Abstract
Centrioles are short microtubule-based organelles with a conserved ninefold symmetry. They are essential for both centrosome formation and cilium biogenesis in most eukaryotes. A core set of five centriolar proteins has been identified and their sequential recruitment to procentrioles has been established. However, structures at atomic resolution for most of the centriolar components were scarce, and the underlying molecular mechanisms for centriole assembly had been a mystery—until recently. In this review, I briefly summarize recent advancements in high-resolution structural characterization of the core centriolar components and discuss perspectives in the field.
Keywords: centriole, protein, structure
1. Introduction
Centrioles are cylindrical cellular structures present in almost all eukaryotic lineages. They are 0.1–0.5 µm long and 0.1–0.2 µm in diameter, and are usually composed of nine microtubule triplets at their outer wall [1,2]. Exceptions to this organization are found in Drosophila embryos (nine doublet microtubules), and in Caenorhabditis elegans sperm cells and early embryos (nine singlets).
Centrioles usually exist in pairs, oriented perpendicularly to one another, and are surrounded by the pericentriolar material (PCM) to form the centrosome [3,4]. The PCM has a layered structure made of fibres and matrix proteins, and is responsible for microtubule nucleation and anchoring [5–8]. As such, the centrosome is the major microtubule organizing centre of animal cells. Centrosome duplication is controlled by centriole replication, which in most animal cells is tightly coordinated with the mitotic cycle. Miscoordination of the two processes often leads to alterations in centrosome number and/or structure that result in chromosome missegregation and severe consequences such as cancerous growth of the cell [9–12]. Besides nucleating centrosomes, centrioles also play a pivotal role in the biogenesis and operation of the cilium, an antenna-like structure projecting out from the cell surface [13]. Cilia are present on virtually every human cell type, and defects in their assembly or function lead to a plethora of human disorders, which are collectively called the ciliopathies [14,15].
Despite cell biologists' great interest in the centriole and extensive cellular and microscopic studies on this organelle for several decades, the molecular composition of the centriole was not fully determined until recently. Because of its favourable features for laboratory study and the availability of powerful genetic and genomic tools, the nematode C. elegans has become an important model organism in studying centrosome duplication and centriole assembly. Studies in several research groups have uncovered five core C. elegans centriolar proteins: the polo-like kinase ZYG-1 [16], and the four coiled-coil-containing proteins, SPD-2 [17,18], SAS-4 [19,20], SAS-5 [21] and SAS-6 [22,23]. All of these proteins contain both structured domains and disordered segments and are conserved in all ciliated cells.
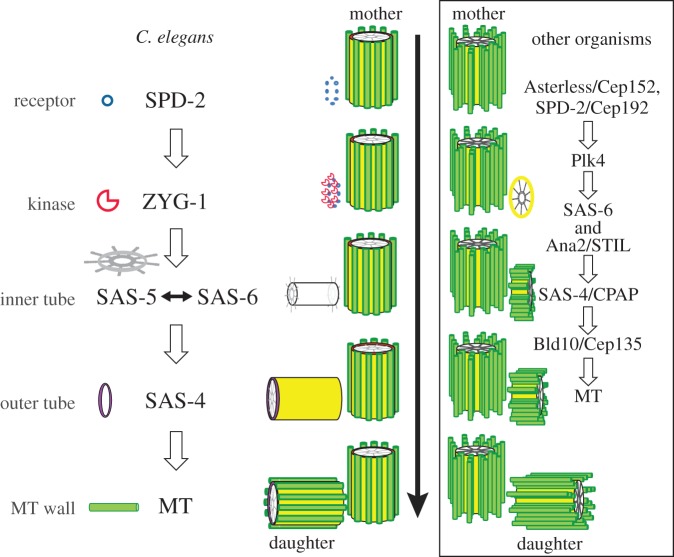
Centriole duplication is a multistep process, with each of the five centriolar proteins being recruited in a hierarchical manner [24,25]. In C. elegans, centriole assembly is initiated by the recruitment of SPD-2 to the proximal side of the mother centriole. The kinase ZYG-1 is then incorporated into the SPD-2 scaffold, and is required for the subsequent recruitment of the SAS-5/SAS-6 complex. SAS-4 is then recruited to add an outer tube around the central tube formed by SAS-5 and SAS-6. Finally, nine singlet microtubules are assembled onto the outer tube to generate a daughter centriole (figure 1). In other organisms, including Drosophila and humans, additional essential proteins besides the five core components are also recruited during centriole assembly (figure 1).
Figure 1.
Centriole assembly in C. elegans. Centriole assembly is initiated by the recruitment of SPD-2 to the vicinity of the mother centriole. ZYG-1 is incorporated and then recruits the SAS-5/SAS-6 complex that forms the inner tube. Incorporation of SAS-5/SAS-6 promotes SAS-4 recruitment, which generates the outer tube. Finally, nine singlet microtubules assemble symmetrically around the outer tube to generate the microtubule wall. For comparison, a schematic of a similar stepwise centriole assembly in other organisms is shown on the right, with the major involved proteins being listed.
Recent years have seen a surge of structural dissections of the centriole, with an increasing number of high-resolution structures reported for most of its core components. Together, these have greatly advanced our understanding of centriole duplication at the atomic level. Nevertheless, a full molecular view of centriolar assembly is a mission that is not yet completed. This review briefly summarizes recent progress in the structural characterization of centriole assembly, which promises a challenging but rewarding future.
2. SPD-2/Cep192 and Asterless/Cep152
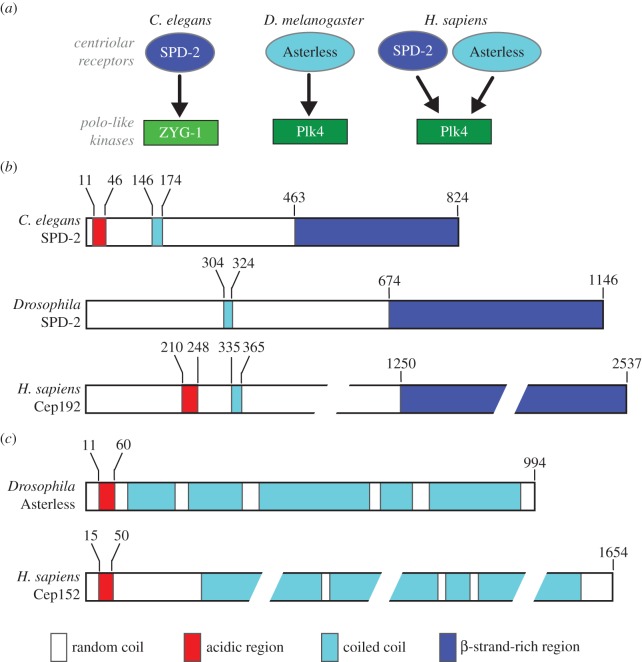
Centriole duplication in animals is initiated by recruiting the centriolar receptors SPD-2 and/or Asterless to the vicinity of the mother centriole. These receptors then recruit polo-like kinase 4 (Plk4) and ZYG-1, depending on which organism is under consideration (figure 2a). SPD-2 is a coiled-coil-containing protein localizing to both the centriole and the PCM in C. elegans [17,18]. It has an N-terminal acidic region that binds to the cryptic polo-box (CPB) domain of ZYG-1 (figure 2b) [26]. The Drosophila orthologue of SPD-2 lacks the acidic region for Plk4 binding, and thus is dispensable for centriole duplication [27,28] (figure 2b). The mammalian SPD-2 orthologue Cep192 also bears an acidic region that interacts with the Plk4 CPB domain, but it is located a bit further downstream in the sequence (figure 2b).
Figure 2.
Recruitment of the polo-like kinases Plk4/ZYG-1 and domain organization of the centriolar receptors. (a) Different recruitment modes of ZYG-1/Plk4 by their respective centriolar receptors SPD-2/Asterless. Caenorhabditis elegans has only one centriolar receptor, SPD-2, to recruit ZYG-1. Drosophila has both SPD-2 and Asterless, but only Asterless can bind and recruit Plk4. In humans, both SPD-2 and Asterless bind Plk4 but in a mutually exclusive manner to temporally and spatially regulate Plk4 recruitment. (b) Domain organization of SPD-2 homologues. Residue numbers of domain boundaries are shown above the schematics. (c) Domain organization of Asterless and Cep152.
Asterless was originally identified in Drosophila as a protein essential for aster formation during male meiosis [29]. The protein was later revealed to be a constitutive centriolar component involved in the initiation of centriole duplication [30–33]. Cep152, the mammalian orthologue of Asterless, is also involved in both centriole duplication and PCM recruitment [34–36]. Both Drosophila Asterless and human Cep152 bind to the CPB domain of Plk4 via their N-terminal acidic region [32,35]. Recent studies demonstrated that Plk4 additionally binds to the C-terminal region of Asterless, which serves to stabilize Plk4 during mitosis [37].
Although it is now well established how SPD-2 and Asterless interact with their kinase partners ZYG-1 and Plk4, they have been structurally less characterized. Bioinformatics analysis suggests that all SPD-2/Cep192 orthologues contain a mostly unstructured N-terminal region with a short coiled-coil domain and a long C-terminal β-strand-rich region (figure 2b). By contrast, all Asterless/Cep152 orthologues contain mostly helical structures that are predicted to form coiled-coils, with no β-strand content (figure 2c). Caenorhabditis elegans and mammalian SPD-2, and Drosophila and mammalian Asterless, also contain a highly acidic region towards their N-termini, which interacts directly and tightly with their kinase partners [26]. This tight interaction creates something of a puzzle however, as the receptors and the kinases do not seem to form a robust complex in the cell until the receptors are targeted to mother centrioles. Recent studies by multiple imaging/biophysical techniques have shown that C. elegans SPD-2 is monomeric in the cytoplasm [38]. It was postulated that the acidic region for kinase binding might be concealed via its interaction with another (probably basic) part of the same protein; receptors targeted to the nascent centriole might then release such auto-inhibition to facilitate interaction with the downstream kinases [26].
Another unresolved issue about the centriolar receptors (SPD-2 and Asterless) in centriole assembly is how they themselves are recruited to the mother centriole. One possibility is that Plk1, another critical polo-like kinase linking centriole biogenesis to cell-cycle progression [39–41], might phosphorylate SPD-2/Asterless to promote their targeting and the initiation of centriole assembly. In human cells, co-localization of another coiled-coil-rich protein Cep63 with Cep152 in a ring-like structure around the proximal end of the mother centriole has been shown [42]. Cep63 knock-out leads to both Cep152 mislocalization and deficiency in procentriole assembly. It was hypothesized that Cep63 might oppose the activity of Plk1 that modifies centrosomal Cep152 [42]. From the structural point of view, it would be interesting to determine the high-resolution structures of the C-terminal β-strand-rich region in SPD-2 and investigate how they are involved in centriole duplication (figure 2b). To determine the high-resolution structures of Asterless and Cep152 might prove challenging because of their high coiled-coil content (figure 2c). Reconstitution of a complex of the coiled-coil with an interaction partner might help to make its structural studies amenable. The MitoCheck consortium has reported that at least nine other proteins interact with Cep152 in mammalian cells [43].
3. Plk4/ZYG-1
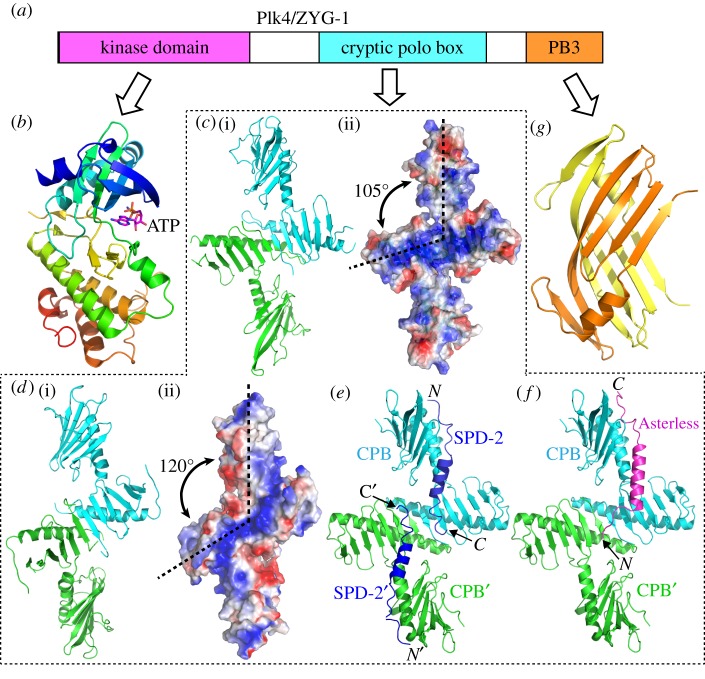
In metazoans, the serine/threonine kinase Plk4 controls daughter centriole assembly and couples centriole duplication with cell-cycle progression [44,45]. As the master regulators of centriole assembly, Plk4 and ZYG-1 are essential for the recruitment of downstream proteins to the procentriole [24,25,46]. In C. elegans, ZYG-1 directly binds SAS-6 in order to recruit it and its interaction partner SAS-5 to the mother centriole [47]. In Drosophila and mammals, Plk4 phosphorylates Ana2/STIL to trigger the direct interaction between Ana2/STIL and SAS-6, which is essential for their recruitment during centriole assembly [48–51]. Plk4 is the most divergent member of the polo-like kinase family, and C. elegans ZYG-1 is one of the most divergent of Plk4 orthologues [52]. Despite the high divergence in their primary sequences, ZYG-1 and Plk4 share a similar structural arrangement consisting of three structural domains: the N-terminal kinase domain, the central CPB domain and the C-terminal single polo-box (PB3) domain (figure 3a).
Figure 3.
Structural dissection of Plk4/ZYG-1. (a) Schematic showing domain organization of Plk4/ZYG-1. (b) Ribbon diagram of the crystal structure of the human Plk4 kinase domain loaded with an ATP analogue (PDB code: 3COK). The structure is colour-ramped from blue (N-terminus) to red (C-terminus). The bound nucleotide is shown as sticks. (c) (i) Ribbon diagram and (ii) corresponding electrostatic surface plot of the crystal structure of the Drosophila Plk4 CPB dimer (PDB code: 4NK7). (d) (i) Ribbon diagram and (ii) corresponding electrostatic surface plot of the crystal structure of the ZYG-1 CPB dimer (PDB code: 4NKB). (e) Crystal structure of human Plk4 CPB in complex with the acidic region of SPD-2 (dark blue) (PDB code: 4N7Z). (f) Crystal structure of human Plk4 CPB in complex with the acidic region of Asterless (pink) (PDB code: 4N7 V). (g) Ribbon diagram of the crystal structure of the mouse Plk4 PB3 (PDB code: 1MBY). Different molecules in all ribbon diagrams are coloured differently. Positive and negative charges in the electrostatic plots are shown in blue and red, respectively.
The kinase domain of Plk4 adopts a similar conformation to that of Plk1, with the nucleotide binding site located in the cleft between two lobe-like modules (figure 3b). The central and C-terminal regions of Plk4 and ZYG-1 are the least similar in terms of protein sequence comparison. It has been debated whether ZYG-1 is a bona fide Plk4 or a structurally distinct functional orthologue [44]. To address this, it was critical to compare their respective CPB and PB3 domains to find out how similar they really are in three-dimensional structure. Initially, a crystal structure of the Drosophila Plk4 CPB was determined, based on which a side-by-side dimeric conformation was proposed [53]. However, this orientation of the Plk4 CPBs was later found to be a crystallographic artefact [26,54]. Other more recent structural studies on the ZYG-1 and Plk4 CPBs revealed that they in fact form a similar Z-shaped dimer, with a basic patch across the dimeric interface for binding by SPD-2 and/or Asterless [26,55]. Interestingly, despite the high similarity in their overall conformation, there are subtle structural variations in the CPBs of ZYG-1 and Plk4 that confer their selective binding to the respective receptors [26]. The CPB of Drosophila Plk4 bears a flat and broad basic patch (figure 3c), whereas the basic patch on the ZYG-1 CPB is narrow and elongated because of the expanded angle between the arms (figure 3d). A crystal structure of the human Plk4 CPB revealed that it is very similar to that of the Drosophila Plk4 CPB [55]. The structure of the human Plk4 CPB in complex with either SPD-2 (figure 3e) or Asterless (figure 3f) showed that the two receptors are bound with distinct modes, suggesting an intricate spatio-temporal regulation of Plk4 recruitment [55].
A crystal structure of the C-terminal PB3 of mouse Plk4 showed that it forms a dimeric structure distinct from the canonical polo-box (figure 3g) [56]. Recent studies suggested that the PB3 of human Plk4 plays an important regulatory role by relieving auto-inhibition of the kinase domain [57]. Similarly, the C-terminal PB3 in ZYG-1 has also been shown to play a regulatory role in its function. ZYG-1 deletion mutations lacking its PB3 make the kinase behave differently in mitosis and meiosis [58]. It was proposed that the ZYG-1 C-terminus may possess two distinct regulatory roles: one for inhibiting the kinase activity and the other for differential regulation of ZYG-1 recruitment in mitosis and meiosis [58]. However, the underlying mechanism for how the PB3 regulates ZYG-1 activity remains elusive. A crystal structure of the mouse Plk4 PB3 dimer revealed a deep interfacial pocket between the two molecules, which was proposed to be a potential ligand-binding site [56]. However, no ligands binding to this pocket of the Plk4 PB3 have yet been identified. Recent studies further demonstrated that the human Plk4 PB3 does not dimerize [57]. Therefore, it remains unclear whether the PB3 of Plk4 in different organisms indeed all forms a dimer and, if so, whether the ZYG-1 PB3 shares a similar conformation to it. Determination of high-resolution structures for the PB3 domains of ZYG-1 and various Plk4s would also help investigate how PB3 mechanistically regulates the function of Plk4 and ZYG-1 in vivo.
4. SAS-5/Ana2/STIL and SAS-6
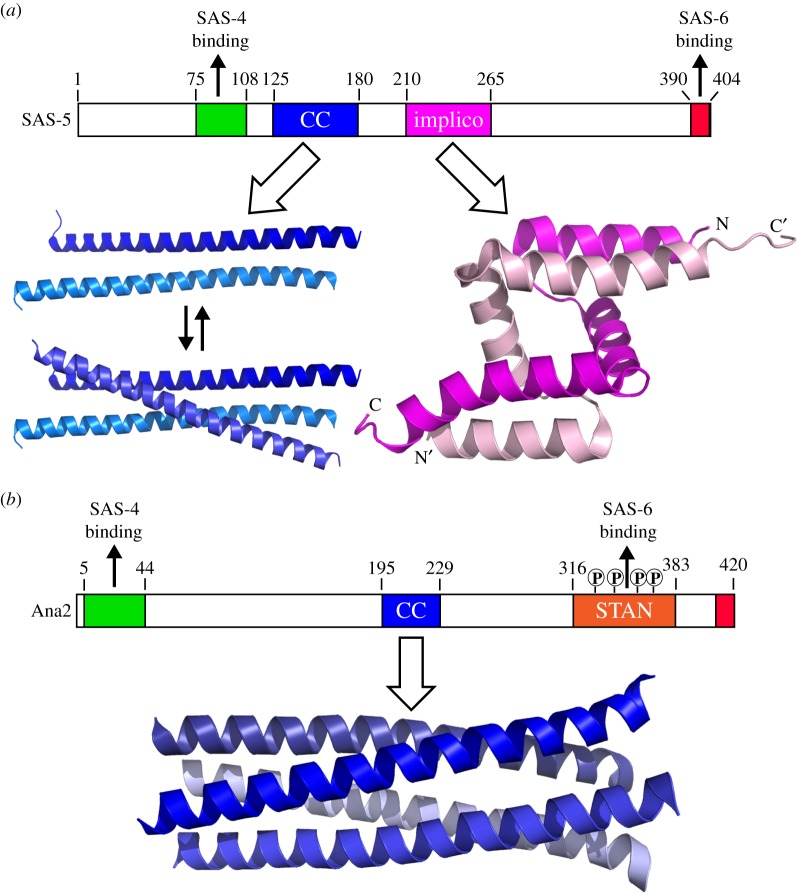
SAS-5 and SAS-6 are co-dependent for their centriolar localization, which is essential for the subsequent recruitment of SAS-4 [23]. SAS-5 is functionally orthologous to Drosophila Ana2 and vertebrate STIL, with a 90-residue STIL/Ana2 (STAN) motif at the C-terminus [59]. Reflecting evolutionary history, this motif is more conserved among vertebrates and flies (greater than 30% identity) than with the corresponding region in C. elegans SAS-5 (12% identity). Likewise, the binding modes of Ana2 and SAS-5 to their respective partners seem to be different. While the binding of SAS-5 to SAS-6 is mediated via synergistic hydrophobic and electrostatic interactions between the C-terminal 15 residues of SAS-5 (which is predicted to form a short α-helix) and a central region of the SAS-6 coiled-coil [60], such an interaction seems to be absent in Drosophila and mammals. This is despite the fact that both Ana2 and STIL are also predicted to possess a short helix towards their C-termini and that two of the four residues at the C-terminus of SAS-5 for SAS-6 binding are also conserved in this helix of Ana2 and STIL. Instead, the interaction between Ana2/STIL and SAS-6 was recently revealed to be mediated by the Plk4-dependent phosphorylation of a number of conserved serine/threonine residues in the STAN motif of Ana2/STIL [48–51]. ZYG-1 was initially reported to phosphorylate SAS-6 at serine 123 residue [61]. However, later studies with mutations of 38 potential phosphorylation sites in C. elegans SAS-6 (including serine123) suggest that SAS-6 is unlikely to be the phosphorylation target of ZYG-1 [47]. It remains to be determined whether ZYG-1 phosphorylates SAS-5 in a similar manner to that of Plk4 on Ana2/STIL.
Studies using various biophysical methods suggested that the central region of SAS-5 forms a tetramer, which may function as a cross-linker to strengthen the higher-order structure formed by SAS-6 [62]. Detailed molecular mechanisms underlying SAS-5 oligomerization have been revealed very recently. Crystal structures show that SAS-5 contains two independent oligomeric domains: a coiled-coil region spanning residues 125–180 and an Implico domain consisting of residues 210–265 (figure 4a) [63]. While the Implico motif forms a stable dimer via the intertwining of the zigzagged triple-helix structures, the coiled-coil can exist as either a dimer or trimer depending on its concentration. Together, these properties allow SAS-5 to form a higher-order assembly that may create a seeding point for SAS-6 oligomerization [63]. In the meantime, a crystal structure of the central coiled-coil domain of Ana2 has been reported, which reveals that it forms a parallel, symmetrical 4-helix bundle (figure 4b) [64]. This tetrameric form, together with its N-terminal SAS-4 binding site and C-terminal phosporylation-dependent SAS-6-binding STAN motif, was believed to actively contribute to centriole assembly by interconnecting neighbouring layers of cartwheels formed by SAS-6 [64]. While auto-assembly of both SAS-5 and Ana2 is essential for centriole biogenesis, their different oligomerization modes might reflect the morphological variations of the respective SAS-6 oligomers that have been observed (see below).
Figure 4.
Domain arrangements and crystal structures of SAS-5 and Ana2. (a) Domain organization and crystal structures of SAS-5. The central coiled-coil may form a dimer or trimer depending on the protein concentration. The Implico domain forms an intertwined homodimer. The C-terminal 15 residues of SAS-5 (shown in red) are predicted to form an α-helix that binds directly to a central region of the SAS-6 coiled-coil. (b) Domain organization of Drosophila Ana2 and crystal structure of its parallel, symmetrical 4-helix bundle. Plk4 phosphorylates four conserved serine residues in the STAN motif to promote the strong interaction between Ana2 and SAS-6.
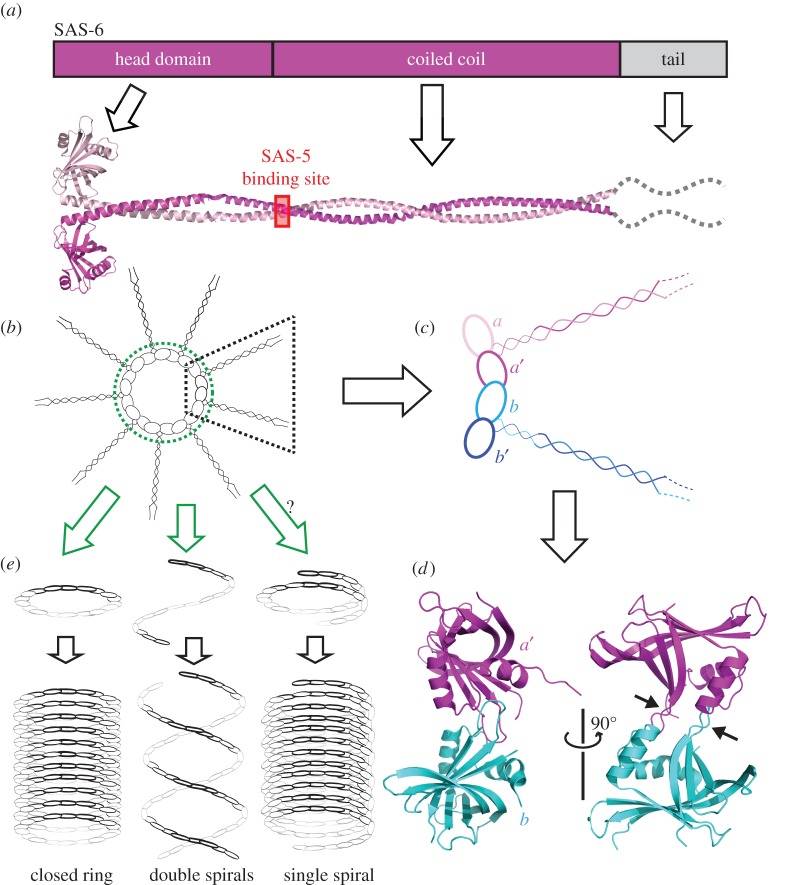
SAS-6 is one of the most thoroughly characterized centriolar proteins at a structural level. It is a modular protein with an N-terminal head domain followed by a long coiled-coil and an intrinsically disordered C-terminal tail (figure 5a). Numerous crystal structures of the head domain of SAS-6 from various organisms have been reported, which show a conserved intermolecular interaction between the head domains [65–68]. Furthermore, a crystal structure of the C. elegans SAS-6 coiled-coil region shows that two SAS-6 molecules form a parallel dimer via a strong interaction between their coiled-coils [60]. Together, these studies suggest that 18 SAS-6 molecules can form nine parallel homodimers that then assemble into a cartwheel-like structure. The cartwheel is stabilized through the interactions between the neighbouring head domains in the closed ring (figure 5b–d). This working model finally provides a molecular explanation for the ninefold cartwheel structure originally identified by electron microscopy [69].
Figure 5.
Structure and assembly of SAS-6. (a) Domain organization and ribbon diagram representation of SAS-6. SAS-6 molecules form a parallel dimer via their coiled-coil domains. The SAS-5 binding site is indicated. (b) Assembly of nine SAS-6 dimers into a cartwheel-like structure via intermolecular interactions of the head domains around the ring. (c) Coloured schematic showing the head–head interaction between two neighbouring SAS-6 dimers. (d) Ribbon diagram of the crystal structure of the C. elegans SAS-6 head domain dimer (PDB code: 3PYI). Arrows point to the loops mediating the intermolecular interaction. (e) Three working models for formation of the cylinder-like structure by SAS-6. The spokes formed by SAS-6 coiled-coils are omitted for clarity.
While the cartwheel formed by SAS-6 is believed to be important for dictating the ninefold symmetry of centrioles, the interaction between the SAS-6 head domains is relatively weak, with a Kd of only 60–100 µM. This argues against the autonomous assembly of the cartwheel by SAS-6 alone. In the cell, this issue might be solved via the cross-linking action of SAS-5/Ana2/STIL as described above. Interestingly, it was shown recently that in human cells cartwheel assembly occurs in the proximal lumen of the mother centriole [70]. This assembly process is initiated by the recruitment of SAS-6 dimers to disengaged centrioles, and mediated by the interaction between the SAS-6 C-terminal tail and SAS-4 located at the luminal wall [70]. However, this seems to be species-specific and might be employed only by cycling cells with strictly controlled centriole number, as de novo assembly of centrioles has been observed in many other species [71–73].
To form a cylindrical centriole, the SAS-6-mediated cartwheel structure apparently has to be assembled longitudinally in the lumen of the centriole. One way to achieve this is by stacking multiple rings on top of one another, as observed in the flagellate Trichonympha (figure 5e, left) [74]. This assembly mechanism might also occur in other organisms [75,76]. The other means of SAS-6 assembly was found in C. elegans where SAS-6 forms a spiral-like structure, and two such spirals wind around each other to form the central tube of the centriole (figure 5e, middle) [65]. It is postulated that SAS-6 might also assemble in a third way, in which SAS-6 forms a single tight spiral formed by the slightly tilted hinge region between the head group and the coiled-coil (figure 5e, right) [77]. However, the single spiral model has not been seen in any real centrioles so far and its existence remains undetermined.
5. SAS-4/CPAP
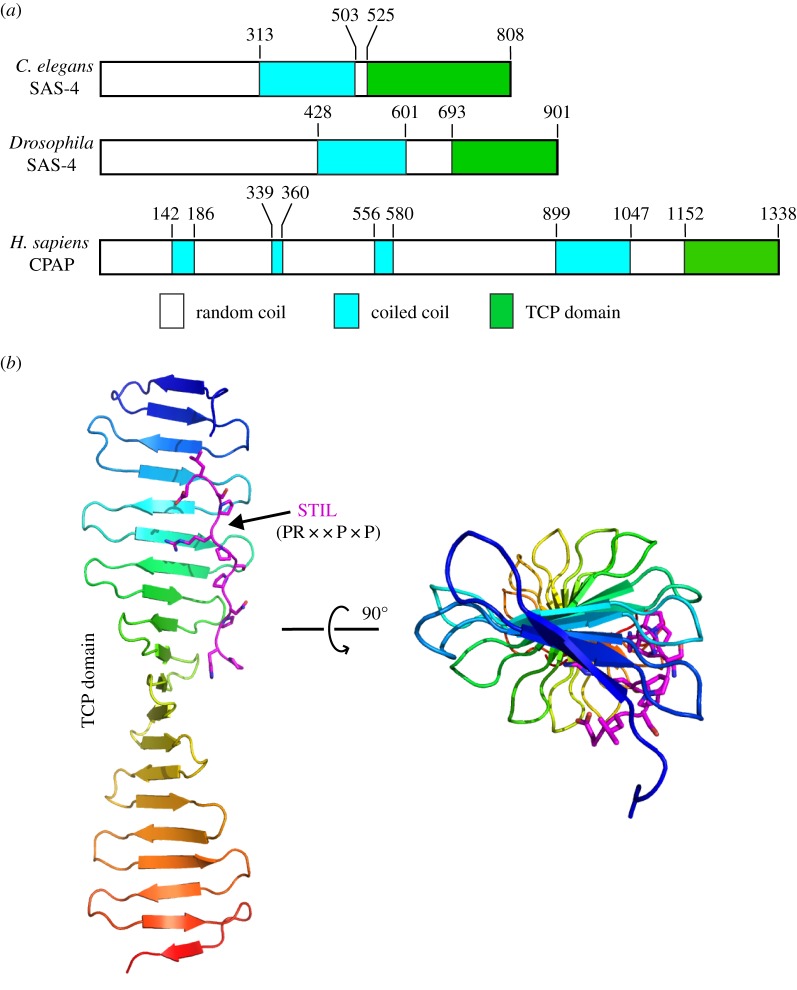
SAS-4/CPAP is the last core centriolar protein to be recruited. It is located at the outer wall of the centriolar barrel and plays an essential role for subsequent recruitment of microtubules [24,25]. SAS-4/CPAP was originally identified in mammals by yeast two-hybrid screens [78]. Homologues of SAS-4/CPAP were also later identified in Drosophila and C. elegans [19,20,79]. All SAS-4/CPAP proteins share a similar overall domain arrangement, with a mostly disordered N-terminus, a central coiled-coil and a globular C-terminal T complex protein 10 (TCP) domain (figure 6a). Crystal structures of the TCP domain of SAS-4 proteins from zebrafish and Drosophila showed that it forms a long β-meander consisting of about 20 β-strands, which interacts directly with a conserved proline-rich region in STIL/Ana2—the consensus sequence of this region is PR××P×P (figure 6b) [80,81]. Interestingly, the seemingly stable TCP structure does not contain a defined hydrophobic core or flanking globular domains on either end.
Figure 6.
Domain organization and structural dissection of SAS-4/CPAP. (a) Domain organization of three representative SAS-4/CPAP homologues. All SAS-4/CPAP homologues share a similar structural arrangement with a mostly disordered N-terminus, a central coiled-coil and a β-strand-rich C-terminal TCP domain. Residue numbers of domain boundaries are shown above the schematics. (b) Ribbon diagram of the crystal structure of the TCP domain of Danio rerio CPAP in complex with the ‘PR××P×P’ motif from STIL. The motif is shown as sticks (PDB code: 4BXR). The complex is shown in two perpendicular orientations.
While the interaction mode between the SAS-4 TCP domain and STIL/Ana2 is conserved in Drosophila and vertebrates, the interaction between C. elegans SAS-4 and SAS-5 may have diverged significantly. For instance, although the TCP domain is required for C. elegans SAS-4 to interact with the N-terminal part of SAS-5, by itself it is not sufficient [80]. Furthermore, SAS-5 does not contain the consensus TCP-binding motif (PR××P×P) present in STIL/Ana2 [80].
The function of SAS-4/CPAP in recruiting microtubules during centriole assembly has been confirmed. However, the mechanism is still unclear. One hypothesis is that the TCP domain of Drosophila CPAP forms monomers in solution, but upon incorporation into procentrioles the TCP domain would bind to STIL towards the interior of the centriole whereas the N-terminal part of SAS-4/CPAP interacts with Asterless/Cep152 and microtubules at the outer wall [80]. Another is that the TCP β-sheet is capable of forming large assemblies via a head-to-tail interaction. This was observed in the crystal structure and was postulated to help CPAP to form continuous connections along the long axis of the centriolar outer wall. Such connections, together with the central coiled-coil of the protein, would then link the pinhead with the microtubules to stabilize the cartwheel stacks in the centriole [81]. Structural and functional analysis of Drosophila SAS-4 TCP domain by another group further confirmed the role of SAS-4 as a mediator to tether PCM complexes to the centriole to facilitate centrosome assembly [82]. However, further studies are needed to find out the exact structural roles of SAS-4/CPAP in centriole assembly.
6. Other proteins involved in centriole assembly
Besides the five core centriolar components, a number of other proteins participating in centriole duplication have recently been identified. Bld10/Cep135 is an approximately 170 kDa protein originally found in Chlamydomonas and essential for cartwheel formation [83]. It consists mostly of coiled-coils and was later found to be a major spoke-tip component, serving to bridge the cartwheel to the triplet microtubules [84]. A homologue of Bld10/Cep135 was subsequently identified in Drosophila, and was reported to play a role in the formation of both the central tube in mature primary spermatocytes [85] and the central pair of microtubules in the flagellar axoneme [86,87]. However, no Bld10/Cep135 homologue has been identified in nematodes.
The localization of Bld10/Cep135 to procentrioles depends on both Cep120 and SPICE1 [88]. Cep120 is an approximately 100 kDa protein participating in centriole assembly by asymmetrically localizing to the daughter centriole [89]. The protein contains an N-terminal C2 domain, a central region for SAS-4/CPAP binding and a C-terminal long coiled-coil for dimerization [90]. It was reported that Cep120 interacts with SPICE1, another large coiled-coil-containing protein, and the two proteins are co-dependent for their localization to procentrioles [88]. Cep120 and SPICE1 are believed to work cooperatively with SAS-4/CPAP to regulate centriole elongation [88,90].
Comparative genomics and proteomics studies of isolated basal bodies from Chlamydomonas reinhardtii have identified 18 proteins (Poc 1–Poc 18) proposed to be centriolar constituents [91]. Poc1 is one of the best-characterized Poc proteins. It contains an N-terminal WD40 domain and a C-terminal coiled-coil and, like Bld10/Cep135, is hypothesized to stabilize microtubules, and thus maintains centriole assembly and integrity [92–95].
In animal cells, a cap structure at the distal end of centrioles is formed to suppress centriole-templated ciliogenesis. The cap complex consists of CP110 and Cep97, two large proteins consisting mostly of coiled-coils [96]. Additionally, several other proteins have been identified to be more specifically involved in assembly of an appendage structure at the distal end of mother centrioles. These proteins include Ccdc41/Cep83, Cep89/Cep123, Sclt1, Fbf1 and Cep164 [97–99]. The distal appendage serves to anchor the mother centriole onto the plasma membrane during ciliogenesis.
7. Conclusion and perspectives
The recent progress in structural studies on centriolar components, with each of the five core components having been at least partially characterized, has greatly advanced our understanding of centriole assembly at the high-resolution level. However, further studies will be necessary to precisely determine their contributions, both physically and temporally, during centriole assembly, and to uncover their interplay with other newly identified constitutive or accessory centriolar proteins and each other. The field faces several challenges in this process including (i) to understand how centriole assembly is initiated, (ii) to uncover how centriole duplication is regulated by both kinases and phosphatases, (iii) to determine the high-resolution structures and the functional roles for those coiled-coil-containing centriolar proteins, and (iv) to precisely assign the relative positions and the interaction network of different centriolar proteins and their structural/functional roles.
Recent studies using various techniques have identified many new centriolar components. It is likely that more centriole-associated proteins will be reported in the future. However, unlike the five core centriolar proteins, at least some of them are organism- or cell-specific. For example, although Bld10/Cep135 is crucial for cartwheel formation in green algae [83,84], Paramecium tetraurelia [100] and mammalian cells [46,101], its homologue in Drosophila is dispensable for cartwheel formation [85,87,102], and no homologue of Bld10/Cep135 seems to exist in nematodes. Another example is CP110 and Cep97, both of which exist only in animals, and are needed for centriole duplication in S-phase-arrested human cells to control centriole length [46,103–105]. Phylogenetic analysis suggests that both CP110 and Cep97 might have been recruited to the centriole-assembly pathway in animals during evolution to coordinate centriole assembly with ciliogenesis and/or cytokinesis [86]. Therefore, it is necessary to carefully examine which of these newly identified centriolar proteins are structural constituents of the centriole and which are accessory proteins, and what their structural and functional roles are in centriole assembly.
It is foreseeable that many more structural characterizations on centriole assembly will be reported in the coming years, which, together with continuous advancements in functional and in vivo studies, will provide a more refined view of one of the most important and fascinating cellular structures that has captivated cell biologists for over a century.
Note added in proof
Arquint et al. [106] just reported high-resolution structures of the PB3 of human Plk4, both alone and in complex with STIL. Their studies demonstrate that the human plk4 PB3, unlike the previously reported dimeric mouse Plk4 PB3 (PDB code: 1MBY), folds into a monomeric canonical polo-box and binds to the central coiled-coil of STIL via a novel polo-box—peptide interaction mode mimicking coiled-coil formation.
Acknowledgements
The author is grateful to Brooke Morriswood (University of Würzburg) for critical reading of the manuscript.
Competing interests
The author declares that there is no conflict of interest.
Funding
This research was supported by institutional support from the Max F. Perutz Laboratories and grant P23440-B20 from the Austrian Science Fund (FWF).
References
- 1.Marshall WF. 2001. Centrioles take center stage. Curr. Biol. 11, R487–R496. (doi:10.1016/S0960-9822(01)00289-5) [DOI] [PubMed] [Google Scholar]
- 2.Preble AM, Giddings TM Jr, Dutcher SK. 2000. Basal bodies and centrioles: their function and structure. Curr. Top. Dev. Biol. 49, 207–233. (doi:10.1016/S0070-2153(99)49010-6) [DOI] [PubMed] [Google Scholar]
- 3.Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M. 1998. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 143, 1575–1589. (doi:10.1083/jcb.143.6.1575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornens M. 2002. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 14, 25–34. (doi:10.1016/S0955-0674(01)00290-3) [DOI] [PubMed] [Google Scholar]
- 5.Conduit PT et al. 2014. The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev. Cell 28, 659–669. (doi:10.1016/j.devcel.2014.02.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conduit PT, et al. 2014. A molecular mechanism of mitotic centrosome assembly in Drosophila. eLife 3, e03399 (doi:10.7554/eLife.03399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mennella V, Agard DA, Huang B, Pelletier L. 2014. Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends Cell Biol. 24, 188–197. (doi:10.1016/j.tcb.2013.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodruff JB, Wueseke O, Hyman AA. 2014. Pericentriolar material structure and dynamics. Phil. Trans. R. Soc. B 369, 20130459 (doi:10.1098/rstb.2013.0459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderhub SJ, Kramer A, Maier B. 2012. Centrosome amplification in tumorigenesis. Cancer Lett. 322, 8–17. (doi:10.1016/j.canlet.2012.02.006) [DOI] [PubMed] [Google Scholar]
- 10.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. 2011. Centrosomes and cilia in human disease. Trends Genet. 27, 307–315. (doi:10.1016/j.tig.2011.05.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinkley BR. 2001. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 11, 18–21. (doi:10.1016/S0962-8924(00)01872-9) [DOI] [PubMed] [Google Scholar]
- 12.Nigg EA. 2006. Origins and consequences of centrosome aberrations in human cancers. Int. J. Cancer 119, 2717–2723. (doi:10.1002/ijc.22245) [DOI] [PubMed] [Google Scholar]
- 13.Satir P, Christensen ST. 2007. Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 69, 377–400. (doi:10.1146/annurev.physiol.69.040705.141236) [DOI] [PubMed] [Google Scholar]
- 14.Adams M, Smith UM, Logan CV, Johnson CA. 2008. Recent advances in the molecular pathology, cell biology and genetics of ciliopathies. J. Med. Genet. 45, 257–267. (doi:10.1136/jmg.2007.054999) [DOI] [PubMed] [Google Scholar]
- 15.Mahjoub MR. 2013. The importance of a single primary cilium. Organogenesis 9, 61–69. (doi:10.4161/org.25144) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. 2001. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105, 547–558. (doi:10.1016/S0092-8674(01)00338-5) [DOI] [PubMed] [Google Scholar]
- 17.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O'Connell KF. 2004. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6, 511–523. (doi:10.1016/S1534-5807(04)00066-8) [DOI] [PubMed] [Google Scholar]
- 18.Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, Ruer M, Muller-Reichert T, Hyman AA. 2004. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14, 863–873. (doi:10.1016/j.cub.2004.04.012) [DOI] [PubMed] [Google Scholar]
- 19.Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA. 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112, 575–587. (doi:10.1016/S0092-8674(03)00117-X) [DOI] [PubMed] [Google Scholar]
- 20.Leidel S, Gonczy P. 2003. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 4, 431–439. (doi:10.1016/S1534-5807(03)00062-5) [DOI] [PubMed] [Google Scholar]
- 21.Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gonczy P. 2004. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 6, 656–664. (doi:10.1038/ncb1146) [DOI] [PubMed] [Google Scholar]
- 22.Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 7, 815–829. (doi:10.1016/j.devcel.2004.10.015) [DOI] [PubMed] [Google Scholar]
- 23.Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7, 115–125. (doi:10.1038/ncb1220) [DOI] [PubMed] [Google Scholar]
- 24.Delattre M, Canard C, Gonczy P. 2006. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16, 1844–1849. (doi:10.1016/j.cub.2006.07.059) [DOI] [PubMed] [Google Scholar]
- 25.Pelletier L, O'Toole E, Schwager A, Hyman AA, Muller-Reichert T. 2006. Centriole assembly in Caenorhabditis elegans. Nature 444, 619–623. (doi:10.1038/nature05318) [DOI] [PubMed] [Google Scholar]
- 26.Shimanovskaya E, Viscardi V, Lesigang J, Lettman MM, Qiao R, Svergun DI, Round A, Oegema K, Dong G. 2014. Structure of the C. elegans ZYG-1 cryptic polo box suggests a conserved mechanism for centriolar docking of Plk4 kinases. Structure 22, 1090–1104. (doi:10.1016/j.str.2014.05.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dix CI, Raff JW. 2007. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 17, 1759–1764. (doi:10.1016/j.cub.2007.08.065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giansanti MG, Bucciarelli E, Bonaccorsi S, Gatti M. 2008. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 18, 303–309. (doi:10.1016/j.cub.2008.01.058) [DOI] [PubMed] [Google Scholar]
- 29.Bonaccorsi S, Giansanti MG, Gatti M. 1998. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J. Cell Biol. 142, 751–761. (doi:10.1083/jcb.142.3.751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, Nicastro D, Malicki J, Avidor-Reiss T. 2008. Drosophila Asterless and vertebrate Cep152 are orthologs essential for centriole duplication. Genetics 180, 2081–2094. (doi:10.1534/genetics.108.095141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobbelaere J, Josue F, Suijkerbuijk S, Baum B, Tapon N, Raff J. 2008. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6, e224 (doi:10.1371/journal.pbio.0060224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dzhindzhev NS, et al. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature 467, 714–718. (doi:10.1038/nature09445) [DOI] [PubMed] [Google Scholar]
- 33.Varmark H, Llamazares S, Rebollo E, Lange B, Reina J, Schwarz H, Gonzalez C. 2007. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 17, 1735–1745. (doi:10.1016/j.cub.2007.09.031) [DOI] [PubMed] [Google Scholar]
- 34.Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191, 731–739. (doi:10.1083/jcb.201007107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Firat-Karalar EN, Rauniyar N, Yates JR 3rd, Stearns T. 2014. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr. Biol. 24, 664–670. (doi:10.1016/j.cub.2014.01.067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191, 721–729. (doi:10.1083/jcb.201006049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klebba JE, Galletta BJ, Nye J, Plevock KM, Buster DW, Hollingsworth NA, Slep KC, Rusan NM, Rogers GC. 2015. Two Polo-like kinase 4 binding domains in Asterless perform distinct roles in regulating kinase stability. J. Cell Biol. 208, 401–414. (doi:10.1083/jcb.201410105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wueseke O et al. 2014. The Caenorhabditis elegans pericentriolar material components SPD-2 and SPD-5 are monomeric in the cytoplasm before incorporation into the PCM matrix. Mol. Biol. Cell 25, 2984–2992. (doi:10.1091/mbc.E13-09-0514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loncarek J, Hergert P, Khodjakov A. 2010. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr. Biol. 20, 1277–1282. (doi:10.1016/j.cub.2010.05.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsou MF, Stearns T. 2006. Mechanism limiting centrosome duplication to once per cell cycle. Nature 442, 947–951. (doi:10.1038/nature04985) [DOI] [PubMed] [Google Scholar]
- 41.Wang WJ, Soni RK, Uryu K, Tsou MF. 2011. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193, 727–739. (doi:10.1083/jcb.201101109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sir JH et al. 2011. A primary microcephaly protein complex forms a ring around parental centrioles. Nat. Genet. 43, 1147–1153. (doi:10.1038/ng.971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutchins JR, et al. 2010. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599. (doi:10.1126/science.1181348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cunha-Ferreira I, et al. 2013. Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr. Biol. 23, 2245–2254. (doi:10.1016/j.cub.2013.09.037) [DOI] [PubMed] [Google Scholar]
- 45.Sillibourne JE, Bornens M. 2010. Polo-like kinase 4: the odd one out of the family. Cell Div. 5, 25 (doi:10.1186/1747-1028-5-25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell 13, 190–202. (doi:10.1016/j.devcel.2007.07.002) [DOI] [PubMed] [Google Scholar]
- 47.Lettman MM, Wong YL, Viscardi V, Niessen S, Chen SH, Shiau AK, Zhou HL, Desai A, Oegema K. 2013. Direct binding of SAS-6 to ZYG-1 recruits SAS-6 to the mother centriole for cartwheel assembly. Dev. Cell 25, 284–298. (doi:10.1016/j.devcel.2013.03.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dzhindzhev NS, Tzolovsky G, Lipinszki Z, Schneider S, Lattao R, Fu J, Debski J, Dadlez M, Glover DM. 2014. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 24, 2526–2532. (doi:10.1016/j.cub.2014.08.061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kratz AS, Barenz F, Richter KT, Hoffmann I. 2015. Plk4-dependent phosphorylation of STIL is required for centriole duplication. Biol. Open 4, 370–377. (doi:10.1242/bio.201411023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moyer TC, Clutario KM, Lambrus BG, Daggubati V, Holland AJ. 2015. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 209, 863–878. (doi:10.1083/jcb.201502088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohta M, Ashikawa T, Nozaki Y, Kozuka-Hata H, Goto H, Inagaki M, Oyama M, Kitagawa D. 2014. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 5, 5267 (doi:10.1038/ncomms6267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jana SC, Bazan JF, Bettencourt-Dias M. 2012. Polo boxes come out of the crypt: a new view of PLK function and evolution. Strucute 20, 1801–1804. (doi:10.1016/j.str.2012.10.008) [DOI] [PubMed] [Google Scholar]
- 53.Slevin LK, Nye J, Pinkerton DC, Buster DW, Rogers GC, Slep KC. 2012. The structure of the plk4 cryptic polo box reveals two tandem polo boxes required for centriole duplication. Structure 20, 1905–1917. (doi:10.1016/j.str.2012.08.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Levine MS, Holland AJ. 2014. Polo-like kinase 4 shapes up. Structure 22, 1071–1073. (doi:10.1016/j.str.2014.07.004) [DOI] [PubMed] [Google Scholar]
- 55.Park SY, et al. 2014. Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis. Nat. Struct. Mol. Biol. 21, 696–703. (doi:10.1038/nsmb.2846) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leung GC, Hudson JW, Kozarova A, Davidson A, Dennis JW, Sicheri F. 2002. The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat. Struct. Biol. 9, 719–724. (doi:10.1038/nsb848) [DOI] [PubMed] [Google Scholar]
- 57.Klebba JE, Buster DW, McLamarrah TA, Rusan NM, Rogers GC. 2015. Autoinhibition and relief mechanism for Polo-like kinase 4. Proc. Natl Acad. Sci. USA 112, E657–E666. (doi:10.1073/pnas.1417967112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peters N, Perez DE, Song MH, Liu Y, Muller-Reichert T, Caron C, Kemphues KJ, O'Connell KF. 2010. Control of mitotic and meiotic centriole duplication by the Plk4-related kinase ZYG-1. J. Cell Sci. 123, 795–805. (doi:10.1242/jcs.050682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stevens NR, Roque H, Raff JW. 2010. DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev. Cell 19, 913–919. (doi:10.1016/j.devcel.2010.11.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiao R, Cabral G, Lettman MM, Dammermann A, Dong G. 2012. SAS-6 coiled-coil structure and interaction with SAS-5 suggest a regulatory mechanism in C. elegans centriole assembly. EMBO J. 31, 4334–4347. (doi:10.1038/emboj.2012.280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kitagawa D, Busso C, Fluckiger I, Gonczy P. 2009. Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryos. Dev. Cell 17, 900–907. (doi:10.1016/j.devcel.2009.11.002) [DOI] [PubMed] [Google Scholar]
- 62.Shimanovskaya E, Qiao R, Lesigang J, Dong G. 2013. The SAS-5 N-terminal domain is a tetramer, with implications for centriole assembly in C. elegans. Worm 2, e25214 (doi:10.4161/worm.25214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rogala KB, Dynes NJ, Hatzopoulos GN, Yan J, Pong SK, Robinson CV, Deane CM, Gonczy P, Vakonakis I. 2015. The Caenorhabditis elegans protein SAS-5 forms large oligomeric assemblies critical for centriole formation. eLife 4, e7410 (doi:10.7554/eLife.07410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cottee MA, Muschalik N, Johnson S, Leveson J, Raff JW, Lea SM. 2015. The homo-oligomerisation of both Sas-6 and Ana2 is required for efficient centriole assembly in flies. eLife 4, e07236 (doi:10.7554/eLife.07236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hilbert M, et al. 2013. Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry. Proc. Natl Acad. Sci. USA 110, 11 373–11 378. (doi:10.1073/pnas.1302721110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kitagawa D, et al. 2011. Structural basis of the 9-fold symmetry of centrioles. Cell 144, 364–375. (doi:10.1016/j.cell.2011.01.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Breugel M, et al. 2011. Structures of SAS-6 suggest its organization in centrioles. Science 331, 1196–1199. (doi:10.1126/science.1199325) [DOI] [PubMed] [Google Scholar]
- 68.van Breugel M, Wilcken R, McLaughlin SH, Rutherford TJ, Johnson CM. 2014. Structure of the SAS-6 cartwheel hub from Leishmania major. eLife 3, e01812 (doi:10.7554/eLife.01812) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anderson RG, Brenner RM. 1971. The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. J. Cell Biol. 50, 10–34. (doi:10.1083/jcb.50.1.10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fong CS, Kim M, Yang TT, Liao JC, Tsou MF. 2014. SAS-6 assembly templated by the lumen of cartwheel-less centrioles precedes centriole duplication. Dev. Cell 30, 238–245. (doi:10.1016/j.devcel.2014.05.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. 2002. De novo formation of centrosomes in vertebrate cells arrested during S phase. J. Cell Biol. 158, 1171–1181. (doi:10.1083/jcb.200205102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szollosi D, Calarco P, Donahue RP. 1972. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J. Cell Sci. 11, 521–541. [DOI] [PubMed] [Google Scholar]
- 73.Vladar EK, Stearns T. 2007. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 178, 31–42. (doi:10.1083/jcb.200703064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guichard P, Desfosses A, Maheshwari A, Hachet V, Dietrich C, Brune A, Ishikawa T, Sachse C, Gonczy P. 2012. Cartwheel architecture of Trichonympha basal body. Science 337, 553 (doi:10.1126/science.1222789) [DOI] [PubMed] [Google Scholar]
- 75.Hirono M. 2014. Cartwheel assembly. Phil. Trans. R. Soc. B 369, 20130458 (doi:10.1098/rstb.2013.0458) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jana SC, Marteil G, Bettencourt-Dias M. 2014. Mapping molecules to structure: unveiling secrets of centriole and cilia assembly with near-atomic resolution. Curr. Opin. Cell Biol. 26, 96–106. (doi:10.1016/j.ceb.2013.12.001) [DOI] [PubMed] [Google Scholar]
- 77.Cottee MA, Raff JW, Lea SM, Roque H. 2011. SAS-6 oligomerization: the key to the centriole? Nat. Chem. Biol. 7, 650–653. (doi:10.1038/nchembio.660) [DOI] [PubMed] [Google Scholar]
- 78.Hung LY, Tang CJ, Tang TK. 2000. Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the gamma-tubulin complex. Mol. Cell Biol. 20, 7813–7825. (doi:10.1128/MCB.20.20.7813-7825.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. 2006. Flies without centrioles. Cell 125, 1375–1386. (doi:10.1016/j.cell.2006.05.025) [DOI] [PubMed] [Google Scholar]
- 80.Cottee MA et al. 2013. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. eLife 2, e01071 (doi:10.7554/eLife.01071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hatzopoulos GN, Erat MC, Cutts E, Rogala KB, Slater LM, Stansfeld PJ, Vakonakis I. 2013. Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture. Structure 21, 2069–2077. (doi:10.1016/j.str.2013.08.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng X, et al. 2014. Conserved TCP domain of Sas-4/CPAP is essential for pericentriolar material tethering during centrosome biogenesis. Proc. Natl Acad. Sci. USA 111, E354–E363. (doi:10.1073/pnas.1317535111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsuura K, Lefebvre PA, Kamiya R, Hirono M. 2004. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J. Cell Biol. 165, 663–671. (doi:10.1083/jcb.200402022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hiraki M, Nakazawa Y, Kamiya R, Hirono M. 2007. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr. Biol. 17, 1778–1783. (doi:10.1016/j.cub.2007.09.021) [DOI] [PubMed] [Google Scholar]
- 85.Roque H, Wainman A, Richens J, Kozyrska K, Franz A, Raff JW. 2012. Drosophila Cep135/Bld10 maintains proper centriole structure but is dispensable for cartwheel formation. J. Cell Sci. 125, 5881–5886. (doi:10.1242/jcs.113506) [DOI] [PubMed] [Google Scholar]
- 86.Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. 2010. Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 123, 1414–1426. (doi:10.1242/jcs.064931) [DOI] [PubMed] [Google Scholar]
- 87.Mottier-Pavie V, Megraw TL. 2009. Drosophila bld10 is a centriolar protein that regulates centriole, basal body, motile cilium assembly. Mol. Biol. Cell 20, 2605–2614. (doi:10.1091/mbc.E08-11-1115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Comartin D, et al. 2013. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr. Biol. 23, 1360–1366. (doi:10.1016/j.cub.2013.06.002) [DOI] [PubMed] [Google Scholar]
- 89.Mahjoub MR, Xie Z, Stearns T. 2010. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J. Cell Biol. 191, 331–346. (doi:10.1083/jcb.201003009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin YN, Wu CT, Lin YC, Hsu WB, Tang CJ, Chang CW, Tang TK. 2013. CEP120 interacts with CPAP and positively regulates centriole elongation. J. Cell Biol. 202, 211–219. (doi:10.1083/jcb.201212060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Keller LC, Romijn EP, Zamora I, Yates JR 3rd, Marshall WF. 2005. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15, 1090–1098. (doi:10.1016/j.cub.2005.05.024) [DOI] [PubMed] [Google Scholar]
- 92.Fourrage C, Chevalier S, Houliston E. 2010. A highly conserved Poc1 protein characterized in embryos of the hydrozoan Clytia hemisphaerica: localization and functional studies. PLoS ONE 5, e13994 (doi:10.1371/journal.pone.0013994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Keller LC, Geimer S, Romijn E, Yates J 3rd, Zamora I, Marshall WF. 2009. Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol. Biol. Cell 20, 1150–1166. (doi:10.1091/mbc.E08-06-0619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pearson CG, Osborn DP, Giddings TH Jr, Beales PL, Winey M. 2009. Basal body stability and ciliogenesis requires the conserved component Poc1. J. Cell Biol. 187, 905–920. (doi:10.1083/jcb.200908019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Venoux M, Tait X, Hames RS, Straatman KR, Woodland HR, Fry AM. 2013. Poc1A and Poc1B act together in human cells to ensure centriole integrity. J. Cell Sci. 126, 163–175. (doi:10.1242/jcs.111203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Spektor A, Tsang WY, Khoo D, Dynlacht BD. 2007. Cep97 and CP110 suppress a cilia assembly program. Cell 130, 678–690. (doi:10.1016/j.cell.2007.06.027) [DOI] [PubMed] [Google Scholar]
- 97.Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179, 321–330. (doi:10.1083/jcb.200707181) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sillibourne JE, Specht CG, Izeddin I, Hurbain I, Tran P, Triller A, Darzacq X, Dahan M, Bornens M. 2011. Assessing the localization of centrosomal proteins by PALM/STORM nanoscopy. Cytoskeleton (Hoboken) 68, 619–627. (doi:10.1002/cm.20536) [DOI] [PubMed] [Google Scholar]
- 99.Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, Asara JM, Tsou MF. 2013. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27, 163–168. (doi:10.1101/gad.207043.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jerka-Dziadosz M, Gogendeau D, Klotz C, Cohen J, Beisson J, Koll F. 2010. Basal body duplication in Paramecium: the key role of Bld10 in assembly and stability of the cartwheel. Cytoskeleton (Hoboken) 67, 161–171. (doi:10.1002/cm.20433). [DOI] [PubMed] [Google Scholar]
- 101.Ohta T, Essner R, Ryu JH, Palazzo RE, Uetake Y, Kuriyama R. 2002. Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J. Cell Biol. 156, 87–99. (doi:10.1083/jcb.200108088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carvalho-Santos Z, et al. 2012. BLD10/CEP135 is a microtubule-associated protein that controls the formation of the flagellum central microtubule pair. Dev. Cell 23, 412–424. (doi:10.1016/j.devcel.2012.06.001) [DOI] [PubMed] [Google Scholar]
- 103.Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. 2002. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell 3, 339–350. (doi:10.1016/S1534-5807(02)00258-7) [DOI] [PubMed] [Google Scholar]
- 104.Kohlmaier G, et al. 2009. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 19, 1012–1018. (doi:10.1016/j.cub.2009.05.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. 2009. Control of centriole length by CPAP and CP110. Curr. Biol. 19, 1005–1011. (doi:10.1016/j.cub.2009.05.016) [DOI] [PubMed] [Google Scholar]
- 106.Arquint C, Gabryjonczyk AM, Imseng S, Bohm R, Sauer E, Hiller S, Nigg EA, Maier T. 2015. STIL binding to Polo-box 3 of PLK4 regulates centriole duplication. eLife. 4 (doi:10.7554/eLife.07888) [DOI] [PMC free article] [PubMed] [Google Scholar]