Abstract
Targeted genome editing mediated by clustered, regularly interspaced, short palindromic repeat (CRISPR)/CRISPR-associated nuclease 9 (Cas9) technology has emerged as one of the most powerful tools to study gene functions, and with potential to treat genetic disorders. Hearing loss is one of the most common sensory disorders, affecting approximately 1 in 500 newborns with no treatment. Mutations of inner ear genes contribute to the largest portion of genetic deafness. The simplicity and robustness of CRISPR/Cas9-directed genome editing in human cells and model organisms such as zebrafish, mice and primates make it a promising technology in hearing research. With CRISPR/Cas9 technology, functions of inner ear genes can be studied efficiently by the disruption of normal gene alleles through non-homologous-end-joining (NHEJ) mechanism. For genetic hearing loss, CRISPR/Cas9 has potential to repair gene mutations by homology-directed-repair (HDR) or to disrupt dominant mutations by NHEJ, which could restore hearing. Our recent work has shown CRISPR/Cas9-mediated genome editing can be efficiently performed in the mammalian inner ear in vivo. Thus, application of CRISPR/Cas9 in hearing research will open up new avenues for understanding the pathology of genetic hearing loss and provide new routes in the development of treatment to restore hearing. In this review, we describe major methodologies currently used for genome editing. We will highlight applications of these technologies in studies of genetic disorders and discuss issues pertaining to applications of CRISPR/Cas9 in auditory systems implicated in genetic hearing loss.
Keywords: Genome editing, CRISPR/Cas9, Hearing loss, Genetics
1. Introduction
Targeted genome editing defined as modification of the genome at a targeted locus, has long been used as a powerful tool to perform genome function studies in biomedical research. Previous methods such as gene knockdown with small interfering RNA (siRNA) and morpholinos have the potential drawbacks of off-targeting (Jackson et al., 2003; Fedorov et al., 2006; Robu et al., 2007) and incomplete editing (Holen et al., 2002; Elbashir et al., 2001; Bill et al., 2009). Conventional gene editing by homologous recombination (HR) can be used to modify genomes in various organisms (Adachi et al., 2006; Meyer et al., 2010; Rong et al., 2000). However, the extremely low efficiency of HR in mammalian cells (ranging from 1 in 108 to 1 in 105) limits its routine use (Reh et al., 2014). To overcome the shortcomings of these earlier methods, genome editing using programmable nucleases has become a promising alternative.
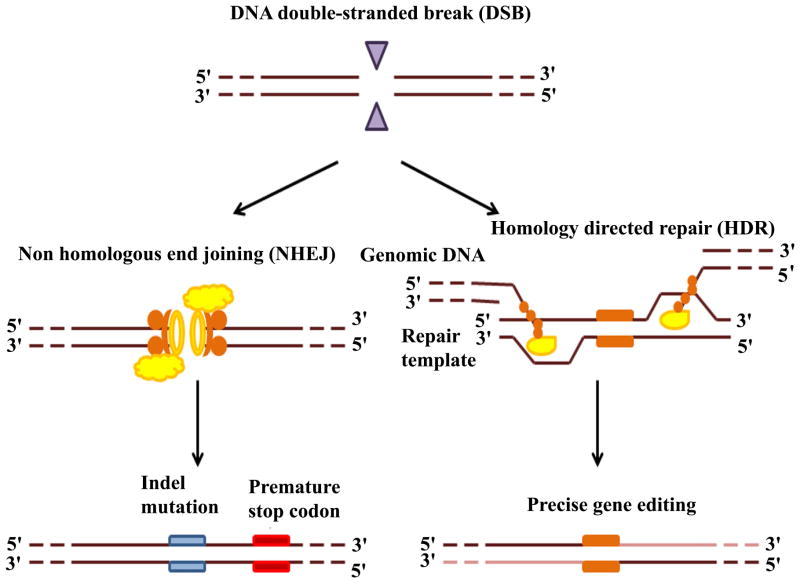
Three major programmable nucleases have been adapted as genome engineering techniques: Zinc finger nucleases (ZFNs), transcriptional activator-like effector nucleases (TALENs), and CRISPR/Cas9. All three nucleases can be guided to induce site-specific DNA double-strand breaks (DSBs), which are repaired by both homologous and non-homologous mechanisms. Homologous repair enhances homologous recombination by at least two orders of magnitude (Rouet et al., 1994), while non-homologous repair leads to targeted frame shift mutations (Bibikova et al., 2002). Applications based on the two DSB repair pathways lead to introduction of different types of mutations at target-specific sites of the genome including gene knock-out, knock-in, and point mutations.
Hearing loss (HL) is the most prevalent sensorineural disorder, affecting approximately 1 in 500 newborns (Mehl et al., 1998). It is estimated that more than two-thirds of prelingual HL cases are found to be inherited, most of which are caused by mutations of a single gene that functions in the inner ear (Liu et al, 2001; Hilgert et al., 2009; Morton et al., 2006). More than 150 chromosomal loci and over 80 genes have been identified to cause non-syndromic as well as syndromic forms of deafness (Yan and Liu 2008; Hilgert et al., 2009; Angel et al, 2012). The strong genetic basis of HL and spectacular advancements in CRISPR/Cas9-based genome editing technologies will surely usher in a new era using genome editing techniques to study HL, as can be glanced from our recent proof-of-principle study in the auditory system (Zuris et al 2015). This review focuses on what we can learn from applications and challenges of CRISPR/Cas9-mediated genome editing and clinical therapeutic potential of CRISPR/Cas9 in the future, with implications in human genome editing in genetic HL. The current state of genome editing technologies will also be presented.
2. Comparisons among three genome editing techniques
2.1 Shared features of three genome editing techniques
All three programmable nucleases contain two functional domains: one is responsible for targeting and binding specific genomic sequence whereas the other is involved in inducing DNA DSBs (Figure 1). For ZFNs, Cys2-His2 zinc fingers are amino termini of ZFNs, each zinc finger can be designed to recognize a three base-pair (bp) DNA sequence, and all zinc fingers (usually 3–6) are joined together to generate a single ZFN unit to target a DNA sequence that is 9-18 bp long (Miller et al., 1985; Nekludova et al., 2000; Urnov et al., 2010; Gaj et al., 2013). For TALENs, transcription activator-like effectors (TALEs) can also be modified to target a predetermined DNA sequence (Figure 2). TALEs are naturally occurring proteins derived from plant pathogenic bacteria Xanthomonas spp., most of which contain 13–28 repeats and 33–35 amino acids per repeat (Boch et al., 2010). Each repeat can recognize a single nucleotide in the target sequence, and the nucleotide specificity is determined by a hypervariable region of two adjacent amino acids at positions 12 and 13 within each amino acid repeat, termed repeat-variable di-residue (RVD) (Moscou et al., 2009; Boch et al., 2009; Morbitzer et al., 2010; Streubel et al., 2012; Cong et al., 2012). Multiple TALEs repeats are joined in tandem to target a specific DNA sequence (Bogdanove et al., 2011). For CRISPR, the repeats are derived from prokaryotic RNA-guided adaptive immune system, which are transcribed as target-specific CRISPR RNA (crRNA) and trans-acting crRNA (tracrRNA). TracrRNA is essential for the maturation of crRNA and can team up with crRNA guiding Cas9 nuclease to cleave invading phages or plasmids by binding to complementary DNA sequence in viruses or plasmids and inducing DSBs (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013; Cho et al., 2013).
Figure 1.
A schematic representation of programmable nucleases demonstrating two functional domains.
Figure 2.
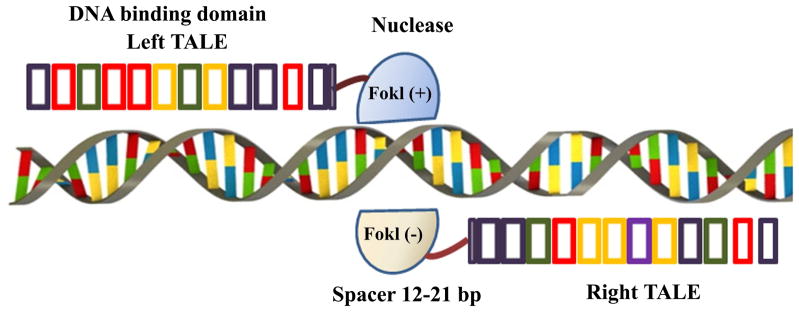
A schematic representation of transcriptional activator-like effector nucleases (TALENs). TALENs are composed of TALE repeats and FokI nuclease. TALENs work in paris, binding two complementary DNA strands across a spacer over which FokI nuclease dimerizes to create a double-stranded break. Each TALE repeat can recognize one single nucleotide, and multiple TALE repeats are joined in tandem to target a specific DNA sequence.
All three systems can induce DNA DSBs: ZFNs have non-specific DNA nuclease – FokI, FokI nucleases must dimerize to be active to induce DNA DSBs (Vanamee et al., 2001). For CRISPR, type II CRISPR-Cas system is adapted as a genome editing tool, in which Cas9 protein acts as an endonuclease, and is guided by dual RNA complex (crRNA and tracrRNA) to cleave a 23-bp target DNA sequence that contains a 20-bp sequence matching the protospacer of crRNA plus a downstream NGG nucleotide motif (protospacer-adjacent motif [PAM]) (Figure 3) (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013; Cho et al., 2013). However, the problem with ZFNs is that there is no well-established way to determine how multiple zinc finger modules can be combined to bind longer sequences without empirical testing. There are still some advantages to the TALEN system in the sense that they can be designed for more sites with potentially more specificity to CRISPR.
Figure 3.
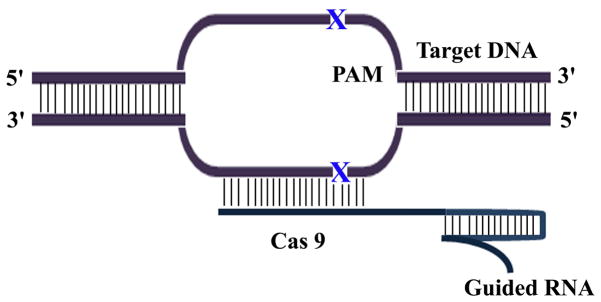
In CRISPR system, guide RNA directs Cas 9 enedonuclease to cleave the target DNA sequence 3′ bp upstream of PAM.
Finally, the DNA DSBs induced by three genome programming nucleases can be repaired by the same two endogenous cellular repair processes: non-homologous end-joining (NHEJ) and homology-directed repair (HDR) (Rouet et al., 1994; Rouet et al., 1994; Jeggo 1998; Van Gent et al., 2001; Bibikova et al., 2002). NHEJ is an error-prone process that can generate small insertions and deletions (indels) of short sequences by quickly ligating DNA DSBs at the target sites (Jeggo 1998). Indels usually cause frameshift mutations in coding regions and eventually lead to gene disruption by knocking out the gene. HDR is a precise process that requires a homologous donor DNA as a template (Rouet et al., 1994). Because of the precision of HDR, point mutations can be corrected or induced to the genome (Bibikova et al., 2001; Bibikova et al., 2003; Porteus et al., 2003; Urnov et al., 2005); additionally, HDR also makes it possible to insert a gene of interest into the genome (Lombardo et al., 2011; Li et al., 2011; Hockemeyer et al., 2011; Doyon et al., 2011; Irion et al. 2014). Induction of two DNA DSBs at the same time can result in chromosomal deletion, duplication or inversion (Lee et al., 2010; Lee et al., 2012; Carlson et al., 2012; Gupta et al., 2013; Cong et al., 2013). Chromosomal translocations can also occur if two DNA DSBs are generated on two different chromosomes (Brunet et al., 2009; Cho et al., 2014).
2.2 Advantages of CRISPR/Cas9 over the other two techniques
The simplicity of design makes CRISPR/Cas9 more promising than the other two genome editing techniques. First of all, as an RNA-guided nuclease, Cas9 can be guided to target any genomic sequence by easily programming the 20-bp targeting sequence of guide RNA (gRNA), which can then be cloned from the gRNA plasmid backbone. However, for both ZFNs and TALENs, large DNA fragments (500–1500 bp) need to be designed for each new target in order to synthesize proteins (zinc fingers or TALEs) that are responsible for targeting specific genomic sequence. In addition, since FokI is an obligate dimer, both ZFNs and TALENs need two novel proteins to function properly, a process that is labor intensive. The successful binding of Cas9 to the genomic sequences requires the PAM sequence immediately adjacent to the target sequence, with the frequency of a PAM motif every 8 bp (NGG) or 4 bp (NGG and NAG) on average (Cong et al., 2013). In contrast, PAM is not a requirement for ZFNs and TALENs. The target density is one in every 100 bp for ZFNs and one per bp for TALENs, and TALENs appears to be advantageous to both ZFNs and CRISPR/Cas9 in terms of high target density (Sander et al., 2011; Gupta et al., 2012; Reyon et al., 2012).
Another advantage of CRISPR/Cas9 is its amenability for multiplexing by delivering multiple gRNAs to target multiple genes in the same cell simultaneously. This approach has been successfully applied to mammalian cells, mice, zebrafish, and monkeys (Mali et al., 2013; Chang et al., 2013; Jao et al., 2013; Li et al., 2013; Niu et al., 2014). Although using multiple ZFNs or TALEs could achieve the similar goal, formation of mismatched dimers, which will increase off-target effects, limits their routine use for multiplexing (Sollu et al., 2010).
The avoidable off-target effects and acceptable cellular toxicity have largely broadened the application of CRISPR/Cas9 in genome editing. Off-target effects of CRISPR/Cas9 reflect the tolerance of multiple mismatches within the protospacer sequence (Fu et al., 2013; Hsu et al., 2013; Pattanayak et al., 2013). However, these off-target effects are all gRNA specific, which can be reduced through multiple strategies (Fu et al., 2013; Hsu et al., 2013; Pattanayak et al., 2013; Cradick et al., 2013; Cho et al., 2014). Well-designed gRNA could significantly reduce off-target effects in comparison with ZFNs and TALENs, as shown by whole-genome analysis of human stem cells (Duan et al., 2014; Kiskinis et al., 2014; Smith et al., 2014; Veres et al., 2014). In addition, modification of CRISPR/Cas9 nuclease specificity by a 5′ truncated gRNA decreases the off-target mutagenesis by over 5000-fold (Fu et al., 2014). An additional strategy is to use a pair of Cas9 nickases, which induce a single-stranded break on each DNA strand, an equivalent to the DSBs generated by normal Cas9. Since the single-stranded break is repaired by base excision pathway with a very low mutation rate, Cas9 nickase can reduce off-target effects (McConnell et al., 2009; Davis et al., 2011). Off-target effects have also been successfully minimized by using a pair of defective Cas9 (dCas9) protein, which lacks the nuclease activity, the dCas9 can then be fused to FokI to induce DNA DSBs (Tsai et al., 2014; Guilinger et al., 2014). As a proof of this concept, we have shown that direct delivery of protein Cas9 with gRNA significantly reduced the off-target effect of genome editing (Zuris et al., 2015).
3. Genetic causes of HL and contribution of CRISPR/Cas9 to hearing research
Most genetic HL cases are caused by monogenic mutations whereas a small portion is resulted from mutations involving more than one gene (Morton et al., 2006; Angeli et al, 2012). The majority of inherited HL is non-syndromic that is often neuroepithelial in origin arising from defects in the function of the organ of Corti – the site of auditory transduction in the inner ear (Yan et al., 2008). Since the discovery of the first nonsyndromic deafness gene in 1993, more than 150 loci for deafness genes have been mapped and more than 80 genes have been implicated in nonsyndromic HL (http://hereditaryhearingloss.org/). These genes belong to very different gene families with various functions, including transcription factors, extracellular matrix molecules, cytoskeletal components, ion channels and transporters (Yan et al., 2008; Hilgert et al, 2010). Various types of mutations have been identified in deaf patients, such as single nucleotide substitution, gene deletion, and gene insertion. These mutations result in missense or/and nonsense mutations of the deafness genes and thus cause HL (http://hereditaryhearingloss.org/). Understanding of functions of human deafness genes has been fueled by the use of vertebrate models for studying hereditary HL.
Progressive HL can initiate at any age, including in the first few years of life. Genes that have been associated with autosomal dominant progressive HL are excellent candidates for age-related hearing loss (ARHL) (Liu et al., 2007). The progressive nature of these diseases could be explained by a gradual increase in the ratio of damaged to normal protein or changes in protein expression. This could result from alterations that influence the transcription, translation and/or degradation of the altered proteins. Developing alternative biological treatments to stop or reverse the progression of HL by repairing these alterations is highly desirable.
The strong genetic basis of HL provides a significant platform to combine CRISPR/Cas9 genome editing technology with model organisms to mimic causal mutations of genetic HL and understand the pathology of the disease. Traditional methods of making transgenic animals to model HL are time-consuming, costly and labor intensive. With CRISPR/Cas9 technology, embryonic stem cells can be efficiently generated through the NHEJ mechanism to create the deletion models or by the HDR mechanism that combines a mutant donor template with Cas9 nickase activity to produce single nucleotide mutation and gene insertion models. We envision the utilization of CRISPR/Cas9 would greatly accelerate the process.
The majority of gene mutations causing HL are associated with dysfunction in auditory hair cells. Hair cell targeted rescue of deleterious gene mutations via expressing or delivering corresponding wild type proteins, or the knock-down of a mutant allele via delivering or expressing silencing RNAs could help protect against the occurring and/or the progression of certain forms of deafness. This makes the cochlear hair cell a preferential target for potential intervention. Adeno-associated virus (AAV) has been the choice in the development of gene therapy for inhereditary HL (Akil et al., 2012). However the size limit of inserts (less than 4.7 kb) severely constrains its use. Advances in methods for delivering in vitro-transcribed mRNA offered a new promising alternative, but still face challenges related to immunogenicity and RNA stability. Thus, CRISPR/Cas9-mediated genome editing could be explored as a new approach to achieve disruption of a dominant mutation or to achieve the repair of recessive mutations for functional recovery of hearing. It is possible that in situations such as the use of CRISPR/Cas9 to edit mRNA where continuous availability of Cas9:gRNA that can be provided by viral delivery will be needed (O’Connell et al., 2014). Researchers have demonstrated that Cas9 can be guided to bind specifically to target RNAs without interfering corresponding DNA sequences by the use of specially designed PAM-presenting oligonucleotides (PAMmers) (O’Connell et al., 2014). This strategy has been proved to isolate GAPDH mRNA from HeLa cells (O’Connell et al., 2014).
CRISPR/Cas9 has been shown to correct genetic disorders including Duchenne muscular dystrophy and liver disorder (Long et al., 2014; Yin et al., 2014). In those studies the corrections were achieved by viral-delivered Cas9/gRNA, resulting in permanent production of the Cas9/gRNA complex. Given the potency of CRISPR/Cas9 in genome editing, transient delivery of Cas9/gRNA complex would be preferred, with permanent editing results while reducing the risk associated with continuous genome editing activities. One emerging route in the development of therapy for genetic HL is the delivery of Cas9/gRNA in protein/nucleic acid complex to achieve CRISPR/Cas9-mediated genome editing for inner ear application.
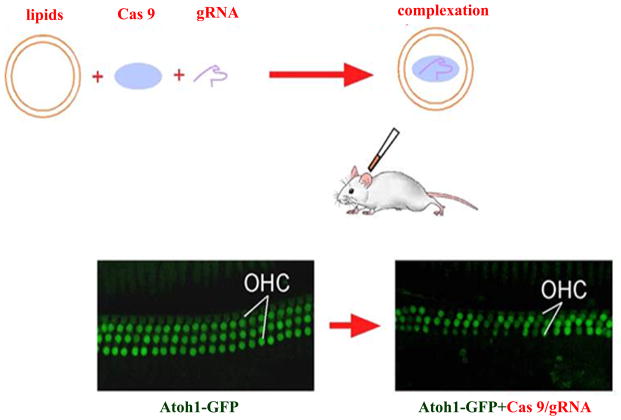
We have shown that a complex formed between protein Cas9 and nucleic acid gRNA by commercially available cationic lipids can be directly delivered to mouse inner ear hair cells in vivo (Zuris et al, 2015) (Figure 4). Further such complexes were shown to induce efficient genome editing by knockdown of GFP signal in the Atoh1-GFP transgenic mice and by detection of indels by the HST sequencing. There was minimum toxicity associated with the delivery of the complex or genome editing. As Cas9 and gRNAs are rapidly degraded upon entering cells, the duration of their activities is short-lived, yet with the permanent genome editing results. However, so far our strategy is only able to target outer hair cells efficiently. We are in the process of developing different methods such as the use of super-charged proteins to carry Cas9 with gRNAs, which is likely to target more inner ear cell types. The immediate implication in the treatment for genetic HL with the approach is to design gRNAs that target dominant hair cell gene mutations such as dominant Myo7a mutations in Usher 1B for disruption by NHEJ. By delivery of Cas9/gRNAs to the mutant inner ear, it is likely that efficient disruption of mutations could be achieved, leading to potential hearing recovery. The easy constitution of Cas9/gRNA complex makes it feasible to study functions of virtually all hair cell genes by guiding RNAs to target the gene of interest. However, we did not test HDR in the inner ear in vivo, primarily due to low efficiency (less than 1%) even for in vitro assay compared to NHEJ where over 85% efficiency can be achieved. It is an area that needs substantial improvement in efficiency. Further studies are warranted to demonstrate that HDR is feasible in the inner ear.
Figure 4.
CRISPR/Cas9-mediated genome editing in hair cells in vivo. Cas9 protein and the GFP gRNA were complexed with the commercial lipids (Lipofectamine 2000), which were then injected into postnatal Atoh1-GFP mouse cochlea in which all outer hair cells were GFP positive. Genome editing was achieved in the outer hair cells by the absence of GFP signal in the injected inner ear, without affecting our hair cell survival. High-throughput sequencing confirmed the disruption of the GFP gene by indels. OHC: Outer Hair Cells
4. Potential challenge and prospect
One major challenge of broad application of CRISPR/Cas9 in hearing research appears to be its requirement of a PAM motif, which lowers its design density and may be a potential problem for small-range precise mutations like single nucleotide substitution because there could not be a PAM motif nearby. The most commonly used Cas9 from S. pyogenes requires a 5′-NGG PAM motif, which limits the design density to one per eight base pairs on average in genome (Jinek et al., 2012; Cong et al., 2013; Mali et al., 2013; Cho et al., 2013). One potential strategy is to combine Cas9 proteins from other bacteria that recognize different PAM motifs, and therefore the target scope of CRISPR/Cas9 can be extended. Some of these Cas9 proteins have already been adapted as genome editing techniques, including Cas9 from Neisseria meningitides (5′-NNNNGATT), Streptococcus thermophilus (5′-NNAGAAW), and Treponema denticola (5′-NAAAAC) (Cong et al., 2013; Esvelt et al., 2013; Hou et al., 2013).
Since its emergence just a few years ago, CRISPR/Cas9 technology has shown great promise to transform biomedical research and to be developed as a new type of treatment-based genome editing for a wide range of genetic disorders. The easy use and high efficiency distinguish it from other existing genome editing technologies, and we fully anticipate its productive applications in hearing research. For the study of inner ear genes, the utilization of reagents including Cre-dependent Cas9 knock-in transgenic mice (Platt et al., 2014) in combination with targeted delivery of gRNAs by AAV in vivo and in vitro could rapidly reveal gene functions by NHEJ-mediated disruptions without the process of germline transmission. The same route can be used to test intervention of mutations to restore hearing in genetic hearing models. However, to develop potential treatment for genetic HL in humans, we consider transient protein/nucleic acid delivery would be a better choice especially in reducing the risk associated with off-target effect. Further development for transient Cas9/gRNA delivery in the inner ear includes the lipid formulations that can target different cell types at different ages. In addition to the disruption of dominant mutations by NHEJ, one major challenge is to improve the efficiency of HDR that can be applied to recessive mutations. Given the rapid progress in the field, it is likely that these obstacles will be overcome in the near future, thus potentiating novel hearing treatments for genetic deafness.
Highlights.
Targeted genome editing using CRISPR/Cas9 is a powerful tool to perform genome function studies.
Applications and challenges of CRISPR/Cas9-mediated genome editing are discussed.
CRISPR/Cas9-mediated technology has implications in human genome editing in genetic hearing loss.
Zebrafish, mice, and primates can serve as model organisms to harness the potential of CRISPR/Cas9-mediated genome editing.
CRISPR/Cas9 genome editing holds great potential to modulate the function of genes involved in hearing loss.
Acknowledgments
This work was supported by grants R01 DC05575, R01 DC01246, 2P50DC000422-Sub-Project 6432, and R01 DC012115 from the National Institutes of Health/National Institute on Deafness and Other Communication Disorders to Xue Zhong Liu; NIH (R01 DC006908), the Bertarelli Foundation, and the David-Shulsky Foundation, and the Frederick and Ines Yeatts Hair Cell Regeneration grants to Zheng-Yi Chen; the University of Miami Provost’s Research Award and College of Arts and Sciences Gabelli Fellowship to Zhongmin Lu; the National Basic Research Program of China (973 program number 2012CB 967904 and 2012CB 967900) to Shiming Yang and Dinghua Xie and the National Nature Science Foundation of China NSFC81300824 to Yilai Shu. We are thankful to Drs. Robert Gerring and Christopher Lisi for the critical reading of the manuscript.
Abbreviations
- AAV
Adeno-associated virus
- ARHL
age-related hearing loss
- Cas9
CRISPR-associated nuclease 9
- CRISPR
clustered, regularly interspaced, short palindromic repeat
- crRNA
CRISPR RNA
- dCas9
defective Cas9
- DSBs
double-strand breaks
- gRNA
guide RNA
- HDR
homology-directed-repair
- HL
hearing loss
- HR
homologous recombination
- NHEJ
non-homologous-end-joining
- PAM
protospacer-adjacent motif
- RVD
repeat-variable di-residue
- siRNA
small interfering RNA
- TALENs
transcriptional activator-like effector nucleases
- TALEs
transcription activator-like effectors
- tracrRNA
trans-acting crRNA
- ZFNs
Zinc finger nucleases
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi N, So S, Iiizumi S, Nomura Y, Murai K, Yamakawa C, Miyagawa K, Koyama H. The human pre-B cell line Nalm-6 is highly proficient in gene targeting by homologous recombination. DNA Cell Biol. 2006;25 (1):19–24. doi: 10.1089/dna.2006.25.19. [DOI] [PubMed] [Google Scholar]
- Akil O, Seal RP, Burke K, Wang C, Alemi A, During M, Edwards RH, Lustig LR. Restoration of hearing in the VGLUT3 knockout mouse using virally mediated gene therapy. Neuron. 2012;75 (2):283–293. doi: 10.1016/j.neuron.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeli S, Lin X, Liu XZ. Genetics of hearing and deafness. Anat Rec. 2012;295 (11):1812–1829. doi: 10.1002/ar.22579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol Cell Biol. 2001;21 (1):289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161 (3):1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300 (5620):764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. A primer for morpholino use in zebrafish. Zebrafish. 2009;6 (1):69–77. doi: 10.1089/zeb.2008.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326 (5959):1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333 (6051):1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci U S A. 2009;106 (26):10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson DF, Tan WF, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CBA, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci U S A. 2012;109 (43):17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23 (4):465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31 (3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off- target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24 (1):132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nat Commun. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41 (20):9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon JB, Zeitler B, Cheng J, Cheng AT, Cherone JM, Santiago Y, Lee AH, Vo TD, Doyon Y, Miller JC, Paschon DE, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Drubin DG. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat Cell Biol. 2011;13 (3):331–337. doi: 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Lu G, Xie Z, Lou M, Luo J, Guo L, Zhang Y. Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Res. 2014;24 (8):1009–1012. doi: 10.1038/cr.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411 (6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10 (11):1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K, Leake D, Marshall WS, Khvorova A. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12 (7):1188–1196. doi: 10.1261/rna.28106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High- frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31 (9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32 (3):279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31 (7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32 (6):577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Christensen RG, Rayla AL, Lakshmanan A, Stormo GD, Wolfe SA. An optimized two-finger archive for ZFN-mediated gene targeting. Nat Methods. 2012;9 (6):588–590. doi: 10.1038/nmeth.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Hall VL, Kok FO, Shin M, McNulty JC, Lawson ND, Wolfe SA. Targeted chromosomal deletions and inversions in zebrafish. Genome Res. 2013;23 (6):1008–1017. doi: 10.1101/gr.154070.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgert N, Smith RJ, Van Camp G. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681 (2–3):189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgert N, Smith RJH, Van Camp G. Function and expression pattern of nonsyndromic deafness genes. Curr Mol Med. 2009;9 (5):546–564. doi: 10.2174/156652409788488775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29 (8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res. 2002;30 (8):1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou ZG, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110 (39):15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31 (9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion U, Krauss J, Nüsslein-Volhard C. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development. 2014;141 (24):1–4. doi: 10.1242/dev.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Gavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21 (6):635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110 (34):13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337 (6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, Han S, Peng T, Thams S, Mikkilineni S, Mellin C, Merkle FT, Davis-Dusenbery BN, Ziller M, Oakley D, Ichida J, Di Costanzo S, Atwater N, Maeder ML, Goodwin MJ, Nemesh J, Handsaker RE, Paull D, Noggie S, McCarroll SA, Joung JK, Woolf CJ, Brown RH, Eggan K. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell. 2014;14 (6):781–795. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20 (1):81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kweon J, Kim E, Kim S, Kim JS. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 2012;22 (3):539–548. doi: 10.1101/gr.129635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Haurigot V, Doyon Y, Li T, Wong SY, Bhagwat AS, Malani N, Anguela XM, Sharma R, Ivanciu L, Murphy SL, Finn JD, Khazi FR, Zhou S, Paschon DE, Rebar EJ, Bushman FD, Gregory PD, Holmes MC, High KA. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475 (7355):217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DL, Qiu ZW, Shao YJ, Chen YT, Guan YT, Liu MZ, Li YM, Gao N, Wang LR, Lu XL, Zhao YX, Liu MY. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31 (8):681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- Liu XZ, Xu LR, Hu Y, Nance WE, Zhang SL, Xu Y. Epidemiological studies on hearing impairment with reference to genetic factors in Sichuan, China. Annals of Otol Rhinol Laryngol. 2001;110 (4):356–363. doi: 10.1177/000348940111000412. [DOI] [PubMed] [Google Scholar]
- Liu XZ, Yan D. Aging and hearing loss. J Pathol. 2007;211 (2):188–197. doi: 10.1002/path.2102. [DOI] [PubMed] [Google Scholar]
- Lombardo A, Cesana D, Genovese P, Di Stefano B, Provasi E, Colombo DF, Neri M, Magnani Z, Cantore A, Lo Riso P, Damo M, Pello OM, Holmes MC, Gregory PD, Gritti A, Broccoli V, Bonini C, Naldini L. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods. 2011;8 (10):861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345 (6201):1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339 (6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell Smith A, Takeuchi R, Pellenz S, Davis L, Maizels N, Monnat RJ, Jr, Stoddard BL. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proc Natl Acad Sci U S A. 2009;106 (13):5099–5104. doi: 10.1073/pnas.0810588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehl AL, Thomson V. Newborn hearing screening: the great omission. Pediatrics. 1998;101 (1):e4. doi: 10.1542/peds.101.1.e4. [DOI] [PubMed] [Google Scholar]
- Miller J, Mclachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor Iiia from Xenopus oocytes. Embo Journal. 1985;4 (6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbitzer R, Romer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci U S A. 2010;107 (50):21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton CC, Nance WE. Current concepts: Newborn hearing screening - A silent revolution. New Engl J M. 2006;354 (20):2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A Simple Cipher Governs DNA Recognition by TAL Effectors. Science. 2009;326 (5959):1501–1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Nance WE. The genetics of deafness. Ment Retard Dev Disabil Res Rev. 2003;9 (2):109–119. doi: 10.1002/mrdd.10067. [DOI] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J. Generation of gene- modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156 (4):836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516 (7530):263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31 (9):839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159 (2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300 (5620):763–763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Reh WA, Vasquez KM. eLS. John Wiley & Sons, Ltd; Chichester: 2014. Gene targeting by homologous recombination. [DOI] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30 (5):460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. Plos Genetics. 2007;3 (5):787–801. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288 (5473):2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14 (12):8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian-cells. Proc Natl Acad Sci U S A. 1994;91 (13):6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, Pierick CJ, Hoffman E, Maeder ML, Khayter C, Reyon D, Dobbs D, Langenau DM, Stupar RM, Giraldez AJ, Voytas DF, Peterson RT, Yeh JR, Joung JK. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat Methods. 2011;8 (1):67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, Ye Z. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014;15 (1):12–13. doi: 10.1016/j.stem.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollu C, Pars K, Cornu TI, Thibodeau-Beganny S, Maeder ML, Joung JK, Heilbronn R, Cathomen T. Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res. 2010;38 (22):8269–8276. doi: 10.1093/nar/gkq720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel J, Blucher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012;30 (7):593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Yu C, Qu J, Li M, Yao X, Yuan T, Goebl A, Tang S, Ren R, Aizawa E, Zhang F, Xu X, Soligalla RD, Chen F, Kim J, Kim NY, Liao HK, Benner C, Esteban CR, Jin Y, Liu GH, Li Y, Izpisua Belmonte JC. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell. 2014;15 (1):31–36. doi: 10.1016/j.stem.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32 (6):569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435 (7042):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11 (9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Van Gent DC, Hoeijmakers JHJ, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nature Reviews Genetics. 2001;2 (3):196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- Vanamee ES, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. J Mol Biol. 2001;309 (1):69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys(2)His(2) zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Yan D, Liu XZ. Cochlear molecules and hereditary deafness. Front Biosci. 2008;1 (13):4972–4983. doi: 10.2741/3056. [DOI] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32 (6):551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33 (1):73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]