown. The average (avg) minimum distance between Aβ42 and CHO or PC across all repeated replicates for the last 50 ns as a function of residue position of Aβ42 in D (avg) (panel D), I (avg) (panel E) and II (avg) (panel F) are shown. The bars indicate standard errors. See Materials and Methods for details in averaging.
In addition to VMD-visualization, quantitative transbilayer density profile analyses were performed on the starting structures as shown in Figure 4. Transbilayer density profile provides information of the spatial distribution of molecules across the bilayer normal. Note that the absolute z value of the density profile depends on the height or z-length of the simulation box and the amount of water above and below the lipid bilayer in any simulation system. Therefore only the widths and peak locations of the density distributions are useful for quantitative comparison.
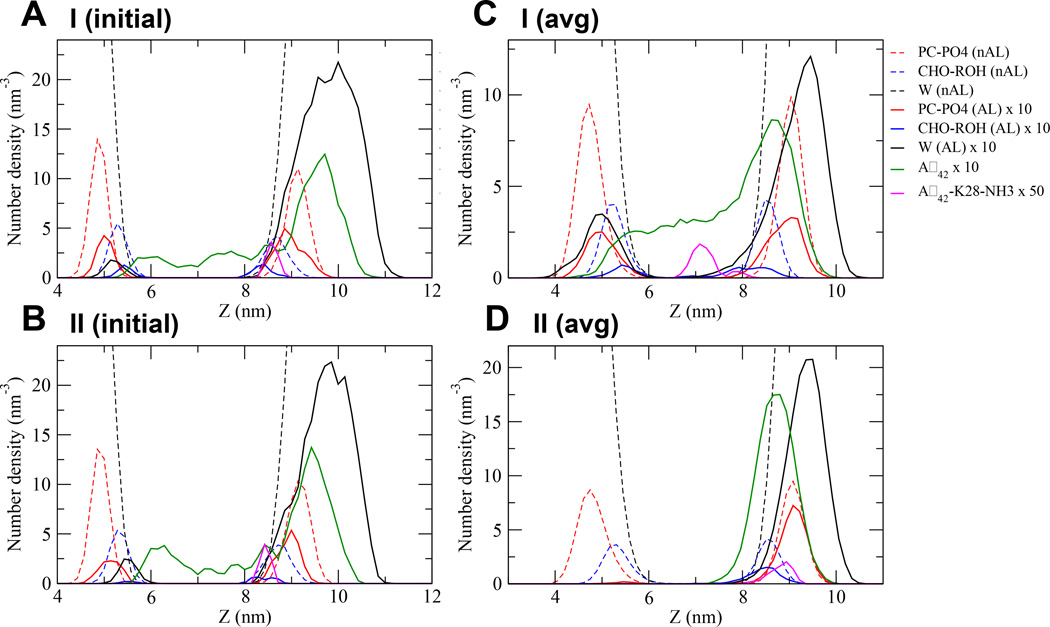
FIG. 4. Transbilayer density profile of molecules in UA Aβ42/lipid/water/ion complexes.
The number density vs. z distance of the I (initial) (A), II (initial) (B), I (avg) (C) and II (avg) (D) complexes are shown. The non-annular (nAL) and annular (AL) lipid and solvent groups are in dashed and solid lines, respectively. The structural groups are PC-PO4 in red, CHO-ROH in blue and solvent or W in black. The entire protein Aβ42 is in green and the K28- NH3 of Aβ42 in pink. Due to the large density differences among AL and nAL molecules, the number densities of protein and all AL molecules are magnified 10 times while K28-NH3 50 times. The average (avg) was across all repeated replicates for the last 50 ns of the I (C) or II (D) complexes. See Materials and Methods for details in averaging.
Figures 4A and 4B illustrate the transbilayer density profile vs. z plots of the protein and lipid polar groups of the I (initial) and II (initial) complexes, respectively. The protein density distribution (green trace) spanned ~ 5 to 10.5 nm along the z-axis and had a width of ~ 5.5 nm in both starting complexes. Note that the density profile peak of K28-NH3, the polar group of the long arm K28 residue, was at ~8.5 nm in both of the I and II complexes. Therefore, the protein distribution from ~8.5 to 10.5 nm with a major peak at ~9.5 nm represented mostly the non-LID distribution of the protein. In comparison, the density distribution of the PC polar groups (red dash trace), represented by PC-PO4 (nAL), had two well-defined peaks of FWHM of ~0.5 nm at ~5 and 9 nm, corresponding to the PC polar group density peak locations of the lower and upper lipid leaflets, respectively. Therefore, a significant positive “peak difference” (= density peak location of protein - density peak location of PC) of ~ 0.5 nm, i.e., 9.5 nm of protein peak - 9.0 nm of PC polar group peak in the upper lipid leaflet, provided evidence that the majority of non-LID was exposed to the aqueous phase. Similar observations were found in all 8 CG starting structures.
2. Deep inserted state for the I Complex
A transmembrane inserted orientation of LID was evident in the I complex after the 1000 ns CG simulation. In addition, both LID and non-LID segments were found to insert deeper towards the center of the lipid bilayer for all 4 replicates (I-rep0, I-rep1, I-rep2 and I-rep3) as illustrated in Figure 1C. After the CG-to-UA mapping and the 200 ns of UA simulations, the transmembrane inserted orientation of the protein and a deeper insertion of both LID and non-LID segments were observed for all replicates as demonstrated in Figure 2C. This new protein orientation is therefore defined as a “deep inserted” state as shown in Table 1.
Figure 3B shows the VMD-visualization of the deep inserted state of a representative replicate (I-rep0-0). It is clear that the non-LID segment descended deeper towards the lipid bilayer center when compared with the starting structure (Figure 3A). Figure 3E shows the average minimum distance from each protein residue to the nearest lipid, CHO or PC for the deep inserted state.
Using the quantitative density profile analysis as shown in Figure 4C, the average transbilayer density profile of the major protein peak was at ~ 8.5 nm relative to the PC peak at ~ 9 nm. The FWHM of either peak was similar to that of the starting structure. Here, a significant negative “peak difference” of ~ −0.5 nm, i.e., 8.5 nm − 9 nm, was found. Therefore, a net ~ −1 nm shift of the protein peak was deducted from the observed change of the “peak difference” from ~ +0.5 to −0.5 nm before and after the multiscale simulations, respectively. Examinations of the transbilayer density profiles of each of the 8 UA starting structures revealed a similar ~ −1 nm negative shift of protein peak (see Figs. S3–S4 of Supporting Information).
3. Deep surface state in the II complex
The inserted orientation of the II complex LID segment moved drastically to a horizontal orientation, after the 1000 ns-CG simulation; the long axis of the peptide backbone was parallel to the bilayer plane, as shown in Figure 1F. Similar to the I complex, the non-LID segment also descended towards the center of the lipid bilayer. This behavior was observed in all 4 replicates (II-rep0, II-rep1, II-rep2 and II-rep3) of the II complex, as demonstrated in Figure 1F. After the CG-to-UA mapping and the 200 ns-UA simulations, the horizontal surface orientation of LID and the insertion of the non-LID segments remained for all 8 replicates, as demonstrated in Figure 2F. This protein orientation is defined as a “deep surface” state as shown in Table 1.
Figure 3C shows the VMD-visualization of the deep surface state of a representative replicate (II-rep0-0). Again, the non-LID descended towards the lipid bilayer center (Figure 3A). Figure 3E shows the average minimum distance from each protein residue to the nearest lipid, CHO or PC for the deep surface state.
Using the quantitative transbilayer density profile analysis as shown in Figure 4D, the average density profile of the major protein peak was also at ~ 8.5 nm relative to the PC peak at ~ 9 nm. Therefore, a similar net ~ −1 nm shift of the protein peak was deducted after the multiscale simulation. Examinations of the transbilayer density profiles of each of the 8 UA starting structures revealed similar observations of the negative shift of protein peak (see Figs. S5–S6 of Supporting Information).
C. Protein Orientational State Transitions
Examination of the starting structures of the I and II complexes revealed that the side chain of K28 of Aβ42 were attached to the polar group of the upper lipid leaflet in the CG (Figures 1B and 1E) representation. This K28-lipid attachment is defined as “K28-snorkeling”. In addition, the C-terminus (A42) of Aβ42 was attached to the polar groups of the lower lipid monolayer in the CG (Figures 1B and 1E) representation. This A42-lipid attachment is defined as “A42-anchoring”.
After the CG 1000 ns simulations, K28-snorkeling was disrupted, as evidenced by the descent of K28 towards the center of the bilayer for all 4 replicates in the I complex (Figure 1C). On the other hand, in the II complex, A42-anchoring was disrupted as evidenced by the ascent of A42 from the lower lipid monolayer to the upper lipid monolayer (Figures 1F). Therefore, disruptions of K28-snorkeling and A42-anchoring signified the protein orientational state transitions in the I and II complexes, respectively.
We have calculated the minimum distances between the positively charged K28 and the negatively charged PO4 group of PC (K28-PO4) and between the negatively charged A42 and the positively charged NC3 of PC (A42-NC3) in both upper and lower lipid leaflets as a function of simulation time. These K28-PO4 and A42-NC3 vs. time plots, Figure 5 and 6, quantify the protein orientational state transition.
FIG. 5. Protein orientational transition in CG Aβ42/lipid/water/ion complexes.
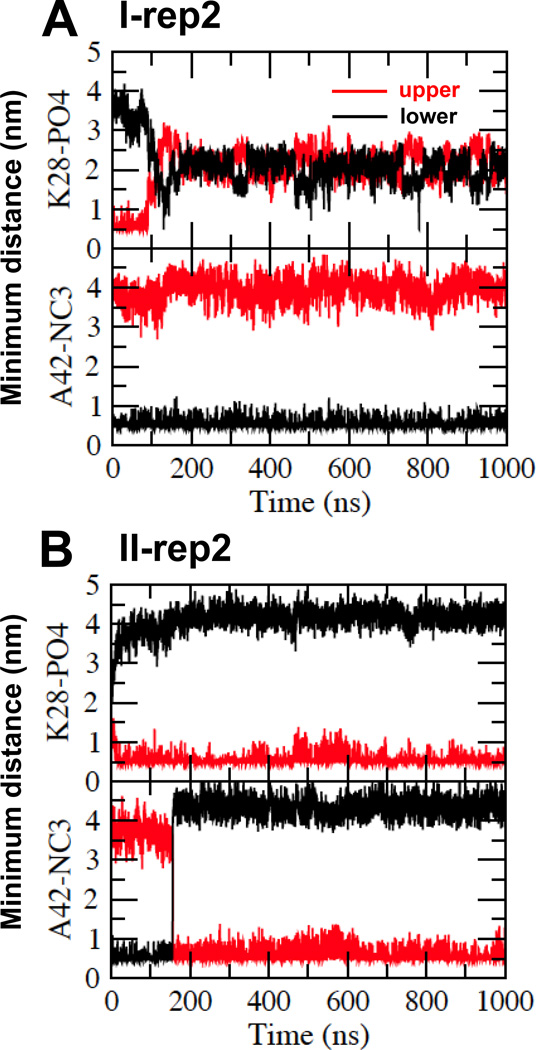
Time evolution of the minimum distance between K28 of Aβ42 and PO4 of PC (K28-PO4) and between A42 of Aβ42 and NC3 of PC (A42-NC3) of representative replicates I-rep2 (A) and II-rep2 (B) as a function of CG simulation time for the upper (red) and lower (black) lipid monolayers are shown.
FIG. 6. Protein orientational transition in UA Aβ42/lipid/water/ion complexes.
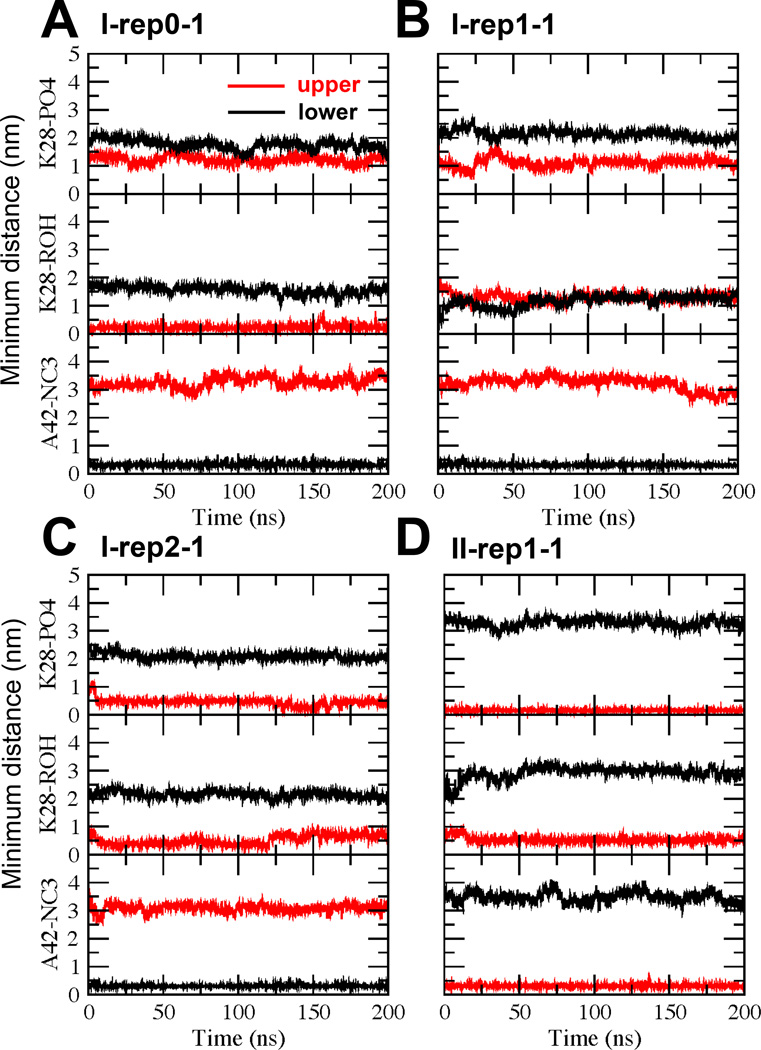
Time evolution of the minimum distanc between K28 of Aβ42 and PO4 of PC (K28-PO4), between K28 of Aβ42 and ROH of CHO (K28-ROH) and between A42 of Aβ42 and NC3 of PC (A42-NC3) of representative replicates I-rep0-1 (A), I-rep1-1 (B), I-rep2-1 (C) and II-rep1-1 (D) as a function of UA simulation time for the upper (red) and lower (black) lipid monolayers.
1. “Inserted” to “deep inserted” state transition in the I complex
For the CG I complex, Figure 5A (upper panel) shows the K28-PO4 distance vs. time plots of a representative replicate (I-rep2) from 0 to 1000 ns with a time-resolution of 0.1 ns. Here, the K28-PO4 distances were approximately 0.7 and 3.5 nm for the upper and lower lipid leaflets, respectively, at 0 ns. At ~ 90 ns, an abrupt increase was seen for the upper monolayer and a concurrent reduction in the lower monolayer K28-PO4 distance. The transition took ~ 30 ns, and from ~ 130–1000 ns, the distances remained stable at ~ 2 nm. Figure 5A (lower panel) shows the representative A42-NC3 distance vs. time plots of the same replicate in both monolayers. The A42-NC3 distance started at approximately 4 and 0.5 nm for the upper and lower lipid monolayers, respectively, at 0 ns. These values remained stable throughout the entire 1000 ns simulation period. Similar observations of K28-PO4 transition and stable A42-NC3 for other three representatives, i.e., I-rep0, I-rep1 and I-rep3 were detected (see Fig. S7 of Supporting Information).
To detect the stability of K28-PO4 and A42-NC3 over time in the relaxed UA representation, similar K28-PO4 and A42-NC3 distance vs. time plots were generated as illustrated in Figure 6. We observed that A42-NC3 maintained a low value of ~ 0.25 nm for the lower leaflet but a high value of ~3 nm for the upper leaflet in all 8 UA replicates indicating the preservation of the A42 anchoring in UA representation. Interestingly, the K28-PO4 distance remained at 1–2 nm for the upper and lower leaflets for 7 out of 8 replicates indicating that K28 snorkeling was disrupted in most replicates. The outlier replicate was I-rep2-1 in which K28-PO4 was closer, 0.5 nm, as illustrated in Figure 6C (middle panel).
Since detachment of K28 with the PC polar group was found in most UA replicates, we also explored the possible “attachment” of K28 with the polar groups of CHO located below the polar PO4 groups of PC (see Figure 4). Interestingly, 2 replicates, I-rep-0-0 and I-rep0–1, of 8 replicates demonstrated attachment of K28 with ROH of CHO as demonstrated in Figure 6A (middle panel), where K28-ROH was 0.25 nm for the upper layer throughout the 200 ns UA simulation in the replicate I-rep0–1. A representative replicate (I-rep1-1) that shows no K28 attachment to PC-PO4 or CHO-ROH is also illustrated in Figure 6B (upper and middle panels). Plots of the other replicates are given in Fig. S8 of Supporting Information.
2. “Inserted” to “deep surface” state transition in the II Complex
For the CG II complex, Figure 5B (upper panel) shows the representative K28-PO4 distance vs. time plots of a representative replicate (II-rep2) in both lipid monolayers. Here, the K28-PO4 distance started at ~1.5 and 2.5 nm for the upper and lower lipid monolayers, respectively, at 0 ns. The values reached steady state of 0.5 and 4 nm, respectively, within the 1000 ns simulation time. Figure 5B (lower panel) shows the representative A42-NC3 distance vs. time plots of the same replicate in both monolayers. Here the A42-NC3 distance started at ~3.5 and 0.5 nm for the upper and lower lipid monolayers, respectively, at 0 ns. At ~150 ns, the K28-NC3 distance decreased abruptly in the upper monolayer and increased in the lower monolayer. The transition was rapid and completed in ~ 1 ns. After the transition, K28-PO4 maintained the steady state values of ~0.5 and 4.5 nm for the upper and lower monolayers throughout the entire 1000 ns simulation. Similar observations were found for the remaining replicates, although the time of transition differed: 300 ns for II-rep0, 220 ns for II-rep1 and 550 ns for II-rep3 (see Fig. S7 of Supporting Information).
To detect the stability of K28-PO4 and A42-NC3 distance over time in the UA representation, similar K28-PO4 and A42-NC3 distance vs. time plots are illustrated in Figure 6. We observed that both K28-PO4 and A42-NC3 distance remains low ~0.25 nm for the upper layer but high of ~3 nm for the lower layer, as illustrated in a representative replicate II-rep1-1 (Figure 6D). Similar observations were found for other replicates.
Similar to the I complex, we also explored possible “attachment” of K28 with the polar groups of CHO. K28-ROH maintained a low value of ~ 0.5 nm in all replicates as illustrated in Figure 6D (middle panel), indicating close interactions between ROH and K28 in the II complex. Plots of all replicates are given in Fig. S8 of Supporting Information.
D. Characterizations of Protein Conformations in Protein Orientational States
The secondary structures of Aβ42 in the I and II complexes before and after the CG simulation were examined by DSSP. The secondary structure as a function of UA simulation time (0 to 200 ns) of each of the 16 reconstructed UA replicates of I and II complexes is given in Fig. S9 of Supporting Information. A summary of the average reduced secondary structures (GHI, BE, T and SC) of each replicate is also presented in Figs. S10 and S11 of Supporting Information. Table 1 summarizes the time-averaged unfolding % or SC % of the protein in both the non-LID and the LID of each replicate over the last 50 ns of the relaxed 200 ns - UA simulations. The unfolding % before the UA simulation relaxation, i.e., after the 1000 ns-CG simulation and the CG-UA mapping, are also shown for comparison. Also shown in Table 1 is the average unfolding % (highlighted in bold) across all replicates of each I or II complex.
The I complex was initially 52% unfolded in the non-LID and 27% unfolded in the LID before the multi-resolution MD simulations. Before the UA simulation relaxation, a diverse range of unfolding of 41 – 63% with an average of 53% in the non-LID but of 20 – 40 % with an average of 28% in the LID was detected among 4 replicates, indicating no significant change in the average unfolding in both domains. After the 200 ns-UA simulation, the level of unfolding relaxed to a range of 32 – 73% with an average of 54% in the non-LID but 28 −68% with an average of 49% in the LID among 8 replicates, indicating a significant increase in the average unfolding of the LID.
The II complex was initially 41% unfolded in the non-LID and 60% unfolded in the LID before the multi-resolution MD simulations. Before the UA simulation relaxation, a range of unfolding of 44 – 85% with an average of 60% in the non-LID and 67 – 100% with an average of 92% in the LID was observed among 4 replicates, indicating significant unfolding in both domains. After the 200 ns-UA simulations, the levels of unfolding relaxed to a range of 37 – 70% with an average of 55% in the non-LID and 53 – 100% with an average of 78% in the LID among 8 replicates, indicating a significant increase in the average unfolding of both non-LID and LID domains.
At the whole chain level, the protein unfolding started at 43% and 53% in the I and II complexes. The corresponding levels became 44%and 71% before the UA simulation relaxation, and finally relaxed to 52 and 64% after the 200 ns-UA simulations. The results indicate a modest 20% increase in the total unfolding for both complexes after the multi-resolution simulations. We noticed a general trend of a decrease in the helix (GHI) and an increase in the turn (T) structures of the protein after the membrane orientational transition of the I or II complex. In addition, beta (BE) structures were evident in several replicates, particularly in the LID, of the II complex as shown in Figs. S9–S11 of Supporting Information.
E. Hydrogen Bonding Profile of K28
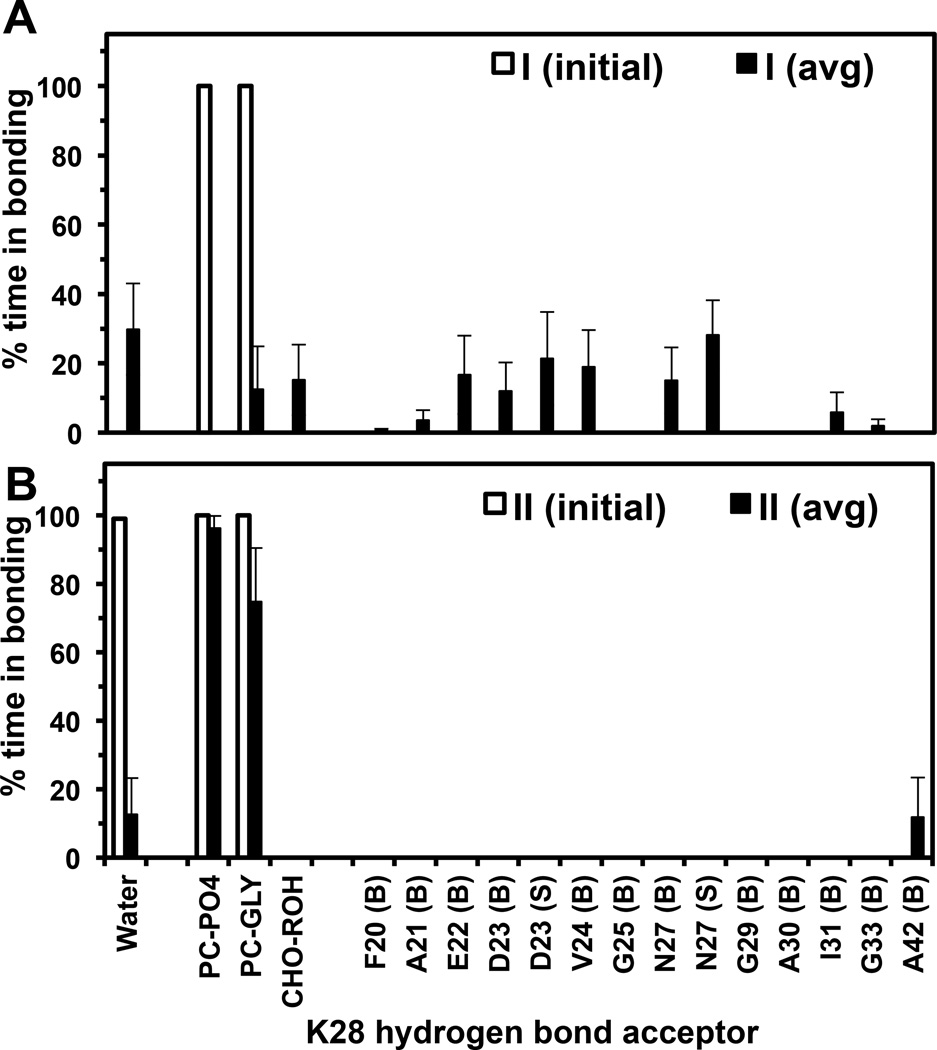
For the I complex, the K28 hydrogen bonding profile of each replicate was determined. In agreement with the minimum distance calculations, I-rep0-0 and I-rep0–1 exhibited K28-ROH hydrogen bonding and I-rep2-1 K28-GLY hydrogen bonding. The other replicates showed only intra-protein hydrogen bonding between K28 and other nearby protein residues. In some cases, K28-solvent hydrogen bonding was also detected. Figure 7A shows the combined K28-hydrogen profile of all the 8 replicates. Individual K28-hydrogen profiles of all replicates are given in Fig. S12 of Supporting Information. Because of its association with PC in K28-hydrogen bonding, I-rep2-1-1 is further identified as “deep inserted-P” state. Replicates I-rep0-0 and I-rep0–1 are denoted “deep inserted-C” state indicating the bonding of K28 with and CHO, respectively. The classification of three sub-states in the I complex is summarized in Table 1. The other replicates, i.e., I-rep1-0, I-rep1-1, I-rep2-0, I-rep3-0 and I-rep3-1, that exhibited no K28-lipid hydrogen bonding are defined as simply “deep inserted” state.
FIG. 7. K28 hydrogen bonding profile in UA Aβ42/lipid/water/ion complexes.
The percentage of time that the NH3 polar group of the side chain of K28 (donor) formed a hydrogen bond with an acceptor group over the last 50 ns of the 200 ns UA simulations of I (initial), II (initial), I (avg) and II (avg) are given. The average (avg) was across all repeated replicates of I or II complex. See Materials and Methods for details in averaging.
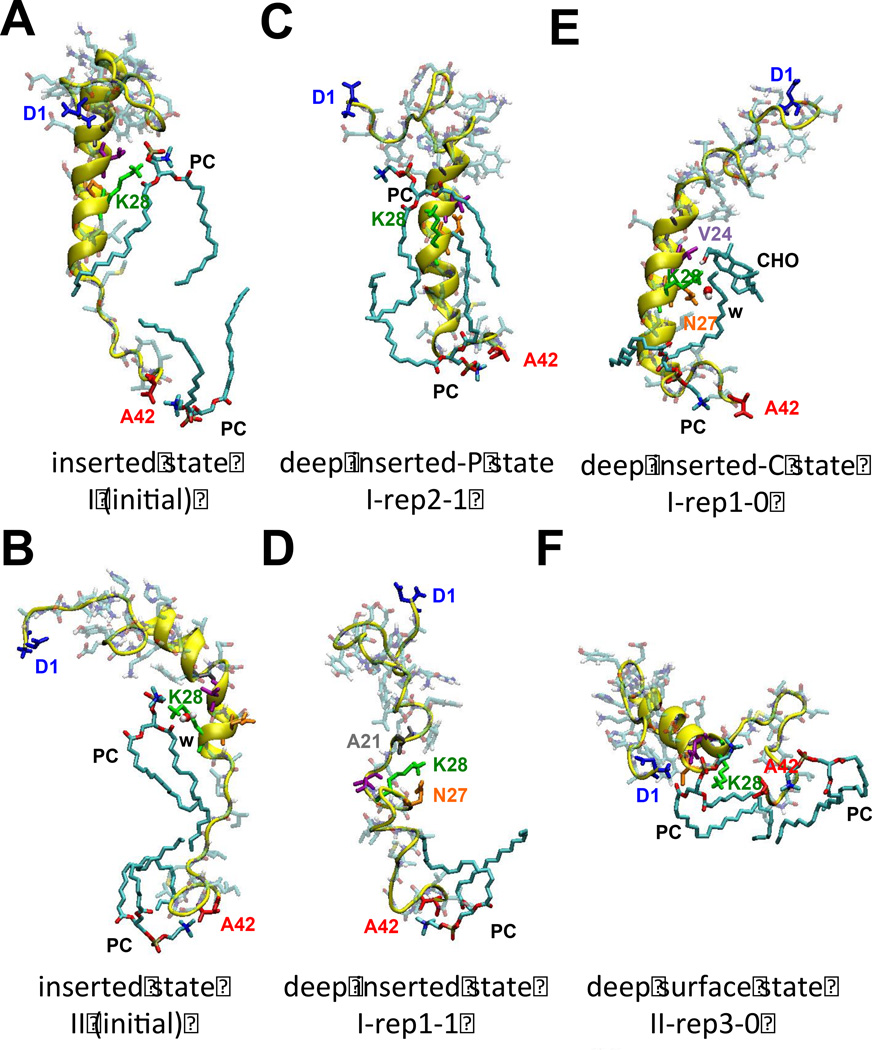
Figure 8 demonstrates the hydrogen bonding partners of K28 with protein residues and nearby lipids. The hydrogen bonding partners of A42 with nearby lipids are also shown for comparison. Note that K28-snorkeling and A42-anchoring with PC were evident in the I (initial) and II (initial) states as shown in Figures 8A and 8B, respectively. Figures 8C and E illustrate the conformation and orientation of K28 and the hydrogen bond acceptors from PC and CHO, of I-rep2-1 and I-rep1-0, respectively. The K28 hydrogen bond partner for I-rep2-1 was only PC, but for replicate I-rep1-0 it formed hydrogen bonds with CHO and other protein residues, N27 and V24, and water.
FIG. 8. K28 and A42 hydrogen bonding partners in UA Aβ42/lipid/water/ion complexes.
Representative hydrogen bonding partners of K28 and A42 of Aβ42 with the hydrogen bonding partners, water (w), polar groups of lipids (PC and CHO), and protein residues (A21, V24, N27) are shown for I (initial) (A) and II (initial) (B) and representative replicates: I-rep2-1 (C), I-rep1-1 (D), I-rep1-0 (E) and II-rep3-0 (F), are shown. The protein orientational state of each structure is identified.
Since K28 snorkeling was preserved in all replicates of the II complex, the hydrogen-bonding profile of all replicates of the II complex was also determined as controls. For the II complex (Figure 7B), K28 formed hydrogen bonds with the PO4 and/or GLY group of PC in all replicates in agreement with minimum distance analysis. Only, one replicate (II-rep3-1) showed an extra K28-A42 hydrogen bond (See Figure S12 of Supporting Information). Figure 8F illustrates the K28 hydrogen bonding lipid partners in a representative replicate II-rep3-0.
F. Protein-Induced Membrane Disruptions
The order parameters of PC acyl chains and bilayer thickness map were determined to assess the protein-induced membrane disruptions of the lipid bilayer.
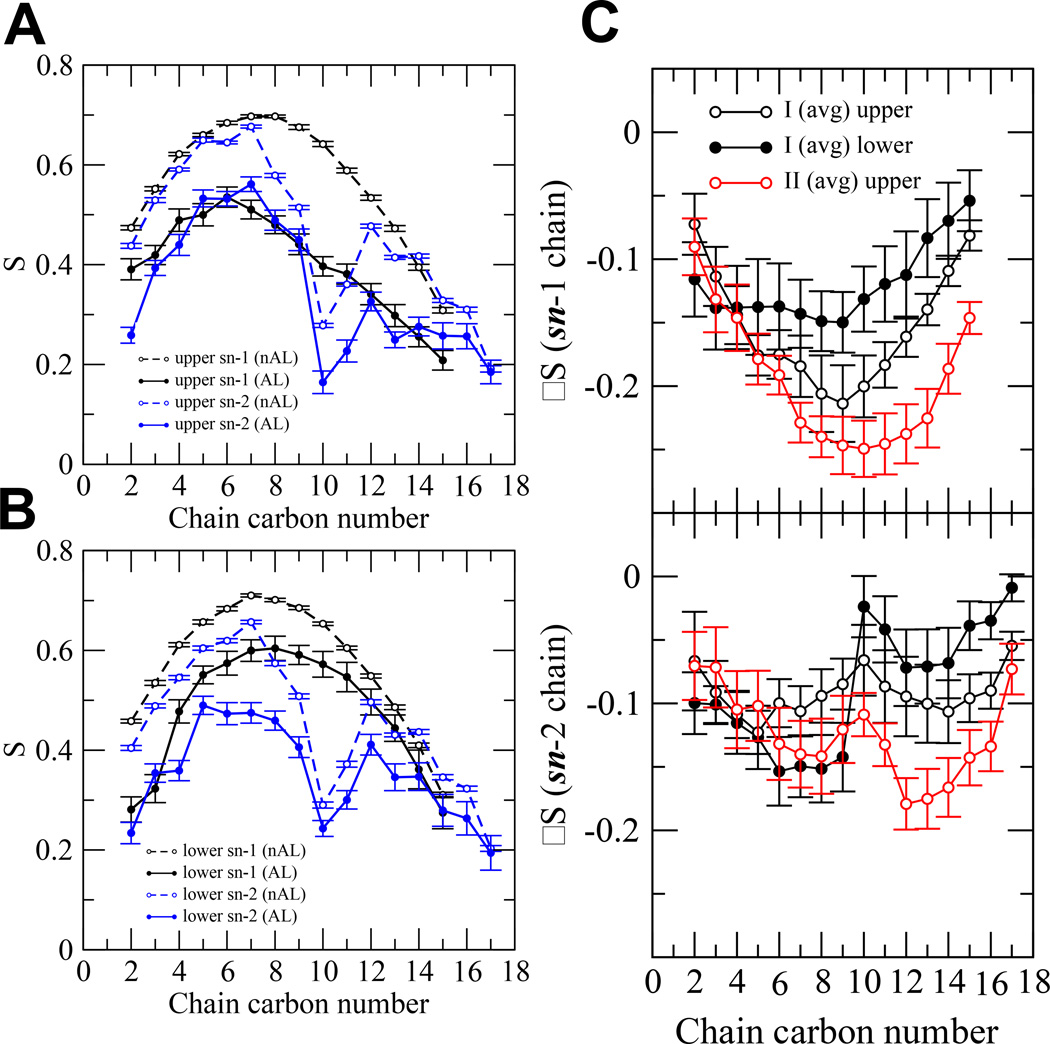
1. Order parameter of PC
The time-averaged order parameters for both the sn-1 and sn-2 chains by carbon number in the AL and nAL regions of both complexes were calculated. Results of all 16 replicates are shown in Figures S13–15 of Supporting Information. The order parameters for a representative replicate I-rep0-0 are shown Figures 9A and B for each carbon number in both acyl chains. It is clear that the order parameters in the AL region were significantly lower than those in the nAL region. The difference of order parameters ΔS (= order parameter of AL – order parameter of nAL) was calculated to evaluate the extent of disruption in the AL compared to the nAL region. As can be seen in Figure 3C, there were no AL lipids in the II complex since the protein was only in the upper lipid leaflet. Hence no ΔS in the lower lipid leaflet was determined.
FIG. 9. Order parameters of PC chains in UA Aβ42/lipid/water/ion complexes.
Order parameters (S) of sn-1 (black) and sn-2 (blue) chains of a representative replicate I-rep-0-0 as a function of chain carbon number in the upper (A) and lower (B) monolayers. Both the annular (AL) (filled circle) and non-annular (nAL) (open circle) lipids are shown. The means and standard errors (error bars) over the last 50 ns of the simulation are shown. Order parameter difference (ΔS = S of AL – S of nAL) of the sn-1 (C) and the sn-2 (D) chains in the upper (black open circle) and lower (black filled circle) monolayers of I (avg) complex and that in the open (red open circle) of II (avg) complex are shown. The calculations were over the last 50 ns of the simulations with the mean and standard errors obtained from the 8 independent replicates of I or II complex.
Figure 9C shows that the average ΔS was more negative for the II complex than for the I complex in the upper layer for both the sn-1 and sn-2. This indicates that the protein in the surface state (II complex) disrupted the acyl chain order more than the protein in the deeper inserted state (I complex) did. The effect was stronger for high carbon number (C10 to C15) of the sn-1 chain but near the middle (C12–14) of the sn-2 chain. Also, the chains in the upper layer were more disrupted by the protein than those in the lower layer in the I complex.
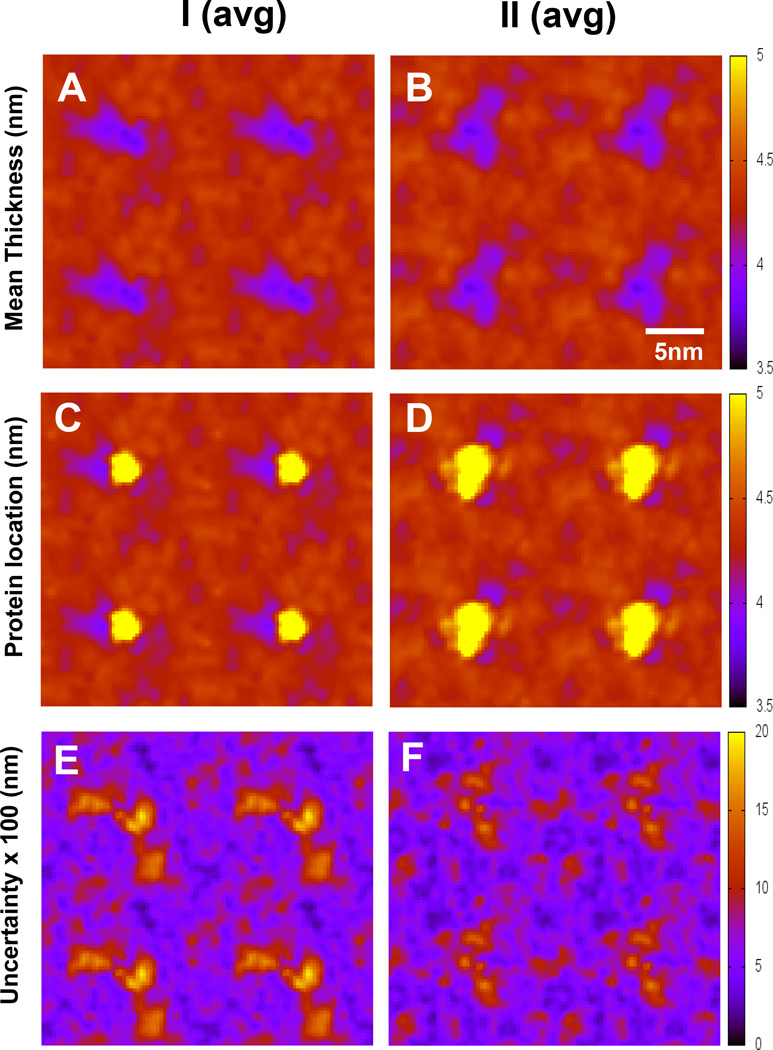
2. Bilayer thickness maps
Figure 10 shows the average bilayer thickness maps over all eight replicates of each I or II complex. The thickness maps indicate that the protein induced similar reduction (~ 1 nm) of the thickness near the vicinity of the protein in the I and II complexes. The protein lateral mobility was lower in the inserted state than in the surface state as evidenced in the protein location map. Also, the variation of the thickness among 8 replicates was larger in the I complex than in the II complex. Time-average bilayer thickness and protein location maps of all replicates are shown in Figures S16–S19 of Supporting Information.
FIG. 10. Lipid bilayer thickness maps of UA Aβ42/lipid/water/ion complexes.
Lipid bilayer thickness maps of I (avg) and II (avg) complexes without (A, B) and with overlays (C, D) of protein locations (yellow) are shown. The uncertainty maps (E, F) of thickness based on the standard errors of the means are also given. The calculations were over the last 50 ns of the simulations with means and standard errors obtained from 8 independent replicates.
IV. DISCUSSION
A multi-resolution simulation approach has been employed to enhance the sampling of protein orientations and to characterize the protein conformations and protein/interaction behavior of a model membrane-active protein in a PC/CHO lipid matrix. Our computational strategy was to sample and characterize new protein orientational states using sequential steps of transformations and simulations.
Starting from a UA structure with a folded (I) or unfolded (II) LID of the protein, a UA-to-CG forward transformation and a long 1000 ns CG simulation allowed sampling protein orientational states of the I or II complex in the CG representation using multiple replicates. With this CG approach, two new protein orientational states: deep inserted and deep surface were discovered in the I and II complexes, respectively. These new CG orientational states were transformed by a CG-to-UA backward transformation followed by a 200 ns UA simulation on multiple replicates. The final UA computational step successfully relaxed the UA reconstructed protein in each replicate. The 200 ns UA simulation step was sufficiently long to sample different protein secondary structures in both non-LID and LID domains and to assess protein/lipid interactions at high spatial resolutions. Based on the K28-hydrogen bonding profile, two new sub-states of the deep inserted state existed: deep inserted-P and deep-inserted-C. These were defined by whether K28-snorkeling targeted PC or CHO.
Hydrogen bonding is an important driving force in driving the membrane orientational transition of the protein. Low levels of hydrogen bonding can stabilize the configuration of membrane-active proteins, since the dielectric constant of the membrane is low. For the deep inserted state with no K28 snorkeling with lipid, the residue K28 no longer fixes the LID to the interface but rather contributes to stabilizing the relatively disordered state of the peptide by hydrogen bonding with several amino acids from what was the non-LID (see Figures 7 and 8) and helps stabilize the compact cytofacial structure of the non-LID. Note that our previous studies [13, 14] on Aβ42 in a PC/CHO bilayer revealed only a fully inserted transition based on the UA simulation alone. This multi-resolution simulation work therefore provides an extension of our previous studies on protein orientational transition of Aβ42 in a PC/CHO bilayer.
Protein orientational states in the Aβ42/lipid/water/ion complex are separated by free energy barriers [55, 56]. If the free energy barriers are too high, identifying or sampling the different orientational states is difficult. Conventional MD simulations are usually biased by the starting structures, limited number of independent simulation replicates, and available computing resources [57, 58]. A number of temperature-based sampling techniques, such as replicate-exchange molecular dynamics (REMD) method [59–61], are commonly used. In the present work, we improve sampling through a CG-reverse mapping approach. The CG representation of Aβ42 is primarily based on the backbone of the peptide and the mapping back to the UA structure through simulated annealing [18] is not unique. Many different reconstructed UA structures of the protein are possible (see Table 1) when it is relaxed in a complex lipid/water/ion environment. A similar reverse mapping procedure has been applied to other amyloid systems [62].
Our previous unconstrained 200 ns-atomistic MD simulations of the same Aβ/PC/CHO/water/ion complex with a partially inserted starting structure revealed “partially inserted”-to-”fully inserted” transition of Aβ42 in all four independent replicates [13]. Depending on the degree of unfolding of the LID, fast and slow relaxation kinetics were detected during the first 50 ns of the simulations [13]. However, studies of model membrane-active peptides [63] have indicated that unfolding of a transbilayer helix structure is enthalpically unfavorable with an energy cost of ~ 4–6 kcal/mol or lower due to a disruption of hydrogen bonding of the peptide backbone in a low dielectric membrane environment. Although, the enthalpy cost could be compensated by the entropy increase from protein unfolding and arrangement of lipid around the disordered protein region ranging from ~ 1.25 to > 6.25 kcal/mol per peptide bond [63, 64]. In addition, the A42-anchoring and K28-snorkeling provide the dual-attachment mechanism for the stabilization of the fully inserted state. However, the important question of the membrane stability for the fully inserted states involving the enthalpically favorable LID folded and enthalpically unfavorable unfolded LID remained unanswered by unconstrained atomistic MD simulation approach alone. The reduced dimensionality of the CG space and the CG forces among molecules ensure that simulation is less likely to be trapped in a local energy minimum [19]. In the CG space, the deep surface state was found in 100 ns or less, and the deep surface state in 550 ns or less (see Supporting Information). Neither of these states was seen in previous multiple-replicate 200 ns UA simulations [14].
Various protein conformations or levels of unfolding were present in the protein orientational states before and after the UA simulation relaxation. In the I complex, no significant change in the average protein unfolding in either LID or non-LID before the UA simulation relaxation, i.e., after the CG simulations and the reverse CG-to-UA mapping, was evident. However, unfolding in the LID over a wide range occurred after the UA simulation relaxation. This result suggests that the unfolding of LID in the deep inserted state was mainly attributed to the interactions of the LID with the lipid matrix during the UA relaxation similar to the observation of the partially inserted -to- fully inserted transition found in earlier studies [14, 65]. In the II complex, significant changes in the average protein unfolding in the LID was found before and after the UA simulation relaxation. This may be explained by the unfavorable interactions of the hydrophobic LID with the relatively polar environment of the polar regions of the lipid bilayer in the deep surface state. This is in contrast to the hydrophobic environment of the LID in the deep inserted state in the I complex.
The choice of force field affects the conformational and orientational behavior of the protein in a lipid matrix. In this multi-resolution MD study, both CG and UA force fields were used. For the CG force field, the non-polarizable Martini FF was used. Recently, the role of polarizability of water in CG simulations of protein/lipid interactions has been raised [66, 67]. In the MARTINI 2.1 and earlier force field [19, 36, 37], four water molecules were represented by a single non-polarizable CG particle. Whether or not the polarizability of water affected the membrane-bound orientations of Aβ42 was examined. We have performed several 1000 ns-CG simulations on the same I (initial) and II (initial) systems using the latest MARTINI 2.2P force field [67] with polarizable water and some adjustments of protein residue interactions. Interestingly, we observed identical transitional behavior from the inserted to the deep inserted state for the I (initial) complex I and from the inserted to the deep surface state for the II (initial) complex (results not shown). Therefore, the protein orientation transition behavior of Aβ42 in PC/CHO was not significantly influenced by the polarizability of water or by better-tuned protein parameters under the MARTINI 2.2P force field [67]. The observed transition from the inserted to the deep surface state is also consistent with the thermodynamics argument that unfolding in the LID is energetically unfavorable [63, 68], and therefore drives the transition towards the surface state.
For the UA simulations the GROMOS87/Berger lipids were used. It should be noted that the GROMOS force field has undergone several revisions since the version employed in this study was released. Kukol [69] found that the secondary structure and integrity of a membrane-active protein was similar for both the GROMOS87/Berger force field and the GROMOS96 53A6 field, and the protein in the GROMOS87/Berger field exhibited greater root mean square deviation, suggesting the structure was less stable. Tieleman et al. [70] found that the strength of lipid-protein interaction was overestimated by the combination of the modified GROMOS87/Berger field. The versions and evolution of the GROMOS force field and its uses have been discussed [69–72]. Therefore, lipid-peptide interactions in this study may be stronger than those in studies using other force fields, leading to a relative increase of unfolded states. This raises the question of how these final protein configurations might be affected. To answer, it should be noted that the deep-inserted and deep surface states occurred during the GG Martini field phase of the simulation so they were not the result of GROMOS87/Berger parameters. The small change in the K28/A42 position (see Figure 6) during the UA simulation suggests stable protein configuration without excessive lipid interaction. In addition, the primary interaction that occurs, snorkeling, has been detected in studies using the GROMOS96 53A6 force field [30, 73]. Therefore, we conclude that there are no appreciable force-field artifacts in the lipid/protein interactions. Since we used the GROMOS87/Berger force field in our previous work with the partially-inserted to fully-inserted transition in PC and PC/CHO, the same force field was used here to complete the work on monomeric protein transition behavior.
With increased recognition that high cholesterol is an important, but poorly understood risk factor for Alzheimer's disease [74–76], there has been a corresponding interest in CHO/Aβ42 interactions. Recent biochemical [77] and molecular docking [78] studies have suggested the existence of a linear cholesterol-binding domain (E22-M35). Note that this cholesterol-binding domain includes K28 and two GXXXG motifs, G25-G29 and G29-G33. K28 [79] has been identified as a key residue involved in downstream self-aggregation pathway and cell toxicity and GXXXG [80, 81] is important in helix-helix association in the lipid membrane. Our finding that the non-LID (1-N27) descended ~ 1 nm toward the center of the bilayer (see Figure 3) to form the deep inserted state allows efficient CHO binding with E22-M35. The minimum distance between the residues of Aβ42 and the lipid for each state (inserted (I (initial), I deep inserted and II deep surface) are shown in Figure 3. It is clear that CHO molecules achieve a minimum distance of 0.25 nm within the linear sub-segments A30-V36 for the I (initial) state, but with E22-M35 and V24-N27 for the I and II states, respectively. Therefore, our results establish that the deep inserted state makes more of the cholesterol-binding domain available for cholesterol to interactions. The presence of three protein orientational states further suggests a molecular “switch” mechanism that controls the cholesterol accessibility to the cholesterol-binding domain in Aβ42 within the membrane. In Figure 3, the proximity of CHO to the peptide backbone in the deep inserted state suggest much stronger lipid/peptide interaction than would be expected from the deep surface state.
The deep inserted state also requires that Aβ42 be more tilted than required for the initial inserted state. This can be seen clearly in Figure 3. It is the result of the extra length of peptide residing within the membrane thickness, which can only be accommodated by tilting. In general, tilted peptides have been shown to trigger membrane instability [82] and in particular, the tilt of Aβ42 has also been proposed to contribute to its cytotoxicity [83].
Insight into the biochemical basis for membrane perturbation and destabilization of both the deep inserted and deep surface states can be seen from Figure 4. The deep inserted state alters the local transverse cholesterol distribution, as shown by the wider cholesterol annular lipid number density peak. In conjunction with the redistribution of CHO, annular POPC was pushed outward from the membrane center, as shown by the shift in its number density peak. This redistribution of annular lipids may be a result of the increased size of the available CHO binding domain [84], or be required to equalize membrane lateral stresses induced by Aβ42 perturbation of the exofacial membrane interface [85, 86]. In either case, the availability of hydrophilic lipid head groups and penetration of polar amino acids into the hydrophobic core of the membrane allowed increased, deeper water penetration of the bilayer.
The deep surface state perturbed the membrane in a different manner. From Figure 4, it is apparent that the peptide mass was much more compact; the number density at the peak maximum increasing about 50%. The implication is that the mass of the peptide is much more planar and concentrated near the surface. Furthermore, by its proximity to the surface, the state provides a scaffold to interact with extracellular Aβ and other exofacial interface peptides. There is also an increase of beta structure of the protein associated with the deep surface state (see Supporting Information). The surface beta structure may serve as the seeding template to promote protein aggregations on membrane surfaces [87].
The lipid order parameters as shown in Figure 9 further verify the perturbation of the membrane annular lipids. While surface associated states have been seen in the previous simulations, they have usually occurred when Aβ was initially placed on or near the water/lipid interface [88–92]. Both the deep surface state and the deep insertion state, Figure 9, show substantial alteration in the AL resulting from the Aβ42 conformational changes. However both Figures 4 and 9 suggest little affect of the conformational change to the nAL.
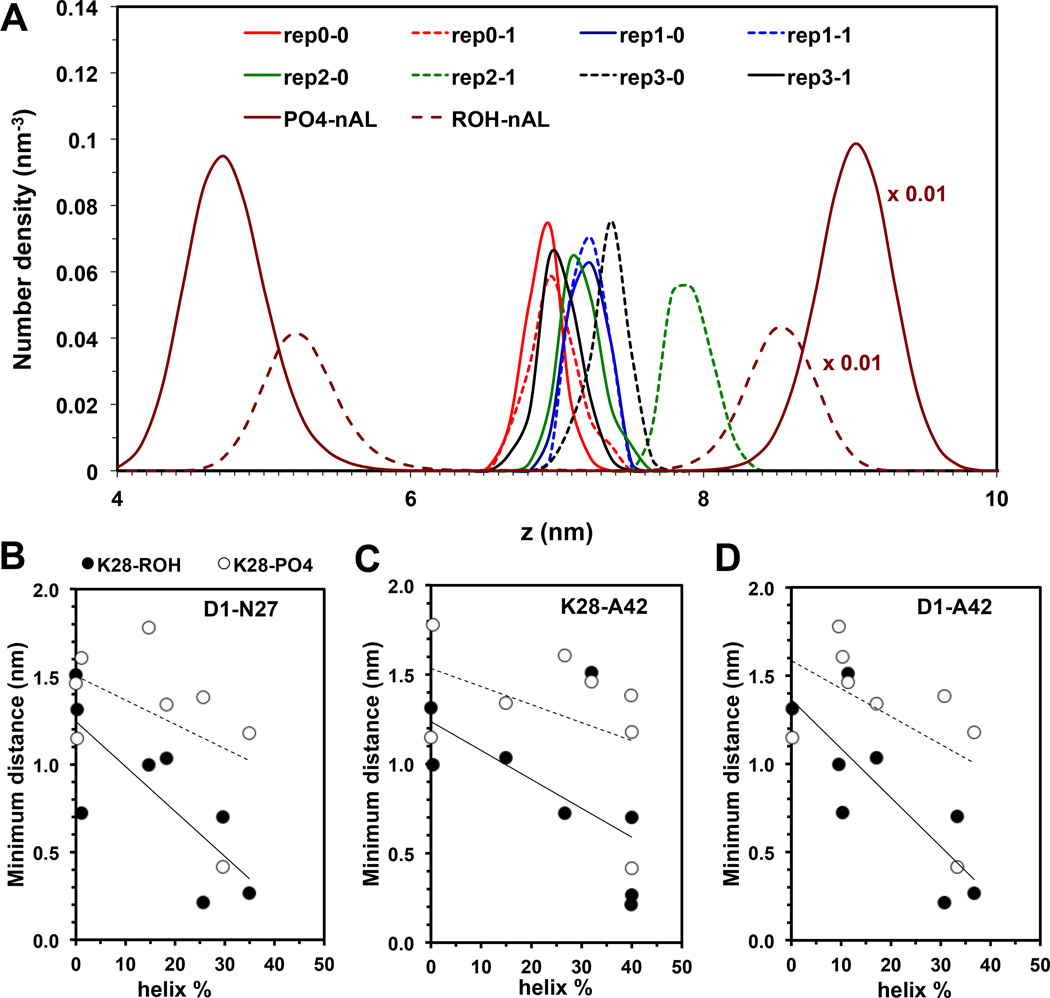
Our results reveal an interesting correlation between K28-lipid interaction and protein conformation in the deep inserted state. Figure 11 illustrates the minimum distance between K28 and ROH of cholesterol (K28-ROH) or between K28 and PO4 of PC (K28-PO4) vs. percent helicity (GHI) of Aβ42 from 8 simulation replicates of the I complex. A general increase of interaction between K28-ROH and K28-PO4 with increasing helicity of the protein was found for the non-LID and LID, suggesting that the secondary structure of the protein in cholesterol-containing membrane may be regulated by the interaction of K28 with the lipid. Specifically, snorkeling of K28 with the polar groups of the lipids in the upper monolayer, PO4 or GLY of PC and K28-ROH of CHO, promotes the helical structure of the protein. As shown in Table 1 and Supporting Information, the two deep inserted sub-states, deep inserted-P and deep inserted-C, possessed less unfolding and higher helicity than the other deep inserted states.
FIG. 11. Proximity of K28 and lipid headgroup in UA Aβ42/lipid/water/ion complexes.
The transbilayer density profiles (A) of PO4 of nAL-PC, ROH of nAL-CHO, and NH3 of K28 of Aβ42 of each simulation replicate of the I complex are shown. Plots of the minimum distance between K28 and ROH of CHO or K28-ROH (black circle) and between K28 and PO4 of PC or K28-PO4 (open circle) as a function of helix % of D1-N27 (B), K28-A42 (C) and D1-A42 (D) segments of Aβ42 are given. The calculations were over the last 50 ns of the simulations with the standard errors (error bars) shown.
Both the deep surface and deep insertion states have pathophysiological importance. For the deep surface state the proximity of the protein molecules to the exofacial interface and the reduced dimensionality of the surface [93] increase the probability of interaction between the deep surface state and membrane-absorbed protein, especially Aβ peptide. Coulombic attraction, hydrophobic, and hydrogen-bonding properties of bilayer-buried and solvent-exposed regions of the protein molecule drive self-association [94]. Alternatively, the deep inserted state could associate with other membrane-active proteins to form a channel [95, 96] that would allow unregulated ion and water flow across the membrane. Both states destabilize the membrane by altering the lipid organization, which alone could result in cytotoxicity.
Supplementary Material
Highlights.
Sampled beta-amyloid membrane orientational states by multiscale MD simulations.
An initial inserted state had a folded (I) or unfolded (II) lipid insertion domain.
Discovered I-to-deep inserted and II-to-deep surface state transitions.
Cholesterol-binding and bilayer structures were altered after the transition.
Lipid insertion domain regulates protein membrane orientational transitions.
ACKNOWLEDGEMENT
This work was supported by the Robert A. Welch Research Foundation grant (D-1158), NIH grant (GM090897-03), Williams Endowment of Trinity University, and Texas Advanced Computing Center (TACC) for the use of Lonestar Cluster under the project (G-803132) “Protein Unfolding in Lipid Membranes”.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE
- 1.Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 2.Groves JT, Kuriyan J. Molecular mechanisms in signal transduction at the membrane. Nat. Struct. Mol. Biol. 2010;17:659–665. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peetla C, Stine A, Labhasetwar V. Biophysical interactions with model lipid membranes: applications in drug discovery and drug delivery. Mol. Pharm. 2009;6:1264–1276. doi: 10.1021/mp9000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang YJ, Shi JM, Bai CJ, Wang H, Li HY, Wu Y, Ji SR. Intra-membrane oligomerization and extra-membrane oligomerization of amyloid-beta peptide are competing processes as a result of distinct patterns of motif interplay. J. Biol. Chem. 2012;287:748–756. doi: 10.1074/jbc.M111.281295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 6.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 7.Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ. Solution structure of amyloid beta-peptide(1–40) in a water-micelle environment. Is the membrane-spanning domain where we think it is? Biochemistry. 1998;37:11064–11077. doi: 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
- 8.Ulmschneider JP, Smith JC, White SH, Ulmschneider MB. In silico partitioning and transmembrane insertion of hydrophobic peptides under equilibrium conditions. J. Am. Chem. Soc. 2011;133:15487–15495. doi: 10.1021/ja204042f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- 10.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 11.Butterfield SM, Lashuel HA. Amyloidogenic protein-membrane interactions: mechanistic insight from model systems. Angew. Chem. Int. Ed. 2010;49:5628–5654. doi: 10.1002/anie.200906670. [DOI] [PubMed] [Google Scholar]
- 12.Stefani M. Structural features and cytotoxicity of amyloid oligomers: implications in Alzheimer's disease and other diseases with amyloid deposits. Prog. in Neurobio. 2012;99:226–245. doi: 10.1016/j.pneurobio.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Qiu L, Buie C, Cheng KH, Vaughn MW. Scaling and alpha-helix regulation of protein relaxation in a lipid bilayer. J. Chem. Phys. 2014;141:225101–225109. doi: 10.1063/1.4902229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu L, Buie C, Reay A, Vaughn MW, Cheng KH. Molecular dynamics simulations reveal the protective role of cholesterol in beta-amyloid protein-induced membrane disruptions in neuronal membrane mimics. J. Phys. Chem. B. 2011;115:9795–9812. doi: 10.1021/jp2012842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji SR, Wu Y, Sui SF. Cholesterol is an important factor affecting the membrane insertion of beta-amyloid peptide (A beta 1–40) which may potentially inhibit the fibril formation. J. Biol. Chem. 2002;277:6273–6279. doi: 10.1074/jbc.M104146200. [DOI] [PubMed] [Google Scholar]
- 16.Marchesi VT. An alternative interpretation of the amyloid Abeta hypothesis with regard to the pathogenesis of Alzheimer's, disease. Proc. Natl. Acad. Sci. USA. 2005;102:9093–9098. doi: 10.1073/pnas.0503181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okada T, Wakabayashi M, Ikeda K, Matsuzaki K. Formation of toxic fibrils of Alzheimer's amyloid beta-protein-(1–40) by nosialoganglioside GM1, a neuronal membrane component. J. Mol. Biol. 2007;371:481–489. doi: 10.1016/j.jmb.2007.05.069. [DOI] [PubMed] [Google Scholar]
- 18.Rzepiela AJ, Schäfer LV, Goga N, Risselada HJ, de Vries AH, Marrink SJ. Reconstruction of Atomistic Details from Coarse-Grained Structures. J. Comp. Chem. 2010;31:1333–1343. doi: 10.1002/jcc.21415. [DOI] [PubMed] [Google Scholar]
- 19.Marrink SJ, Tieleman DP. Perspective on the Martini model. Chem. Soc. Rev. 2013;42:6801–6822. doi: 10.1039/c3cs60093a. [DOI] [PubMed] [Google Scholar]
- 20.Wassenaar TA, Pluhackova K, Böckmann RA, Marrink SJ, Tieleman DP. Going Backward: A Flexible Geometric Approach to Reverse Transformation from Coarse Grained to Atomistic Models. J. Chem. Theory Comput. 2014;10:676–690. doi: 10.1021/ct400617g. [DOI] [PubMed] [Google Scholar]
- 21.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 22.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 23.Williams TL, Serpell LC. Membrane and surface interactions of Alzheimer's Abeta peptide--insights into the mechanism of cytotoxicity. FEBS J. 2011;278:3905–3917. doi: 10.1111/j.1742-4658.2011.08228.x. [DOI] [PubMed] [Google Scholar]
- 24.Gadad BS, Britton GB, Rao KS. Targeting oligomers in neurodegenerative disorders: lessons from α-synuclein, tau, and amyloid-β peptide. J. Alzheimers Dis. 2011;24:223–232. doi: 10.3233/JAD-2011-110182. [DOI] [PubMed] [Google Scholar]
- 25.Urbanc B, Betnel M, Cruz L, Bitan G, Teplow DB. Elucidation of amyloid beta-protein oligomerization mechanisms: discrete molecular dynamics study. J. Am. Chem. Soc. 2010;132:4266–4280. doi: 10.1021/ja9096303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 27.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 28.Nasica-Labouze J, Nguyen PH, Sterpone F, Berthoumieu O, Buchete NV, Cote S, De Simone A, Doig AJ, Faller P, Garcia A, Laio A, Li MS, Melchionna S, Mousseau N, Mu Y, Paravastu A, Pasquali S, Rosenman DJ, Strodel B, Tarus B, Viles JH, Zhang T, Wang C, Derreumaux P. Amyloid beta Protein and Alzheimer's Disease: When Computer Simulations Complement Experimental Studies. Chem. Rev. 2015;115:3518–3563. doi: 10.1021/cr500638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis CH, Berkowitz ML. A molecular dynamics study of the early stages of amyloid-beta (1–42) oligomerization: the role of lipid membranes. Proteins. 2010;78:2533–2545. doi: 10.1002/prot.22763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemkul JA, Bevan DR. Lipid composition influences the release of Alzheimer's amyloid beta-peptide from membranes. Protein Sci. 2011;20:1530–1545. doi: 10.1002/pro.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, Shen J, Luo X, Zhu W, Chen K, Ma J, Jiang H. Conformational transition of amyloid beta-peptide. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5403–5407. doi: 10.1073/pnas.0501218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X, Wang Q, Pan Q, Zhou F, Zheng J. Molecular interactions of Alzheimer amyloid-beta oligomers with neutral and negatively charged lipid bilayers. Phys. Chem. Chem. Phys. 2013;15:8878–8889. doi: 10.1039/c3cp44448a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao X, Xu J. Free cholesterol induces higher β-sheet content in Aβ peptide oligomers by aromatic interaction with Phe19. PLoS One. 2012;7:e46245. doi: 10.1371/journal.pone.0046245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 35.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 36.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 37.Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ. The MARTINI Coarse-Grained Force Field: Extension to Proteins. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 38.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction Models for Water in Relation to Protein Hydration. In: Pullman B, editor. Intermolecular Forces. Dordrecht, The Netherlands: Reidel; 1981. pp. 331–342. [Google Scholar]
- 39.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity-rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 40.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 41.Bekker H, Berendsen HJC, Dijkstra EJ, Achterop S, van Drunen R, van der Spoel D, Sijbers A, Keegstra H, Reitsma B, Renardus MKR. Physics Computing. Vol. 92. Singapore: World Scientific; 1993. Gromacs: A Parallel Computer for Molecular Dynamics Simulations. [Google Scholar]
- 42.Berendsen HD, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comp. Phys. Comm. 1995;91:43–56. [Google Scholar]
- 43.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Mod. 2001;7:306–317. [Google Scholar]
- 44.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast flexible and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 45.Berger O, Edholm O, Jahnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holtje M, Forster T, Brandt B, Engels T, von Rybinski W, Holtje HD. Molecular dynamics simulations of stratum corneum lipid models: fatty acids and cholesterol. Biochim. Biophys. Acta. 2001;1511:156–167. doi: 10.1016/s0005-2736(01)00270-x. [DOI] [PubMed] [Google Scholar]
- 47.van Buuren AR, Marrink SJ, Berendsen HJC. A molecular dynamics study of the decane/water interface. J. Phys. Chem. 1993;97:9206–9212. [Google Scholar]
- 48.van der Spoel D, van Buuren AR, Tieleman DP, Berendsen HJC. Molecular dynamics simulations of peptides from BPTI: a closer look at amide-aromatic interactions. J. Biomol. NMR. 1996;8:229–238. doi: 10.1007/BF00410322. [DOI] [PubMed] [Google Scholar]
- 49.Darden T, York D, Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 50.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen L. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 51.Hess B, Bekke H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J. Comp. Chem. 1997;13:952–962. [Google Scholar]
- 52.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 53.Hess B, Kutzner C, van der Spoel D, Lindahl E. Gromacs 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 54.Allen WJ, Lemkul JA, Bevan DR. GridMAT-MD: a grid-based membrane analysis tool for use with molecular dynamics. J. Comput. Chem. 2009;30:1952–1958. doi: 10.1002/jcc.21172. [DOI] [PubMed] [Google Scholar]
- 55.Stansfeld PJ, Sansom MSP. From Coarse Grained to Atomistic: A Serial Multiscale Approach to Membrane Protein Simulations. J. Chem. Theory Comput. 2011;7:1157–1166. doi: 10.1021/ct100569y. [DOI] [PubMed] [Google Scholar]
- 56.Qin SS, Yu YX, Li QK, Yu ZW. Interaction of human synovial phospholipase A2 with mixed lipid bilayers: a coarse-grain and all-atom molecular dynamics simulation study. Biochemistry. 2013;52:1477–1489. doi: 10.1021/bi3012687. [DOI] [PubMed] [Google Scholar]
- 57.Meier K, Choutko A, Dolenc J, Eichenberger AP, Riniker S, van Gunsteren WF. Multi-resolution simulation of biomolecular systems: a review of methodological issues. Angew Chem. Int. Ed. Engl. 2013;52:2820–2834. doi: 10.1002/anie.201205408. [DOI] [PubMed] [Google Scholar]
- 58.Ayton GS, Voth GA. Systematic multiscale simulation of membrane protein systems. Curr. Opin. Struct. Biol. 2009;19:138–144. doi: 10.1016/j.sbi.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou R. Replica exchange molecular dynamics method for protein folding simulation. Methods Mol. Biol. 2007;350:205–223. doi: 10.1385/1-59745-189-4:205. [DOI] [PubMed] [Google Scholar]
- 60.Kubitzki MB, de Groot BL. Molecular dynamics simulations using temperature-enhanced essential dynamics replica exchange. Biophys. J. 2007;92:4262–4270. doi: 10.1529/biophysj.106.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999;314:141–151. [Google Scholar]
- 62.Pannuzzo M, Raudino A, Milardi D, La Rosa C, Karttunen M. alpha-helical structures drive early stages of self-assembly of amyloidogenic amyloid polypeptide aggregate formation in membranes. Sci. Rep. 2013;3:2781. doi: 10.1038/srep02781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Engelman DM, Steitz TA, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 64.Nymeyer H, Woolf TB, Garcia AE. Folding is not required for bilayer insertion: replica exchange simulations of an alpha-helical peptide with an explicit lipid bilayer. Proteins. 2005;59:783–790. doi: 10.1002/prot.20460. [DOI] [PubMed] [Google Scholar]
- 65.Qiu L, Buie C, Cheng KH, Vaughn MW. Scaling and alpha-helix regulation of protein relaxation in a lipid bilayer. J. Chem. Phys. 2014;141:225101. doi: 10.1063/1.4902229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu Z, Cui Q, Yethiraj A. A New Coarse-Grained Force Field for Membrane-Peptide Simulations. J. Chem. Theory Comp. 2011;7:3793–3802. doi: 10.1021/ct200593t. [DOI] [PubMed] [Google Scholar]
- 67.de Jong DH, Singh G, Bennett WFD, Arnarez C, Wassenaar TA, Schäfer LV, Periole X, Tieleman DP, Marrink SJ. Improved Parameters for the Martini Coarse-Grained Protein Force Field. J. Chem. Theory Comp. 2012;9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 68.Jaud S, Fernandez-Vidal M, Nilsson I, Meindl-Beinker, Hubner NC, Tobias DJ, von Heijne G, White SH. Insertion of sort transmembrane helices by the Sec61 translocon. Proc Natl Acad Sci U.S.A. 2009;106:11588–11593. doi: 10.1073/pnas.0900638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kukol A. Lipid models for united-atom molecular dynamics simulations of proteins. J. Chem. Theory Comput. 2009;5:615–626. doi: 10.1021/ct8003468. [DOI] [PubMed] [Google Scholar]
- 70.Tieleman DP, Justin LM, Walter LA, Christian K, Zhitao X, Luca M. Membrane protein simulations with a united-atom lipid all-atom protein model: lipid-protein interactions side chain transfer free energies and model proteins. J. Phys.: Condens. Matter. 2006;18:1221–1234. doi: 10.1088/0953-8984/18/28/S07. [DOI] [PubMed] [Google Scholar]
- 71.Poger D, van Gunsteren WF, Mark AE. A new force field for simulationg phosphatidylcholine bilayers. J. Comp. Chem. 2010;31:1117–1125. doi: 10.1002/jcc.21396. [DOI] [PubMed] [Google Scholar]
- 72.Poger D, Mark AE. Lipid bilayers: The effec of force field on ordering and dynamics. J. Chem. Theory Comp. 2012;8:4807–4817. doi: 10.1021/ct300675z. [DOI] [PubMed] [Google Scholar]
- 73.Lemkul JA, Bevan DR. A comparative molecular dynamics analysis of the amyloid beta-peptide in a lipid bilayer. Arch. Biochem. Biophys. 2008;470:54–63. doi: 10.1016/j.abb.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 74.Kivipelto M, Helkala EL, Laakso MP, Hanninen T, Hallikainen M, Alhainen K, Soininen H, Tuomilehto J, Nissinen A. Midlife vascular risk factors and Alzheimer's disease in later life: longitudinal, population based study. BMJ (Clinical research ed.) 2001;322:1447–1451. doi: 10.1136/bmj.322.7300.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pappolla MA, Bryant-Thomas TK, Herbert D, Pacheco J, Fabra Garcia M, Manjon M, Girones X, Henry TL, Matsubara E, Zambon D, Wolozin B, Sano M, Cruz-Sanchez FF, Thal LJ, Petanceska SS, Refolo LM. Mild hypercholesterolemia is an early risk factor for the development of Alzheimer amyloid pathology. Neurology. 2003;61:199–205. doi: 10.1212/01.wnl.0000070182.02537.84. [DOI] [PubMed] [Google Scholar]
- 76.Simons M, Keller P, Dichgans J, Schulz JB. Cholesterol and Alzheimer's disease: is there a link? Neurology. 2001;57:1089–1093. doi: 10.1212/wnl.57.6.1089. [DOI] [PubMed] [Google Scholar]
- 77.Barrett PJ, Song Y, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336:1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Di Scala C, Yahi N, Lelievre C, Garmy N, Chahinian H, Fantini J. Biochemical identification of a linear cholesterol-binding domain within Alzheimer's beta amyloid peptide. ACS Chem. Neurosci. 2013;4:509–517. doi: 10.1021/cn300203a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sinha S, Lopes DH, Bitan G. A key role for lysine residues in amyloid beta-protein folding, assembly, and toxicity. ACS Chem. Neurosci. 2012;3:473–481. doi: 10.1021/cn3000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 81.Curran AR, Engelman DM. Sequence motifs polar interactions and conformational changes in helical membrane proteins. Curr. Opin. Struct. Biol. 2003;13:412–417. doi: 10.1016/s0959-440x(03)00102-7. [DOI] [PubMed] [Google Scholar]
- 82.Thomas A, Brasseur R. Tilted peptides: the history. Curr. Protein Pept. Sci. 2006;7:523–527. doi: 10.2174/138920306779025594. [DOI] [PubMed] [Google Scholar]
- 83.Pillot T, Goethals M, Vanloo B, Talussot C, Brasseur R, Vandekerckhove J, Rosseneu M, Lins L. Fusogenic properties of the C-terminal domain of the Alzheimer beta-amyloid peptide. J. Biol. Chem. 1996;271:28757–28765. doi: 10.1074/jbc.271.46.28757. [DOI] [PubMed] [Google Scholar]
- 84.Di Scala C, Chahinian H, Yahi N, Garmy N, Fantini J. Interaction of Alzheimer's beta-amyloid peptides with cholesterol: mechanistic insights into amyloid pore formation. Biochemistry. 2014;53:4489–4502. doi: 10.1021/bi500373k. [DOI] [PubMed] [Google Scholar]
- 85.Cantor RS. The influence of membrane lateral pressures on simple geometric models of protein conformational equilibria. Chem. Phys. Lipids. 1999;101:45–56. doi: 10.1016/s0009-3084(99)00054-7. [DOI] [PubMed] [Google Scholar]
- 86.Cantor RS. Lipid composition and the lateral pressure profile in bilayers. Biophys. J. 1999;76:2625–2639. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burke KA, Yates EA, Legleiter J. Biophysical insights into how surfaces, including lipid membranes, modulate protein aggregation related to neurodegeneration. Front. Neurol. 2013;4:17. doi: 10.3389/fneur.2013.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davis CH, Berkowitz ML. Structure of the amyloid-beta (1–42) monomer absorbed to model phospholipid bilayers: a molecular dynamics study. J. Phys. Chem. B. 2009;113:14480–14486. doi: 10.1021/jp905889z. [DOI] [PubMed] [Google Scholar]
- 89.Davis CH, Berkowitz ML. Interaction between amyloid-beta (1–42) peptide and phospholipid bilayers: a molecular dynamics study. Biophys. J. 2009;96:785–797. doi: 10.1016/j.bpj.2008.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu X, Wang Q, Pan Q, Zhou F, Zheng J. Molecular interactions of Alzheimer amyloid-beta oligomers with neutral and negatively charged lipid bilayers. Phys. Chem. Chem. Phys. 2013;15:8878–8889. doi: 10.1039/c3cp44448a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu X, Zheng J. Polymorphic structures of Alzheimer's beta-amyloid globulomers. PLoS One. 2011;6:e20575. doi: 10.1371/journal.pone.0020575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu X, Zheng J. Cholesterol promotes the interaction of Alzheimer beta-amyloid monomer with lipid bilayer. J. Mol. Biol. 2012;421:561–571. doi: 10.1016/j.jmb.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 93.Aisenbrey C, Borowik T, Bystrom R, Bokvist M, Lindstrom F, Misiak H, Sani MA, Grobner G. How is protein aggregation in amyloidogenic diseases modulated by biological membranes? Eur. Biophys. J. 2008;37:247–55. doi: 10.1007/s00249-007-0237-0. [DOI] [PubMed] [Google Scholar]
- 94.Gorbenko GP, Kinnunen PK. The role of lipid-protein interactions in amyloid-type protein fibril formation. Chem. Phys. Lipids. 2006;141:72–82. doi: 10.1016/j.chemphyslip.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 95.Jang H, Zheng J, Nussinov R. Models of beta-amyloid ion channels in the membrane suggest that channel formation in the bilayer is a dynamic process. Biophys. J. 2007;93:1938–1949. doi: 10.1529/biophysj.107.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Arispe N, Diaz J, Durell SR, Shafrir Y, Guy HR. Polyhistidine peptide inhibitor of the Abeta calcium channel potently blocks the Abeta-induced calcium response in cells. Theoretical modeling suggests a cooperative binding process. Biochemistry. 2010;49:7847–7853. doi: 10.1021/bi1006833. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.