Abstract
Background and Objective
Epidermal growth factor receptor (EGFR) inhibitors are not equally effective in all cancer patients. One potential clinical factor that could help in selecting patients who may benefit from treatment with cetuximab is acneiform rash, which correlates with the clinical response to EGFR inhibitors. Some previous studies have suggested that the tendency to develop rash may depend on polymorphisms in the EGFR gene. In this investigation, the association of degree of CA dinucleotide polymorphism with skin rash and cetuximab therapy outcome was examined.
Methods
The study included 60 patients treated with cetuximab. For each patient, the severity of acneiform rash was assessed, and the type of polymorphism was determined by genotyping. Associations between genotypes, the acneiform rash, and response to treatment were determined by using the chi-square test and Spearman’s rank correlation. The cutoffs S ≤ 17(CA), L > 17(CA), n(CA) ≤ 35, and n(CA) > 35 were tested, as well as the sum of the two allele repetitions.
Results
A correlation was found between body surface area covered by rash and the sum of the two allele repetitions (p = 0.030). No statistically significant relationship between genotype and response to treatment was observed. However, in patients who have had partial remission, we noticed a higher incidence of polymorphism, with less CA dinucleotide repetitions and early onset of rash.
Conclusion
A correlation between genotype and severity of rash was observed. That is, the severity of rash decreased with an increased number of CA repetitions.
Key Points
| Skin rash is an adverse reaction typical for anti-epidermal growth factor receptor (EGFR) therapeutics that is independent of tumor type. |
| Severe skin rash is associated with clinical benefit in cancer patients treated with cetuximab. The aim of this study was to verify whether CA simple sequence repeat in intron 1 (CA-SSR1) polymorphism is a predictive factor for skin rash and possibly for clinical response to cetuximab. |
| This study found that an increased number of CA repetitions was inversely associated with severity of skin rash as determined by body area covered; however, the association with clinical response was unclear. |
Introduction
The epidermal growth factor receptor (EGFR, ErbB-1, HER1) is a membrane receptor in the receptor tyrosine kinase family (ErbB). The activation of EGFR is an important factor in metastasis formation and progression. Signaling pathways activated by EGFR interact with such processes as cell growth, proliferation, apoptosis, differentiation, and migration, as well as the secretion of some proteins [1–4]. Overexpression of EGFR, up to 106 receptors per cell [5], often occurs in tumor cells of epithelial origin (head and neck cancer, stomach cancer, colorectal cancer, ovarian cancer, prostate cancer, and glioma) and correlates with poor prognosis [1, 2, 4, 6].
Targeted drugs that inhibit signaling through the EGFR are currently widely used. Such drugs include monoclonal antibodies (mAbs), such as cetuximab and panitumumab, and small-molecule tyrosine kinase inhibitors, such as gefitinib and erlotinib. Cetuximab (IMC-225, Erbitux®) is a human–mouse chimeric mAb. It was first approved for use in metastatic colorectal cancer manifesting EGFR expression, as monotherapy or in combination with cytotoxic chemotherapy [7, 8]. However, EGFR inhibitors are not equally effective in all cancer patients [9–12]. The efficacy of cetuximab with concurrent use of chemotherapy in colorectal cancer patients is about 23 % [13]. This poor efficacy may be explained by the discovery of additional mutations in the RAS gene in patients with colorectal cancer. Cetuximab and panitumumab do not work in patients with mutations of KRAS, NRAS, or BRAF [14–17]. However, cetuximab also has poor efficacy in approximately 40 % of patients with wild-type KRAS and other types of cancers with overexpression of EGFR receptors, the reasons for which are poorly understood [18].
There is a lack of information about predictive factors other than RAS somatic mutation for the selection of patients who will benefit the most from treatment with cetuximab. The one potential clinical factor is acneiform rash (rash), which correlates with the response to EGFR inhibitors [19–22]. Generally, EGFR inhibitors are well-tolerated by patients; however, acneiform rash, a characteristic dermatological adverse effect, occurs in over 50 % of patients [3, 20, 23]. Studies carried out with the use of anti-EGFR mAbs have also shown that the severity of the rash is dependent on the dose of the drug [21, 24, 25]. The severity is similar in patients receiving cetuximab alone or in combination with irinotecan chemotherapy [13]. The rash is most common in areas rich in sebaceous glands [3]. It usually presents a few days after the start of treatment, reaches maximum severity up to 3 weeks later, and then disappears over several weeks after the end of treatment [3].
Numerous studies have shown that the rash correlates with better response to treatment with anti-EGFR mAbs [13, 23, 26]. The occurrence of the rash is thought to be associated with a genetic variation in the human population. One factor that may influence the occurrence of the rash is CA dinucleotide repeat polymorphism in intron 1 [CA simple sequence repeat in intron1 (CA-SSR1)]. This variation is present in a highly polymorphic genomic DNA region of the EGFR gene [9, 17, 27], the 5′ end of intron 1. This location is considered to be important because it is in the immediate neighborhood of the second enhancer [5, 28, 29] and is believed to influence the expression of the EGFR gene.
Studies performed in patients with non-small cell lung cancer and pancreatic cancer have shown that a smaller number of CA dinucleotide repeats in intron 1 of the EGFR gene is associated with worse survival [29, 30]. On the other hand, the positive therapeutic effect of EGFR inhibitors may be correlated with a smaller number of CA-SSR1 repeats [9, 31].
Materials and Methods
Patients
Sixty patients treated with cetuximab for colorectal, lung, and head and neck cancer were enrolled in this study. All colorectal cancer patients (n = 52) in this study had a wild-type KRAS genotype. Cetuximab was given intravenously at an initial dose of 400 mg/m2 and then at 250 mg/m2 every 2 weeks.
The severity of skin reactions was evaluated at weekly intervals according to the National Cancer Institute Common Toxicity Criteria for Adverse Events (NCI CTCAE) version 3.0. Grade 0 is defined as no rash and no additional symptoms associated with rash; grade 1 as macular or papular eruptions or erythema without additional symptoms associated with rash; grade 2 as macular or papular eruptions or erythema pruritus or other associated symptoms, such as localized desquamation or other lesion covering less than 50 % of the body surface area; grade 3 as severe generalized erythoderma or macular, papular, or vesicular eruptions, desquamation covering more than 50 % of the body surface area, plus additional adverse effects; and grade 4 as generalized exfoliative, ulcerative, or bullous dermatitis. In this investigation, the degree of rash coverage of the body surface area was estimated on the basis of nomograms.
The study was multicentered and carried out with the approval of the local ethics committee of the University of Warmia-Mazury in Olsztyn, Poland (No. 6/2010 dated 25 February 2010 and No. 18/2010 dated 27 May 2010). The patients were informed about the subject and aim of the study before the start of the investigation.
Laboratory Analysis
For the genotype analysis, DNA was isolated from peripheral blood by using the Blood Mini kit (A&A Biotechnology, Gdynia, Poland). The isolated DNA was re-suspended in 100 µL of TE buffer and stored at −20 °C until further analysis.
To determine the degree of polymorphism in the CA-SSR1 region, a PCR reaction was carried out, followed by genotyping with the use of the GenScan method. The primers used in the PCR were designed by using Primer3 on the basis of the sequence obtained from the National Center for Biotechnology Information (NCBI) database (reference sequence: NG_007726). The length of the PCR product was 298 bp, with 16 repetitions of the CA dinucleotide. To analyze the GenScan, one primer was end-labeled with the fluorescent dye FAM (forward primer 5-FAMGGACTCTTGAGCGGAAGC-3 and reverse primer 5-CCATAAACCCACTTGACAGG-3). The PCR reaction was performed under the following conditions: 3 min initial denaturation at 94 °C, followed by 30 cycles of denaturation for 60 s at 94 °C, annealing for 60 s at 61 °C, and elongation for 60 s at 72 °C; the final extension was carried out for 7 min at 72 °C.
The fragment length was determined by analysis carried out with the GeneScan ABI Prism® 3700 DNA Analyzer (Life Technologies, Carlsbad, CA, USA). GS500 ROX (-250 LIZ) size marker was added to each sample. To validate the first method, GenScan direct sequencing was carried out for several randomly selected homozygote samples.
Grouping by Allele, Genotype, and Grade of Rash
Samples were grouped arbitrarily depending on the number of CA dinucleotide repeats. The first criterion was based on the division of the first and second alleles into two groups: the short allele, labeled S, and the long allele, labeled L. The cutoffs used were S ≤ 17 and L > 17. The division created three kinds of genotypes: SS, SL, and LL. The second criterion was based on the sum of repetitions for the first and second alleles, with the sum divided into two groups: long genotype [n(CA) > 35] and short [n(CA) ≤ 35] genotype. The third criterion used the sum of CA repetitions for the first and second alleles.
In the assessment of the severity of rash, two criteria were used. In the first criterion, the severity of rash, was evaluated according to the NCI CTCAE version 3.0 scale. Due to the relatively small sample size, the patients were grouped into insignificant rash (grade 0–1) and significant rash (grade 2–3). In the second criterion, the percentage of body surface area covered by rash was used.
Statistical Analysis
Statistical analysis was performed with the Statistica software version 10 (Statsoft, Inc., 2011; http://www.statsoft.com). Due to the relatively small group of patients and the lack of normal distributions in the studied variables, we used non-parametric statistical methods.
The clinical response to cetuximab [Response Evaluation Criteria In Solid Tumors (RECIST)] was determined for 49 patients. The association between response to treatment and rash or genotype was determined by using Spearman’s rank correlation. The association between rash severity (NCI CTCAE v3.0) and CA-SSR1 genotype subgroup [cutoffs: S ≤ 17(CA), L > 17(CA), n(CA) ≤ 35, and n(CA) > 35] was determined using Pearson’s chi-square test. The relationship between body surface area covered by rash and CA-SSR1 genotype was determined by using Spearman’s rank correlation, Mann Whitney U test, and Kruskal–Wallis test. Two quantitative variables, the percentage of body surface area covered by rash and the sum of CA dinucleotide repeats in the EGRF gene, were analyzed using Pearson’s correlation. The significance level for all tests was p < 0.05.
Results
Data from 60 patients were analyzed. The sample consisted of 70 % males and 30 % females, ages 38–79 years, with a mean age of 62 years. The majority of patients had colorectal cancer. All patients received cetuximab in two treatment regimens: alone or in combination with chemotherapy. No relationship was found between severity of rash and cetuximab administration scheme.
For 49 patients, the response to cetuximab treatment was determined. Stable disease was observed in 57 % of patients and partial remission in 29 %; 14 % of patients progressed. Table 1 presents the characteristics of the patients and their treatment schemes.
Table 1.
General characteristic of patients and treatment
| Characteristic | n (%)a |
|---|---|
| Sex | |
| Men | 42 (70) |
| Women | 18 (30) |
| Age (n = 60) | |
| Average | 60 years |
| Range | 38–79 years |
| Median age | 61 years |
| Type of cancer (n = 60) | |
| Head and neck | 5 (8.06) |
| Colorectal | 52 (86.67) |
| Small cell lung | 1 (1.66) |
| Stomach | 2 (3.33) |
| Treatment regimen (n = 60) | |
| Monotherapy | 27 (45) |
| Chemotherapy | 33 (55) |
| Response to treatment (RECIST scale) [n = 49] | |
| Partial remission | 14 (29) |
| Stabilization | 28 (57) |
| Progression | 7 (14) |
| Body surface area covered by the rash (n = 60) | |
| Average | 26 % |
| Range | 0–98 % |
| Median | 22 % |
| Acneiform rash severity (scale NCI CTCAE v.3.0) (n = 60) | |
| No rash and grade 1 rash | 16 (27) |
| Grades 2 and 3 rash | 44 (73) |
| Time of appearance the rash (week of treatment) [n = 59] | |
| First and second | 33 (16) |
| Third and fourth | 19 (32) |
| After the fourth | 7 (12) |
| Time of maximum severity of rash (week of treatment) [n = 59] | |
| First and second | 10 (17) |
| Third and fourth | 34 (57) |
| After the fourth | 15 (25) |
| Additional symptoms associated with acneiform (n = 60) | |
| Itching | 35 (58) |
| Dry skin (xerosis) | 35 (58) |
| Exfoliation skin | 21 (35) |
| Hair changes | 3 (5) |
| Nail changes | 12 (20) |
| Symptomatic treatment of acneiform rash (n = 60) | |
| No treatment | 20 (33) |
| Antibacterials | 20 (33) |
| Corticosteroids | 9 (15) |
| Antibacterials + corticosteroids | 11 (18.33) |
NCI CTCAE National Cancer Institute Common Toxicity Criteria for Adverse Events, RECIST Response evaluation criteria in solid tumors
aUnless otherwise stated
The Rash and Associated Symptoms
For analysis, the patients were divided into two subgroups: those with absence of rash or grade 1 rash (16 patients; 27 %) and those with grade 2 or 3 rash (44; 73 %).
The rash covered, on average, 26 % of the body surface of patients, with the median being 22 % coverage. Most frequently, the rash appeared on the head, especially on the face (54 patients; 90 %), and on the neck (33; 55 %), chest (14; 23 %), back (28; 47 %), abdomen (11; 18 %), hips, thighs, and buttocks (9; 15 %), arms (8; 13 %), and hands (7; 12 %). The rash also appeared, although rarely, in the vicinity of the lower legs and feet (less than 10 % of patients).
The majority of patients (67 %) received symptomatic treatment for the rash, such as antibacterials and corticosteroids, separately or in combination. The rash usually appeared in the second or third week after initiation of cetuximab administration and reached maximum intensity on the third or fifth week. Table 1 presents detailed data on the appearance, severity, and duration of the rash.
CA Simple Sequence Repeat in Intron 1 (CA-SSR1) Polymorphism of the EGFR Gene
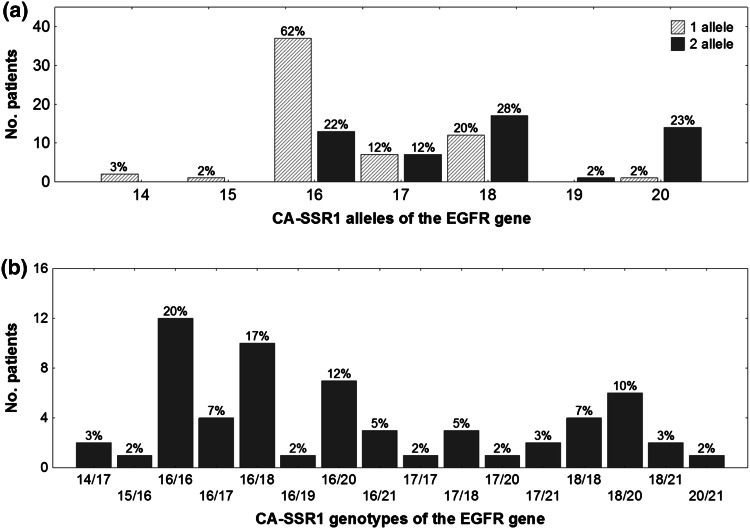
The number of CA dinucleotide repeats present in patients was between 14 and 21. The distribution of polymorphisms was trimodal; the dominant alleles had 16, 18, and 20 repeats. The most common allele had 16 CA dinucleotide repeats, which occurred in 62 % of patients, followed by 18 CA repetitions in 28 %, and 20 allele repeats in 23 %. The median length of alleles was 17 CA dinucleotide repeats; Fig. 1a shows the distribution of the repeat lengths.
Fig. 1.
Distribution polymorphic CA-SSR1 of the EGFR gene. a Distribution of alleles of the EGFR gene and b genotypes of polymorphic CA-SSR1 fragment of the EGFR gene. CA-SSR1 CA simple sequence repeat in intron 1, EGFR epidermal growth factor receptor
The most common genotype was 16/16 CA repeats, which occurred in 20 % of patients, followed by 16/18 CA repeats in 17 %, 16/20 CA repeats in 12 %, and 18/20 repeats in 10 %. Figure 1b shows the CA genotype distribution.
Association Between Rash and CA-SSR1 Polymorphism
An evaluation of the association between rash and CA-SSR1 polymorphism of the EGFR gene was performed for the described grouping criteria (Sect. 2.3). Due to the lack of clear examples in the scientific literature for this type of analysis, three possibilities were tested. Table 2 provides detailed information on the analysis.
Table 2.
Examination of the relationship between the acneiform rash and CA simple sequence repeat in intron 1 (CA-SSR1) polymorphism of the epidermal growth factor receptor (EGFR) gene
| Criterion | Subgroup [n (%)] | Test | Test value | p value | ||||
|---|---|---|---|---|---|---|---|---|
| SS | SL | LL | Short n(CA) ≤ 35 | Long n(CA) > 35 | ||||
| Grouped NCI CTCAE v3.0 scale vs. sum of CA repetitions of a single allele [cutoff S ≤ 17(CA), L > 17(CA)] | Chi-squared Pearson’s test |
χ 2 = 8.09 df = 2 |
p = 0.059 | |||||
| Grade 0–1 | 2 (3) | 11 (18) | 3 (5) | |||||
| Grade 2–3 | 18 (30) | 16 (27) | 10 (17) | |||||
| Grouped NCI CTCAE v3.0 scale vs. sum of CA repetitions of two alleles [cutoff n(CA) ≤ 35, n(CA) > 35] | Chi-squared Pearson’s (Yeats’) test |
χ 2 = 0.85 df = 1 |
p = 0.356 | |||||
| Grade 0–1 | 7 (12) | 9 (15) | ||||||
| Grade 2–3 | 27 (45) | 17 (28) | ||||||
| Body surface area covered by the rash (%) vs. sum of CA repetitions of a single allele [cutoff S ≤ 17(CA), L > 17(CA)] | 20 (33) | 27 (45) | 13 (22) | Kruskal–Wallis test | H = 4.781 | p = 0.092 | ||
| Spearman’s rank correlation |
R = −0.258 t = −2.035 |
p = 0.046 | ||||||
| Body surface area covered by the rash (%) vs. sum of CA repetitions of two alleles and the cutoff n(CA) ≤ 35, n(CA) > 35 | 34 (57) | 26 (43) | Mann–Whitney U test | Z = 2.039 | p = 0.042 | |||
| Spearman’s rank correlation |
R = −0.266 t = −2.105 |
p = 0.040 | ||||||
| Body surface area covered by the rash (%) vs. sum of CA repetitions of two alleles | The whole group of patients | Pearson’s correlation (Fig. 2) |
r = −0.3251 r 2 = 0.106 |
p = 0.011 | ||||
χ 2 chi-squared, df degrees of freedom, L long allele, NCI CTCAE National Cancer Institute Common Toxicity Criteria for Adverse Events, S short allele
An insignificant association was found for “the grouped NCI CTCAE version 3.0 scale versus the sum of CA repetitions of a single allele [cutoffs: S ≤ 17(CA) and L > 17(CA)]” (p = 0.059). An insignificant statistical result was also obtained for “the grouped NCI CTCAE version 3.0 scale versus the sum of CA repetitions of two alleles [cutoffs: n(CA) ≤ 35 and n(CA) > 35]” (p = 0.356).
The correlation between body surface area covered by rash and genotype was also analyzed. A positive correlation between surface area covered by rash (%) and the sum of CA repetitions of a single allele [cutoffs: S ≤ 17(CA) and L > 17(CA)] was found (p = 0.046). The body surface area covered by rash (%) and the sum of CA repetitions of the two alleles [cutoffs: n(CA) ≤ 35 and n(CA) > 35] were also found to be associated (p = 0.040).
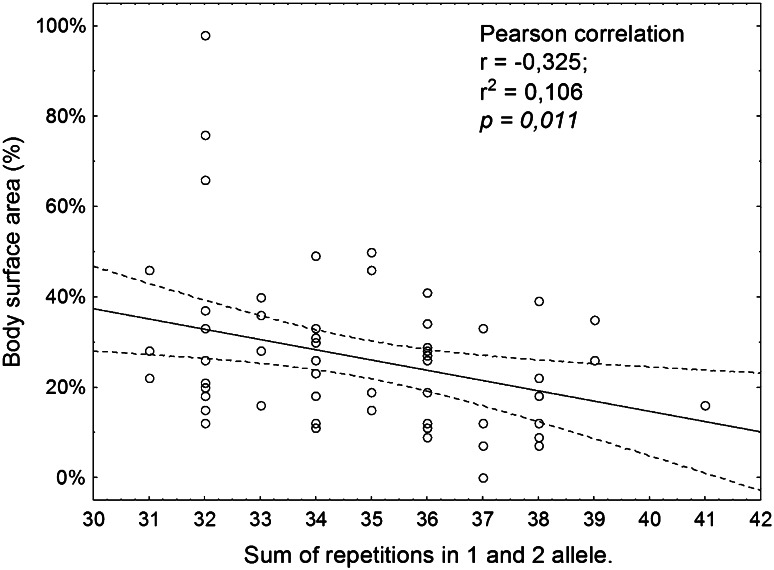
A negative correlation (r = −0.3251) was observed between percentage of body surface area covered by rash and the sum of CA dinucleotide repeats in the EGFR gene (p = 0.011). Figure 2 shows a scatter diagram of this correlation. A decrease in the body surface area covered by rash was found with increasing sum of CA repetitions of the two alleles.
Fig. 2.
Examination of the relationship between the rash and CA-SSR1 polymorphism of the EGFR gene. Scatterplot of total repetitions two alleles CA-SSR1 vs. body surface area covered by acneiform rash. CA-SSR1 CA simple sequence repeat in intron 1, EGFR epidermal growth factor receptor
Response to Treatment Versus Severity of Rash
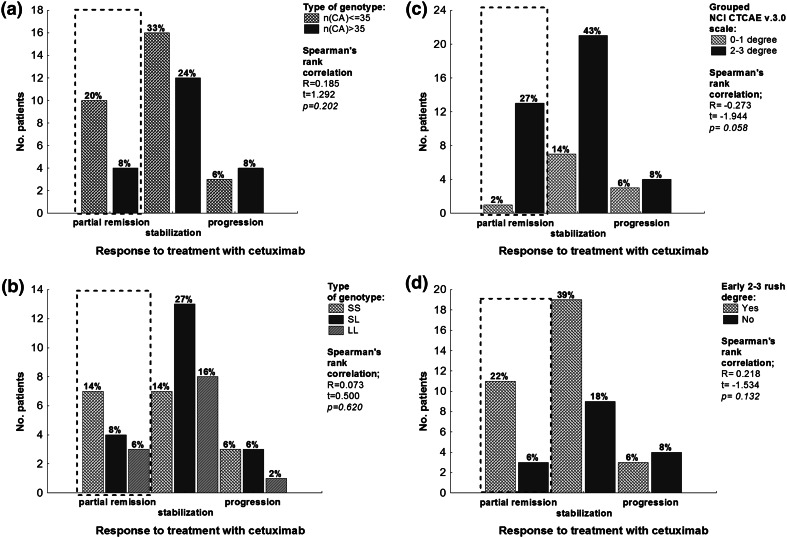
A weak correlation was found between severity of rash and response to treatment (p = 0.058), as shown in Fig. 3c. In patients with partial remission and stabilization, better response to treatment with cetuximab and early occurrence of rash were observed.
Fig. 3.
Histogram presenting examination of the relationship between the response to treatment with cetuximab and CA-SSR1 genotype and the patient’s acneiform rash. a Response to treatment with cetuximab (RECIST scale) vs. the sum of CA repetitions of two alleles, cutoff n(CA) ≤ 35, n(CA) > 35. b Response to treatment with cetuximab (RECIST scale) vs. the sum of CA repetitions of a single allele cutoff S ≤ 17(CA), L > 17(CA). c Response to treatment with cetuximab (RECIST scale) vs. grouped NCI CTCAE v3.0 scale. d Response to treatment with cetuximab (RECIST scale) vs. the early grade 2–3 rash. A Spearman’s rank correlation was calculated for each relationship. In all cases, we did not obtain a statistically significant result. Dotted lines indicate the group characterized by the highest coefficient of variation. In patients with partial remission we observed a better response to cetuximab treatment when there was a smaller number of repetitions in the polymorphic region of CA-SSR1 of the EGFR gene and early grade 2–3 rash. CA-SSR1 CA simple sequence repeat in intron 1, L long allele, NCI CTCAE National Cancer Institute Common Toxicity Criteria for Adverse Events, RECIST Response Evaluation Criteria In Solid Tumors, S short allele
Response to Treatment Versus CA-SSR1 Polymorphism of the EGFR Gene
The relationship between CA-SSR1 polymorphism and response to treatment with cetuximab was evaluated based on the RECIST criteria. In all cases [cutoffs: S ≤ 17(CA) and L > 17(CA), as shown in Fig. 3a; cutoffs: S ≤ 17(CA) and L > 17(CA), as shown Fig. 3b], there was no statistically significant result. However, among patients with partial remission, better response to treatment with cetuximab was observed in those with a smaller number of repetitions in the CA-SSR1 polymorphic region of the EGFR gene.
Discussion
EGFR inhibitors are not equally effective in all cancer patients, even in patients with overexpression of the EGFR gene receptor [13]. Cunningham et al. reported that the level of response to treatment is approximately 23 % for patients receiving cetuximab plus chemotherapy and about 11 % for patients receiving cetuximab as monotherapy [13]. Among patients tested for mutation in the KRAS gene, the level of response to wild-type genotypes is still not more than 40 % [20].
Many studies have indicated that there are molecular factors that may modulate the response to treatment with EGFR inhibitors, such as KRAS status [8], the rash [23, 26], and polymorphism of the EGFR gene [32, 33].
Acneiform Rash
Acneiform skin rash is a characteristic adverse effect of EGFR inhibitor therapy, occurring in 50–95 % of patients [3, 20, 23]. There seems to be greater severity of rash in patients undergoing anti-EGFR mAb treatment [34, 35]. In this study, the rash occurred in nearly all patients (59; 98 %), which is a similar finding to that of other studies [12, 20, 26, 33, 36]. The patients in this study were not homogeneous with regard to type of tumor. However, based on the available literature, the occurrence of rash is assumed to be independent of cancer type: colorectal cancer [26], small cell lung cancer [25, 37], or head and neck cancer [38]. Moreover, the genotype of the patient, and not the genotype of the tumor, determines the occurrence of rash. Usually, the rash starts after the 7th to 10th day of treatment with EGFR inhibitors [39]. In this investigation, the highest frequency of occurrence of rash was observed during the second week after initiation of treatment (38 % of patients). The maximum intensity of rash was observed most often in the third week (35 %). The study by Stintzing et al. showed that the appearance of rash during the first cycle of treatment (up to 21 days of treatment with cetuximab) was associated with a significantly longer overall survival (30.7 vs. 20.2 months; p = 0.007) [20].
In this analysis, the body surface area covered by rash was assumed to be one of the most important factors in evaluating the intensity of rash. Because some patients with skin rash (67 %) were treated with antibacterials and corticosteroids, which could bias the estimation of rash severity, it is likely that the rash severity was underestimated in some patients. A meta-analysis by Ocvirk et al. [40] showed that the administration of antibacterials could reduce the risk of grade 3 or 4 rash by about 42–77 %.
Many previous studies have confirmed the positive effect of rash on the prediction of treatment outcome and survival of patients treated with EGFR inhibitors [13, 20–22, 41]. In this study, a relationship between degree of rash and response to treatment was found (p = 0.058; n = 49), but it was not statistically significant.
CA-SSR1 Polymorphism of the EGFR Gene
One of the factors associated with the occurrence of rash and better response to EGFR inhibitors is CA-SSR1 polymorphism of the EGFR gene. However, genotypic predictors are more desirable than symptoms because of the possibility of easy testing before initiation of therapy.
The CA-SSR1 polymorphism of the EGFR gene has a relatively high number of alleles (8–9) in the range of 14–22 CA dinucleotide repeats [11]. In this study, we found eight types of alleles in the range of 14–21 CA dinucleotide repetitions, for a total of 16 kinds of genotypes. The most common alleles had 16 and 18 CA dinucleotide repeats. Alleles with 16 or 18 CA dinucleotide repeats are the most characteristic for the European population. According to the analysis by Kharrat et al., the CA-SSR1 polymorphism varies geographically [42]. The most common allele in the European population has 16 repeats [43]; in the population of Turkey and Kuwait it has 17 repeats [42]; and in the Asian population it has 20 repeats [33, 44].
Relationship Between CA-SSR1 Polymorphism and Skin Rash
The large number of genotypes present in the CA-SSR1 region of the EGFR gene causes problems in the evaluation of the relationship between the polymorphism and the severity of rash. In the scientific literature, it is not yet clearly resolved how the grouping of genotypes should be carried out. An analysis of the available literature suggests that researchers investigating patients from the Asian population often use cutoff criteria with a higher number of repetitions than those studying patients from the European population. Asian and European populations also have different distributions of alleles [42]. The difference in the choice of cutoff is probably due to an attempt to split the study sample into subgroups of a similar size. In small groups of patients, the cutoff selection is very important. This is well illustrated in the analysis performed here and explains why different cutoff criteria for analysis were chosen (Table 2).
A weak association between intensity of rash and CA-SSR1 polymorphism was found for the cutoff S ≤ 17 < L, but it was not statistically significant. The most significant results were obtained when we did not use any grouping criteria for CA-SSR1 polymorphism but instead applied the sum of CA repetitions of the two alleles. To evaluate the severity of rash, we did not use the NCI CTCAE version 3 scale but rather the body surface area covered by the rash at maximum intensity, calculated as percentage of body surface. Through this approach, a significant statistical correlation (p = 0.011) was found, in which the decrease in body surface area covered by rash was inversely correlated with the increasing number of CA dinucleotide repeats for the two alleles. A statistical relationship was also found when the grouping criteria of n(CA) ≤ 35, n(CA) > 35, S ≤ 17(CA), and L > 17(CA) were compared with the percentage of body surface area covered by rash (p = 0.046 and p = 0.040, respectively).
Other researchers have noted that the use of the NCI CTCAE version 3 scale can be problematic due to the possibility of different evaluation criteria, particularly for grades 1 and 2 rash [45]. In our observation, the treatment was used prophylactically without consideration of the intensity of rash. In grades 0–1 rash, the treatment was used in 12 % of patients. It also seems important that the NCI CTCAE version 3 scale contains only two criteria for the evaluation of body surface area covered by rash: <50 or >50 % of the body surface area. An area of <50 % occurs in grades 1 and 2, whereas an area of >50 % occurs in grades 3 and 4. It should be noted that grades 3 and 4 are relatively rare. In this study, 82 % of patients were classified in the 0–1 subgroup, whereas only 13 % of the patients belonged to the grade 3 subgroup. A better criterion seems to be the percentage of body surface area covered by rash or a scale that would better define the difference between grades 1 and 2, as well as the criteria for medical intervention.
In other publications, an association between type of CA dinucleotide polymorphism and response to treatment has been reported [32, 44]. The present investigation did not find a statistically significant association between treatment response and type of polymorphism (Fig. 3a, b). Loupakis et al. obtained a similar result when considering progression-free survival and overall survival in colorectal cancer [46]. However, among the patients with partial remission involved in this study, there was better response to cetuximab in cases of smaller number of repetitions, which seems to be consistent with other studies [32, 44]. The inconclusive results for the subgroup of patients with stabilization and progression may be caused by too short an observation time from the start of treatment until the time the medical questionnaire was filled in by the oncologist. In general, the analysis of tumor response and CA genotype has limited value in this study because of the heterogeneity of tumors and treatment types.
Molecular Mechanism
The molecular mechanism, which explains the relationship between CASSR1 polymorphism and response to treatment with EGFR inhibitors, is poorly understood. One factor that may contribute to this relationship is an increased expression of the EGFR gene with varying CA repeat lengths.
An in vitro study by Gebhardt et al. [28] showed that in the case of CASSR1 alleles containing 21 repeats of the CA dinucleotide EGFR gene, transcription is inhibited by approximately 80 % compared with genes containing a smaller numbers of repeats. Buerger et al. [47] found that Japanese patients with breast cancer, having much longer CA-SSR1, showed a significantly lower expression of the EGFR gene. Further evidence supporting the concept that CA-SSR1 polymorphism is directly linked to the expression of the EGFR gene is provided by Lee et al. In their study, the correlation between survival and number of repetitions in the CA-SSR1 polymorphic region in esophageal cancer patients was examined. Patients with homozygous genotypes in which alleles had fewer than 20 CA dinucleotide repeats were characterized by shorter survival than patients with genotypes in which two long alleles had more than 20 CA repetitions [31]. From the above studies, it can be deduced that one of the factors that determine the efficacy of EGFR inhibitors is the number of EGFR receptors on the cell. Amador et al. [32] noted that cells with a smaller number of CA dinucleotide repeats in the CA-SSR region have higher expression of the EGFR gene; thus, they have a greater amount of EGFR receptors and are more sensitive to the inhibitory effects of erlotinib.
In contrast, other studies have indicated that the protein expression level of EGFR determined by immunohistochemistry does not correlate with the response to treatment with cetuximab [13, 26, 48]. These observations show that the relationship between gene polymorphism and response to treatment may be more complicated. The rash observed during treatment with EGFR inhibitors is probably the final phenotypic effect created by the interaction between the EGFR genotype [28, 31, 32, 47] of the patients and their immune system [2, 48–50]. Perhaps this is why the search for relationships between patient genotype and response to treatment results in observations in which such a relationship is weak and not entirely clear.
Conclusions
The data provided in this study show that there is a correlation between CA-SSR1 polymorphism and body surface area covered by rash in response to cetuximab treatment. However, the relationship between CA-SSR1 polymorphism and response to cetuximab treatment is unclear. For a better evaluation of the association between the rash and polymorphism, a study on a larger group of patients should be performed, and the evaluation criteria for both the rash and the CA-SSR1 polymorphism should be better clarified.
Conflict of interest
TJ, MR, WR, ML, JT, MH, AK, AB, RD, and SN declare no conflicts of interest.
The study was funded by a grant from the Polish Ministry of Science and Higher Education No. N N402 453539.
Author contribution
Study concept—SN; collection of biological samples and clinical data—MR, WR, ML, JT, MH, AK, AB, and RD; laboratory analysis—TJ, ML, and SN; statistical analysis of clinical and molecular data and interpretation—TJ, ML, and SN; manuscript preparation and final redaction—TJ, MR, WR, ML, JT, MH, AK, AB, RD, and SN; responsibility for overall content—TJ and SN (equally contributed).
References
- 1.Herbst RS. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 2004;59:21–26. doi: 10.1016/j.ijrobp.2003.11.041. [DOI] [PubMed] [Google Scholar]
- 2.Kawaguchi Y, Kono K, Mimura K, Sugai H, Akaike H, Fujii H. Cetuximab induce antibody-dependent cellular cytotoxicity against EGFR-expressing esophageal squamous cell carcinoma. Int J Cancer J Int Cancer. 2007;120:781–787. doi: 10.1002/ijc.22370. [DOI] [PubMed] [Google Scholar]
- 3.Segaert S, Van Cutsem E. Clinical signs, pathophysiology and management of skin toxicity during therapy with epidermal growth factor receptor inhibitors. Ann Oncol. 2005;16:1425–1433. doi: 10.1093/annonc/mdi279. [DOI] [PubMed] [Google Scholar]
- 4.Yarden Y. The EGFR family and its ligands in human cancer. Signalling mechanisms and therapeutic opportunities. Eur J Cancer 1990. 2001;37(Suppl 4):S3–S8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 5.Brandt B, Meyer-Staeckling S, Schmidt H, Agelopoulos K, Buerger H. Mechanisms of egfr gene transcription modulation: relationship to cancer risk and therapy response. Clin Cancer Res. 2006;12:7252–7260. doi: 10.1158/1078-0432.CCR-06-0626. [DOI] [PubMed] [Google Scholar]
- 6.Vlahovic G, Crawford J. Activation of tyrosine kinases in cancer. Oncologist. 2003;8:531–538. doi: 10.1634/theoncologist.8-6-531. [DOI] [PubMed] [Google Scholar]
- 7.Behl AS, Goddard KAB, Flottemesch TJ, Veenstra D, Meenan RT, Lin JS, et al. Cost-effectiveness analysis of screening for KRAS and BRAF mutations in metastatic colorectal cancer. J Natl Cancer Inst. 2012;104:1785–1795. doi: 10.1093/jnci/djs433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonçalves A, Esteyries S, Taylor-Smedra B, Lagarde A, Ayadi M, Monges G, et al. A polymorphism of EGFR extracellular domain is associated with progression free-survival in metastatic colorectal cancer patients receiving cetuximab-based treatment. BMC Cancer. 2008;8:169. doi: 10.1186/1471-2407-8-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graziano F, Ruzzo A, Loupakis F, Canestrari E, Santini D, Catalano V, et al. Pharmacogenetic profiling for cetuximab plus irinotecan therapy in patients with refractory advanced colorectal cancer. J Clin Oncol. 2008;26:1427–1434. doi: 10.1200/JCO.2007.12.4602. [DOI] [PubMed] [Google Scholar]
- 10.Lurje G, Nagashima F, Zhang W, Yang D, Chang HM, Gordon MA, et al. Polymorphisms in cyclooxygenase-2 and epidermal growth factor receptor are associated with progression-free survival independent of K-ras in metastatic colorectal cancer patients treated with single-agent cetuximab. Clin Cancer Res. 2008;14:7884–7895. doi: 10.1158/1078-0432.CCR-07-5165. [DOI] [PubMed] [Google Scholar]
- 11.Han S-W, Oh D-Y, Im S-A, Park SR, Lee K-W, Song HS, et al. Epidermal growth factor receptor intron 1 CA dinucleotide repeat polymorphism and survival of advanced gastric cancer patients treated with cetuximab plus modified FOLFOX6. Cancer Sci. 2010;101:793–799. doi: 10.1111/j.1349-7006.2009.01447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boccia RV, Cosgriff TM, Headley DL, Badarinath S, Dakhil SR. A phase II trial of FOLFOX6 and cetuximab in the first-line treatment of patients with metastatic colorectal cancer. Clin Colorectal Cancer. 2010;9:102–107. doi: 10.3816/CCC.2010.n.014. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 14.Douillard J-Y, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 15.Lewandowska MA, Jóźwicki W, Żurawski B. KRAS and BRAF mutation analysis in colorectal adenocarcinoma specimens with a low percentage of tumor cells. Mol Diagn Ther. 2013;17:193–203. doi: 10.1007/s40291-013-0025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lièvre A, Bachet J-B, Le Corre D, Boige V, Landi B, Emile J-F, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 17.Dahan L, Norguet E, Etienne-Grimaldi M-C, Formento J-L, Gasmi M, Nanni I, et al. Pharmacogenetic profiling and cetuximab outcome in patients with advanced colorectal cancer. BMC Cancer. 2011;11:496. doi: 10.1186/1471-2407-11-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Cutsem E, Köhne C-H, Hitre E, Zaluski J, Chang Chien C-R, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 19.Jaka A, Gutiérrez-Rivera A, Ormaechea N, Blanco J, La Casta A, Sarasqueta C, et al. Association between EGFR gene polymorphisms, skin rash and response to anti-EGFR therapy in metastatic colorectal cancer patients. Exp Dermatol. 2014;23:751–753. doi: 10.1111/exd.12510. [DOI] [PubMed] [Google Scholar]
- 20.Stintzing S, Kapaun C, Laubender RP, Jung A, Neumann J, Modest DP, et al. Prognostic value of cetuximab-related skin toxicity in metastatic colorectal cancer patients and its correlation with parameters of the epidermal growth factor receptor signal transduction pathway: results from a randomized trial of the GERMAN AIO CRC Study Group. Int J Cancer. 2013;132:236–245. doi: 10.1002/ijc.27654. [DOI] [PubMed] [Google Scholar]
- 21.Petrelli F, Borgonovo K, Barni S. The predictive role of skin rash with cetuximab and panitumumab in colorectal cancer patients: a systematic review and meta-analysis of published trials. Target Oncol. 2013;8(3):173–181. doi: 10.1007/s11523-013-0257-x. [DOI] [PubMed] [Google Scholar]
- 22.Liu H, Wu Y, Lv T, Yao Y, Xiao Y, Yuan D, et al. Skin rash could predict the response to EGFR tyrosine kinase inhibitor and the prognosis for patients with non-small cell lung cancer: a systematic review and meta-analysis. PloS One. 2013;8:e55128. doi: 10.1371/journal.pone.0055128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fracasso PM, Burris H, 3rd, Arquette MA, Govindan R, Gao F, Wright LP, et al. A phase 1 escalating single-dose and weekly fixed-dose study of cetuximab: pharmacokinetic and pharmacodynamic rationale for dosing. Clin Cancer Res. 2007;13:986–993. doi: 10.1158/1078-0432.CCR-06-1542. [DOI] [PubMed] [Google Scholar]
- 24.Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard J-Y, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (the IDEAL 1 Trial) [corrected] J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Soler R. Rash as a surrogate marker for efficacy of epidermal growth factor receptor inhibitors in lung cancer. Clin Lung Cancer. 2006;8(Suppl 1):S7–S14. doi: 10.3816/CLC.2006.s.008. [DOI] [PubMed] [Google Scholar]
- 26.Saltz LB, Meropol NJ, Loehrer PJ, Sr, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201–1208. doi: 10.1200/JCO.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 27.Park JH, Han S-W, Oh D-Y, Im S-A, Jeong S-Y, Park KJ, et al. Analysis of KRAS, BRAF, PTEN, IGF1R, EGFR intron 1 CA status in both primary tumors and paired metastases in determining benefit from cetuximab therapy in colon cancer. Cancer Chemother Pharmacol. 2011;68:1045–1055. doi: 10.1007/s00280-011-1586-z. [DOI] [PubMed] [Google Scholar]
- 28.Gebhardt F, Zänker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem. 1999;274:13176–13180. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- 29.Nomura M, Shigematsu H, Li L, Suzuki M, Takahashi T, Estess P, et al. Polymorphisms, mutations, and amplification of the EGFR gene in non-small cell lung cancers. PLoS Med. 2007;4:e125. doi: 10.1371/journal.pmed.0040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzeng C-WD, Frolov A, Frolova N, Jhala NC, Howard JH, Vickers SM, et al. Pancreatic cancer epidermal growth factor receptor (EGFR) intron 1 polymorphism influences postoperative patient survival and in vitro erlotinib response. Ann Surg Oncol. 2007;14:2150–2158. doi: 10.1245/s10434-007-9409-5. [DOI] [PubMed] [Google Scholar]
- 31.Lee J-M, Yang S-Y, Yang P-W, Shun C-T, Wu M-T, Hsu C-H, et al. Polymorphism in epidermal growth factor receptor intron 1 predicts prognosis of patients with esophageal cancer after chemoradiation and surgery. Ann Surg Oncol. 2011;18:2066–2073. doi: 10.1245/s10434-011-1559-9. [DOI] [PubMed] [Google Scholar]
- 32.Amador ML, Oppenheimer D, Perea S, Maitra A, Cusatis G, Cusati G, et al. An epidermal growth factor receptor intron 1 polymorphism mediates response to epidermal growth factor receptor inhibitors. Cancer Res. 2004;64:9139–9143. doi: 10.1158/0008-5472.CAN-04-1036. [DOI] [PubMed] [Google Scholar]
- 33.Huang C-L, Yang C-H, Yeh K-H, Hu F-C, Chen K-Y, Shih J-Y, et al. EGFR intron 1 dinucleotide repeat polymorphism is associated with the occurrence of skin rash with gefitinib treatment. Lung Cancer. 2009;64:346–351. doi: 10.1016/j.lungcan.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 34.Abdullah SE, Haigentz M, Jr, Piperdi B. Dermatologic toxicities from monoclonal antibodies and tyrosine kinase inhibitors against EGFR: pathophysiology and management. Chemother Res Pract. 2012;2012:351210. doi: 10.1155/2012/351210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacot W, Bessis D, Jorda E, Ychou M, Fabbro M, Pujol J-L, et al. Acneiform eruption induced by epidermal growth factor receptor inhibitors in patients with solid tumours. Br J Dermatol. 2004;151:238–241. doi: 10.1111/j.1365-2133.2004.06026.x. [DOI] [PubMed] [Google Scholar]
- 36.Wacker B, Nagrani T, Weinberg J, Witt K, Clark G, Cagnoni PJ. Correlation between development of rash and efficacy in patients treated with the epidermal growth factor receptor tyrosine kinase inhibitor erlotinib in two large phase III studies. Clin Cancer Res. 2007;13:3913–3921. doi: 10.1158/1078-0432.CCR-06-2610. [DOI] [PubMed] [Google Scholar]
- 37.Rosell R, Robinet G, Szczesna A, Ramlau R, Constenla M, Mennecier BC, et al. Randomized phase II study of cetuximab plus cisplatin/vinorelbine compared with cisplatin/vinorelbine alone as first-line therapy in EGFR-expressing advanced non-small-cell lung cancer. Ann Oncol. 2008;19:362–369. doi: 10.1093/annonc/mdm474. [DOI] [PubMed] [Google Scholar]
- 38.Klinghammer K, Knödler M, Schmittel A, Budach V, Keilholz U, Tinhofer I. Association of epidermal growth factor receptor polymorphism, skin toxicity, and outcome in patients with squamous cell carcinoma of the head and neck receiving cetuximab-docetaxel treatment. Clin Cancer Res. 2010;16:304–310. doi: 10.1158/1078-0432.CCR-09-1928. [DOI] [PubMed] [Google Scholar]
- 39.Robert C, Soria J-C, Spatz A, Le Cesne A, Malka D, Pautier P, et al. Cutaneous side-effects of kinase inhibitors and blocking antibodies. Lancet Oncol. 2005;6:491–500. doi: 10.1016/S1470-2045(05)70243-6. [DOI] [PubMed] [Google Scholar]
- 40.Ocvirk J, Heeger S, McCloud P, Hofheinz R-D. A review of the treatment options for skin rash induced by EGFR-targeted therapies: evidence from randomized clinical trials and a meta-analysis. Radiol Oncol. 2013;47:166–175. doi: 10.2478/raon-2013-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L, Cao Y, Tan A, Liao C, Mo Z, Gao F. Cetuximab-based therapy vs noncetuximab therapy in advanced or metastatic colorectal cancer: a meta-analysis of seven randomized controlled trials. Colorectal Dis. 2010;12:399–406. doi: 10.1111/j.1463-1318.2009.01916.x. [DOI] [PubMed] [Google Scholar]
- 42.Kharrat N, Al’Fadhli S, Rebai M, Aifa MS, Kallel I, Khabir A, et al. (AC) dinucleotide repeat polymorphism in intron 1 of human EGFR shows ethnic specificities and high evidence for association with breast cancer. Int J Biol Markers. 2007;22:258–264. doi: 10.5301/jbm.2008.1479. [DOI] [PubMed] [Google Scholar]
- 43.Buisine M-P, Wacrenier A, Mariette C, Leteurtre E, Escande F, Aissi S, et al. Frequent mutations of the CA simple sequence repeat in intron 1 of EGFR in mismatch repair-deficient colorectal cancers. World J Gastroenterol. 2008;14:1053–1059. doi: 10.3748/wjg.14.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han S-W, Jeon YK, Lee K-H, Keam B, Hwang PG, Oh D-Y, et al. Intron 1 CA dinucleotide repeat polymorphism and mutations of epidermal growth factor receptor and gefitinib responsiveness in non-small-cell lung cancer. Pharmacogenet Genomics. 2007;17:313–319. doi: 10.1097/FPC.0b013e328011abc0. [DOI] [PubMed] [Google Scholar]
- 45.Agero ALC, Dusza SW, Benvenuto-Andrade C, Busam KJ, Myskowski P, Halpern AC. Dermatologic side effects associated with the epidermal growth factor receptor inhibitors. J Am Acad Dermatol. 2006;55:657–670. doi: 10.1016/j.jaad.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 46.Loupakis F, Antoniotti C, Cremolini C, Zhang W, Yang D, Wakatsuki T, et al. Prospective study of EGFR intron 1 (CA)n repeats variants as predictors of benefit from cetuximab and irinotecan in chemo-refractory metastatic colorectal cancer (mCRC) patients. Pharmacogenomics J. 2014;14:322–327. doi: 10.1038/tpj.2014.1. [DOI] [PubMed] [Google Scholar]
- 47.Buerger H, Packeisen J, Boecker A, Tidow N, Kersting C, Bielawski K, et al. Allelic length of a CA dinucleotide repeat in the egfr gene correlates with the frequency of amplifications of this sequence—first results of an inter-ethnic breast cancer study. J. Pathol. 2004;203:545–550. doi: 10.1002/path.1542. [DOI] [PubMed] [Google Scholar]
- 48.Vallböhmer D, Zhang W, Gordon M, Yang DY, Yun J, Press OA, et al. Molecular determinants of cetuximab efficacy. J Clin Oncol. 2005;23:3536–3544. doi: 10.1200/JCO.2005.09.100. [DOI] [PubMed] [Google Scholar]
- 49.Argiris A, Lee SC, Feinstein T, Thomas S, Branstetter BF, IV, Seethala R, et al. Serum biomarkers as potential predictors of antitumor activity of cetuximab-containing therapy for locally advanced head and neck cancer. Oral Oncol. 2011;47:961–966. doi: 10.1016/j.oraloncology.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takata T, Tarutani M, Zouboulis CC, Sano S. Sebaceous glands as the primary target of EGFR-inhibitors in the development of papulopustular eruption. J Dermatol Sci. 2012;66:165–168. doi: 10.1016/j.jdermsci.2012.02.012. [DOI] [PubMed] [Google Scholar]