Abstract
The objective of this study was to explore the molecular epidemiology and the genetic support of clinical multidrug resistant (MDR) Acinetobacter baumannii (A. baumannii) isolates in an ICU ward of a comprehensive hospital. A total of 102 non-duplicate drug-resistant A. baumannii isolates were identified and 93 (91.1%) of them were MDR strains. Molecular analysis demonstrated that carbapenemase genes blaOXA-23 and blaOXA-51 were presented in all 93 MDR isolates (100%), but other carbapenemase genes, including blaOXA-24, blaOXA-58, blaIMP-1, blaIMP-4, blaSIM, and blaVIM genes were completely absent in all isolates. In addition, genes of AdeABC efflux system were detected in 88.2% (90/102) isolates. Interestingly, an addition to efflux pump inhibitor, reserpine could significantly enhance the susceptibility of MDR isolates to moxifloxacin, cefotaxime, and imipenem (p < 0.01). Clonal relationship analysis further grouped these clinical drug-resistant isolates into nine clusters, and the MDR strains were mainly in clusters A, B, C, and D, which include 16, 13, 25, and 15 isolates, respectively. This study demonstrated that clinical isolates carrying carbapenemase-encoding genes blaOXA-23 and AdeABC efflux pump genes are the main prevalent MDR A. baumannii, and the co-expression of oxacillinase and efflux pump proteins are thus considered to be the important reason for the prevalence of this organism in the ICU of this hospital.
Keywords: Acinetobacter baumannii, multidrug resistance, nosocomial infections, oxacillinase, efflux pump
1. Introduction
The emergence of multidrug-resistant (MDR) bacterial strains has been recognized as a main challenge for treatment of clinical infection with broad-spectrum antibiotics. Acinetobacter baumannii (A. baumannii) is an emerging opportunistic nosocomial pathogen with great concern worldwide, which is the most common clinically isolated Acinetobacter species and a cause of severe infections in intensive care units (ICU) of hospitals, owing to its remarkable ability to acquire resistance to most antimicrobials [1,2]. Carbapenem, aminoglycosides, and quinolone antibiotics are often efficient in the treatment of A. baumannii infection. However, MDR A. baumannii isolates have recently been increasingly reported in many countries, particularly in Asia-Pacific countries including China [3,4,5,6,7,8,9,10,11,12]. A recent surveillance report from CHINET for the resistance rates of Acinetobacter species shows that A. baumannii isolates accounted for 89.6% of the resistance, and their resistances to imipenem and meropenem are up to 62.8% and 59.4% in China, respectively (http://narin.minke.cn) [12].
The molecular basis of multidrug resistance in this species has been attributed to combined mechanisms of an increased expression of oxacillinase (OXA)-type carbapenemases and non-enzymatic mechanisms, such as increased cell membrane impermeability, expression of multidrug efflux pump proteins, and/or and alterations in penicillin-binding proteins, due to a high level of genomic plasticity and mutation of endogenous genes associated with antimicrobial resistance [13,14,15,16]. An analysis of the molecular epidemiology of nosocomial infection MDR A. baumannii, particularly in the ICU ward of a hospital thus may help to develop efficient guidelines to control the spread of these bugs [17].
To date, there are only few reports demonstrating combinations of different mechanisms of resistance in MDR bacterial pathogens, despite this there is an apparent correlation of antimicrobial resistance with the carbapenemase production and an expression of multidrug efflux pumps in A. baumannii has been reported [18,19,20,21]. In the present study, we interrogated the molecular mechanism with a focus in the genetic linkage of OXA-type carbapenemases and multidrug efflux pumps in drug-resistant A. baumannii isolates recovered from the ICU ward in the General Hospital of Ningxia Medical University, a national comprehensive hospital in Northwestern China from January 2013 to July 2014.
2. Experimental Section
2.1. Bacterial Isolation
The Clinical Research Ethics Committee at the General Hospital of Ningxia Medical University approved this study with a waiver for informed consents. All of the non-duplicate clinical isolates were routinely collected from patients who were not received an antibiotic therapy at the General Hospital of Ningxia Medical University (Yinchuan, China) in the ICU from January 2013 to July 2014. Isolated bacteria were stored at −80 °C prior to be used in this study. All clinical strains of Acinetobacter spp. were identified with a ViteK-2 Compact automated microbiological system (Biomerieux, France). The Microseq 500 16S rDNA bacterial identification kit was used to identify isolates for Acinetobacter species (Applied Biosystem, Foster City, CA, USA).
2.2. Test of Antimicrobial Susceptibility
The susceptibility of an antimicrobial agent was determined using E-test strips (AB Biodisk, Slona, Sweden) per manufacturer’s instruction. The susceptibility was interpreted according to the guidelines of the Clinical Laboratory Standards Institute (CLSI) [22,23], and defined as previously described [24]. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 strains were used as references for antimicrobial susceptibility testing. A total of 102 drug-resistant A. baumannii isolates were recovered from patients hospitalized in the ICU ward of this hospital, among which 93 were multi-drug resistant strains and 9 were resistant to less than three classes of the following 11 tested antibiotics or a synergistic combination. The tested antibiotics in this study were ampicillin-sulbactam, cefepime, ceftazidime, ceftriaxome, imipenem, levofloxacin, piperacillin, polymyxin B, ticarcillin/clavulanicac, tobramycin and trimethoprim/sulfamethoazole. A multidrug resistant A. baumannii was defined as an isolate resistant to at least three classes of antibiotics, and isolates resistant to less than three classes of antibiotics were designated as antibiotic unsusceptible isolates in this study. In order to examine the effect of efflux pumps in antimicrobial resistance, the MICs of cefotaxime, moxifloxacin and imipenem for A. baumannii were measured in the presence of an efflux pump inhibitor (EPI) reserpine (Dalian Meilun Biology Technology Co., Ltd., Dalian, China) at a concentration of 25 mg/L with an agar dilution method as described previously [25]
2.3. Identification of the Drug Resistance Genes
All isolates were subjected to detect genes of drug resistance, including the carbapenem-resistance genes (blaNDM-1, blaSIM, blaVIM, blaIMP-1, blaIMP-4, blaOXA-23, blaOXA-24, blaOXA-58, blaOXA-51) and the efflux pump genes (AdeA, AdeB, AdeC, AdeR, AdeS) by a polymerase chain reaction (PCR) assay as described elsewhere [26]. All primers used for PCR in this study were listed in the Table 1. Genomic DNA of A. baumannii isolates was extracted using TIANamp Bacteria DNA Kit (Tiangen, Beijing, China). PCR was performed using Taq PCR Master Mix (TaKaRa, Dalian, China). The cycling parameters of PCR were as follows: an initial denaturation at 94 °C for 5 minutes, followed by 30 cycles of 94 °C denature for 30 seconds, 55 °C annealing for 30 seconds and 72 °C extension for 90 seconds. Then the PCR products were resolved in Ethidium Bromide (EM) agarose gels and visualized under an Ultraviolet (UV) light.
Table 1.
Sequences of PCR primer sets for genes of carbapenemase and ABC efflux pumps of A. baumannii used in this study.
| Target genes | Primer sets | Primer sequence (5′→3′) | Amplicon Size (bp) |
|---|---|---|---|
| Carbapenemase | blaNDM-1 F | GCATTGGCGGCGAAAGTCA | 921 |
| blaNDM-1 R | CTCGCACCGAATGTCTGGC | ||
| blaSIM F | TACAAGGGATTCGGCATCG | 741 | |
| blaSIM R | TAATGGCCTGTTCCCATGTG | ||
| blaVIM F | TTATGGAGCAACCGATGT | 920 | |
| blaVIM R | CAAAAGTCCCGCTCCAACGA | ||
| blaIMP-1 F | ATCCAAGCAGCAAGCGCGTTA | 474 | |
| blaIMP-1 R | AGGCGTGCTGCTGCAACGACTTGT | ||
| blaIMP-4 F | CTACCGCAGCAGAGTCTTTG | 879 | |
| blaIMP-4 R | AACCAGTTTTGCCTTACCAT | ||
| blaOXA-23 F | GATGTGTCATAGTATTCGTCG | 774 | |
| blaOXA-23 R | TCACAACAACTAAAAGCACTG | ||
| blaOXA-24 F | TTCCCCTAACATGAATTTGT | 828 | |
| blaOXA-24 R | GTACTAATCAAAGTTGTGAA | ||
| blaOXA-58 F | AAGTATTGGGGCTTGTGCTG | 800 | |
| blaOXA-58 R | CCCCTCTGCGCTCTACATAC | ||
| blaOXA-51 F | TAATGCTTTGATCGGCCTTG | 760 | |
| blaOXA-51 R | TGGATTGCACTTCATCTTGG | ||
| Multidrug resistance efflux pumps | AdeA F | GGCGTATTGGGCAATCTTTTGT | 1157 |
| AdeA R | GTCACCGACTTTCAAGCCTTTG | ||
| AdeB F | TGGCGGAATGAAGTATGT | 1323 | |
| AdeB R | GCAGTGCGGCAGGTTAG | ||
| AdeC F | GACAATCGTATCTCGTGGACTC | 1331 | |
| AdeC R | AGCAATTTTCTGGTCAGTTTCC | ||
| AdeR F | TCACATGGCTATCTACGGTTGG | 538 | |
| AdeR R | TGAAGGCATGAGTGTTATTCGG | ||
| AdeS F | GTGGACGTTAGGTCAAGTTCTG | 949 | |
| AdeS R | TGTTATCTTTTGCGGCTGTATT |
2.4. Genetic Relationship among A. Baumannii Isolates Determined by Pulse Field Gel Electrophoresis (PFGE)
Pulsed-field gel electrophoresis (PFGE) was performed as previously described [27]. Briefly, the purified bacterial genomic DNA was digested by the restriction enzyme ApaI (TaKaRa, Dalain, China), and the digested product was separated in a Bio-Rad CHEF-Mapper apparatus with parameters of pulses ranging from 5 to 20 seconds at a voltage of 5 V/cm and switch angle of 120° at 14 °C for 19 h (Bio-Rad Laboratories, Hercules, CA, USA). The gel was then stained with ethidium bromide and the restricted pattern of DNAs was acquired using Bio-Rad Vilber Lourmat. The BioNumerics 6.6 software (Applied Maths, Kortrijk, Belgium) was employed for analyzing the similarities between the digitized PFGE profiles. The similarity between DNA restriction profiles were interpreted according to the criteria described by Seifert et al. [28] using the Dice correlation coefficient F. F = 2Nxy/(Nx + Ny), where Nxy represents the number of identical bands between isolate x and y, and the Nx and Ny are total numbers of bands acquired from the digestion in isolates x and y, respectively. Isolates with a similarity of >85% following dendrogram analysis were considered to represent an identical PFGE genotype (pulsotype) and categorized into the same group [28].
2.5. Statistical Analysis
All data were managed using the WHONET version 5.6 software. The statistical analysis was processed with the Statistical Package for the Social Sciences (SPSS) software version 18.0 (SPSS, Chicago, IL, USA). The changes of MICs for MDR A. baumanniican between the presence and absence of reserpine were compared with a t-test analysis. A p < 0.05 was defined as a statistical significance.
3. Results
3.1. Epidemiological Data of Drug-Resistant A. Baumannii Infection in ICU
During the study period, a total of 102 non-duplicated drug-resistant A. baumannii isolates to tested antibiotics were collected from the ICU ward of this hospital. The tested antibiotics or synergistic combinations in this study include ampicillin-sulbactam, cefepime, ceftazidime, ceftriaxome, imipenem, levofloxacin, piperacillin, polymyxin B, ticarcillin/clavulanicac, tobramycin, and trimethoprim/sulfamethoazole. The age of patients ranged from 1 to 92 years (median, 55 years); 78 (76.47%) patients were males and 24 (23.53%) were females. The majority of drug-resistant strains were recovered from respiratory specimens (72/102), followed by samples from body secretions (9/102), bloodstream (6/102), urine (5/102), chest drainage fluid (4/102), pus (4/102), and cerebrospinal fluid (2/102). Among the 102 drug-resistant isolates, 93 out of the 102 isolates were categorized as MDR A. baumannii which were resistant to at least three classes of antimicrobials; nine of them were identified as drug-resistance to less than three classes of the tested antibiotics, which were designated as antibiotic unsusceptible isolates in this study (Table 2). All of the 102 drug-resistant clinical isolates were resistant to trimethoprim/sulfamethoazole (102/102, 100%); high resistant rates were also observed to cefepime (94/102, 92.2%), ceftazidime (94/102, 92.2%), ceftriaxome (99/102, 97.1%), imipenem (83/102, 81.4%), levofloxacin 92/102, 90.2%), piperacillin (99/102, 97.1%), ticarcillin/clavulanicac (98/102, 96.1%), and tobramycin (90/102, 88.2%) (Table 2). Noteworthy, in spite of 13 out of the 102 (12.7%) drug-resistant clinical isolates were resistant to polymyxin B, A. baumannii isolates showed most susceptible to this drug of “last resort” in this study [6] (Table 2).
Table 2.
The susceptibilities of clinical A. baumannii isolates to tested antibiotics (N = 102).
| Antibiotics | Drug-resistant isolates | MIC ranges (mg/mL) | Drug-susceptible isolates |
|---|---|---|---|
| Trimethoprim/sulfamethoazole | 102/102, (100%) | 4–32/64 | 0/102, (0.0%) |
| Piperacillin | 99/102, (97.1%) | 128–512 | 3/102, (2.9%) |
| Ceftriaxome | 99/102, (97.1%) | 16–256 | 3/102, (2.9%) |
| Ampicillin-sulbactam | 98/102, (96.1%) | 8–256/8 | 4/102, (3.9%) |
| Ticarcillin-clavulanicac | 98/102, (96.1%) | 32–1224/1 | 4/102, (3.9%) |
| Cefepime | 94/102, (92.2%) | 8–512 | 8/102, (7.8%) |
| Ceftazidime | 94/102, (92.2%) | 4–128 | 8/102, (7.8%) |
| Levofloxacin | 92/102, (90.2%) | 2–64 | 10/102, (9.8%) |
| Tobramycin | 90/102, (88.2%) | 4–256 | 12/102, (9.8%) |
| Imipenem | 83/102, (81.4%) | 2–64 | 19/102, (18.6%) |
| Polymyxin B | 13/102, (12.7%) | 1–16 | 89/102, (87.3%) |
3.2. Genes of Antimicrobial Resistance Identified by Multiplex PCR Assays
To uncover the underlying mechanism involved in the drug-resistance of these 102 A. baumannii isolates, genes encoding carbapenemase and efflux pumps were determined by multiplex PCR assays. The distribution of carbapenemase and efflux pump genotypes in drug-resistant strains was shown in Table 3. There were no carbapenemase genes blaOXA-23, blaOXA-24, blaOXA-51, blaOXA-58, blaVIM, blaIMP-1, blaIMP-4, blaSIM, blaNDM-1 detected in all nine antibiotic unsusceptible isolates, while efflux pump AdeA, AdeB, AdeC genes and their regulator AdeR and AdeS genes were found in these isolate (data not shown). Of note, blaOXA-23, blaOXA-51 genes were present in all 93 MDR A. baumannii isolates, while the other carbapenemase genes, blaoxa-24, blaOXA-58, blaVIM, blaIMP-1, blaIMP-4, blaSIM, blaNDM-1 were completely absent (Table 3). Among efflux genes, the majority of the MDR isolates were found to harbor AdeABC efflux pump genes, and only 11.8% (12/102) of the isolates were undetectable for AdeABC and AdeRS genes (Table 3). The AdeA, AdeB, AdeC, AdeR and AdeS genes were detected in 79.6% (74/93), 77.4% (72/93), 86.0% (80/93), 81.7% (76/93), and 80.6% (75/93) of the 93 MDR isolates, respectively. Equally noteworthy, there were 73 and 68 out of the 93 MDR isolates harbored AdeABC and AdeABCRS multiple genes, respectively. The AdeA, AdeB, AdeC, AdeR, and AdeS genes were detected in 4/9, 1/9, 3/9, 2/9, and 1/9 of the 9 of the antibiotic unsusceptible isolates, respectively (Table 3).
Table 3.
Distribution of Carbapenemase genes and Ade efflux pump genes in the 102 clinical drug-resistant A. baumannii isolates in this study (Number of isolates harboring Carbapenemase gene/number of isolates containing efflux pump gene).
| Genes | AdeA | AdeB | AdeC | AdeS | AdeR |
|---|---|---|---|---|---|
| blaOXA-23 | 93/78 | 93/73 | 93/83 | 93/78 | 93/76 |
| blaOXA-24 | 0/78 | 0/73 | 0/83 | 0/78 | 0/76 |
| blaOXA-51 | 93/78 | 93/73 | 93/83 | 93/78 | 93/76 |
| blaOXA-58 | 0/78 | 0/73 | 0/83 | 0/78 | 0/76 |
| BlaNDM-1 | 0/78 | 0/73 | 0/83 | 0/78 | 0/76 |
| BlaSIM | 0/78 | 0/73 | 0/83 | 0/78 | 0/76 |
| BlaVIM | 0/78 | 0/73 | 0/83 | 0/78 | 0/76 |
| BlaIMP-1 | 0/78 | 0/73 | 0/83 | 0/78 | 0/76 |
| BlaIMP-4 | 0/78 | 0/73 | 0/83 | 0/78 | 0/76 |
3.3. Impact of Efflux Pumps on the Susceptibility of Clinical MDR A. Baumannii Isolates to Antibiotics
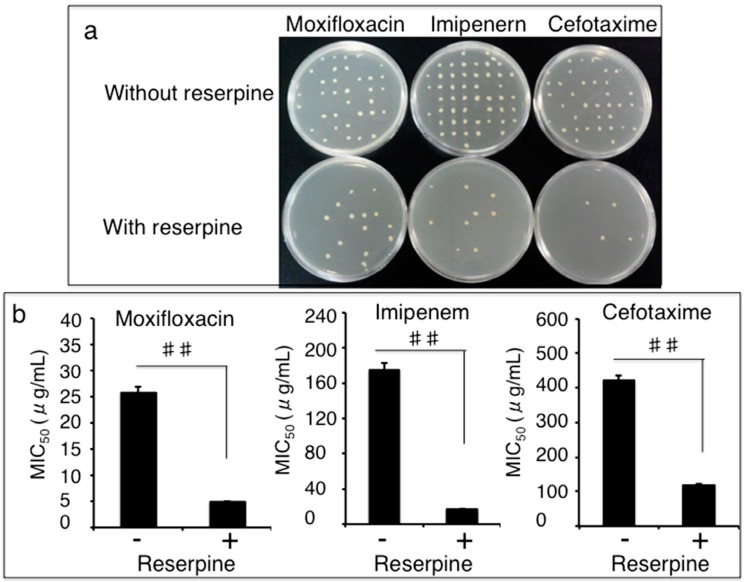
We next sought to explore whether the efflux pumps played a role in the drug-resistance of these 102 clinical isolates of A. baumannii to antibiotics by an active efflux inhibition test on the M-H agar plates with or without 25 mg/L of efflux pump inhibitor (EPI) reserpine. The presence of reserpine showed a dramatically enhanced inhibitory capacity of moxifloxacin, cefotaxime, and imipenem to the growth of the clinical MDR A. baumannii isolates (Figure 1a). In the presence of reserpine, the susceptibilities of 80 out of the 93 MDR isolates (86.0%, 80/93) to moxifloxacin, 75 of the 93 isolates to cefotaxime (80.6%, 75/93), and 88 of the 93 MDR isolates (94.6%, 88/93) to imipenem were increased by 2–8, 2–4, and 2–32 folds as determined by an MIC assay, respectively (Figure 1b). The reserpine-mediated decreases of MICs of these MDR isolates were statistically different in comparison with they were treated with the antibiotics alone (p < 0.01) (Figure 1b), indicating that a bacterial antibiotic efflux mechanism was involved in the clinical MDR isolates. Intriguingly, the presence of reserpine had no inhibitory effect on the growth and MICs of the nine antibiotic unsusceptible A. baumannii isolates (data not shown).
Figure 1.
Impact of reserpine on the susceptibility of MDR A. baumannii to antibiotics. The clinical drug-resistant A. baumannii isolates were culture in the presence of cefotaxime, moxifloxacin, or imipenem with or without 25 mg/L of efflux pump inhibitor reserpine, the MICs were determined by agar dilution method. (a) Representative images showed an enhanced inhibitory activity of indicated antibiotics in the presence of reserpine. (b) Effect of reserpine on the susceptibility of MDR A. baumannii to antibiotics. An addition of reserpine resulted in a significantly reduction of MICs of indicated antibiotics the clinical isolates (p < 0.01), indicative of an enhanced susceptibility to these antimicrobials. Compared to the corresponding reserpine absent group, ##: p < 0.01. Data represented the mean ± SD from three independent triplicated experiments (N = 102).
3.4. Clonal Relationship of Drug-Resistant A. Baumannii Isolates Determined by a Pulsed-Field Gel Electrophoresis (PFGE) Method
In order to identify clonal relationship of the 102 clinical drug-resistant A. baumannii isolates, PFGE analysis was employed using Apa I-digested A. baumannii genomic DNA. The 102 clinical isolates gave 47 reproducible ApaI-digested DNA profiles (PFGE genotypes) with a Dice coefficient, F ranging from 0.85 to 1.00 [28]. Cluster analysis of the pulsotypes grouped the 102 clinical drug-resistant A. baumannii isolates into nine clusters with a cutoff point at 85% similarity (Table 4). The MDR isolates were grouped into four main clusters, A, B, C, and D, which had respective 16, 13, 25 and 15 strains, while none of the nine antibiotic unsusceptible isolates was in the clusters A–D. Interestingly, isolates in cluster B and E exhibited a tendency of AdeABC efflux pump genotypes (Table 4). This result suggested that the majority of the MDR isolates had the diversity with multivariate clones.
Table 4.
Distribution of AdeABC efflux pump genes in different groups of clinical drug-resistant A. baumannii isolates.
| Efflux pump AdeABC genes | PFGE groups | Constituent ratio | |||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | Other | ||
| AdeABC, AdeRS | 11 | 12 | 18 | 5 | 3 | 19 | 68/102 (66.7%) |
| AdeABC | 0 | 1 | 0 | 0 | 0 | 3 | 4/102 (3.9%) |
| AdeABC, AdeR | 0 | 0 | 0 | 0 | 0 | 2 | 2/102 (1.9%) |
| AdeABC, AdeS | 2 | 0 | 2 | 0 | 0 | 0 | 4/102 (3.9%) |
| Other genotypes | 3 | 0 | 1 | 3 | 0 | 5 | 12/102 (11.8%) |
| Negative | 0 | 0 | 4 | 7 | 0 | 1 | 12/102 (11.7%) |
| Sum | 16 | 13 | 25 | 15 | 3 | 30 | 102/102 (100%) |
4. Discussion
An increasing emergence of antibiotic resistance in microbes has had significant impact on the patient outcomes and challenges treatments of clinical infection using broad-spectrum antibiotics. Moreover, A. baumannii recently emerged as an important pathogen responsible for epidemics of nosocomial infections, particularly in the ICU ward of a hospital [1]. Therefore, an identification of molecular mechanisms of drug resistance in A. baumannii will improve treatments of hospital-acquired infections and help for developing appropriate control measures to prevent further spread of multidrug-resistant organism [10]. In the present study, we investigated possible molecular epidemic mechanisms of MDR A. baumannii in the ICU ward of the General Hospital of Ningxia Medical University in Northwestern China, and found that most common MDR A. baumannii strains identified in the ICU of this hospital were isolates harboring genes of class D oxacillinases blaOXA-23/blaOXA-51 and drug-resistant efflux pump AdeABC in the period of January 2013 to July 2014. Total of 102 clinical drug-resistant A. baumannii isolates were recovered, and PFGE analysis further revealed that the MDR isolates were mainly grouped into A, B, C, and D clusters. Furthermore, these drug-resistant isolates displayed a relative low resistance to polymyxin B (12.5%) but high resistance to trimethoprim/sulfamethoazole (100%); an addition of efflux pump inhibitor reserpine could significantly enhance the susceptibility of these MDR A. baumannii strains to moxifloxacin, cefotaxime, and imipenem.
Mechanisms of MDR A. baumannii are complex. In addition to a remarkable ability of this organism to horizontally acquire resistance determinants, intrinsic resistance mechanisms include production of enzyme, change of outer membrane permeability, expression of drug resistance and efflux pump genes [15]. For instance, A. baumannii is able to gain its resistance to carbapenems mainly through a mechanism of producing different carbapenemase enzymes including class B metallo-b-lactamases (MBLs) and class D oxacillinases (OXAs) [29]. blaOXA-23, blaOXA-24, blaOXA-51 and blaOXA-58 are most common class D blaOXAs reported in clinical MDR A. baumannii isolates, particularly in Asia-Pacific countries [4,9,30,31], where MDR A. baumannii isolates harboring blaOXA-23 gene were more prevalent than any other gene type, and the blaOXA-58 gene was rarely detected in these strains [31]; while MBLs IMP, VIM, and SIM-producing A. baumannii isolates have also been often reported worldwide [29].
In line with the findings from other studies of Asia-Pacific countries including China and Korea [4,8,9,10,30,31,32], the majority of prevalent MDR A. baumannii in the ICU of this hospital were strains carrying blaOXA-23 and blaOXA-51 genes, of which were detected in all of the 93 MDR A. baumannii isolates, but none of the blaOXA-24, blaOXA-58, blaVIM, blaIMP-1, blaIMP-4, blaSIM, blaNDM-1 genes were detected in these drug-resistant isolates. Of note, since blaOXA-51 gene is an intrinsic, chromosomal carbapenemase naturally present in all A. baumannii strains regardless of drug susceptibility, it is an intrinsic cambapenem resistance mechanism. Therefore, it has been used as a target gene for identification of A. baumannii species using PCR, which was correlated well with 16S rRNA and rpoB sequencing [33]. Interestingly, all the 93 MDR isolates were A. baumannii strains, but the nine antibiotic unsusceptible isolates lacking blaOXA-51 gene might be Acinetobacter spp. strains. Such a high frequency of blaOXA-51 gene detected in these MDR isolates may imply that an intrinsic drug resistance mechanism also contribute the multidrug resistance. For instance, the porin deficiency is another intrinsic carbapenem resistance mechanism in A. baumannii [15]. Porins are outer membrane proteins (OMPs) able to form transport channels for molecules crossing membranes. The carbapenem-associated OMP (CarO) is the most characterized porin and the best characterized causes of intrinsic carbapenem resistance in A. baumannii [15,34]. An alteration of CarO gene expression could contribute imipenem resistance by reducing the penetration of drug into the cells [15,34,35].
In terms of the genetic basis of blaOXA-23 dissemination in A. baumannii in China, Liu et al. recently uncovered that the plasmid pAZJ221 and Tn2009 might effectively contribute the broad dissemination of blaOXA-23 gene in Acinetobacter spp. in China, suggesting that the mechanism of horizontal gene transfer may play a key role in the dissemination of blaOXA-23 gene in this country [30]. Of interest, despite a remarkably increased proportion of MDR A. baumannii isolates carrying the blaOXA-23 gene has been reported in Asia-Pacific region since last decade, while the prevalence of MDR A. baumannii harboring blaOXA-51 gene has been decreased [4,10,31]. Differing from these observations, the blaOXA-51 gene was determined in all of the 93 MDR A. baumannii isolates in this study. However, the contribution of blaOXA-51 gene in the MDR of these isolates needs to be further identified by a quantitative assay. Together with other findings, this finding thus further supports a notion of that the prevalent blaOXA genes are significantly varied depending on the time, place, and even hospital ward of isolation [36,37,38].
Apart from the production of carbapenemase, MDR efflux pumps also have displayed multifactorial roles in the resistance of Acinetobacter spp to antibiotics [15,19,39,40,41,42]. There are three resistance-nodulation-cell division (RND) systems, AdeFGH, AdeIJK, and AdeABC have been characterized in A. baumannii, among which the AdeABC was the most frequently involved in MDR A. baumannii, which was found in approximately 80% of clinical isolates [43]. A. baumannii overexpressing AdeABC has been reported to be significantly correlated with resistance to tigecycline, minocycline, and gentamicin and other biological functions [15].
Using a set of isogenic mutants of A. baumannii strains, Yoon et al. recently demonstrated that the expression of RND-efflux systems, particularly the AdeABC contributed to the drug resistance and biofilm formation in A. baumannii [44]. An A. baumannii mutant overproduced AdeABC could confer a clinical resistance to aminoglycosides. More importantly, the AdeABC pump showed a synergistic effect of the level of resistance of the host when it was in combination with enzymatic resistance to carbapenems and aminoglycosides, indicative of a synergistic effect between the expression of an efflux pump and a resistance gene on MDR of A. baumannii strains [44]. In this study, AdeABC genes were detected in the majority of clinical MDR isolates of those also harboring blaOXA-23 gene, and an addition of EPI reserpine led to an enhanced susceptibility of MDR isolates to antibiotics (p < 0.01). A. baumannii isolates carrying both of oxacillinase and efflux pump genes were also reported in several previous studies [19,41]. For instance, the expression both of AdeABC efflux pump and blaOXA-23 played a role in acquiring carbapenem resistant A. baumannii isolates in a hospital of Korea [41]. In addition, results from a study in imipenem resistant A. baumannii harboring blaOXA-66/blaOXA-51 genes by Hu et al. suggested that the production of carbapenemase could account for the intrinsic resistance to imipenem, but drug export by an efflux pump might contribute more in the prevalence of imipenem-resistant A. baumannii [19]. These studies and ours clearly suggest that the RND-type efflux systems and oxacillinase may play a synergistic role in multidrug resistance of A. baumannii in this hospital.
5. Conclusions
Collectively, 102 drug-resistant A. baumannii isolates were identified from the ICU ward of the General Hospital of Ningxia Medical University, China. The majority (93/102) of these isolates were MDR strains. Genotyping analysis revealed that 100% (93/93) of the MDR isolates carried carbapenemase genes blaOXA-23/blaOXA-51 but were absent other carbapenemase genes blaOXA-24, blaOXA-58, blaVIM, blaIMP-1, blaIMP-4, blaSIM, blaNDM-1. Importantly, most of the MDR A. baumannii isolates also carried genes of AdeABC efflux pumps, and a presence of efflux pump inhibitor reserpine could significantly enhance their susceptibility to antibiotics (p < 0.01). These clinical drug-resistant isolates could be grouped into nine clusters as determined by a PFGE analysis, and the MDR were mainly in clusters A, B, C, and D. These findings illustrate a challenge of increasing MDR A. baumannii isolates in the ICU ward. The high distribution of multiple genes, mainly the genes of blaOXA-23/blaOXA-51 carbapenemase and RND AdeABC efflux pump contributes to distinct drug-resistant mechanisms, which also indicates an emerging threat in this hospital. Therefore, local molecular detection of genes accounting for drug resistance in a hospital ward, such as the ICU is essential to limit the spread of nosocomial infections caused MDR A. baumannii. This result thus may be useful for developing an effective guidance to prevent nosocomial infections caused by A. baumannii in this hospital.
Acknowledgments
This work was supported by grants from the Ningxia Key Laboratory of Clinical and Pathogenic Microbiology for Wei Jia and Jun Wei.
List of abbreviations
- A. baumannii
Acinetobacter baumannii
- Ade
Acinetobacter drug efflux
- ICU
Intensive care unit
- IMP
metallo-β-lactamase resistant to imipenem
- MDR
multidrug-resistant
- NDM
New Delhi metallo-β-lactamase
- MIC
Minimal inhibitory concentration
- OXA
oxacillinase
- PCR
polymerase chain reaction
- PFGE
pulsed-field gel electrophoresis
- SIM
seoul imipeneamase
- VIM
verona integron encoded metallo-β-lactamase
Author Contributions
Wei Jia, Jun Wei and Xiaoming Liu conceived and designed the experiments; Jun Wei, Caiyun Li, and Xiaoming Liu analyzed the data and drafted the manuscript; Caiyun Li, Haiyun Zhang, and Gang Li performed experiments and acquired data; Wei Jia and Gang Li collected samples; Jun Wei and Xiaoming Liu interpreted data and critically revised the manuscript. All authors read and approved the final version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai X.T., Sun F.J., Chen Z.H., Luo G.M., Feng W., Xiong W., Xia P.Y. The epidemiology and resistance mechanisms of Acinetobacter baumannii isolates from the respiratory department ICU of a hospital in China. Microb. Drug Resist. 2014;20:618–622. doi: 10.1089/mdr.2014.0005. [DOI] [PubMed] [Google Scholar]
- 3.Hong S.B., Shin K.S., Ha J., Han K. Co-existence of blaOXA-23 and armA in multidrug-resistant Acinetobacter baumannii isolated from a hospital in South Korea. J. Med. Microbiol. 2013;62:836–844. doi: 10.1099/jmm.0.055384-0. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y., Kim Y.R., Kim J., Park Y.J., Song W., Shin J.H., Uh Y., Lee K., Lee S.H., Cho J.H., et al. Increasing prevalence of blaOXA-23-carrying Acinetobacter baumannii and the emergence of blaOXA-182-carrying Acinetobacter nosocomialis in Korea. Diagn. Microbiol. Infect. Dis. 2013;77:160–163. doi: 10.1016/j.diagmicrobio.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Sung J.Y., Koo S.H., Cho H.H., Kwon K.C. Nosocomial infection by sequence type 357 multidrug-resistant Acinetobacter baumannii isolates in a neonatal intensive care unit in Daejeon, Korea. Ann Lab Med. 2013;33:279–282. doi: 10.3343/alm.2013.33.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lean S.S., Suhaili Z., Ismail S., Rahman N.I., Othman N., Abdullah F.H., Jusoh Z., Yeo C.C., Thong K.L. Prevalence and genetic characterization of carbapenem- and polymyxin-resistant Acinetobacter baumannii isolated from a tertiary hospital in Terengganu, Malaysia. ISRN Microbiol. 2014 doi: 10.1155/2014/953417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hasan B., Perveen K., Olsen B., Zahra R. Emergence of carbapenem-resistant Acinetobacter baumannii in hospitals in Pakistan. J. Med. Microbiol. 2014;63:50–55. doi: 10.1099/jmm.0.063925-0. [DOI] [PubMed] [Google Scholar]
- 8.Peymani A., Higgins P.G., Nahaei M.R., Farajnia S., Seifert H. Characterisation and clonal dissemination of OXA-23-producing Acinetobacter baumannii in Tabriz, northwest Iran. Int. J. Antimicrob. Agents. 2012;39:526–528. doi: 10.1016/j.ijantimicag.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Aly M., Tayeb H.T., Al Johani S.M., Alyamani E.J., Aldughaishem F., Alabdulkarim I., Balkhy H.H. Genetic diversity of OXA-51-like genes among multidrug-resistant Acinetobacter baumannii in Riyadh, Saudi Arabia. Euro. J. Clin. Microbiol. Infect. Dis. 2014;33:1223–1228. doi: 10.1007/s10096-014-2068-0. [DOI] [PubMed] [Google Scholar]
- 10.Lee M.H., Chen T.L., Lee Y.T., Huang L., Kuo S.C., Yu K.W., Hsueh P.R., Dou H.Y., Su I.J., Fung C.P. Dissemination of multidrug-resistant Acinetobacter baumannii carrying blaOXA-23 from hospitals in central Taiwan. J. Microbiol. Immunol. Infect. 2013;46:419–424. doi: 10.1016/j.jmii.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Apisarnthanarak A., Pinitchai U., Warachan B., Warren D.K., Khawcharoenporn T., Hayden M.K. Effectiveness of infection prevention measures featuring advanced source control and environmental cleaning to limit transmission of extremely-drug resistant Acinetobacter baumannii in a Thai intensive care unit: An analysis before and after extensive flooding. Am. J. Infect. Control. 2014;42:116–121. doi: 10.1016/j.ajic.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 12.Xiao Y., Wei Z., Shen P., Ji J., Sun Z., Yu H., Zhang T., Ji P., Ni Y., Hu Z., et al. Bacterial-resistance among outpatients of county hospitals in China: Significant geographic distinctions and minor differences between central cities. Microbes Infect. 2015 doi: 10.1016/j.micinf.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Perez F., Hujer A.M., Hujer K.M., Decker B.K., Rather P.N., Bonomo R.A. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007;51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X.Z., Nikaido H. Efflux-mediated drug resistance in bacteria: An update. Drugs. 2009;69:1555–1623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rumbo C., Gato E., Lopez M., Ruiz de Alegria C., Fernandez-Cuenca F., Martinez-Martinez L., Vila J., Pachon J., Cisneros J.M., Rodriguez-Bano J., et al. Contribution of efflux pumps, porins, and beta-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 2013;57:5247–5257. doi: 10.1128/AAC.00730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siroy A., Cosette P., Seyer D., Lemaitre-Guillier C., Vallenet D., Van Dorsselaer A., Boyer-Mariotte S., Jouenne T., De E. Global comparison of the membrane subproteomes between a multidrug-resistant Acinetobacter baumannii strain and a reference strain. J. Proteome Res. 2006;5:3385–3398. doi: 10.1021/pr060372s. [DOI] [PubMed] [Google Scholar]
- 17.Toledo P.V.M., Arend L.N., Pilonetto M., Costa Oliveira J.C., Luhm K.R. Surveillance programme for multidrug-resistant bacteria in healthcare-associated infections: An urban perspective in South Brazil. J. Hosp. Infect. 2012;80:351–353. doi: 10.1016/j.jhin.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Dolejska M., Villa L., Poirel L., Nordmann P., Carattoli A. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16s RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J. Antimicrob. Chemother. 2013;68:34–39. doi: 10.1093/jac/dks357. [DOI] [PubMed] [Google Scholar]
- 19.Hu W.S., Yao S.M., Fung C.P., Hsieh Y.P., Liu C.P., Lin J.F. An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2007;51:3844–3852. doi: 10.1128/AAC.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koh T.H., Sng L.H., Wang G.C., Hsu L.Y., Zhao Y. Carbapenemase and efflux pump genes in Acinetobacter calcoaceticus—Acinetobacter baumannii complex strains from Singapore. J. Antimicrob. Chemother. 2007;60:1173–1174. doi: 10.1093/jac/dkm334. [DOI] [PubMed] [Google Scholar]
- 21.Seecoomar G.D., Marmol B.C., Kwon D.H. Promoter deletions of Klebsiella pneumoniae carbapenemase (KPC)-encoding genes (blaKPC-2) and efflux pump (AcrAB) on β-lactam susceptibility in KPC-producing Enterobacteriaceae. FEMS Microbiol. Lett. 2013;348:120–126. doi: 10.1111/1574-6968.12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute . Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard-9th Edition. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2012. [Google Scholar]
- 23.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2014. [Google Scholar]
- 24.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 25.Shi W.F., Jiang J.P., Xu N., Huang Z.M., Wang Y.Y. Inhibitory effects of reserpine and carbonyl cyanide m-chloro-phenylhydrazone on fluoroquinolone resistance of Acinetobacter baumannii. Chin. Med. J. 2005;118:340–343. [PubMed] [Google Scholar]
- 26.Jia W., Li G., Wang W. Prevalence and antimicrobial resistance of Enterococcus species: A hospital-based study in China. Int. J. Environ. Res. Public Health. 2014;11:3424–3442. doi: 10.3390/ijerph110303424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu H., Manos J. Pulsed-field gel electrophoresis of Pseudomonas aeruginosa. Methods Mol. Biol. 2015;1301:157–170. doi: 10.1007/978-1-4939-2599-5_14. [DOI] [PubMed] [Google Scholar]
- 28.Seifert H., Dolzani L., Bressan R., van der Reijden T., van Strijen B., Stefanik D., Heersma H., Dijkshoorn L. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 2005;43:4328–4335. doi: 10.1128/JCM.43.9.4328-4335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh T.R. Emerging carbapenemases: A global perspective. Int. J. Antimicrob. Agents. 2010;36:S8–S14. doi: 10.1016/S0924-8579(10)70004-2. [DOI] [PubMed] [Google Scholar]
- 30.Liu L.L., Ji S.J., Ruan Z., Fu Y., Fu Y.Q., Wang Y.F., Yu Y.S. Dissemination of blaOXA-23 in Acinetobacter spp. in China: Main roles of conjugative plasmid pAZJ221 and transposon TN2009. Antimicrob. Agents Chemother. 2015;59:1998–2005. doi: 10.1128/AAC.04574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendes R.E., Bell J.M., Turnidge J.D., Castanheira M., Jones R.N. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: Report from the SENTRY Surveillance Program. J. Antimicrob. Chemother. 2009;63:55–59. doi: 10.1093/jac/dkn434. [DOI] [PubMed] [Google Scholar]
- 32.Chen T.L., Lee Y.T., Kuo S.C., Hsueh P.R., Chang F.Y., Siu L.K., Ko W.C., Fung C.P. Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob. Agents Chemother. 2010;54:4575–4581. doi: 10.1128/AAC.00764-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J., Ruan Z., Feng Y., Fu Y., Jiang Y., Wang H., Yu Y. Species distribution of clinical Acinetobacter isolates revealed by different identification techniques. PloS ONE. 2014;9:e104882. doi: 10.1371/journal.pone.0104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbott I., Cerqueira G.M., Bhuiyan S., Peleg A.Y. Carbapenem resistance in Acinetobacter baumannii: Laboratory challenges, mechanistic insights and therapeutic strategies. Expert Rev. Anti Infect. Ther. 2013;11:395–409. doi: 10.1586/eri.13.21. [DOI] [PubMed] [Google Scholar]
- 35.Novovic K., Mihajlovic S., Vasiljevic Z., Filipic B., Begovic J., Jovcic B. Carbapenem-resistant Acinetobacter baumannii from Serbia: Revision of CarO classification. PloS ONE. 2015;10:e0122793. doi: 10.1371/journal.pone.0122793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao S.Y., Jiang D.Y., Xu P.C., Zhang Y.K., Shi H.F., Cao H.L., Wu Q. An investigation of drug-resistant Acinetobacter baumannii infections in a comprehensive hospital of East China. Ann. Clin. Microbiol. Antimicrob. 2015 doi: 10.1186/s12941-015-0066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higgins P.G., Poirel L., Lehmann M., Nordmann P., Seifert H. OXA-143, a novel carbapenem-hydrolyzing class D β-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2009;53:5035–5038. doi: 10.1128/AAC.00856-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coelho J.M., Turton J.F., Kaufmann M.E., Glover J., Woodford N., Warner M., Palepou M.F., Pike R., Pitt T.L., Patel B.C., et al. Occurrence of carbapenem-resistant Acinetobacter baumannii clones at multiple hospitals in London and Southeast England. J. Clin. Microbiol. 2006;44:3623–3627. doi: 10.1128/JCM.00699-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon E.J., Courvalin P., Grillot-Courvalin C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: Major role for adeabc overexpression and adeRS mutations. Antimicrob. Agents Chemother. 2013;57:2989–2995. doi: 10.1128/AAC.02556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou P.F., Chen X.Y., Yan G.F., Wang Y.P., Ying C.M. Study of the correlation of imipenem resistance with efflux pumps AdeABC, AdeIJK, AdeDE and AbeM in clinical isolates of Acinetobacter baumannii. Chemotherapy. 2012;58:152–158. doi: 10.1159/000335599. [DOI] [PubMed] [Google Scholar]
- 41.Lee Y., Yum J.H., Kim C.K., Yong D., Jeon E.H., Jeong S.H., Ahn J.Y., Lee K. Role of OXA-23 and AdeABC efflux pump for acquiring carbapenem resistance in an Acinetobacter baumannii strain carrying the blaOXA-66 gene. Ann. Clin. Lab. Sci. 2010;40:43–48. [PubMed] [Google Scholar]
- 42.Ruzin A., Keeney D., Bradford P.A. AdeABC multidrug efflux pump is associated with decreased susceptibility to tigecycline in Acinetobacter calcoaceticus—Acinetobacter baumannii complex. J. Antimicrob. Chemother. 2007;59:1001–1004. doi: 10.1093/jac/dkm058. [DOI] [PubMed] [Google Scholar]
- 43.Coyne S., Courvalin P., Perichon B. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob. Agents Chemother. 2011;55:947–953. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoon E.J., Nait Chabane Y., Goussard S., Snesrud E., Courvalin P., De E., Grillot-Courvalin C. Contribution of resistance-nodulation-cell division efflux systems to antibiotic resistance and biofilm formation in Acinetobacter baumannii. mBio. 2015 doi: 10.1128/mBio.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]