Abstract
Almost all proteins contain charged residues, and their chain distribution is tailored to fulfill essential ionic interactions for folding, binding and catalysis. Among proteins, the hinged two-domain chain of the cellular prion protein (PrPC) exhibits a peculiar charge structure with unclear consequences in its structural malleability. To decipher the charge design role, we generated charge-reverted mutants for each domain and analyzed their effect on conformational and metabolic features. We found that charges contain the information for interdomain interactions. Use of dynamic light scattering and thermal denaturation experiments delineates the compaction of the α-fold by an electrostatic compensation between the polybasic 23–30 region and the α3 electronegative surface. This interaction increases stability and disfavors fibrillation. Independently of this structural effect, the N-terminal electropositive clusters regulate the α-cleavage efficiency. In the fibrillar state, use of circular dichroism, atomic-force and fluorescence microscopies reveal that the N-terminal positive clusters and the α3 electronegative surface dictate the secondary structure, the assembly hierarchy and the growth length of the fibril state. These findings show that the PrP charge structure functions as a code set up to ensure function and reduce pathogenic routes.
The charge organization in proteins defines the inter- and intramolecular ionic interactions essential for folding, binding and catalysis1,2,3. Altered charges modify the structural dynamics, the aggregation, and amyloid formation propensity, among others, of elementary proteins processes in all protein conformational diseases4,5,6,7,8,9,10,11,12,13,14. In prion disorders, the prion protein (PrP), a two-domain chain with an N-terminal effector tail (FT) hinged to a C-terminal globular domain (GD), forms transmissible amyloids15,16. Although the information for both folding and misfolding is contained in the 90–231 sequence region, the regulatory role exerted by the charged FT and the pathogenicity of mutations related to exposed charges underline an intramolecular code that remains to be elucidated10,17,18,19,20,21,22,23,24,25,26,27,28,29,30.
PrP displays a peculiar charge pattern in its two-domain chain, with a polybasic FT and all acid residues located at the GD. The FT is composed of repeats flanked at either side by positively charged clusters, known as CC1 (residues 23–30) and CC2 (residues 101–110). Despite its intrinsically disordered tail, the protein undergoes ligand-induced folding and can wrap around the GD facing the α2–α3 exposed surface, which induces compaction of the α-fold19,27,31,32. Removing the CC1 cluster alters the α-fold stability, early nucleation steps, the polymer shape and the prion propagation efficiency17,18,20,21,22,23,24,25,26,28,29,30. CC2 plays a very active metabolic role, participating in both biogenesis and processing33,34. Importantly, its charge abrogation yields amyloids with PrPSc-like features35. The spacing between CC1 and CC2 affects disease onset, GD stability and the FT effector function28,30,36,37. On the contrary, the GD contains all the acid residues of the chain. Among them, D144, D147, D178, E196 and E211 (numbered according to the human sequence) are involved in salt bridges that either stabilize (α1/α3) or link structural elements (β2 to α2 and α1 to α2/α3)38,39,40,41. Other residues, such as E146 and E152 in α1, D167 in β2-α2, and E200, D202, E207, E221 and E228 in α3, expose their side chains to solvent, thus defining electronegative surface clusters39. Of these charges, the structural D144 and D147 residues and their respective salt bridges stabilize PrPC, preventing conversion to protease resistance forms, whereas D178, E196 and E211 are prone sites for pathogenic mutations upon charge alteration38,42. Moreover, pathogenic mutations, such as E200K and Q217R, and the dominant negative E219K polymorphism alter the α3 surface electrostatic potential, inducing minor folding effects39.
To unveil the information encoded in the complementary solvent-exposed charge structure, we constructed several mutants consisting of charge reversions and inclusions and tested their effects on the properties of both the α-folded and fibrillar states. These modifications avoid the formation of non-polar surface patches resulting from abrogating charges and it solubility effect43. At the FT, both the CC1 and CC2 regions were modified by substituting their K residues with E (K2: K24EK27E, K4:K101EK104EK106EK110E and K6: K2–K4). At the GD, the α3 electronegative surface was perturbed by replacing E200 and E221 with K and by replacing Q217 and Q219 with charged R and K, respectively. We found that charges dictate a variety of structural and metabolic traits mostly through communication between domains. Effects such as the stabilization of the native α-fold, dictating the efficiency of the α-cleavage, attenuating the fibrillation propensity and yielding the most benign amyloids suggest that the charge design ensures PrPC functions.
Results
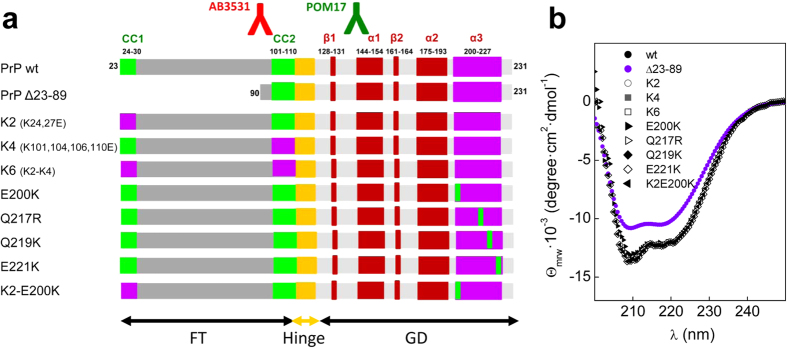
To gain insight into the charge design of the PrP chain, we utilized reversion and insertion approaches using the rHaPrP (23–230) (PrP wt) as template (Fig. 1a). This choice preserved surface charge avoiding solubility effects resulting from charge abrogation43. At the FT, both the CC1 and CC2 charges were reversed (K2: K24EK27E, K4:K101EK104EK106EK110E and K6: K2-K4). At the GD, the α3 charge surface was modified by independent E200K, Q217R, Q219K, and E221K substitutions. Of these chains, K4 is equivalent to MoFBOM122, E200K and Q217R are pathogenic mutations found in humans44,45, and Q219K represents the dominant-negative variant of MoPrP Q218K46. All of these chains were produced recombinantly, yielding cooperative folds with a predominantly α-helical secondary structure (Fig. 1b).
Figure 1. Charge structure of the PrP chain.
(a) Modular organization of the PrP chain into an N-terminal domain (FT) hinged to a C-terminal globular domain (GD), displaying the location of the charged regions and their mutations that were considered in this study. Epitopes for Ab3531 and POM17 are depicted using the color codes of fluorescence microscopy. (b) Far-UV CD spectra in 10 mM MES pH 6.5 of PrP wt and of its charge and length mutants due to their α-folding.
PrP charge structure encodes an interdomain interaction promoting the α-fold compaction
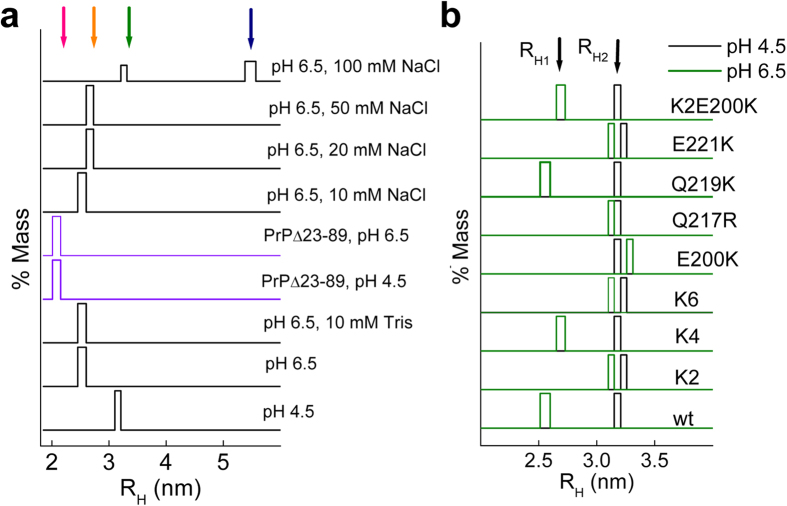
Because charges are fundamental for intra- and intermolecular interactions, we probed the fold hydrodynamic properties using dynamic light scattering (DLS) (Fig. 2). To minimize insolubility interferences, measurements were performed using 15 μM protein concentrations at pH values of 4.5 and 6.5, for which open and closed PrP wt states have been reported, respectively27,40. Figure 2a shows that PrP wt yielded monodisperse species with RH of 3.2 ± 0.1 (pH 4.5) and 2.6 ± 0.1 (pH 6.5) nm, whereas its GD as PrPΔ23–89 yielded an RH of 2.1 ± 0.1 nm at both pH values47,48. Of these values, only the 2.6 nm for the PrP wt and 2.1 nm for the PrPΔ23–89 were comparable to the theoretical RH values of globular protein spheres with the same molecular weight (2.25 and 1.96 nm, respectively), whereas the value of 3.2 nm for the PrP wt at pH 4.5 deviated from the ideal behavior, agreeing with the open state indicated via NMR. Importantly, the RH values at pH 6.5 remained constant in the presence of 10 mM Tris used as a cation quencher, ruling out the effects of cation traces (Fig. 2a).
Figure 2. Effect of charges on the PrP hydrodynamic features.
(a) Mass size distributions of PrP wt and PrPΔ23–89 as a function of pH and NaCl concentration. The theoretical values of RH for spheres with similar MWs to PrP wt and PrPΔ23–89 are 2.25 and 1.96 nm, respectively. (b) Mass size distributions of PrP mutants in 10 mM NaAc pH 4.5 (black) and 10 mM Mes pH 6.5 (green). The measurements were performed at 25 °C using at least two different protein batches. Column widths show the standard deviation among measurements. Arrows at the top indicate the different RH values.
At pH 4.5 all charge mutants yielded an RH (approximately 3.2 ± 0.1 nm) similar to the PrP wt, ruling out perturbations in the open state (Fig. 2b). On the contrary, at pH 6.5, the RH of K4 and Q219K decreased to 2.6 ± 0.1 nm, whereas the RH values of K2 (both independently or combined with K4 as K6), E200K, Q217R and E221K remained unaltered. The lack of an RH difference for these mutants suggests an impaired compaction and a structural role of CC1, E200, Q217 and E221 in such process. Given the hydrodynamic invariability of the GD and the pH-induced electropositive charge changes (His residues in octarepeats) in the FT, the impaired RH reduction for the CC1, E200, Q217 and E221 mutants suggests that compaction involves an electrostatic interaction between the electropositive CC1 and the electronegative α3 surface, which is altered in Q217R. To further test this possibility, we constructed a K2-E200K mutant containing a double reversion and, after verifying its cooperative folding (Figs 1 and 3a), we tested its hydrodynamic properties. Figure 2b shows that the K2-E200K mutant behaved similar to the PrP wt, supporting the idea that the α-fold undergoes a compaction due to a long-range electrostatic interdomain interaction between the CC1 and α3 surface charges. The interaction driving the interdomain lock in monomeric PrP wt is weak, and NaCl concentrations above 50 mM provoke the emergence of species corresponding to the open (3.2 ± 0.1) and oligomeric (5.6 ± 0.2 nm) states (Fig. 2a), agreeing with the relevance of stabilizing factors such as coordinated cations27,49.
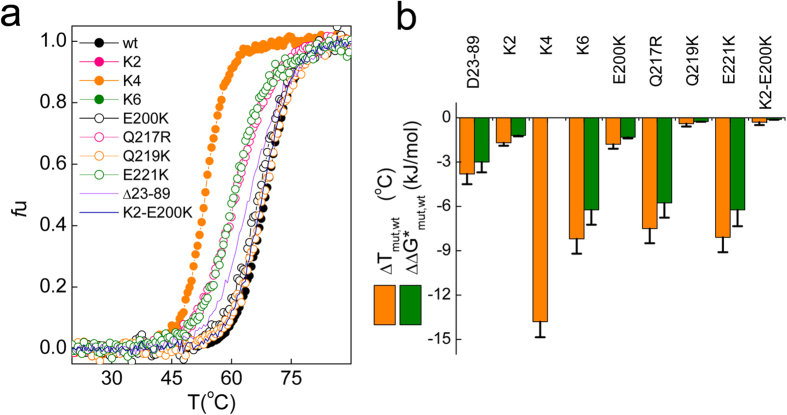
Figure 3. Effect of domain charges on the PrP stability.
(a) Thermal denaturation of PrP wt and mutants. The unfolded fraction was calculated using the ϴ222 temperature function according to a two-state transition, as described48. (b) Differences in the unfolding temperature (ΔTm) and in the free energy of unfolding (ΔΔG*) induced by the charge mutations. ΔTm is the difference between the denaturation temperature of PrP mutant (PrPmut or PrP90−231 wild type) and full length PrPwt (Tmmut −Tmwt). Tm values were obtained as the midpoints of the temperature denaturation curves. ΔΔG* was obtained as ΔH,wt × (1−Tmwt/Tmmut), where ΔHvH is the van´t Hoff enthalpy of PrP wt denaturation (255 kJ/mol). ΔΔG* < 0 indicates a destabilization of the mutant chain compared to the wt. Displayed data are the average of three independent measurements, performed with at least two different protein batches. Error bar represents the standard deviation (s.d.). Calculations for K4 were omitted given its irreversible denaturation.
Charge structure regulates α-fold stability through interdomain interactions in the native and denature states
To gain insights on the effect of the PrP charge structure on its thermodynamic stability, we analyzed the thermal denaturation curves, as shown in Fig. 3. It must be noted that measurements were performed in the absence of described interdomain cation stabilizers to avoid effects other than their complexation with His residues of the N-terminal octarepeats27,49. With the exception of K4, all of the thermal denaturations were reversible, as indicated by the recovery of at least 90% of the initial signal after cooling from the highest temperature. K4 denaturation was irreversible, and insoluble aggregates were detected upon cooling from the highest temperature (data not shown). Figure 3a,b show that the mutants with impaired compaction, such as K2, K6, E200K, Q217R, E221K and Δ23–89, unfolded with Tm values lower than that of wt PrP. This effect varied according to K6 ≈ E221K ≈ Q217R >Δ23–89> E200K ≈ K2, leading to decreases in the free energy of unfolding of about 1.2–6 kJ/mol. On the contrary, Q219K and K2-E200K, which undergo compaction, exhibited a PrP wt-like denaturation profile. Notably, the K4 mutant, exhibiting PrP wt-like hydrodynamic properties, yielded the most thermally labile fold, possibly due to its singular irreversible denaturation. Thus, with the exception of K4, these results indicate that altering the charges that mediate the interdomain lock decrease the native fold stability.
The destabilization pattern of the mutants precluding compaction agree with the effects described for the chains containing N-terminal truncations at pH > 6.0 and slightly differ from that described for PrP chains consisting in mutant GD moieties20,21,37,50,51,52 (Fig. 3b). By virtue of its additivity, the ΔG of folding of a two-domain protein (TD or full length) can be expressed as the sum of the contributions arising from the folding of each of domain (ΔGFT and ΔGGD) and from their interaction (ΔGFTGD). Since the ΔGFT and ΔGFTGD are linked in PrP, their sum (ΔG*FT) can be calculated as ΔG*FT = ΔG TD - ΔGGD. For the PrP wt and a GD consisting in PrPΔ23–89, ΔG*FT corresponded to 3 ± 0.7 kJ/mol (−ΔΔG* in Fig. 3b), which agree with the 3.2 ± 1 kJ/mol value that can be calculated for GD consisting in PrPΔ32–89 chains20. For mutants exhibiting either identical GD as K2 or negligible effects on its free energy of unfolding as E200K50,51, which increasing GD breathing facilitates M213 sulfoxidation53, ΔG*FT can be approximated to ΔGTDwt-ΔGTDmutant difference (−ΔΔG* in Fig. 3b). This approach yielded values of 1.2 ± 0.2 and 1.5 ± 0.7 kJ/mol for K2 and E200K respectively. Taken together the values of PrPΔ32–89, K2 and E200K yielded an averaged estimation of ΔG*FT of approximately 2 kJ/mol. On the hand, for Q217R the ΔΔG*Q217R reported for the GD chains was −8.9 ± 2 kJ/mol50,51 and the calculated for the TD amounted to −5.8 ± 1 kJ/moly. As with Q217R, K6 reversing the charge of FT flanks provoked a similar ΔΔG* of −6.2 ± 1 kJ/mol. The higher destabilization of Q217R and K6 compare to PrP Δ23–89 suggested that in these mutants their charge changes affected interdomain interactions not only in the native state but also in their denature state54.
Charges are gatekeepers of PrP fibrillation
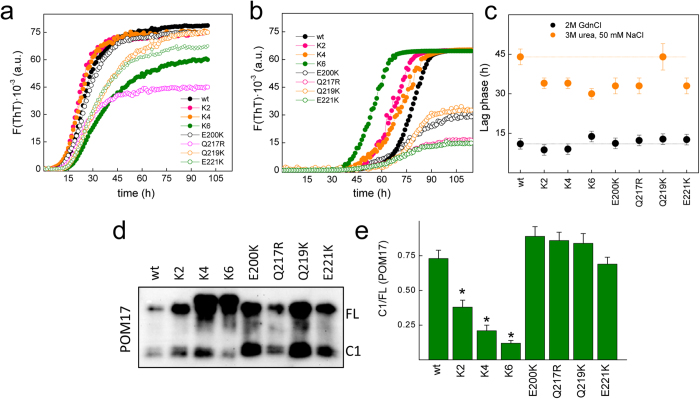
Although most PrP amyloids generated in vitro lack the infectivity and proteolytic signatures of PrPSc, their formation models the chain propensity and the conformational changes required for GD self-assembly55,56. To test whether the charge structure, through either the α-fold stabilization described above or the initial self-assembly step, impacts the fibrillation, we performed time-dependent Thioflavin T (ThT) binding experiments using the various PrP chains at pH 6.5 and calculated the lag-phase as indicator of propensity (Fig. 4). To induce the required mild denaturation, we used either 2 M GdnCl (conventional ionic media) or 3 M urea containing 50 mM NaCl (low salt). As anticipated from the exposed character of the mutated charges and the high ionic strength of the media, fibrillation in 2 M GdnCl (Fig. 4a) yielded lag-phases that were roughly similar for all chains, with minor reductions (K2,K4) or enhancements (K6) (Fig. 4c). On the contrary, reactions in the low salt media containing 3 M urea and 50 mM NaCl revealed that with the exception of Q219K, all mutants form fibrils faster than the PrP wt as indicated by their higher lag-phases (Fig. 4b,c). Kinetic curves also showed that charge changes in the GD (E200K, Q217R, Q219K and E221K) decrease the ThT fluorescence increment linked to fibrillation predominantly under low salt (Fig. 4a,b), suggesting off-pathway events.
Figure 4. Charge changes modify the fibrillation propensity and processing of PrP.
Time-dependence of ThT binding of the PrP wt and mutants (40 μM protein concentrations) in 50 mM MES pH 6.5 at 37 °C containing (a) 2 M GdnCl and (b) 3 M urea and 50 mM NaCl. The curves represent the average of three independent measurements, performed in triplicate. (c) Lag-phases of the fibrillation reactions of the PrP wt and mutants in 50 mM Mes pH 6.5 containing either 2 M GdnCl or 3 M urea with 50 mM NaCl. The depicted values correspond to three independent experiments, each performed in triplicate. (d) Western blot of PNGase-treated cell lysates of CHO cells transfected with PrP wt and the charged mutants. Detection was performed using POM17, and the positions of the full-length (FL) and N-terminal-truncated (C1 fragment) chains are depicted. (e) Variations in the C1/FL ratio of the PrP chains. Quantifications are the average of two independent transfection assays. Error bar represents the standard deviation (s.d.). *p < 0.01.
The observed reduction in the lag-phase in the low salt reaction media in K2, K4, K6, E200K, Q217R and E221K variants indicated that CC1, CC2 and the α3 electronegative caps function as fibrillation gatekeepers. Since K2, K6, E200K, Q217R and E221K impeded domain compaction and K4, with preserved compaction, lack the charges preventing the PrP fibril N-terminal packing35, together their effects suggest that charges regulate fibrillation propensity through their role in both the native fold compaction and the packing of critical regions for the seed stability. This charge effect agree with the increased propensity of PrP Δ23–89 compared to the full length chain57,58 and the dependence of anionic cofactors for functional fibrillation59.
FT charges regulate the efficiency of C1 production
PrP processing into C1 chains is essential for the abrogation of prion formation and propagation, involving a complex cleavage within the interdomain hinge (around residue 110)60,61,62. To test whether the PrP charge structure plays a role in this processing, CHO cells were transfected with plasmids coding for PrP wt or its mutants, and the expressed chains were analyzed using POM 17 after PNGaseF digestion. Figure 4d,e shows that among the different PrP chains, only K2, K4 and their combination as K6 drastically reduced the efficiency of C1 production compared to wt PrP, which is in agreement with previous reports that showed impaired C1 production for mouse variants of K4 and PrPΔ23–31 in both Hpl cells and transgenic mice22,25. On the contrary, the mutants including modifications of the α3 surface charge impairing the interdomain lock, such as E200K, Q217R and E221K, yielded C1 levels similar to the PrP wt (Fig. 4d,e). Differences in the dependence of C1 production on the charge structure of either domain indicated that processing through the α-cleavage site is highly dependent on the FT charge structure but fairly independent of their role in driving the α-fold compaction. Thus, C1 production may require the recognition of the FT through its charge clusters by an unknown anionic ligand52,63,64,65, implying conditions with unlocked α-fold. These results suggest that the PrP charge structure regulates processing through its role in interactions.
Charges impact the secondary to quaternary structure of the fibrils
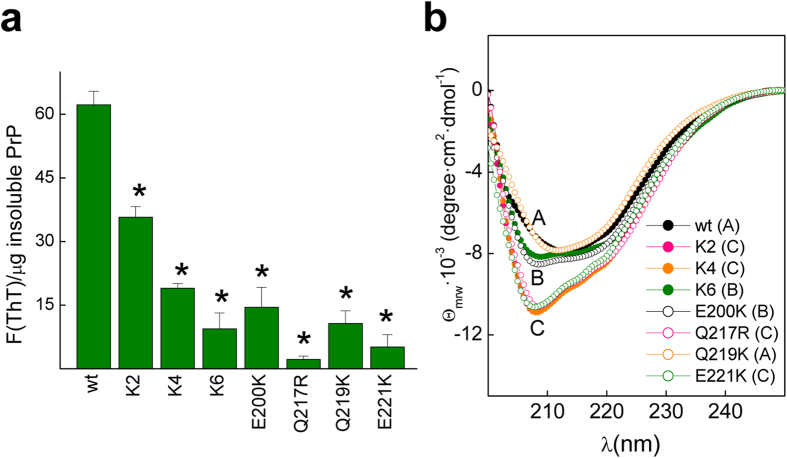
Similar to the α-folded wt PrP chain, variants with altered charge structure form fibrils that differ in their shape and proteinase-resistant core21,35. To gain insight into the effects of the charged design on the fibril structure and its properties, we used the reaction conditions yielding PrP wt S-fibrils with resolution for imaging analysis. S-fibrils, produced under slow orbital agitation as opposed to the fast rotation leading to the R-polymorph, are featured by a β-sheet-like far-UV CD spectrum along with a thin and curvy fibril topology that expose the 90–102 region while partially shield the POM17 epitope66 (Figs 5, 6, 7). It must be stressed that this fibrillation reaction was performed in the presence of 2 M GndCl, which abrogates the charge effects on the native α-fold and permits the assignation of the deviations from the wt behavior to differences in the assembly process. Fibrils formed by all PrP mutants displayed a reduced specific ThT binding, suggesting major structural differences (Fig. 5a).
Figure 5. Properties of the PrP wt and mutant fibrils.
(a) Specific ThT binding of the PrP wt and mutant fibrils. Typically 50 μM of fibrils in 10 mM ammonium acetate pH 5 were incubated for 10 min with ThT (15 μM) before fluorescence determination. ThT fluorescence intensities were corrected for the background (absence of fibrils) and divided by the protein amount in the pellet of a 12000 rpm 20 min centrifugation. The depicted data represent the average of two independent experiments performed in duplicate (*p < 0.01). (b) Far-UV CD spectra of the PrP wt and mutant assemblies in 10 mM ammonium acetate pH 5. The fibrils were formed in 50 mM MES pH 6.5 containing 2 M GdnCl at 37 °C under continuous rotation at 24 rpm, and then dialyzed against 10 mM ammonium acetate pH 5.
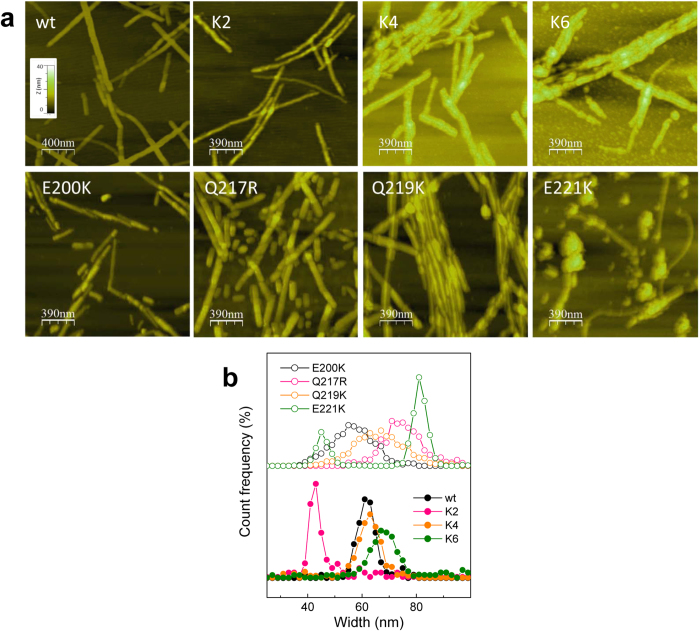
Figure 6. Topology images of the PrP wt and mutant fibrils.
(a) AFM images of the PrP wt and mutant fibrils corresponding to the topology mode. The z-axis was fixed for all graphs, and the color scale is displayed as an insert in the PrP wt panel. (b) Histogram of the width distribution of the distinct PrP assemblies.
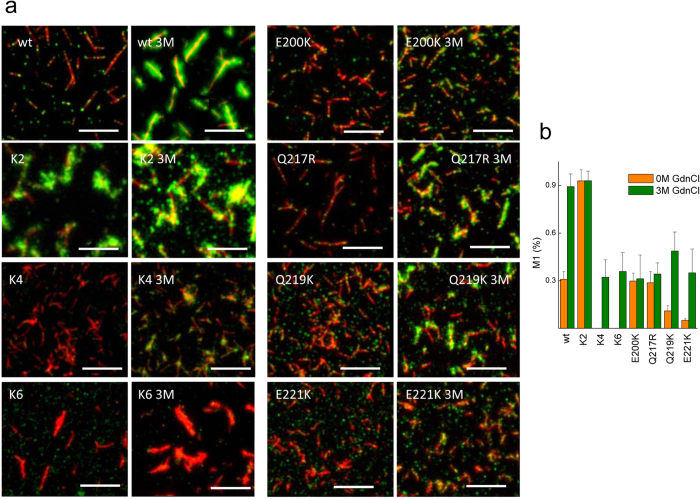
Figure 7. Surface reactivity of PrP wt and mutant fibrils.
(a) Representative fluorescence images of the fibrils formed by the PrP wt and mutant fibrils stained with AB3531 (red) and POM17 (green). Pretreatment with 3 M GdnCl is indicated as 3 M. The white bar represents 5 μm. (b) Effect of charges on the variation of the POM17 overlap with AB3531 (Manders overlap coefficient) based on the denaturant concentration. Error bar represents the standard deviation (s.d.).
Among the fibrils, only those formed by Q219K exhibited an S-like CD spectrum (Fig. 5b), whereas the assemblies formed by K2, K4, K6, E200K, Q217R and E221K displayed R-like CD spectra featuring a higher negative ellipticity, a minimum at 207 nm, and a shoulder at 217 nm of the β-sheet and turn structures (Fig. 5b). These data suggest a strict dependence of the S-fibril on CC1, CC2, and α3 surface charges. Because only the R-type fibril exhibited in vivo infectivity, the PrP charge structure appears to drive the fibrillation path to the less pathogenic S-type state67. The blockage of S-fibril type formation in K2, K4, K6, E200K, Q217R and E221K mutants also suggest that these charges may behave as sites for cofactor binding that, through their shielding, may shift the conversion route to the most infectious path35,68,69. Accordingly, in addition to known anionic cofactors which may function recognizing CC1 and CC2 regions may be molecules that exhibiting a basic charge would produce similar effects shielding the α3 surface charges , which would amplify the group of molecules functioning as cofactor in conversion35,59,68,69.
The Q219K substitution forms S-type-fibrils as PrP wt, but their assembly exhibited several differences. First, AFM imaging revealed that the Q219K fibrils, which were similar in dimension to those formed by the PrP wt, displayed a high degree of lateral association, yielding fibril layers rarely observed in PrP wt (Fig. 6). Second, the immunofluorescence analysis resolving fibrils in the μm range showed that in the wt and the mutant fibrils, the 90–102 (red, Ab3531 epitope) region is exposed along the fibril axis, whereas POM17 epitopes (140/145 region) were detected as discrete regularly spaced dots (green), which after treatment with 3 M GdnCl become disrupted in the wt fibrils but persist in the Q219K fibrils (Fig. 7a). These differences in the POM17/Ab3531 overlap both in the absence and presence of denaturant treatment are depicted in Fig. 7b.Thus, the lateral association and the attenuated surface reactivity of the Q219K fibrils may presumably contribute to the dominant-negative property of this charge insertion.
The K2 fibrils were thinner than the wt fibrils and tending to arrange into Y-shapes (Fig. 6). The immunofluorescence analysis revealed that the POM17 epitope was fully exposed in the absence of denaturant treatment (Fig. 7). On the contrary, K4 and K6 formed thick fibrils, indicating a higher assembly complexity. Of them, the peculiar stacking of the K6 assemblies resembled the aggregates of PrP27–30 rods35,70. Immunostaining revealed that the POM17 epitope was highly secluded in the K4 and K6 assemblies, resisting treatments, with distinct GdnCl concentrations (Fig. 7). These results indicate that CC1 and CC2 charges, in addition to allowing for S-type fibrillation, govern the evolution of the hierarchical assembly and the surface exposure of the region 140–145. CC1 appeared to work during the initial assembly step involving the shielding of the 140–145 region, whereas CC2 in a dominant fashion functioned during later assembly steps, compromising the discrete POM17 epitope exposure. These effects suggest that the FT charges modulate properties of the fibril state of the C-terminal domain.
Changes in the α3 charge structure resulted in R-type (Fig. 5b). AFM imaging revealed thicker fibrils compared to those of the PrP wt and images that were rich in short structures, rod-shaped in E200K and Q217R and spherical in E221K (Fig. 6). Interestingly, the E221K mutant revealed extremely thin, curvy fibrils emerging from the spherical aggregates (Fig. 6). The E200K and Q217R fibrils displayed a surface reactivity similar to that of the PrP wt but with green dots at reduced spacing, suggesting POM17 epitope location at the joint of the short rod-shaped structures (Fig. 7). In both assemblies, mild denaturing treatment increased POM17 staining but to a lesser extent that in the wt fibrils, suggesting differences in the folding of the 140–145 region. On the contrary, the E221K fibrils resembled those formed by K4, with a complete seclusion of the POM17 epitope and a discrete exposure upon treatment with 3 M GdnCl (Fig. 7). Taken together, these features suggest that the charges of the α3 primarily regulate the fibril length by either limiting growth or promoting fragmentation and the exposure of the POM17 epitope.
Discussion
Here, we show that the PrP charge structure considered as solvent exposed contains information regarding interdomain interactions in both the PrPC-like conformation and the fibrillar state. In the PrPC-like formation, FT wraps the GD via the interaction between the N-terminal polybasic motive (CC1) and the α3 electronegative surface. This complementary charge interaction is relatively weak, and in the absence of stabilizers such bound Zn2+ the modifications provoked by the ionic strength or diminished surface electronegativity (single charge reversion or inclusions along the helix) impaired it. Notwithstanding, the interdomain interaction adds to the global fold a stabilization lower threshold of approximately 2 kJ/mol which interferes with fibrillation. Independent of this structural effect, the FT charge design also dictates the efficiency of cleavage between domains through the α-site indicating that the production of neuroprotecting fragments involves conditions with unlocked domains. In the fibrillar state, the PrP charge structure regulates the assembly process, dictating the secondary structure of the scaffold, the allowance of different (initial, intermediate and late) assembly steps, and the polymer length.
The sequence of proteins contains the protein’s functional information, including signaling mechanisms for maturation, sorting, covalent modification, and folding regulation, whereas other functions are encrypted and operate metabolically. The PrP sequence exhibits an unusual abundance of encrypted codes. The abundance of Met residues in the PrP chain allows for the translation of a minor chain that segregates out of the major secretory route, a structural regulation through a redox process, and a metabolically controlled substitution by SeMet71,72,73. Similarly, the His distribution entails a complex pH-regulated, cation binding trait in the FT that provides a diverse structural landscape with functional consequences27,32,63,74. Regarding charges, the complementary polybasic stretches in the FT and electronegative surfaces from the GD folding provide the basis for molecular compaction and its regulation by FT and GD ligands. Charge structures also participate in the fibrillation process, bestowing interdomain-dependent and domain-specific growth and assembly features.
The PrP charge structure seems to encode information for latent states with balanced ligand binding functions. The interdomain lock, functionally in non-transmembrane PrP formations, may sense changes in the length and flexibility of the FT, which depend on the number of repeats, the pH and cation binding27,28,30,36,37,75. Indeed, the interaction can be isolated either under partial protonation of the octarepeat His, as described here, or stabilized by Zn2+ binding27 to ensure ionic strength resistance. FT ligands that recognize its charge clusters regulate the cleavage of domains, suggesting that this process that monitors the dose of prion precursors uses as substrate the unlocked domain molecule. On the other hand, GD ligands precluding the lock by shielding the interacting motives (POM 4,10,19 and α-219–232) or by providing steric impediments (POM1) allow for FT toxic signaling28,76, suggesting that at the cell surface PrPC may exit under the compact state. As in the GD the α3 surface constitutes the prion binding site, its shielding by the interdomain lock may also interfere with prion amplification by competing with seed binding76. Moreover, the interdomain lock prevents fibrillation of PrP by stabilizing the α-fold, which could be utilized in therapeutic intervention using the lock state as a target.
The charge structure code also operates in the fibrillation path, dictating properties that compromise propagation and toxicity. Despite the differences with PrPSc, only R-type PrP fibrils exhibit in vivo propagation, which are only found in mutants with a modified charge structure66. The PrP charges appear to drive the chain into the fibril structures with attenuated pathogenicity and provide sites for cofactor action. Therefore, cofactors that bind to CC1 may regulate the initial steps, whereas cofactors that shield CC2, singly or combined with CC1, may drive the progression to the advanced assembly steps. These mechanistic hints agree with the mode of action of polyanions in previous conversion studies35,59. Moreover, the limits of assembly evolution due to FT charges are inversely correlated with the POM17 epitope exposure. Because this region contains the DWED sequence, forming part of the α1 in the α-fold and stabilized by salt bridges, it is tempting to postulate that the assembly involves its interaction with CC1. On the other hand, charged residues at either end of the α3, which are part of the β-core but not engaged in β-sheets, mainly govern the fibril length. Changing these charges reduces the length, either impeding growth or favoring fragmentation, which increases the seed concentration and consequently the propagation efficiency77. Thus, α3 mutations may strengthen their pathogenicity by optimizing their propagation through the seed concentration.
In conclusion, PrP charge design encodes several of structural and metabolic traits set up for ensuring function and diminishing pathogenic routes. These traits are: 1) shaping an interdomain lock, which prevents the FT from its toxic signaling and the GD from its prion receptor activity, 2) promoting the α-cleavage yielding anti-prion C1 chains and reducing the dose of chains with pro-prion activity, and 3) stabilizing the α-fold against conversion and if so allowing the less pathogenic assembly.
Methods
Production of PrP chains
rHaPrP (23–231) chains were produced from their pET11a constructs as described48,78, with minor modifications. Briefly, inclusion bodies were solubilized in 20 mM Tris pH 7.5 containing 6 M GdnCl, 05 M NaCl and 2.6 mM imidazole, and after column loading 6 M GdnCl was replaced by 8 M urea. rHaPrP(90–231) was produced from a pET15b construct using thrombin digestion to cleave the His-tag. The various mutants were produced via site-directed mutagenesis using the primers listed in Table S1. Before use proteins were equilibrated in the desired buffer (10 mM NaAc pH 4.5 or 10 mM Mes at pH 6.5, unless stated) by extensive dialysis and cleared by centrifugation before concentration determinations48.
Dynamic Light Scattering
Dynamic light scattering (DLS) measurements were performed using a DynaPro spectroscatter (Wyatt Technology) with a 1.5-mm path length and a 12 μl quartz cuvette. The average of 20–25 acquisitions of buffers and protein solutions (15 μM protein concentrations) were filtered using a 0.1 μm Whatman Anodisc-3 filters. The hydrodynamic radii (RH) and mass proportions (%) of the species were derived from the autocorrelation data assuming a model of n-monodisperse globular proteins and using the software provided by the manufacturer13,48. Measurements were performed in triplicate using two different protein batches. The theoretical hydrodynamic radius (RHT) for PrP wt chain was calculated using 0.73 cm3 g−1 and 0.35 g H2O (g protein)−1 for the particle specific volume and the hydration, as described47.
CD Spectroscopy
CD spectra were recorded in a Jasco-810 CD spectrometer with 15 μM protein solutions in either 10 mM Mes at pH 6.5 or 10 mM ammonium acetate pH 5.0 using a 0.1-cm cuvette48. Thermal denaturation experiments were performed following the ellipticity changes at 222 nm upon heating from 15 °C to 90 °C at a 1 degree/min heating rate and analyzed as a two-state transition as described48. Briefly, the changes in ϴ222 with temperatures were normalized to the fraction of unfolded protein (fU) using fU(T) = (ϴ222(T) − ϴ222N(T))/(ϴ222U(T)− ϴ222N(T)), where N and U refer to the native and unfolded states, respectively. The value of fU was plotted as a function of temperature for the calculation of Tm and ΔHm. The experimental Tm values of mutants were converted into the apparent relative changes in free energy with respect to the wt protein (ΔΔGomut/wt) using the equation ΔΔGmut/wt = ΔHwt x (1 – Tmwt / Tmmut), where Tmwt and Tmmut are the Tm values for the wt and mutant protein, respectively, and ΔHwt is the denaturation van´t Hoff enthalpy of the wt protein.
Fibril formation
The proteins in 5 mM MES pH 6.5 were cleared by centrifuging at 13200 rpm for 20 min at 4 °C and placed at 40 μM protein concentrations in 50 mM MES at pH 6.5 containing either 2 M GdnCl and 3 M urea with 50 mM NaCl. For kinetic analyses, 0.2-ml samples containing 15 μM ThT were placed in wells with a 3 mm glass ball. ThT binding was monitored at 37 °C, as described13,73. All measurements were performed in triplicate, and the experiments were repeated at least with two different protein batches. For imaging, fibrils were formed in 50 mM MES pH 6.0 containing 2 M GdnCl at 37 °C with continuous rotation at 24 rpm. After 100 h, the products were dialyzed against 10 mM ammonium acetate at pH 5.
Atomic force microscopy
First, 2-μM fibril solutions were deposited onto freshly cleaved mica surfaces. After 10 min of adsorption, the samples were washed with H2O and dried with a stream of N2. AFM studies were performed using a MultiMode Veeco microscope with a 125-μm lateral range and a 5-μm vertical range equipped with a J-scanner and a NanoScope IIIa controller, using rectangular cantilevers with tetrahedral tips for the dynamic mode in air (Olympus, OMCL-AC240TS)13. The analysis was performed using WSxM (Nanotec).
Fluorescence Microscopy
PrP fibrils were deposited onto Permanox 8-well Lab-Teks chamber slides and immunostained with rabbit anti-PrP Ab 3531 (1:1000, recognizes 90–102) and mouse POM17 anti-PrP Ab (1:1000, recognizes 140–145), followed by goat anti-rabbit and goat anti-mouse antibodies labeled with Alexa-488 and Alexa-546, respectively (Invitrogen/Molecular Probes, 1:1000). Fluorescence images were captured using an Axioplan Universal Microscope (Zeiss) equipped with a DFC 350 FX digital camera (Leica) and a 100x objective. Processing and analyses were performed using WCIF ImageJ software79. Manders overlap coefficients were determined as the amount of Ab3531 signal overlapped with the POM17 stain with at least 20 independent fibrils.
Cell culture, transfections and processing analysis
Mutants were generated using pcDNA4.1-HaPrP(1–254) as a template (Table 1S)80. Transient transfections were performed using Chinese hamster ovary (CHO) cells and Fugene 6 as a transfection reagent (Roche), as described80. After 40 h, the cells were harvested via in situ lysis in a cold RIPA buffer (10 mM Tris-HCl pH 7.5, 100 mM NaCl 10 mM EDTA, 0.5% Triton X-100, 0.5% deoxycholate). The lysates were cleared via 5 min of centrifugation at 500 g, supplemented with 0.5 mM Pefabloc, and precipitated with 5 volumes of methanol at −20 °C. Samples were then centrifuged at 10000 × g for 30 min, and the pellets were re-dissolved in TNE buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM EDTA). Enzymatic digestions with PNGaseF (New England Biolabs) were performed for 1 h at 37 °C80. After digestion, the reactions were stopped by the addition of Laemmli buffer and PrP analyzed via immunoblotting using the POM17 antibody.
Western blot analysis
The samples were resolved using SDS-PAGE (13.5% acrylamide gels) and electrophoretically transferred onto PVDF. The membranes were blocked for 1 h in 5% (w/v) non-fat dried skimmed milk powder in Tris-buffered saline containing 0.05% Tween 20. After incubation with mouse anti-PrP POM17 and HRP (horseradish peroxidase)-conjugated goat-anti mouse antibody (Sigma-Aldrich, 1:5000), the signals were developed using an ECL-Western-blotting reagent (Bio-Rad) and detected using ChemiDoc XRS equipment.
Statistical analysis of experiments
Statistical analysis of experiments was performed either using paired Student’s t-test, for comparing two samples, or one-way ANOVA with Dunnett’s post-hoc test, for comparison of all columns to a control column. Results are displayed as the average of replicates ± s.d.
Additional Information
How to cite this article: Martínez, J. et al. PrP charge structure encodes interdomain interactions. Sci. Rep. 5, 13623; doi: 10.1038/srep13623 (2015).
Supplementary Material
Acknowledgments
We thank Prof. A. Estepa, V. Muñoz, and S. Zorrilla for contributing reagents and equipment and for critically reading the manuscript. This work was partially supported by the Ministerio de Economía y Competitividad (BFU2009-07971 and SAF2014-52661 to MG, BIO2011-28092 and CSD2009-00088 to MC), Fundación CIEN (MG), Raman Health (MG) and the National Institutes of Health (grants NS045585 and NS074998 to IVB). JM was supported by an FPI-research contract and an FPI-short staying grant. MC was supported by a Juan de la Cierva Postdoctoral contract.
Footnotes
The authors declare no competing financial interests.
Author Contributions M.G. and J.M. conceived the experiments; J.M., R.S., M.C. and N.M. conducted the experiments; J.M., M.G., I.B. and A.A. analyzed the results. M.G. and J.M. draft the manuscript. All authors reviewed the manuscript
References
- Nakamura H. Roles of electrostatic interaction in proteins. Quarterly reviews of biophysics 29, 1–90 (1996). [DOI] [PubMed] [Google Scholar]
- Bosshard H. R., Marti D. N. & Jelesarov I. Protein stabilization by salt bridges: concepts, experimental approaches and clarification of some misunderstandings. Journal of molecular recognition: JMR 17, 1–16, 10.1002/jmr.657 (2004). [DOI] [PubMed] [Google Scholar]
- Gitlin I., Carbeck J. D. & Whitesides G. M. Why are proteins charged? Networks of charge-charge interactions in proteins measured by charge ladders and capillary electrophoresis. Angew Chem Int Ed Engl 45, 3022–3060, 10.1002/anie.200502530 (2006). [DOI] [PubMed] [Google Scholar]
- Uversky V. N., Gillespie J. R. & Fink A. L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41, 415–427 (2000). [DOI] [PubMed] [Google Scholar]
- Chiti F. et al. Studies of the aggregation of mutant proteins in vitro provide insights into the genetics of amyloid diseases. Proceedings of the National Academy of Sciences of the United States of America 99 Suppl 4, 16419–16426, 10.1073/pnas.212527999 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F., Stefani M., Taddei N., Ramponi G. & Dobson C. M. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature 424, 805–808, 10.1038/nature01891 (2003). [DOI] [PubMed] [Google Scholar]
- Schmittschmitt J. P. & Scholtz J. M. The role of protein stability, solubility, and net charge in amyloid fibril formation. Protein science : a publication of the Protein Society 12, 2374–2378, 10.1110/ps.03152903 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E., Kim S., Schafer K. N. & Kuret J. Pseudophosphorylation of tau protein directly modulates its aggregation kinetics. Biochimica et biophysica acta 1814, 388–395, 10.1016/j.bbapap.2010.10.005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dima R. I. & Thirumalai D. Proteins associated with diseases show enhanced sequence correlation between charged residues. Bioinformatics 20, 2345–2354, 10.1093/bioinformatics/bth245 (2004). [DOI] [PubMed] [Google Scholar]
- Guest W. C., Cashman N. R. & Plotkin S. S. Electrostatics in the stability and misfolding of the prion protein: salt bridges, self energy, and solvation. Biochemistry and cell biology = Biochimie et biologie cellulaire 88, 371–381, 10.1139/o09-180 (2010). [DOI] [PubMed] [Google Scholar]
- Funk K. E. et al. Lysine methylation is an endogenous post-translational modification of tau protein in human brain and a modulator of aggregation propensity. The Biochemical journal 462, 77–88, 10.1042/BJ20140372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant’Anna R. et al. The importance of a gatekeeper residue on the aggregation of transthyretin: implications for transthyretin-related amyloidoses. The Journal of biological chemistry 289, 28324–28337, 10.1074/jbc.M114.563981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez J. et al. Fish β-parvalbumin acquires allergenic properties by amyloid assembly. Swiss medical weekly (2015). [DOI] [PubMed] [Google Scholar]
- Mao Y. et al. Charge and charge-pair mutations alter the rate of assembly and structural properties of apolipoprotein C-II amyloid fibrils. Biochemistry 54, 1421–1428, 10.1021/bi5014535 (2015). [DOI] [PubMed] [Google Scholar]
- Aguzzi A. & Calella A. M. Prions: protein aggregation and infectious diseases. Physiological reviews 89, 1105–1152, 10.1152/physrev.00006.2009 (2009). [DOI] [PubMed] [Google Scholar]
- Soto C. Transmissible proteins: expanding the prion heresy. Cell 149, 968–977, 10.1016/j.cell.2012.05.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillas M., Swietnicki W., Gambetti P. & Surewicz W. K. Membrane environment alters the conformational structure of the recombinant human prion protein. The Journal of biological chemistry 274, 36859–36865 (1999). [DOI] [PubMed] [Google Scholar]
- Li R. et al. Identification of an epitope in the C terminus of normal prion protein whose expression is modulated by binding events in the N terminus. Journal of molecular biology 301, 567–573, 10.1006/jmbi.2000.3986 (2000). [DOI] [PubMed] [Google Scholar]
- Wuthrich K. & Riek R. Three-dimensional structures of prion proteins. Advances in protein chemistry 57, 55–82 (2001). [DOI] [PubMed] [Google Scholar]
- Cordeiro Y. et al. The amino-terminal PrP domain is crucial to modulate prion misfolding and aggregation. Biophysical journal 89, 2667–2676, 10.1529/biophysj.105.067603 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostapchenko V. G., Makarava N., Savtchenko R. & Baskakov I. V. The polybasic N-terminal region of the prion protein controls the physical properties of both the cellular and fibrillar forms of PrP. Journal of molecular biology 383, 1210–1224, 10.1016/j.jmb.2008.08.073 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Martins J. B. et al. Unexpected tolerance of alpha-cleavage of the prion protein to sequence variations. PloS one 5, e9107, 10.1371/journal.pone.0009107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland M. P. et al. Anionic phospholipid interactions of the prion protein N terminus are minimally perturbing and not driven solely by the octapeptide repeat domain. The Journal of biological chemistry 285, 32282–32292, 10.1074/jbc.M110.123398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. B., Geoghegan J. C. & Supattapone S. Dissociation of infectivity from seeding ability in prions with alternate docking mechanism. PLoS pathogens 7, e1002128, 10.1371/journal.ppat.1002128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh J. A. et al. The N-terminal, polybasic region of PrP(C) dictates the efficiency of prion propagation by binding to PrP(Sc). The Journal of neuroscience: the official journal of the Society for Neuroscience 32, 8817–8830, 10.1523/JNEUROSCI.1103-12.2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh J. A., Westergard L., Unterberger U., Biasini E. & Harris D. A. The N-terminal, polybasic region is critical for prion protein neuroprotective activity. PloS one 6, e25675, 10.1371/journal.pone.0025675 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spevacek A. R. et al. Zinc drives a tertiary fold in the prion protein with familial disease mutation sites at the interface. Structure 21, 236–246, 10.1016/j.str.2012.12.002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonati T. et al. The toxicity of antiprion antibodies is mediated by the flexible tail of the prion protein. Nature 501, 102–106, 10.1038/nature12402 (2013). [DOI] [PubMed] [Google Scholar]
- Zurawel A. A. et al. Prion nucleation site unmasked by transient interaction with phospholipid cofactor. Biochemistry 53, 68–76, 10.1021/bi4014825 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benetti F. et al. Structural determinants in prion protein folding and stability. Journal of molecular biology 426, 3796–3810, 10.1016/j.jmb.2014.09.017 (2014). [DOI] [PubMed] [Google Scholar]
- Liu A. et al. NMR experiments for resonance assignments of 13C, 15N doubly-labeled flexible polypeptides: application to the human prion protein hPrP(23-230). Journal of biomolecular NMR 16, 127–138 (2000). [DOI] [PubMed] [Google Scholar]
- Morante S. et al. Inter- and intra-octarepeat Cu(II) site geometries in the prion protein: implications in Cu(II) binding cooperativity and Cu(II)-mediated assemblies. The Journal of biological chemistry 279, 11753–11759, 10.1074/jbc.M312860200 (2004). [DOI] [PubMed] [Google Scholar]
- Kim S. J., Rahbar R. & Hegde R. S. Combinatorial control of prion protein biogenesis by the signal sequence and transmembrane domain. The Journal of biological chemistry 276, 26132–26140, 10.1074/jbc.M101638200 (2001). [DOI] [PubMed] [Google Scholar]
- Johanssen V. A. et al. C-terminal peptides modelling constitutive PrPC processing demonstrate ameliorated toxicity predisposition consequent to alpha-cleavage. The Biochemical journal 459, 103–115, 10.1042/BJ20131378 (2014). [DOI] [PubMed] [Google Scholar]
- Groveman B. R. et al. Charge neutralization of the central lysine cluster in prion protein (PrP) promotes PrP(Sc)-like folding of recombinant PrP amyloids. The Journal of biological chemistry 290, 1119–1128, 10.1074/jbc.M114.619627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiesa R., Piccardo P., Ghetti B. & Harris D. A. Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron 21, 1339–1351 (1998). [DOI] [PubMed] [Google Scholar]
- Leliveld S. R., Stitz L. & Korth C. Expansion of the octarepeat domain alters the misfolding pathway but not the folding pathway of the prion protein. Biochemistry 47, 6267–6278, 10.1021/bi800253c (2008). [DOI] [PubMed] [Google Scholar]
- Speare J. O., Rush T. S. 3rd, Bloom M. E. & Caughey B. The role of helix 1 aspartates and salt bridges in the stability and conversion of prion protein. The Journal of biological chemistry 278, 12522–12529, 10.1074/jbc.M211599200 (2003). [DOI] [PubMed] [Google Scholar]
- Zahn R. et al. NMR solution structure of the human prion protein. Proceedings of the National Academy of Sciences of the United States of America 97, 145–150 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzolai L. & Zahn R. Influence of pH on NMR structure and stability of the human prion protein globular domain. The Journal of biological chemistry 278, 35592–35596, 10.1074/jbc.M303005200 (2003). [DOI] [PubMed] [Google Scholar]
- Zuegg J. & Gready J. E. Molecular dynamics simulations of human prion protein: importance of correct treatment of electrostatic interactions. Biochemistry 38, 13862–13876 (1999). [DOI] [PubMed] [Google Scholar]
- Norstrom E. M. & Mastrianni J. A. The charge structure of helix 1 in the prion protein regulates conversion to pathogenic PrPSc. Journal of virology 80, 8521–8529, 10.1128/JVI.00366-06 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumetz A. C., Chockla A. M., Kaler E. W. & Lenhoff A. M. Effects of pH on protein-protein interactions and implications for protein phase behavior. Biochimica et biophysica acta 1784, 600–610, 10.1016/j.bbapap.2007.12.016 (2008). [DOI] [PubMed] [Google Scholar]
- Gabizon R. et al. Insoluble wild-type and protease-resistant mutant prion protein in brains of patients with inherited prion disease. Nature medicine 2, 59–64 (1996). [DOI] [PubMed] [Google Scholar]
- Hsiao K. et al. Mutant prion proteins in Gerstmann-Straussler-Scheinker disease with neurofibrillary tangles. Nature genetics 1, 68–71, 10.1038/ng0492-68 (1992). [DOI] [PubMed] [Google Scholar]
- Lee C. I., Yang Q., Perrier V. & Baskakov I. V. The dominant-negative effect of the Q218K variant of the prion protein does not require protein X. Protein science: a publication of the Protein Society 16, 2166–2173, 10.1110/ps.072954607 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva D. et al. Oligomerization of the proteolytic products is an intrinsic property of prion proteins. Biochemical and biophysical research communications 323, 1278–1286, 10.1016/j.bbrc.2004.08.230 (2004). [DOI] [PubMed] [Google Scholar]
- Lisa S. et al. The structural intolerance of the PrP alpha-fold for polar substitution of the helix-3 methionines. Cellular and molecular life sciences: CMLS 67, 2825–2838, 10.1007/s00018-010-0363-1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A. K., Srivastava A. K., Srinivas V., Chary K. V. & Rao C. M. Copper alters aggregation behavior of prion protein and induces novel interactions between its N- and C-terminal regions. The Journal of biological chemistry 286, 38533–38545, 10.1074/jbc.M111.265645 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liemann S. & Glockshuber R. Influence of amino acid substitutions related to inherited human prion diseases on the thermodynamic stability of the cellular prion protein. Biochemistry 38, 3258–3267, 10.1021/bi982714g (1999). [DOI] [PubMed] [Google Scholar]
- Apetri A. C., Surewicz K. & Surewicz W. K. The effect of disease-associated mutations on the folding pathway of human prion protein. The Journal of biological chemistry 279, 18008–18014, 10.1074/jbc.M313581200 (2004). [DOI] [PubMed] [Google Scholar]
- Bera A. & Nandi P. K. Nucleic acid induced unfolding of recombinant prion protein globular fragment is pH dependent. Protein science : a publication of the Protein Society 23, 1780–1788, 10.1002/pro.2573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canello T. et al. Oxidation of Helix-3 methionines precedes the formation of PK resistant PrP. PLoS pathogens 6, e1000977, 10.1371/journal.ppat.1000977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xiao S. et al. Rational modification of protein stability by targeting surface sites leads to complicated results. Proceedings of the National Academy of Sciences of the United States of America 110, 11337–11342, 10.1073/pnas.1222245110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus A., Groveman B. R. & Caughey B. Prions and the potential transmissibility of protein misfolding diseases. Annual review of microbiology 67, 543–564, 10.1146/annurev-micro-092412-155735 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. B. Biology and genetics of prions causing neurodegeneration. Annual review of genetics 47, 601–623, 10.1146/annurev-genet-110711-155524 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb N. J., Sonnichsen F. D., McHaourab H. & Surewicz W. K. Molecular architecture of human prion protein amyloid: a parallel, in-register beta-structure. Proceedings of the National Academy of Sciences of the United States of America 104, 18946–18951, 10.1073/pnas.0706522104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Moore R. A. & McGuirl M. A. Dissociation of recombinant prion protein fibrils into short protofilaments: implications for the endocytic pathway and involvement of the N-terminal domain. Biochemistry 51, 4600–4608, 10.1021/bi300201e (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supattapone S. Synthesis of high titer infectious prions with cofactor molecules. The Journal of biological chemistry 289, 19850–19854, 10.1074/jbc.R113.511329 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. G. et al. Truncated forms of the human prion protein in normal brain and in prion diseases. The Journal of biological chemistry 270, 19173–19180 (1995). [DOI] [PubMed] [Google Scholar]
- Walmsley A. R., Watt N. T., Taylor D. R., Perera W. S. & Hooper N. M. alpha-cleavage of the prion protein occurs in a late compartment of the secretory pathway and is independent of lipid rafts. Molecular and cellular neurosciences 40, 242–248, 10.1016/j.mcn.2008.10.012 (2009). [DOI] [PubMed] [Google Scholar]
- McDonald A. J., Dibble J. P., Evans E. G. & Millhauser G. L. A new paradigm for enzymatic control of alpha-cleavage and beta-cleavage of the prion protein. The Journal of biological chemistry 289, 803–813, 10.1074/jbc.M113.502351 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Iglesias R. et al. Prion protein interaction with glycosaminoglycan occurs with the formation of oligomeric complexes stabilized by Cu(II) bridges. Journal of molecular biology 319, 527–540, 10.1016/S0022-2836(02)00341-8 (2002). [DOI] [PubMed] [Google Scholar]
- Leucht C. et al. The 37 kDa/67 kDa laminin receptor is required for PrP(Sc) propagation in scrapie-infected neuronal cells. EMBO reports 4, 290–295, 10.1038/sj.embor.embor768 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghera N. et al. Deciphering the molecular details for the binding of the prion protein to main ganglioside GM1 of neuronal membranes. Chemistry & biology 18, 1422–1431, 10.1016/j.chembiol.2011.08.016 (2011). [DOI] [PubMed] [Google Scholar]
- Ostapchenko V. G. et al. Two amyloid States of the prion protein display significantly different folding patterns. Journal of molecular biology 400, 908–921, 10.1016/j.jmb.2010.05.051 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimova N., Makarava N. & Baskakov I. V. The diversity and relationship of prion protein self-replicating states. Virus research . 10.1016/j.virusres.2014.10.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. B. et al. Cofactor molecules induce structural transformation during infectious prion formation. Structure 21, 2061–2068, 10.1016/j.str.2013.08.025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble G. P., Walsh D. J., Miller M. B., Jackson W. S. & Supattapone S. Requirements for mutant and wild-type prion protein misfolding in vitro. Biochemistry 54, 1180–1187, 10.1021/bi501495j (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley M. P. & Prusiner S. B. Ultrastructural studies of prions. Current topics in microbiology and immunology 172, 75–91 (1991). [DOI] [PubMed] [Google Scholar]
- Juanes M. E., Elvira G., Garcia-Grande A., Calero M. & Gasset M. Biosynthesis of prion protein nucleocytoplasmic isoforms by alternative initiation of translation. The Journal of biological chemistry 284, 2787–2794, 10.1074/jbc.M804051200 (2009). [DOI] [PubMed] [Google Scholar]
- Colombo G., Meli M., Morra G., Gabizon R. & Gasset M. Methionine sulfoxides on prion protein Helix-3 switch on the alpha-fold destabilization required for conversion. PloS one 4, e4296, 10.1371/journal.pone.0004296 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. et al. Selenomethionine incorporation into amyloid sequences regulates fibrillogenesis and toxicity. PloS one 6, e27999, 10.1371/journal.pone.0027999 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millhauser G. L. Copper and the prion protein: methods, structures, function, and disease. Annual review of physical chemistry 58, 299–320, 10.1146/annurev.physchem.58.032806.104657 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A. et al. Octarepeat region flexibility impacts prion function, endoproteolysis and disease manifestation. EMBO molecular medicine 7, 339–356, 10.15252/emmm.201404588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi M. & Caughey B. Specific binding of normal prion protein to the scrapie form via a localized domain initiates its conversion to the protease-resistant state. The EMBO journal 18, 3193–3203, 10.1093/emboj/18.12.3193 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R., Savtchenko R., Ostapchenko V. G., Makarava N. & Baskakov I. V. The alpha-helical C-terminal domain of full-length recombinant PrP converts to an in-register parallel beta-sheet structure in PrP fibrils: evidence from solid state nuclear magnetic resonance. Biochemistry 49, 9488–9497, 10.1021/bi1013134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaei H. et al. High yield purification and physico-chemical properties of full-length recombinant allelic variants of sheep prion protein linked to scrapie susceptibility. European journal of biochemistry/FEBS 267, 2833–2839 (2000). [DOI] [PubMed] [Google Scholar]
- Ostapchenko V., Gasset M. & Baskakov I. V. Atomic force fluorescence microscopy in the characterization of amyloid fibril assembly and oligomeric intermediates. Methods Mol Biol 849, 157–167, 10.1007/978-1-61779-551-0_11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisa S. et al. Failure of prion protein oxidative folding guides the formation of toxic transmembrane forms. The Journal of biological chemistry 287, 36693–36701, 10.1074/jbc.M112.398776 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.