Abstract
Introduction
Anal squamous cell carcinoma is preceded by persistent infection with high-risk human papillomavirus (HPV) and the cancer precursor, high-grade squamous intraepithelial lesion (HSIL). Detection of specific HPV genotypes and HPV-related biomarkers may be an option for primary anal screening. However, more data on the natural history of HPV-related anal lesions are required. The outcomes from this study will enhance our understanding of the clinical and biological behaviour of HPV-related anal lesions and inform the development of future HPV genotype and/or biomarker screening tests.
Methods and analysis
HIV-negative and HIV-positive men who have sex with men, aged 35 years and over, recruited from community-based settings in Sydney, Australia, attend 6 clinic visits over 3 years. At the first 5 visits, participants undergo a digital anorectal examination, an anal swab for HPV genotyping and anal cytology, and high-resolution anoscopy with directed biopsy of any visible abnormalities that are suggestive of any abnormality suspicious of SIL. Tissue sections from participants diagnosed with histologically confirmed HSIL at the baseline clinic visit will undergo laser capture microdissection, HPV detection and genotyping, and quantitation of CpG methylation in baseline and follow-up biopsies. Histological and cytological findings in combination with HPV genotyping data will be used to identify persistent HSIL. HSIL will be stratified as non-persistent and persistent based on their status at 12 months. The performance of HPV genotype and methylation status in predicting disease persistence at 12 months will be assessed, along with associations with HIV status and other covariates such as age.
Ethics and dissemination
The St Vincent's Hospital Ethics Committee granted ethics approval for the study. Written informed consent is obtained from all individuals before any study-specific procedures are performed. Findings from this study will be disseminated to participants and the community through study newsletters, and through peer-reviewed publications and international conferences.
Keywords: Human papillomavirus, Anal cancer, Biomarkers, Precancerous conditions, Laser capture microdissection, HSIL/HGAIN
Strengths and limitations of this study.
One of only a very small number of cohorts with prospectively collected longitudinal anal cytology and histology samples from HIV-positive and HIV-negative participants.
Participants are not routinely treated for high grade squamous intraepithelial lesion, allowing for longitudinal follow-up of lesions.
Use of laser capture microdissection to isolate lesion tissue.
Challenges relating to sampling and diagnostic uncertainty and establishing whether or not a lesion is persistent from one visit to the next.
Some lesions will be missed during high resolution anoscopy; therefore, the study uses both histological diagnoses and cytological findings.
Introduction
Human papillomavirus (HPV)-associated anal squamous cell carcinoma (ASCC), like cervical cancer, is preceded by persistent infection with high-risk HPV (HR-HPV) and precancerous high-grade squamous intraepithelial lesions (HSILs). In developed nations, rates of ASCC in both genders have been increasing for decades.1 Subpopulations at greatest risk of ASCC include men who have sex with men (MSM), HIV-positive men and women, solid organ transplant recipients on immunosuppressant drugs, and women with previous HPV-related cervical, vulvar or vaginal disease.2 MSM have a 20-fold increased incidence. HIV-positive people have at least a 30-fold increased incidence, and anal cancer has become the most commonly occurring non-AIDS-defining cancer in developed nations. HIV-positive MSM are the group at highest risk, with 50-fold to 150-fold increased incidence.3–6 Women with previous HPV-related anogenital disease have a 5-fold to 20-fold increased risk of anal cancer,7 and solid organ transplant recipients have a 5-fold increased incidence.8
Vaccination against HPV infection is likely to reduce anal cancer rates in the long term, but not within the next few decades. The quadrivalent HPV (qHPV) vaccine (targeting HPV 6, 11, 16 and 18) has been shown in phase III clinical trials to greatly reduce the risk of development of anal cancer precursor lesions in MSM.9 10 The introduction of prophylactic qHPV vaccination of young Australian females in 2007 led to a 77% reduction in prevalence of vaccine preventable HPV types,11 as well as a reduction in the incidence of cervical precancerous lesions in young women.12 13 Vaccination of Australian high school-aged boys with the qHPV vaccine began in 2013.14 A new nonavalent HPV vaccine, targeting an extended range of genotypes (HPV 6, 11, 16, 18, 31, 33, 45, 52 and 58) has recently completed clinical trials.15 This vaccine has been approved for use by the US Food and Drug Administration (FDA),16 and decisions about the use of this vaccine are yet to be made for general and at-risk populations, and ongoing studies are generating data to inform these decisions.
Currently available HPV vaccines are not therapeutic and do not protect against disease development in individuals already HPV DNA-positive for vaccine genotypes,17 18 although individuals who are seropositive and DNA-negative are protected from future disease.19 Because vaccination does not protect against existing ongoing infections, it may not be as effective in adults, and consequently widespread vaccination of adults over the age of 26 years has not been adopted by any country to date. To reduce ASCC morbidity in the unvaccinated populations, other approaches are necessary. Organised population-based cytology screening programmes have led to a continuing decline in incidence and mortality from cervical cancer in Australia and other industrialised nations, where high standards of quality assurance and quality control are in place and there is high coverage of the target population.20 Epidemiological, aetiological and pathological similarities between squamous cancers of the cervix and anus have led to suggestions that similar screening programmes in high-risk groups could also reduce the incidence of ASCC;21 22 however, it has been established that anal cytology has not been shown to be as effective as cervical cytology in terms of screening to prevent cancer.21 It has also been determined that genotyping of anal swabs is not predictive for lesion-specific genotypes.23 Existing data indicate that approximately 20–40% of MSM have anal HSIL at any one time,4 with only a very small proportion progressing to cancer even in the absence of treatment.24 Therefore, the specificity of anal cytological screening is likely to be low. More specific screening targets, such as DNA or RNA from the most high-risk genotypes, and/or other biomarkers of cancer risk, may be a necessary part of any screening programme.
Between 70% and 90% of ASCC are associated with HPV type 16 in single or mixed infections; the contributions of other HPV genotypes are not well established, but may predominantly be restricted to a small subset of causative genotypes.25 26 In line with ongoing changes to cervical screening practices in Australia and the USA,27 28 detection of specific HPV genotypes and HPV-related biomarkers, such as methylation, HPV mRNA transcript patterns and protein markers, may be options for primary anal screening. Host-specific biomarkers such as increased methylation in regulatory regions of tumour suppressor genes have also shown promise as a potential triage marker for cervical high-grade disease,29 and there are some data to support this finding in the anal canal.30
HPV DNA-based anal screening algorithms and vaccination policy for anal cancer prevention in high-risk populations will rely on more accurate HPV genotype data than are currently available. Genotype data for anal cancers are limited; however, they suggest that the distribution of HPV genotypes associated with anal cancers is different to that observed in cervical cancers, with a higher proportion of HPV16-positive ASCC and a higher, but still low, proportion of low-risk HPV (LR-HPV) and possible/probable HR-HPV genotypes.25 31 32 Reliable genotype data for anal HSIL are even more limited than for cancers. In addition to the limited quantity of data,33–36 the interpretation of these data are confounded by the common occurrence of multiple HPV genotypes and multiple squamous lesions in the anal canals of MSM37 and, to a lesser extent, women.38 While each individual lesion or cancer is caused by a single genotype,23 39 up to a quarter of anal cancer specimens from men and women25 40 41 and up to half of anal HSIL biopsies contain multiple detectable genotypes,23 most of which will be incidental to the lesion of interest. Consequently, attributions of genotype to lesions, based on swab or biopsy samples, are indirect and often rely on the use of mathematical algorithms to determine the most likely causative genotype.35 42
A more direct method, laser capture microdissection (LCM) combined with sensitive genotype testing, is a highly effective approach to assign HPV genotypes to individual lesions.23 39 LCM utilises lasers to precisely isolate and capture specifically targeted tissue, from a single cell to a selected segment of an intraepithelial lesion, from which nucleic acid can be extracted and tested. We describe here the use of LCM to study the natural history of longitudinally collected HPV-associated anal lesions. The data generated using the methods described below will enable us to accurately determine which HPV genotypes present the highest risk of anal HSIL persistence and/or progression to cancer, investigate patterns of HPV-related biomarkers related to persistence of HSIL, and inform the development of screening and vaccine strategies for the prevention of anal cancer in at-risk populations.
Methods and analysis
Participant recruitment
Approximately 600 MSM, 35 years and older, recruited from community-based settings in Sydney, Australia, are being enrolled into this 3-year cohort study. Participant recruitment and sample collection have been previously described in detail.43 Once recruited, participants attend a baseline clinic visit and four follow-up visits at 6, 12, 24 and 36 months after baseline. LCM is performed on lesions collected at baseline and at follow-up visits from consenting participants with biopsy-confirmed HSIL diagnosed at baseline.
Sampling of anal lesions
At each clinic visit, participants undergo a digital anorectal examination, intra-anal swabbing for HPV DNA genotyping as well as anal cytology, which is followed by high-resolution anoscopy (HRA) and biopsy of any lesions suspicious of SIL as described elsewhere.43 Clinical impressions of intra-anal abnormalities and their anatomical positions observed during HRA are recorded on a preformatted form by the physician and digital photographs of the area are taken. All clinical reporting by the anoscopist is carried out with the knowledge of the participants’ HIV status, cytology and HRA results from previous visits. Biopsies are placed into a 10% formalin solution and sent to Douglass Hanly Moir Pathology (Sydney, Australia) where they are routinely processed and embedded in paraffin. Formalin-fixed paraffin-embedded (FFPE) biopsies are sectioned and stained with H&E, and an initial diagnosis obtained, as described.43 Reporting of the biopsies is performed in accordance with criteria, terminology and recommendations of the Lower Anogenital Squamous Terminology (LAST) Project.44 45 In addition to the LAST criteria, anal low-grade squamous intraepithelial lesions (LSILs) are routinely subdivided into exophytic (eLSIL) and flat (fLSIL), based on architectural criteria (eLSIL corresponding to condyloma acuminatum).43 46 47
HPV genotyping of anal cytology samples
A 1 ml aliquot of intra-anal swab material in ThinPrep PreservCyt (Hologic, Inc, Marlborough, Massachusetts, USA) is centrifuged at 13 000×g for 20 min and the cell pellet resuspended in 200 µL of sterile phosphate-buffered saline. DNA is extracted using the MagNAPure LC (Roche Diagnostics GmbH, Penzberg, Germany) using the DNA-I kit (blood cells high-performance protocol) or on the MagNAPure 96 using the DNA and Viral Nucleic Acid Small Volume Kit (Pathogen Universal 200 protocol). Extracted DNA is eluted into a final volume of 100 µL. HPV genotyping is performed on all samples using the Linear Array HPV Genotyping Test (Roche Diagnostics), which detects and identifies 37 of the most common mucosal HPV genotypes. Samples which return an unassessable result on Linear Array are retested using DNA diluted 1 in 2 in nuclease-free water, to dilute any substances which may be inhibiting PCR amplification. Samples which test positive for HPV52/33/35/58 and one or more of HPV33, 35 and/or 58 are further tested on a quantitative PCR (qPCR) assay to determine whether HPV52 is also present, as previously described.48 Samples that are negative for HPV16 or 18 will be tested on a type-specific qPCR assay for these genotypes, as previously described.49
Selection of biopsies
Participants diagnosed with biopsy-confirmed HSIL at the baseline visit and a further 50 participants diagnosed with LSIL are included for comparison. Participants are excluded if no tissue remains in the biopsy block, the biopsy is negative for any SIL on review, or if the participant did not give consent for their tissue to be stored and used for procedures other than histological diagnosis. Participants who have had ablative treatment are also excluded. More than one biopsy may be selected from a single participant.
Follow-up of HSILs
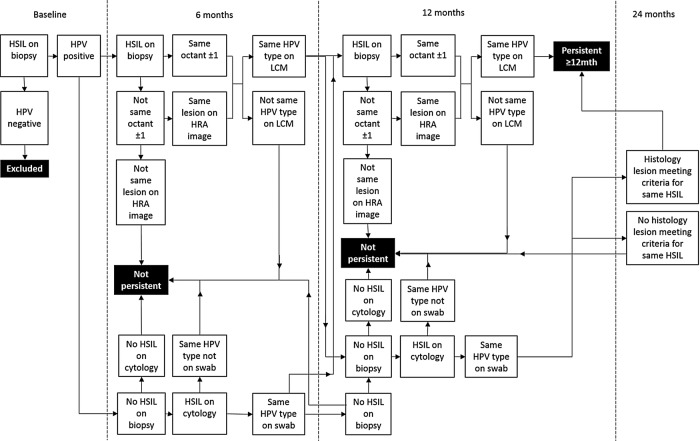
By reference to clinical notes and photographs taken during HRA (figure 1A), biopsies from follow-up visits where available are selected for sectioning. A HSIL is considered persistent if it is (1) detected at least 12 months after the baseline visit within the same octant or up to one octant either side of the baseline lesion (figure 1B); (2) where possible confirmed by reference to photographs taken during HRA and annotated by a clinician; and (3) if it contains the same HPV genotype following LCM and genotyping. An anal intraepithelial neoplasia grade 2 (AIN2) could be considered the same lesion as an AIN3 diagnosed at a subsequent visit, and vice versa, as both are HSIL. An LSIL diagnosed on follow-up is not necessarily considered to be the same lesion as a HSIL diagnosed at a previous visit; however, exceptions may include low-grade lesions that are p16-positive and positive for the same HPV genotype, if an eligible HSIL is detected at the next visit. In cases where it is not clear whether a HSIL is still present at a follow-up visit, additional data including cytology results and reference to HRA photographs will be used in accordance with the algorithm in figure 2. HSIL will be defined as persistent for <6 or <12 (not persistent), or ≥12 months (persistent).
Figure 1.

(A) Photograph of the anal transformation zone taken through a speculum during high-resolution anoscopy. A digital photograph at 10× magnification of an area treated with 3% acetic acid. An area suspicious for high-grade disease is outlined; this was confirmed by biopsy to be anal intraepithelial neoplasia grade 3 (AIN3). (B) Clock face diagram of anal transformation zone segmented into eight quadrants. A high-grade squamous intraepithelial lesion is considered to be potentially persistent if it was biopsied from within the same quadrant or up to one quadrant either side of the baseline lesion at follow-up clinic visits.
Figure 2.
Flow chart for defining probable persistence of high-grade squamous intraepithelial lesion (HSIL) detected at baseline. A HSIL biopsy detected at consecutive visits which tests positive for the same human papillomavirus (HPV) genotype following laser capture microdissection (LCM) and is located in the same octant or an octant either side of the baseline biopsy is considered persistent. In the event that not all of these criteria are met, additional information including the cytology result and swab HPV genotype result and extent of disease on high-resolution anoscopy (HRA) will be considered. A HSIL lesion is considered to be not persistent if there were no follow-up biopsy taken at 6 months meeting the criteria for persistence, and if the cytology and swab results also do not support persistence. If there is doubt, 12-month samples will be taken into consideration and if not found to support the existence of a persistent HSIL, the result will be ‘not persistent’.
Sandwich sectioning and diagnosis of FFPE samples for LCM
For each biopsy, a series of nine sections are cut and processed for histological analysis using a sandwich sectioning method modified from refs 39 and 49 (figure 3). To minimise cross-contamination, the microtome blade is changed and the microtome surface is cleaned with xylene after sectioning of each sample. Where possible without resetting the blocks, biopsies are cut from the basal side towards the epithelial/superficial side to minimise carryover of unrelated viral DNA to lesional tissue. The two outer sections (sections 1 and 9) are cut to 3 µm thickness and stained with H&E. The first inner section (section 2) is also cut to 3 µm thickness and automatically stained with CINtec anti-p16 (Roche, Tucson, Arizona, USA) and counterstained with Gills No. 2 haematoxylin (Australian Biostain, Traralgon, Australia) on the Ventana Benchmark Ultra instrument (Ventana Medical Systems, Inc, Oro Valley, Arizona, USA). Sections 1, 2 and 9 are reviewed by a histopathologist experienced in reading anal biopsy specimens (JMR), to assess p16 positivity and lesion grade. Biopsies are classified as negative for intraepithelial neoplasia, eLSIL, fLSIL, HSIL-AIN2, HSIL-AIN3 or ASCC, and assessment of p16 staining is performed, both in accordance with accepted international criteria.45 Hence, p16 staining is reported as positive if there is continuous strong nuclear or nuclear plus cytoplasmic staining of the basal cell layer with extension upwards involving at least one-third of the epithelial thickness. Other patterns are regarded as negative. The second inner section (section 3) is cut to 9 µm thickness and placed onto an Arcturus polyethylene naphthalate (PEN) membrane glass slide (Applied Biosystems, Foster City, California, USA) for LCM and allowed to dry. The next two sections (sections 4 and 5, designated whole tissue sections (WTS)) are cut to 9 µm thickness, placed into sterile 1.5 mL microfuge tubes as scrolls and stored away from light. An additional three sections are cut to 3 µm thickness, mounted and stored for future immunohistochemical staining.
Figure 3.

Sandwich sectioning of formalin-fixed, paraffin-embedded anal biopsy tissue samples and downstream testing of each section. (1) First H&E slide, 3 µm section; (2) p16-ink4a slide, 3 µm section; (3) membrane slide to be used for LCM, 9 µm section; (4 and 5) whole tissue sections, unmounted, 9 µm sections; (6–8) unstained tissue sections mounted on slides, 3 µm sections; (9) last H&E slide, 3 µm section. HPV, human papillomavirus; LCM, laser capture microdissection.
Digestion and DNA extraction of WTS
Section 5 is archived at 4°C and section 4 is digested with a mixture of histolene (800 µL) and ethanol (400 µL). The tissue is pelleted at 14 000×g for 10 min, and the resultant pellet washed twice with 500 µL of 70% ethanol. The tissue is air dried to ensure that no residual ethanol is present and is subsequently incubated with 400 µg of proteinase K in 200 µL of tissue lysis buffer (Roche Molecular Systems) on a 55°C heat block for 1 h followed by 37°C overnight. Samples which are not fully digested are vortexed briefly and incubated for a further hour following the addition of a further 10 µL of proteinase K (10 mg/mL in tissue lysis buffer). The digested tissue is then transferred onto an automated MagnaPure 96 (Roche) isolation and purification system, using the DNA and Viral NA small volume kit, Universal Pathogen 200 protocol. The final volume of eluted DNA in solution is 100 µL.
Dewaxing, LCM and DNA extraction of lesions
Immediately prior to tissue capture by LCM, PEN membrane slides are de-paraffinised via sequential 5 min incubations in 100% xylene (twice) and 100% ethanol (twice) and then allowed to dry thoroughly prior to laser capture. An annotated photograph of the H&E-stained section is used to guide selection of abnormal tissue to be captured. Tissue sections are visualised and dissected with an ultraviolet cutting laser on a Veritas 704 Laser Capture Microdissection System (Arcturus Bioscience, Mountain View, California, USA) and a single lesion is captured onto each thermoplastic CapSure Macro LCM Cap (Applied Biosystems) with an infrared capture laser (figure 4). Care is taken to avoid capturing the superficial layers of the epithelium (figure 5). For any lesions which test positive for more than one HPV genotype (genotype testing described below), additional sections will be captured by LCM from the basal layer at either end of the originally dissection area of tissue to test for multiple colliding lesions or contamination (figure 6). DNA is extracted from each cap with the Arcturus PicoPure DNA Extraction Kit (Applied Biosystems) as per protocol F of the PicoPure DNA Extraction Kit User guide.
Figure 4.
Laser capture microdissection of high-grade squamous intraepithelial lesion (HSIL). (A) Adjacent H&E stained and p16 stained biopsy section with HSIL annotated by a histopathologist in green. (B) Dewaxed, unstained biopsy section on polyethylene naphthalate membrane slide ready for laser capture microdissection. (C) Lesion excised with ultraviolet laser, thermoplastic cap heated with infrared laser and fragments lifted off. (D) Excised fragment ready for DNA extraction.
Figure 5.

Laser capture microdissection (LCM) of the basal layer to avoid superficial contamination with incidental human papillomavirus (HPV) genotypes. H&E section of an anal canal biopsy containing an anal intraepithelial neoplasia grade 3 (AIN3). The basal and superficial layers of the AIN3 were dissected separately by LCM. The basal layers of the lesion were positive for HPV16 and the biopsy also had superficial contamination with HPV33 DNA.
Figure 6.
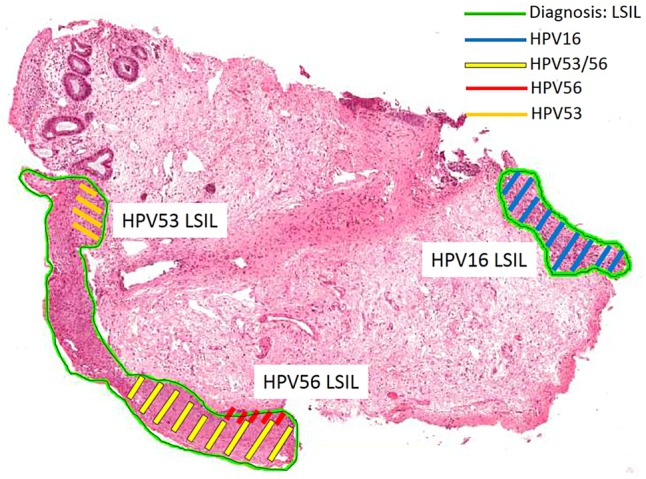
Example of a biopsy containing colliding lesions each positive for different HPV genotypes. H&E section of an anal canal biopsy with three lesions positive for HPV16, 53 and 56, respectively. Shaded areas indicate tissue dissected by LCM and genotyped. The bottom two lesions collide, and the original LCM section tested positive for both HPV53 and 56. Subsequent sections each tested positive singly for either HPV53 or 56. HPV, human papillomavirus; LCM, laser capture microdissection; LSIL, low-grade squamous intraepithelial lesion.
Genotyping of DNA extracted from WTS and LCM samples
A 110 bp region of the human β-globin gene is detected by quantitative real-time PCR to confirm successful DNA extraction, as previously described.49 DNA from WTS-extracted and LCM-extracted tissue sections are both tested for HPV genotype on the RHA kit HPV SPF10-LiPA25, V.1 (Labo Bio-medical Products BV, Rijswijk, The Netherlands) according to the manufacturer's instructions. Because of an inability to discriminate between HPV68 and 73 by the SPF10-LiPA25 assay, samples which are positive for HPV68/73 are tested on HPV68 and 73 type-specific qPCR assays targeting the E6 region as previously described,49 using the following primers and probes: HPV68 (forward primer 5′-CTGTGCAGGACATTGGACAC-3′; reverse primer 5′-TTGGCATGCAGCAAATGGTA-3′; probe 5′-LC610-TGCAGAAGGCAACTACAACGGACA-BHQ2–3′); HPV73 (forward primer 5′-CCAATTCAGAAGAACGACCA-3′; reverse primer 5′-CGTTGGCAAAACACACAGTC-3′; probe 5′-FAM-CTTCGTCACATAACGCTTGTAGCTTG-BHQ1-3′). Confirmed lesions that are positive for β-globin, but not detectable on the SPF10-LiPA25, are tested for HPV DNA on the DNA ELISA kit HPV SPF10, V.1 (Labo Bio-medical Products BV) which is validated for detection of 44 HPV genotypes. Samples which are ELISA-positive are genotyped on the RHA Kit HPV SPF+ (Labo Bio-medical Products BV) which is able to identify HPV26, 30, 55, 61, 62, c64, 67, 69, 71, 71sub, 82, 83, 84, 85, 87, 89, 90 and 91. ELISA-negative samples are tested on a qPCR assay targeting the E6 region of HPV16 and 18 as previously described,49 in addition to HPV45 (forward primer 5′-CTACAAGACGTATCTATTGCCTGTG-3′; reverse primer 5′-CCAGTGTCTCTCCATATACAGAGTTT-3′; probe 5′-Cy5-CCTCTGTGCGTTCCAATGTTGCTT-BHQ3-3′). Confirmed lesions which are still negative on ELISA and/or all previous HPV tests are tested for the presence of β-papillomavirus (cutaneous or skin) genotypes as follows. Samples are PCR amplified using degenerate PCR primers CP65 (5′-CARGGTCAYAAYAATGGYAT-3′) and CP70 (5′-AAYTTTCGTCCYARAGRAWATTGRTC-3′)50 as follows: 5 µL of DNA template is included in a final reaction volume of 50 µL (2.5U AmpliTaq Gold and 1× Buffer II (Applied Biosystems, Foster City, California, USA); 3.6 mM MgCl2; 0.2 mM of each dNTP; 1 µM of each primer). Amplicons are visualised on a 2% agarose gel. These samples are also genotyped using the RHA Kit Skin (β) HPV (Diassay B.V., Rijswijk, The Netherlands) which is able to identify HPV5, 8(I), 8(II), 9, 12, 14, 15, 17, 19, 20, 21, 22, 23, 24, 25 36, 37, 38, 47, 49, 75, 80, 92, 93 and 96, or by Sanger sequencing of the CP65/CP70 amplicon and comparison with the (National Center for Biotechnology Information) NCBI Blast nucleotide database (http://www.ncbi.nlm.nih.gov/Blast.cgi) and/or HPV-QUEST database (http://www.ijbcb.org/HPV/)51 to identify the closest matching known sequence. Samples that are negative for both β-globin and HPV, after re-extraction by LCM if possible, are denoted ‘unassessable’. Samples that are positive for β-globin but negative for all HPV tests are denoted ‘HPV not detected’. Samples that are positive for HPV DNA ELISA but not able to be genotyped are denoted ‘HPV X’. Samples that are negative for mucosal HPV types (negative on SPF10 LiPA, SPF+ LiPA and E6 qPCR) and positive for β-papillomavirus DNA gel electrophoresis assay but not able to be genotyped are denoted ‘HPV β’. Additionally, all WTS samples that are negative for HPV16 or 18 will be tested on a type-specific qPCR assay for these genotypes.
Viral genome methylation sequence analysis
DNA from HPV16-positive HSIL is subjected to methylation sequencing of viral genomic regions associated with regulation of oncogene expression using modifications of published protocols52 53 on anal swab, WTS and LCM-extracted tissue. DNA from the second WTS sample (section 5) is extracted using the QIAmp DNA FFPE Tissue Kit (Qiagen GmbH, Hilden, Germany). The same DNA sample used for HPV genotyping of swab and LCM samples is also used for methylation analysis. Known controls for low (SiHa cell line, ATCC HTB35; American Type Culture Collection (ATCC), Manassas, Virginia, USA) and high (CaSki cell line, ATCC CRL1550; ATCC) methylation at the target sites are included in each experiment as well as no-template controls. For each HPV16-positive HSIL, 24 µL of extracted DNA is subjected to bisulfite conversion and DNA cleanup using the Methylamp DNA Modification Kit (Epigentek, Farmingdale, New York, USA), according to the manufacturer's instructions. This procedure converts unmethylated cytosine (C) residues into tyrosine (T) residues. Following conversion, two targeted sections of the HPV16 genome are amplified by nested PCR; samples are amplified in a first round of PCR using the primary pair of sequences, and a 1 µL aliquot is then amplified in a second round of PCR using the secondary (nested) PCR primers, as listed in table 1. The amplified sections are the E2BS1 region from nucleotide position 7348 to 7489 (final amplicon size 142 bp), and E2BS2-4 region from nucleotide position 7839 to 117 (final amplicon size 184 bp). Primary PCR amplifications are performed in reaction mixes with a final volume of 25 μL (12.5 μL of Hot Start Taq Master mix (Qiagen); 0.25 μmol of each primer; 5 μL of bisulfite modified template DNA; nuclease-free water to final volume). Thermocycler parameters are as follows: 15 min at 95°C, followed by 50 cycles of (40 s at 95°C, 30 s at 50°C and 40 s at 72°C), followed by 6 min at 72°C. Amplicons are visualised on a 2% agarose gel. The secondary (nested) PCR amplification for the E2BS1 region is performed in reaction mixes with a final volume of 25 μL (12.5 μL of Hot Start Taq Master mix (Qiagen); 0.25 μmol of each nested primer; 1 μL of amplicon from the primary PCR; nuclease-free water to final volume). Thermocycler parameters are as follows: 15 min at 95°C, followed by 50 cycles of (40 s at 95°C, 30 s at 56°C and 40 s at 72°C), followed by 6 min at 72°C. The secondary PCR amplification for the E2BS2-4 region is performed in reaction mixes with a final volume of 25 μL (12.5 μL of Hot Start Taq Master mix (Qiagen); 0.25 μmol of each nested primer; 1 μL of amplicon from the primary PCR; nuclease-free water to final volume). Thermocycler parameters are as follows: 15 min at 95°C, followed by 50 cycles of (40 s at 95°C, 30 s at 50°C and 40 s at 72°C), followed by 6 min at 72°C. CpG sequence analysis and quantitation of methylation at nucleotide positions 7426, 7432, 7453, 7459, 7860, 31, 37, 43, 52 and 68 on the HPV16 viral genome52 is performed using pyrosequencing technology on the PyroMark Q24 system (Qiagen) at the Australian Genome Research Facility (AGRF). Assay setup, pyrosequence run and analysis are performed by PyroMark Q24 Software. Sequencing primers for E2BS1, E2BS2 and E2BS3,4 regions are listed in table 1.
Table 1.
Primer sets for determination of human papillomavirus 16 CpG methylation using pyrosequencing
| Primer name | Primer sequence | Amplicon size (bp) |
|---|---|---|
| E2BS1 amplicon | ||
| Primary amplification forward primer Ampy E2BS1 F2 bio | 7348 5′-Biotin-ATTGTGTTGTGGTTATTTATTGTA-3′ 7371 | 175 |
| Primary amplification reverse primer Ampy E2BS1 R3 | 7522 5′-AACCATAATTACTAACATAAAACT-3′ 7499 | |
| Nested amplification forward primer Ampy E2BS1 F2 bio | 7348 5′-Biotin-ATTGTGTTGTGGTTATTTATTGTA-3′ 7371 | 142 |
| Nested amplification reverse primer Ampy E2BS1 R2 | 7489 5′-AACACATTTTATACCAAAAAAC-3′ 7468 | |
| Sequencing primer Sempy E2BS1 R2 |
7486 5′-ACATTTTATACCAAAAAACAT-3′ 7466 | – |
| E2BS2-4 amplicon | ||
| Primary amplification forward primer Ampy E2BS2,3,4 F | 7830 5′-TTGTAAAATTGTATATGGGTGTG-3′ 7852 | 193 |
| Primary amplification reverse primer Ampy E2BS2,3,4 R bio | 117 5′-Biotin-AAATCCTAAAACATTACAATTCTC-3′ 94 | |
| Nested amplification forward primer Ampy E2BS2 F | 7839 5′-TGTATATGGGTGTGTGTAAAT-3′ 7859 | 184 |
| Nested amplification reverse primer Ampy E2BS2,3,4 R bio | 117 5′-Biotin-AAATCCTAAAACATTACAATTCTC-3′ 94 | |
| Sequencing primer Sempy E2BS2 F |
7839 5′-TGTATATGGGTGTGTGTAAAT-3′ 7859 | – |
| Sequencing primer Sempy E2BS3,4 F |
9 5′-AATTTATGTATAAAATTAAGGG-3′ 30 | – |
Other biomarkers
Additional biomarker analyses may include host genome marker methylation sequencing, viral genome integration frequency and relative transcription levels of HPV mRNA species, contingent on sample availability and assay performance.
Statistical considerations
In total, about 600 participants (30–40% of whom will be HIV-positive) will be recruited by June 2015. As of the end of 2014, 480 participants have been recruited with 125 (26%, 95% CI 22% to 30%) men diagnosed with biopsy-confirmed LSIL and 143 (30%, 95% CI 26% to 34%) men with biopsy-confirmed anal HSIL. Therefore, we anticipate between 130 and 180 of the total 600 participants will be diagnosed with LSIL, and between 150 and 200 will be diagnosed with biopsy-confirmed HSIL at baseline.
HPV prevalence in LCM-isolated lesions, including low-risk (including β-papillomavirus or HPV X), high-risk and individual genotypes will be calculated overall and stratified by HIV status as the proportion of lesions that tested positive for a specific HPV type or risk group. The exact binomial method will be used to calculate the corresponding 95% CIs for prevalence values. Biomarker positivity associated with lesion type will be calculated using the same method. Univariate and multivariate logistic regression models will be carried out to measure the strength of association between HPV status and each biomarker in lesions at baseline.
To determine the performance of biomarkers in predicting disease persistence at 12 months, univariate and multivariate Cox regressions will be performed to assess the strength of the association between biomarker positivity and anal HSIL persistence at 12 months in those who are diagnosed with anal HSIL at baseline. The sensitivity and specificity of each biomarker detected on anal swab to predict persistent HSIL at 12 months will be calculated alone and in combination with HPV genotyping results. In addition, univariate logistic regression models will be carried out to measure the strength of each test in relation to a diagnosis of HSIL. A multivariate logistic regression model will be constructed to determine a combination of these biomarkers that best predict persistent HSIL and to assess the association with HIV status and other covariates such as age.
Percentages (%) agreement and κ statistic will be used to determine HPV concordance in anal swab, WTS and LCM sample (poor if κ≤0.20; fair if 0.21≤κ≤0.40; moderate if 0.41≤κ≤0.60; substantial if 0.61≤κ≤0.80; good if κ>0.80). Differences in HPV genotypes prevalence between samples will be assessed by the McNemar test.
Currently available algorithms to determine the most likely lesion-associated genotype based on swab samples42 will be compared with actual lesion-associated genotypes determined using LCM. Percentage detection and 95% CIs will be used to calculate the performance of hierarchical and proportional attribution methods against LCM-determined lesion-associated genotypes.
Ethics and dissemination
Written informed consent is obtained from all individuals before any study-specific procedures are performed. As clinical guidelines do not indicate treatment and there is currently no scientific evidence to support routine treatment of anal HSIL,54 55 participants diagnosed with HSIL are not routinely treated, except where deemed clinically necessary; this may include severely abnormal blood vessels, persistent large lesions or other risk factors for ASCC, such as prolonged immunosuppression, low CD4 nadir and history of smoking.
Findings from this study will be disseminated to participants and the community through study newsletters, and through peer-reviewed publications and international conferences.
Discussion
Importance of this study
With continuing increases in ASCC incidence,3 56–58 future prevention of this disease will rely on effective vaccination and screening measures and these in turn will depend on accurate data pertaining to the association of individual HPV genotypes with ASCC and with anal HSIL. Persistent anal HSIL is widely accepted to be the precursor to ASCC, and is the anal counterpart to cervical HSIL, the main disease end point in cervical screening programmes. The end point for anal screening will likely differ from cervical screening as rates of progression from anal HPV infection and/or HSIL to anal cancer are very low,4 24 and the populations that are likely to be most frequently screened contain a high proportion of individuals with HR-HPV and/or HSIL.4 59 Effective anal screening programmes will therefore require very high specificity for an appropriate end point, to limit unnecessary procedures on a large number of low-risk individuals, as well as high sensitivity to identify the small population of high-risk individuals. This research will contribute to the determination of appropriate end points for anal screening.
Approximately 70–90% of cases of ASCC are attributed to infection with HPV16.25 26 33 58 60 However, the exact contribution of HPV16, and perhaps more importantly other HPV genotypes, to the load of ASCC cases is not known, as up to a quarter of anal cancer specimens test positive for multiple HPV genotypes.25 40 41 Where HPV16 is present, it is usually assumed to be the causative genotype; given that HPV16 is a commonly found genotype in any population of sexually active adults, it is highly likely to be incidental to the cancer in a number of cases and therefore its contribution to cancer development overestimated. Conversely, an increasing quantity of evidence suggests that so-called LR-HPV genotypes—particularly HPV6 and 11—contribute to a larger proportion than expected based on cervical cancer data sets25 31 49 60 61 and present data collection methods probably lead to under-estimation of their contribution to anal cancer rates.
In addition to HPV genotype detection, methylation patterns also have potential utility as biomarkers in cancer screening,52 62 particularly with rapid advances in sequencing technology and the increasing recognition in other medical fields of the diagnostic potential of epigenetic markers.63 Currently available data on methylation of viral DNA targets are insufficient to inform development of methylation as a screening marker for HPV-related cancers, and are derived predominantly from cervical tissue and cervical cancer cell lines with only a single study presenting any methylation sequence data for the HPV genome in anal lesions.64
Because it is the persistence of HR-HPV and HSIL that lead to increased risk of anal cancer development, longitudinal studies provide more useful information than cross-sectional data, particularly for rare events such as anal cancer incidence. Longitudinal or natural history studies provide data to allow more accurate risk categories to be assigned to single screening results, so that individual patients can be triaged appropriately. This prospective, longitudinal study is designed to provide data on risk factors associated with persistent HSIL—and by extension increased risk of anal cancer development—to inform future decisions on anal cancer prevention in at-risk populations.
Strengths of this study
This participant cohort is a unique resource, as one of only a very small number of cohorts with prospectively collected longitudinal cytology and histology samples from HIV-positive and HIV-negative participants diagnosed with the full spectrum of HPV-associated anal lesions. A team of intensively trained anoscopists have their diagnostic skills monitored regularly with extensive quality control metrics, ensuring high-quality clinical data. Participants in this cohort are not routinely treated when HSIL is detected unless it is determined to be clinically necessary, for example, severely abnormal blood vessels, persistent large lesions or other risk factors for ASCC, such as prolonged immunosuppression, low CD4 nadir and history of smoking. This approach allows for longitudinal follow-up of lesions, and therefore allows assessment of when treatment is likely to be most beneficial.
A major strength of the present study is the use of LCM to distinguish lesions from adjacent normal tissue, which allows for precise genotype and biomarker testing from which to infer the natural history of HPV-associated anal lesions. Methylation patterns and the behaviour of other biomarkers are also often tissue-specific or lesion-specific, and can be heterogeneous within even a small tissue sample.53 The use of LCM generates extremely low-concentration DNA samples, and the assays being used in this study are very sensitive for low copies of DNA targets, and have a small amplicon size for the detection of fragmented DNA, which is partially degraded in the fixation process. These methods have been proven sensitive enough for HPV genotyping and methylation sequence analysis in published studies23 39 53 and in our own laboratory.
In routine diagnostic work, p16 immunostaining is utilised, as per LAST criteria, for confirming a diagnosis of HSIL-AIN2 and for distinguishing true HSIL-AIN3 from atypical immature metaplasia. This latter differential diagnosis is more challenging in the anal canal that in the cervix (T Darragh, personal communication, 2013). Routine p16 staining is not indicated for diagnostic samples due to the uncertain significance of p16-positive LSIL,45 but it is a useful research tool for further exploring the natural history of p16-positive LSIL in conjunction with HPV genotype and/or biomarker analysis and potentially clarifying the significance of these lesions.
Anticipated challenges
The major challenges of this study relate to diagnostic uncertainty and establishing whether or not a lesion is persistent from one visit to the next. It is inevitable that some lesions will be missed during HRA, and therefore multiple sources of data will be used to define the persistence of individual lesions, as indicated in figure 2. The study uses both cytology and histology (based on HRA-guided biopsy collection) to establish disease grade in participants at each visit. It is known that anal cytology and HRA/histology each have their limitations,21 65 and therefore both methods will be used in a complementary fashion to derive a more accurate diagnosis. Longitudinal sampling and identification of the same individual lesion has not been attempted before in a systematic manner. This adds an extra dimension of complexity to the project. Defining persistent lesions will not be straightforward due to the difficulties inherent in sampling the anal canal, the possibility of missed lesions, the probability of multiple lesions and the uncertainties in locating and identifying the same lesion at consecutive visits.23 The presence of multiple distinct lesions caused by the same genotype cannot be ruled out. All anoscopists are aware of each participant's previous clinic results and have access to HRA photographs; this improves the likelihood of sampling the same lesion during subsequent HRA visits. It is known that the anal canal may rotate up to 90° between clinic visits (personal communication R J Hillman, 2012), making precise location of anatomical features difficult; this has been acknowledged and follow-up biopsies will be considered if they are taken up to one octant either side of the baseline biopsy to reduce the chance of missing a persistent lesion. The use of features in HRA photographs will also aid in determining lesion location, and in conjunction with anoscopists’ notes will be used to indicate the extent of disease (ie, across multiple octants). Each original biopsy diagnosis is made by one of a team of three experienced anogenital pathologists, and this is the basis on which tissues are selected for inclusion. The review diagnoses, however, are all made by one of these three pathologists (JMR) based on the new H&E and p16 slides as described in the Methods section. The main issue resulting from this will be confirming persistence of HSIL when the review diagnosis at follow-up has been downgraded to LSIL or negative for SIL. In these cases, the study investigators will make a decision based on the original diagnosis, the HPV genotype detected by LCM, and the histological diagnosis at the next clinic visit.
This study will use precise molecular biology techniques and international expertise to characterise the natural history and biological markers of individual anal lesions in a population at high risk of anal cancer. This is an ambitious and unique study which will enhance our understanding of the clinical and biological behaviour of HPV-related anal lesions and the potential prognostic implications of future HPV genotype or biomarker screening tests.
Footnotes
Collaborators: The SPANC study team includes: AEG, IMP, FJ, Brian Acraman, Garrett Prestage, Leonie Crampton, Patrick McGrath, Kathy Petoumenos, Matthew O'Dwyer, Robert Mellor, Piero Pezzopane, Matthew Law (Kirby Institute, University of New South Wales); Carmella Law, Winnie Tong, Daniel Seeds, Eddie Fraissard and Andrew Carr (St Vincent's Hospital, Sydney); DJT and Rick Varma (RPA Sexual Health, Sydney); Kit Fairley (Melbourne Sexual Health Centre); RJH, Kirsten McCaffery and Kirsten Howard (University of Sydney); Annabelle Farnsworth, JMR, Adele Richards and Julia Thurloe (Douglass Hanly Moir Pathology, Sydney); SMG, SNT, AMC and SP (The Royal Women's Hospital, Melbourne); and Lance Feeney and Russ Gluyas (community representatives).
Contributors: All authors contributed to study design. AMC drafted the manuscript. JMR provided histopathological expertise. SP and MM provided molecular biology expertise. DAM provided statistical expertise. RJH provided clinical expertise. AEG, FJ, IMP, DJT, SMG and SNT collectively conceived of the study and provided epidemiological and molecular biology expertise. All authors read and approved of the final manuscript.
Funding: The SPANC study is funded by Australian Commonwealth Government National Health and Medical Research Council Program grants (#568971 and #1071269) and a Cancer Council New South Wales Strategic Research Partnership Program grant (#13-11).
Competing interests: None declared.
Ethics approval: The St Vincent's Hospital Ethics Committee.
Provenance and peer review: Not commissioned; externally peer reviewed.
Contributor Information
Collaborators: on behalf of the SPANC Study Team, Brian Acraman, Garrett Prestage, Leonie Crampton, Patrick McGrath, Kathy Petoumenos, Matthew O'Dwyer, Robert Mellor, Piero Pezzopane, Matthew Law, Carmella Law, Winnie Tong, Daniel Seeds, Eddie Fraissard, Andrew Carr, Rick Varma, Kit Fairley, Kirsten McCaffery, Kirsten Howard, Annabelle Farnsworth, Adele Richards, Julia Thurloe, Lance Feeney, and Russ Gluyas
References
- 1.Grulich AE, Poynten IM, Machalek DA et al. The epidemiology of anal cancer. Sex Health 2012;9:504–8. 10.1071/SH12070 [DOI] [PubMed] [Google Scholar]
- 2.Grulich AE, Jin F, Conway L et al. Cancers attributable to human papillomavirus infection. Sex Health 2010;7:244–52. 10.1071/SH10020 [DOI] [PubMed] [Google Scholar]
- 3.Jin F, Stein AN, Conway EL et al. Trends in anal cancer in Australia, 1982–2005. Vaccine 2011;29:2322–7. 10.1016/j.vaccine.2011.01.015 [DOI] [PubMed] [Google Scholar]
- 4.Machalek DA, Poynten M, Jin F et al. Anal human papillomavirus infection and associated neoplastic lesions in men who have sex with men: a systematic review and meta-analysis. Lancet 2012;13:487–500. 10.1016/S1470-2045(12)70080-3 [DOI] [PubMed] [Google Scholar]
- 5.van Leeuwen MT, Vajdic CM, Middleton MG et al. Continuing declines in some but not all HIV-associated cancers in Australia after widespread use of antiretroviral therapy. AIDS 2009;23:2183–90. 10.1097/QAD.0b013e328331d384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daling JR, Weiss NS, Hislop TG et al. Sexual practices, sexually transmitted diseases, and the incidence of anal cancer. N Engl J Med 1987;317:973–7. 10.1056/NEJM198710153171601 [DOI] [PubMed] [Google Scholar]
- 7.van der Zee RP, Richel O, de Vries HJC et al. The increasing incidence of anal cancer: can it be explained by trends in risk groups? Neth J Med 2013;71:401–11. [PubMed] [Google Scholar]
- 8.Grulich AE, van Leeuwen MT, Falster MO et al. Incidence of cancers in people with HIV/AIDS compared with immunosuppressed transplant recipients: a meta-analysis. Lancet 2007;370:59–67. 10.1016/S0140-6736(07)61050-2 [DOI] [PubMed] [Google Scholar]
- 9.Giuliano AR, Palefsky JM, Goldstone S et al. Efficacy of quadrivalent HPV vaccine against HPV infection and disease in males. N Engl J Med 2011;364:401–11. 10.1056/NEJMoa0909537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palefsky JM, Giuliano AR, Goldstone S et al. HPV vaccine against anal HPV infection and anal intraepithelial neoplasia. N Engl J Med 2011;365:1576–85. 10.1056/NEJMoa1010971 [DOI] [PubMed] [Google Scholar]
- 11.Tabrizi SN, Brotherton JML, Kaldor JM et al. Fall in human papillomavirus prevalence following a national vaccination program. J Infect Dis 2012;206:1645–51. 10.1093/infdis/jis590 [DOI] [PubMed] [Google Scholar]
- 12.Brotherton JML, Fridman M, May CL et al. Early effect of the HPV vaccination programme on cervical abnormalities in Victoria, Australia: an ecological study. Lancet 2011;377:2085–92. 10.1016/S0140-6736(11)60551-5 [DOI] [PubMed] [Google Scholar]
- 13.Gertig DM, Brotherton JM, Budd AC et al. Impact of a population-based HPV vaccination program on cervical abnormalities: a data linkage study. BMC Med 2013;11:227 10.1186/1741-7015-11-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson E. Australia leads way on HPV vaccination in boys. Lancet Infect Dis 2012;12:831–2. 10.1016/S1473-3099(12)70282-7 [DOI] [PubMed] [Google Scholar]
- 15.Luxembourg A; On behalf of the V503 Program Team. An overview of the 9-valent HPV L1 virus-like particle vaccine clinical development program. Florence, Italy: EUROGIN International Congress, 2013. [Google Scholar]
- 16.U.S. Food and Drug Administration. News & Events. http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm426485.htm (accessed 18 Feb 2015).
- 17.Ault KA, The Future II Study Group. Effect of prophylactic human papillomavirus L1 virus-like-particle vaccine on risk of cervical intraepithelial neoplasia grade 2, grade 3, and adenocarcinoma in situ: a combined analysis of four randomised clinical trials. Lancet 2007;369:1861–8. 10.1016/S0140-6736(07)60852-6 [DOI] [PubMed] [Google Scholar]
- 18.Haupt RM, Wheeler CM, Brown DR et al. Impact of an HPV6/11/16/18 L1 virus-like particle vaccine on progression to cervical intraepithelial neoplasia in seropositive women with HPV16/18 infection. Int J Cancer 2011;129:2632–42. 10.1002/ijc.25940 [DOI] [PubMed] [Google Scholar]
- 19.Joura EA, Garland SM, Paavonen J et al. Effect of the human papillomavirus (HPV) quadrivalent vaccine in a subgroup of women with cervical and vulvar disease: retrospective pooled analysis of trial data. BMJ 2012;344:e1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farnsworth A. Screening for the prevention of cervical cancer in the era of human papillomavirus vaccination: an Australian perspective. Acta Cytol 2011;55:307–12. 10.1159/000326956 [DOI] [PubMed] [Google Scholar]
- 21.Darragh TM, Winkler B. Anal cancer and cervical cancer screening: key differences. Cancer Cytopathol 2011;119:5–19. 10.1002/cncy.20126 [DOI] [PubMed] [Google Scholar]
- 22.Palefsky JM, Holly EA, Hogeboom CJ et al. Anal cytology as a screening tool for anal squamous intraepithelial lesions. J Acquir Immune Defic Syndr 1997;14:415–22. 10.1097/00042560-199704150-00004 [DOI] [PubMed] [Google Scholar]
- 23.Richel O, Quint KD, Lindeman J et al. One lesion, one virus: individual components of high grade anal intraepithelial neoplasia in HIV+ men contain a single HPV type. J Infect Dis 2014;210:111–20. 10.1093/infdis/jiu052 [DOI] [PubMed] [Google Scholar]
- 24.Cachay E, Agmas W, Mathews C. Five-year cumulative incidence of invasive anal cancer among HIV-infected patients according to baseline anal cytology results: an inception cohort analysis. HIV Med 2014;16:191–5. 10.1111/hiv.12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hillman RJ, Garland SM, Gunathilake MPW et al. Human papillomavirus (HPV) genotypes in an Australian sample of anal cancers. Int J Cancer 2014;135:996–1001. 10.1002/ijc.28730 [DOI] [PubMed] [Google Scholar]
- 26.Ouhoummane N, Steben M, Coutlee F et al. Squamous anal cancer: patient characteristics and HPV type distribution. Cancer Epidemiol 2013;37:807–12. 10.1016/j.canep.2013.09.015 [DOI] [PubMed] [Google Scholar]
- 27.Australian Government Department of Health. National Cervical Screening Program: Overview of the Renewal. http://www.cancerscreening.gov.au/internet/screening/publishing.nsf/Content/overview-of-the-renewal (accessed 18 Feb 2015).
- 28.Huh WK, Ault KA, Chelmow D et al. Use of primary high-risk human papillomavirus testing for cervical cancer screening: interim clinical guidance. J Low Genit Tract Dis 2015;19:91–6. 10.1097/LGT.0000000000000103 [DOI] [PubMed] [Google Scholar]
- 29.Bierkens M, Hesselink AT, Meijer CJLM et al. CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer 2013;133:1293–300. 10.1002/ijc.28138 [DOI] [PubMed] [Google Scholar]
- 30.Zhang J, Martins CR, Fansler ZB et al. DNA methylation in anal Intraepithelial lesions and anal squamous cell carcinoma. Clin Cancer Res 2005;11:6544–9. 10.1158/1078-0432.CCR-05-0374 [DOI] [PubMed] [Google Scholar]
- 31.Alemany L, Saunier M, Alvarado-Cabrero I et al. HPV DNA prevalence and type distribution in anal carcinomas worldwide. Int J Cancer 2015;136:98–107. 10.1002/ijc.28963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walboomers JMM, Jacobs MV, Manos MM et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 1999;189:12–19. [DOI] [PubMed] [Google Scholar]
- 33.De Vuyst H, Clifford GM, Nascimento MC et al. Prevalence and type distribution of human papillomavirus in carcinoma and intraepithelial neoplasia of the vulva, vagina and anus: a meta-analysis. Int J Cancer 2009;124:1626–39. 10.1002/ijc.24116 [DOI] [PubMed] [Google Scholar]
- 34.Gohy L, Gorska I, Rouleau D et al. Genotyping of human papillomavirus DNA in anal biopsies and anal swabs collected from HIV-seropositive men with anal dysplasia. J Acquir Immune Defic Syndr 2008;49:32–9. 10.1097/QAI.0b013e318183a905 [DOI] [PubMed] [Google Scholar]
- 35.Sahasrabuddhe VV, Castle PE, Follansbee S et al. Human papillomavirus genotype attribution and estimation of preventable fraction of anal intraepithelial neoplasia cases among HIV-infected men who have sex with men. J Infect Dis 2013;207:392–401. 10.1093/infdis/jis694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong AK, Chan RC, Aggarwal N et al. Human papillomavirus genotypes in anal intraepithelial neoplasia and anal carcinoma as detected in tissue biopsies. Mod Pathol 2010;23:144–50. 10.1038/modpathol.2009.143 [DOI] [PubMed] [Google Scholar]
- 37.Machalek DA, Grulich AE, Jin F et al. The epidemiology and natural history of anal human papillomavirus infection in men who have sex with men. Sex Health 2013;9:527–37. 10.1071/SH12043 [DOI] [PubMed] [Google Scholar]
- 38.Nyitray AG. The epidemiology of anal human papillomavirus infection among women and men having sex with women. Sex Health 2012;9:538–46. 10.1071/SH12021 [DOI] [PubMed] [Google Scholar]
- 39.Quint W, Jenkins D, Molijn A et al. One virus, one lesion—individual components of CIN lesions contain a specific HPV type. J Pathol 2012;227:62–71. 10.1002/path.3970 [DOI] [PubMed] [Google Scholar]
- 40.Abramowitz L, Jacquard A-C, Jaroud F et al. Human papillomavirus genotype distribution in anal cancer in France: the EDiTH V study. Int J Cancer 2010;129:433–9. 10.1002/ijc.25671 [DOI] [PubMed] [Google Scholar]
- 41.Steinau M, Unger ER, Hernandez BY et al. Human papillomavirus prevalence in invasive anal cancers in the United States prior to vaccine introduction. J Low Genit Tract Dis 2013;17:397–403. 10.1097/LGT.0b013e31827ed372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wentzensen N, Schiffman M, Dunn ST et al. Multiple HPV genotype infections in cervical cancer progression in the Study to Understand Cervical Cancer Early Endpoints and Determinants (SUCCEED). Int J Cancer 2009;125:2151–8. 10.1002/ijc.24528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Machalek DA, Grulich AE, Hillman RJ et al. The Study of the Prevention of Anal Cancer (SPANC) design and methods of a three-year prospective cohort study. BMC Public Health 2013;13:946 10.1186/1471-2458-13-946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darragh TM, Colgan TJ, Cox JT et al. The Lower Anogenital Squamous Terminology Standardization Project for HPV-associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. J Low Genit Tract Dis 2012;16:205–42. 10.1097/LGT.0b013e31825c31dd [DOI] [PubMed] [Google Scholar]
- 45.Darragh TM, Colgan TJ, Cox JT et al. The Lower Anogenital Squamous Terminology Standardization Project for HPV-associated lesions: background and consensus recommendations from the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology. Arch Pathol Lab Med 2012;136:1266–97. 10.5858/arpa.LGT200570 [DOI] [PubMed] [Google Scholar]
- 46.Pirog EC, Quint KD, Yantiss RK. P16/CDKN2A and Ki-67 enhance the detection of anal intraepithelial neoplasia and condyloma and correlate with human papillomavirus detection by polymerase chain reaction. Am J Surg Pathol 2010;34:1449–55. 10.1097/PAS.0b013e3181f0f52a [DOI] [PubMed] [Google Scholar]
- 47.Oh K-T, Palefsky J. Diseases of the anus. In: CP C, KR L, eds. Diagnostic gynecologic and obstetric pathology. Philadelphia: Elsevier Saunders, 2006:199–227. [Google Scholar]
- 48.Stevens MP, Garland SM, Tabrizi SN. Development and validation of a real-time PCR assay specifically detecting human papillomavirus 52 using the Roche LightCycler® 480 system. J Virol Methods 2008;147:290–6. 10.1016/j.jviromet.2007.09.018 [DOI] [PubMed] [Google Scholar]
- 49.Cornall AM, Roberts J, Garland SM et al. Anal and perianal cancers exclusively associated with low-risk HPV genotypes 6 and 11. Int J Cancer 2013;133:2253–8. 10.1002/ijc.28228 [DOI] [PubMed] [Google Scholar]
- 50.Berkhout RJM, Tieben LM, Smits HL et al. Nested PCR approach for detection and typing of epidermodysplasia verruciformis-associated human papillomavirus types in cutaneous cancers from renal transplant recipients. J Clin Microbiol 1995;33:690–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin L, Yao J, Gardner BP et al. HPV-QUEST: a highly customized system for automated HPV sequence analysis capable of processing next generation sequencing data set. Bioinformation 2012;8:388–90. 10.6026/97320630008388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaiwongkot A, Vinokurova S, Pientong C et al. Differential methylation of E2 binding sites in episomal and integrated HPV 16 genomes in preinvasive and invasive cervical lesions. Int J Cancer 2013;132:2087–94. 10.1002/ijc.27906 [DOI] [PubMed] [Google Scholar]
- 53.Vinokurova S, von Knebel Doeberitz M. Differential methylation of the HPV 16 upstream regulatory region during epithelial differentiation and neoplastic transformation. PLoS ONE 2011;6:e24451 10.1371/journal.pone.0024451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Macaya A, Munoz-Santos C, Balaguer A et al. Interventions for anal canal intraepithelial neoplasia. Cochrane Database Syst Rev 2012(10):CD009244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richel O, de Vries HJC, van Noesel CJM et al. Comparison of imiquimod, topical fluorouracil, and electrocautery for the treatment of anal intraepithelial neoplasia in HIV-positive men who have sex with men: an open-label, randomised controlled trial. Lancet Oncol 2013;14:346–53. 10.1016/S1470-2045(13)70067-6 [DOI] [PubMed] [Google Scholar]
- 56.Joseph DA, Miller JW, Wu X et al. Understanding the burden of human papillomavirus-associated anal cancers in the US. Cancer 2008;113(S10):2892–900. 10.1002/cncr.23744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brewster DH, Bhatti LA. Increasing incidence of squamous cell carcinoma of the anus in Scotland, 1975–2002. Br J Cancer 2006;95:87–90. 10.1038/sj.bjc.6603175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frisch M, Glimelius B, van den Brule AJC et al. Sexually transmitted infection as a cause of anal cancer. N Engl J Med 1997;337:1350–8. 10.1056/NEJM199711063371904 [DOI] [PubMed] [Google Scholar]
- 59.Coutlee F, de Pokomandy A, Franco EL. Epidemiology, natural history and risk factors for anal intraepithelial neoplasia. Sex Health 2012;9:547–55. 10.1071/SH11167 [DOI] [PubMed] [Google Scholar]
- 60.Daling JR, Madeleine MM, Johnson LG et al. Human papillomavirus, smoking, and sexual practices in the etiology of anal cancer. Cancer 2004;101:270–80. 10.1002/cncr.20365 [DOI] [PubMed] [Google Scholar]
- 61.Guimera N, Lloveras B, Lindeman J et al. The occasional role of low-risk human papillomaviruses 6, 11, 42, 44, and 70 in anogenital carcinoma defined by laser capture microdissection/PCR methodology: results from a global study. Am J Surg Pathol 2013;37:1299–310. 10.1097/PAS.0b013e31828b6be4 [DOI] [PubMed] [Google Scholar]
- 62.Kalantari M, Calleja-Macias IE, Tewari D et al. Conserved methylation patterns of human papillomavirus type 16 DNA in asymptomatic infection and cervical neoplasia. J Virol 2004;78:12762–72. 10.1128/JVI.78.23.12762-12772.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Partin AW, Van Neste L, Klein EA et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J Urol 2014;192:1081–7. 10.1016/j.juro.2014.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiley DJ, Huh J, Rao JY et al. Methylation of human papillomavirus genomes in cells of anal epithelia of HIV-infected men. J Acquir Immune Defic Syndr 2005;39:143–51. [PubMed] [Google Scholar]
- 65.Berry JM, Palefsky JM, Jay N et al. Performance characteristics of anal cytology and human papillomavirus testing in patients with high-resolution anoscopy-guided biopsy of high-grade anal intraepithelial neoplasia. Dis Colon Rectum 2009;52: 239–47. 10.1007/DCR.0b013e31819793d9 [DOI] [PubMed] [Google Scholar]