In contrast to other similar compounds, [FeCl2(C14H30N4)]PF6 is a monomer. Comparison with the mononuclear Fe2+ complex of the same ligand shows that the smaller Fe3+ ion is more fully engulfed by the cavity of the bicyclic ligand. Comparison with the μ-oxo dinuclear complex of an unsubstituted ligand of the same size demonstrates that the methyl groups of 4,11-dimethyl-1,4,8,11-tetraazabicyclo[6.6.2]hexadecane prevent dimerization upon oxidation.
Keywords: crystal structure, macrocycle, cross bridge, iron, cylam
Abstract
The title compound, [FeCl2(C14H30N4)]PF6, contains Fe3+ coordinated by the four nitrogen atoms of an ethylene cross-bridged cyclam macrocycle and two cis chloride ligands in a distorted octahedral environment. In contrast to other similar compounds this is a monomer. Intermolecular C—H⋯Cl interactions exist in the structure between the complex ions. Comparison with the mononuclear Fe2+ complex of the same ligand shows that the smaller Fe3+ ion is more fully engulfed by the cavity of the bicyclic ligand. Comparison with the μ-oxido dinuclear complex of an unsubstituted ligand of the same size demonstrates that the methyl groups of 4,11-dimethyl-1,4,8,11-tetraazabicyclo[6.6.2]hexadecane prevent dimerization upon oxidation.
Chemical context
The tendency for iron complexes to form oxido-bridged iron(III) species and ultimately hydrated iron oxides limits their utility, especially in aqueous media, as functional catalysts based on common ligands (Ortiz de Montellano, 1986 ▸). Even so, iron is one of the predominant metal ions found in biological catalytic systems (Jang et al., 1991 ▸; Wallar & Lipscomp, 1996 ▸; Boyington et al., 1993 ▸). A major feature of numerous synthetic catalysts having nitrogen donors and vacant coordination sites is their propensity to form dimers in which higher valent metal ions are present. One of us has produced iron(II) (Hubin et al., 2000 ▸) and iron(III) (Hubin et al., 2001 ▸) complexes of ethylene cross-bridged tetraazamacrocyclic ligands that are remarkably resistant to oxidative hydrolysis while still having available sites for binding of the metal ion to either a terminal oxidant or substrate. The ability of the complex to remain mononuclear, and thus catalytically useful, appears to hinge on the substitution pattern of the non-bridgehead nitrogen atoms of the bicyclic ligands (Hubin et al., 2001 ▸). Methyl or benzyl substitution results only in mononuclear complexes, even in the M 3+ (Hubin et al., 2001 ▸, 2003 ▸) or M 4+ (Yin et al., 2006 ▸) oxidation state, while oxidation of the unsubstituted-ligand complexes results in μ-oxido iron(III) dimers (Hubin et al., 2003 ▸).
Recently, the iron(II) triflate complex of this same ligand, 4,11-dimethyl-1,4,8,11-tetraazabicyclo[6.6.2]hexadecane, has been investigated as a catalyst for olefin oxidation by H2O2 and was found to be an active catalyst with reactivity properties similar to [Fe(TPA)(OTf)2] [TPA is tris(2-pyridylmethyl)amine; OTf is trifluoromethanesulfonate; Feng et al., 2011 ▸]. A key result of this study was that the location of two available cis binding sites on the metal ion is crucial for maximum catalytic activity. Very recently an FeIV analogue has been reported, but no crystal structure data were given (England et al., 2015 ▸).
Structural commentary
The asymmetric unit of the title compound contains one complete Fe3+ mononuclear cross-bridged cyclam complex and a single PF6 − anion. The metal is hexacoordinate in the so-called cis-V geometry common for macrocycles of this type. It is coordinated by four nitrogen atoms of the macrocycle and two cis chloride ions, as shown in Fig. 1 ▸. The Fe—Cl bond lengths are similar to those of other comparable Fe3+ complexes. The relatively long Fe—N bonds strongly suggest the Fe3+ present is in a high-spin configuration.
Figure 1.
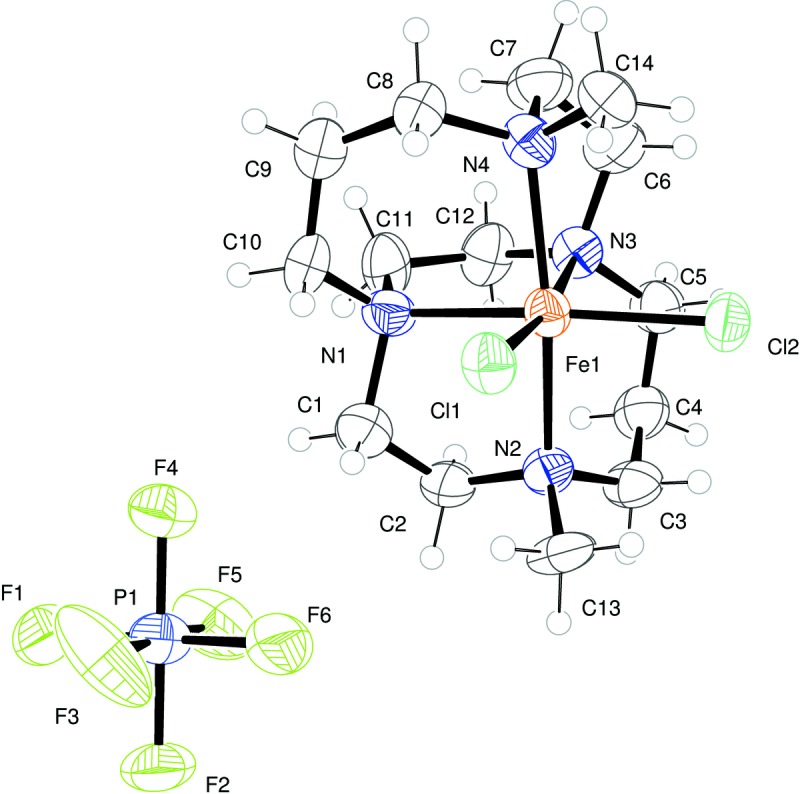
ORTEP representation of the asymmetric unit with atoms drawn as 50% probability ellipsoids.
The Fe3+ resides within a pocket in the rigid macrocycle, slightly displaced from the centre. The N2—Fe1—N4 bond angle is 166.8 (3) Å and the N1—Fe1—N3 bite angle is 79.8 (3) Å.
Comparison with related structures
Structural characterization of an Fe3+ mononuclear cross-bridged cyclam complex has not been achieved prior to the present study. In the present case, even upon oxidation of the iron from Fe(II) to Fe(III), the methyl-substituted ligand does not allow dimerization to occur. We will now compare the observed structure with that of the lower valent analogue and to the iron(III) μ-oxido dimer of the unsubstituted analogue.
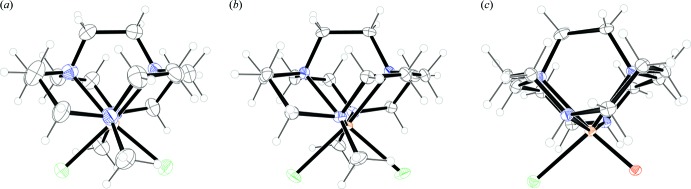
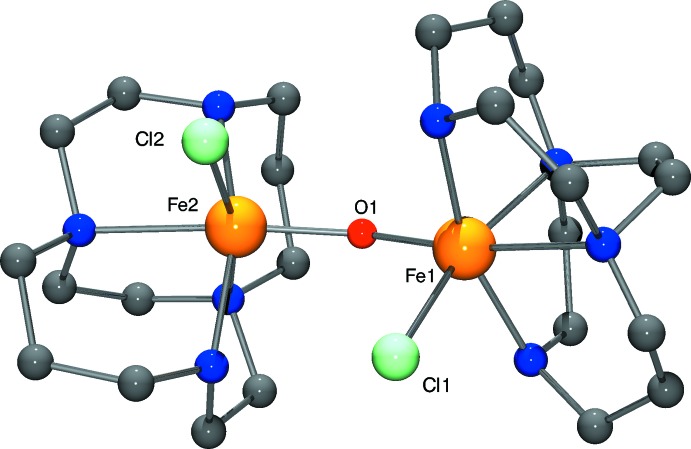
From a comparison of the Fe3+ 4,11-dimethyl-1,4,8,11-tetraazabicyclo[6.6.2]hexaadecane dichloride complex hexafluoridophosphate with the Fe2+ analogue, the reduction in ionic radius of the iron ion upon oxidation is clear (Table 1 ▸). Nax—Fe3+—Nax is 166.8 (3)° in the present structure, while Nax—Fe2+—Nax is 161.88 (5)° in the reduced complex (Hubin et al., 2000 ▸). The smaller Fe3+ ion is pulled further into the ligand cavity as the favored octahedral geometry is approached, as can be seen by viewing each complex down the Nax—Fen+—Nax axis (Fig. 2 ▸). Fe—N bond lengths are also affected, going from a mean of 2.255 Å in the Fe2+ complex, to 2.209 Å in the Fe3+ complex. Comparison of the present Fe3+ monomer with the μ-oxido dimer complex is also informative. The crystal structure of the dimeric Fe3+ complex (Hubin, 2003 ▸) is represented in Fig. 3 ▸. The Fe3+ ion of this complex is also found in a pseudo-octahedral, six-coordinate geometry. Usually, these dimers contain five-coordinate metal cations, although dimers with six- and seven-coordinate cations are known (Murray, 1974 ▸). However, one monodentate chlorido ligand is maintained in this structure. Since the macrocyclic ligand is uncharged, the attractive Coulombic forces between the halide and the Fe3+ ion may be enough to keep it bound. Also, the folded ligand conformation helps separate the ligands from each other, easing the steric interactions that might favor lower coordination numbers with more nearly planar ligands. The secondary amine/Fe3+ bond lengths in the dimer are somewhat shorter than the tertiary amine/Fe3+ bond lengths: the Fe—N(secondary) mean length is 2.153 Å while the Fe—N(tertiary) mean length is 2.239 Å. In the monomer, with all tertiary amines, the mean Fe—N bond length is 2.209 Å, shorter but not quite matching that of the shortest, secondary amine bonds in the dimer. The Nax—Fe—Nax mean bond angle is 161.6° in the dimer, while this value is 166.85 (19)° in the monomer. Clearly, dimerization, and its associated steric consequences, pulls the Fe3+ ion further out of the ligand cavity than it is in the Fe3+ monomer. In fact, the dimer Nax—Fe—Nax bond angle is much closer to that of the Fe2+ monomer at 161.88 (5)° than that of the Fe3+ monomer at 166.85 (19)°. This steric consequence is consistent with the observation that the more sterically demanding methyl-substituted ligand prevents dimerization altogether.
Table 1. Geometric parameters (, ) for the macrocyclic cavity in Fe2+ and Fe3+ macrocyclesa .
| Parameter | Fe3+Me2 Lb | Fe2+Me2 Lc | Fe3+H2 L dimerd |
|---|---|---|---|
| FeN1 | 2.195(5) | 2.2574(13) | 2.151 |
| FeN2 | 2.229(5) | 2.2866(14) | 2.155 |
| FeN3 | 2.220(5) | 2.2634(13) | 2.234 |
| FeN4 | 2.190(5) | 2.2748(13) | 2.245 |
| N2axFeN4ax | 166.8(3) | 161.88(5) | 161.6 |
| N1eqFeN3eq | 79.8(3) | 78.36(5) | 78.9 |
Figure 2.
Comparison of Fe2+ complex (Hubin et al., 2000 ▸) labelled (a), with Fe3+ monomer complex (b), and the one half of the dimer complex (Hubin et al., 2003 ▸) (c), in each case viewed perpendicular to the Cl–Fe–Cl or Cl–Fe–O plane. Atoms are drawn as 50% probability ellipsoids.
Figure 3.
Molecular structure of μ-oxidobis[chlorido(1,4,8,11-tetraazabicyclo[6.6.2]hexadecane)iron(III)] (Hubin et al., 2003 ▸).
Supramolecular features
There are no classical hydrogen bonds within the structure, but many C—H⋯Cl and C—H⋯F intermolecular interactions exist (Table 2 ▸). Pairs of complexes form dimers sustained by C—H⋯Cl interactions (H⋯Cl distances lie in the range 2.76 to 2.97 Å) and further C—H⋯Cl interactions link these into tapes that run parallel to the b-axis. These tapes are stacked along the a and c axes. Between them lie PF6 − anions, forming C—H⋯F interactions to generate a three-dimensional array of intermolecular contacts.
Table 2. Hydrogen-bond geometry (, ).
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| C1H1AF4 | 0.99 | 2.59 | 3.244(13) | 124 |
| C2H2AF1i | 0.99 | 2.36 | 3.249(12) | 148 |
| C3H3BCl2 | 0.99 | 2.73 | 3.304(11) | 117 |
| C5H5BCl2 | 0.99 | 2.67 | 3.277(11) | 120 |
| C6H6BCl2 | 0.99 | 2.91 | 3.453(10) | 115 |
| C8H8ACl1 | 0.99 | 2.78 | 3.337(10) | 116 |
| C8H8BCl2ii | 0.99 | 2.78 | 3.726(11) | 159 |
| C9H9ACl2iii | 0.99 | 2.76 | 3.571(12) | 140 |
| C10H10ACl1 | 0.99 | 2.69 | 3.311(11) | 121 |
| C11H11AF2iv | 0.99 | 2.51 | 3.097(13) | 118 |
| C11H11BF5i | 0.99 | 2.41 | 3.360(15) | 161 |
| C13H13ACl1 | 0.98 | 2.63 | 3.198(10) | 117 |
| C13H13BF1v | 0.98 | 2.59 | 3.323(11) | 131 |
| C14H14ACl1 | 0.98 | 2.97 | 3.554(10) | 119 |
| C14H14BCl1vi | 0.98 | 2.89 | 3.772(11) | 150 |
| C14H14BCl2 | 0.98 | 2.76 | 3.265(11) | 113 |
Symmetry codes: (i) 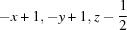 ; (ii)
; (ii) 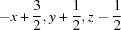 ; (iii)
; (iii) 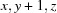 ; (iv)
; (iv) 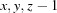 ; (v)
; (v)  ; (vi)
; (vi) 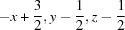 .
.
Database survey
For coordination chemistry of cross-bridged tetraazamacrocycle derivatives and their applications, see: Hubin (2003 ▸); Jones et al. (2015 ▸); Springborg (2003 ▸); Wong et al. (2000 ▸). For related structures involving iron complexes of 4,11-dimethyl-1,4,8,11-tetraazabicyclo[6.6.2]hexadecane derivatives, see: Hubin et al. (2000 ▸, 2001 ▸, 2003 ▸); McClain et al. (2006 ▸); Feng et al. (2011 ▸).
Synthesis and crystallization
The title complex was prepared by a procedure slightly modified from those found in Hubin et al. (2000 ▸, 2001 ▸). In an inert atmosphere glovebox, 0.381 g (0.001 mol) of the iron(II) dichloride complex of 4,11-dimethyl-1,4,8,11-tetraazabicyclo[6.6.2]hexadecane (Hubin et al., 2000 ▸) was dissolved in 20 ml of methanol in a round-bottom flask. Five equivalents of NH4PF6 (0.005 mol, 0.815 g) were dissolved in the solution. The flask was removed from the glovebox with a stopper to protect it from air. In a fume hood, a stream of nitrogen gas was directed over the surface of the solution. Four to six drops of Br2 were added and the reaction was stirred for 15 minutes. Care must be taken when adding the bromine drops, as its vapor pressure and density tend to cause it to spurt out of the pipette. Practicing dispensing drops back into the bromine bottle (in the hood) can allow for successful dispensing.
A bright yellow–orange precipitate formed immediately. The nitrogen gas was then allowed to bubble through the solution for 15 minutes to remove excess Br2. The flask was then stoppered and placed in a freezer for 30 minutes to complete the precipitation. The yellow–orange solid product was collected by vacuum filtration on a glass frit and washed with methanol and then ether. The product (0.428 g, 80% yield) was analytically pure as calculated with one-half molar equivalent of water of crystallization. Crystals suitable for X-ray diffraction were grown from ether diffusion into a dichloromethane solution.
Refinement
Standard data collection and refinement procedures were adopted. Crystal data, data collection and structure refinement details are summarized in Table 3 ▸.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | [FeCl2(C14H30N4)]PF6 |
| M r | 526.14 |
| Crystal system, space group | Orthorhombic, P n a21 |
| Temperature (K) | 150 |
| a, b, c () | 26.002(4), 8.5752(15), 9.3829(16) |
| V (3) | 2092.1(6) |
| Z | 4 |
| Radiation type | Mo K |
| (mm1) | 1.11 |
| Crystal size (mm) | 0.12 0.09 0.07 |
| Data collection | |
| Diffractometer | Stoe IPDS2 |
| No. of measured, independent and observed [I > 2(I)] reflections | 21858, 4186, 2249 |
| R int | 0.143 |
| (sin /)max (1) | 0.620 |
| Refinement | |
| R[F 2 > 2(F 2)], wR(F 2), S | 0.050, 0.109, 0.84 |
| No. of reflections | 4186 |
| No. of parameters | 254 |
| No. of restraints | 1 |
| H-atom treatment | H-atom parameters constrained |
| max, min (e 3) | 0.39, 0.33 |
| Absolute structure | Refined as a two-component inversion twin |
| Absolute structure parameter | 0.03(5) |
Hydrogen atoms were placed using a riding model with fixed bond lengths and angles. For methylene groups U iso (H) was set at 1.2U iso(C) and for methyl groups U iso (H) was set at 1.5U iso(C).
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015015340/zl2640sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015015340/zl2640Isup2.hkl
CCDC reference: 1419250
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| [FeCl2(C14H30N4)]PF6 | Dx = 1.670 Mg m−3 |
| Mr = 526.14 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pna21 | Cell parameters from 8849 reflections |
| a = 26.002 (4) Å | θ = 1.7–24.1° |
| b = 8.5752 (15) Å | µ = 1.11 mm−1 |
| c = 9.3829 (16) Å | T = 150 K |
| V = 2092.1 (6) Å3 | Block, orange |
| Z = 4 | 0.12 × 0.09 × 0.07 mm |
| F(000) = 1084 |
Data collection
| Stoe IPDS2 diffractometer | 2249 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.143 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 26.1°, θmin = 1.6° |
| ω–scans | h = −32→32 |
| 21858 measured reflections | k = −10→8 |
| 4186 independent reflections | l = −11→11 |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.050 | H-atom parameters constrained |
| wR(F2) = 0.109 | w = 1/[σ2(Fo2) + (0.0378P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 0.84 | (Δ/σ)max = 0.001 |
| 4186 reflections | Δρmax = 0.39 e Å−3 |
| 254 parameters | Δρmin = −0.33 e Å−3 |
| 1 restraint | Absolute structure: Refined as a two-component inversion twin |
| Primary atom site location: structure-invariant direct methods | Absolute structure parameter: 0.03 (5) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refined as a two-component inversion twin. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Fe1 | 0.66232 (4) | 0.14982 (13) | 0.20386 (14) | 0.0394 (3) | |
| Cl1 | 0.71279 (10) | 0.2523 (4) | 0.3803 (3) | 0.0471 (6) | |
| Cl2 | 0.69285 (9) | −0.0987 (3) | 0.2182 (3) | 0.0493 (6) | |
| N1 | 0.6182 (3) | 0.3664 (9) | 0.1775 (7) | 0.0479 (19) | |
| N2 | 0.5950 (3) | 0.0889 (10) | 0.3401 (7) | 0.043 (2) | |
| N3 | 0.6144 (3) | 0.0869 (10) | 0.0162 (8) | 0.042 (2) | |
| N4 | 0.7148 (3) | 0.2294 (12) | 0.0366 (8) | 0.045 (2) | |
| C1 | 0.5844 (5) | 0.3692 (15) | 0.3148 (11) | 0.058 (3) | |
| H1A | 0.6066 | 0.3876 | 0.3990 | 0.069* | |
| H1B | 0.5593 | 0.4558 | 0.3087 | 0.069* | |
| C2 | 0.5569 (4) | 0.2208 (13) | 0.3320 (11) | 0.057 (3) | |
| H2A | 0.5335 | 0.2044 | 0.2502 | 0.069* | |
| H2B | 0.5360 | 0.2236 | 0.4201 | 0.069* | |
| C3 | 0.5698 (4) | −0.0588 (14) | 0.2979 (10) | 0.056 (3) | |
| H3A | 0.5409 | −0.0765 | 0.3645 | 0.067* | |
| H3B | 0.5949 | −0.1440 | 0.3138 | 0.067* | |
| C4 | 0.5490 (4) | −0.0768 (14) | 0.1479 (10) | 0.059 (3) | |
| H4A | 0.5320 | −0.1799 | 0.1407 | 0.071* | |
| H4B | 0.5222 | 0.0036 | 0.1328 | 0.071* | |
| C5 | 0.5867 (4) | −0.0642 (13) | 0.0324 (11) | 0.055 (3) | |
| H5A | 0.5689 | −0.0866 | −0.0585 | 0.066* | |
| H5B | 0.6128 | −0.1471 | 0.0463 | 0.066* | |
| C6 | 0.6538 (4) | 0.0671 (13) | −0.0993 (10) | 0.060 (3) | |
| H6A | 0.6364 | 0.0548 | −0.1924 | 0.072* | |
| H6B | 0.6743 | −0.0283 | −0.0810 | 0.072* | |
| C7 | 0.6900 (5) | 0.2105 (14) | −0.1046 (11) | 0.067 (3) | |
| H7A | 0.7166 | 0.1952 | −0.1789 | 0.081* | |
| H7B | 0.6701 | 0.3053 | −0.1286 | 0.081* | |
| C8 | 0.7315 (4) | 0.4001 (13) | 0.0555 (11) | 0.059 (3) | |
| H8A | 0.7520 | 0.4078 | 0.1442 | 0.071* | |
| H8B | 0.7545 | 0.4280 | −0.0247 | 0.071* | |
| C9 | 0.6882 (5) | 0.5214 (14) | 0.0628 (13) | 0.073 (4) | |
| H9A | 0.7043 | 0.6258 | 0.0699 | 0.088* | |
| H9B | 0.6693 | 0.5179 | −0.0287 | 0.088* | |
| C10 | 0.6499 (4) | 0.5070 (11) | 0.1805 (13) | 0.067 (3) | |
| H10A | 0.6685 | 0.5103 | 0.2724 | 0.080* | |
| H10B | 0.6269 | 0.5990 | 0.1773 | 0.080* | |
| C11 | 0.5873 (5) | 0.3684 (14) | 0.0505 (12) | 0.064 (4) | |
| H11A | 0.6044 | 0.4357 | −0.0209 | 0.077* | |
| H11B | 0.5537 | 0.4166 | 0.0735 | 0.077* | |
| C12 | 0.5775 (5) | 0.2089 (12) | −0.0168 (11) | 0.060 (3) | |
| H12A | 0.5430 | 0.1728 | 0.0135 | 0.072* | |
| H12B | 0.5765 | 0.2222 | −0.1216 | 0.072* | |
| C13 | 0.6132 (4) | 0.0676 (13) | 0.4876 (9) | 0.058 (3) | |
| H13A | 0.6302 | 0.1632 | 0.5200 | 0.087* | |
| H13B | 0.5838 | 0.0450 | 0.5497 | 0.087* | |
| H13C | 0.6376 | −0.0194 | 0.4911 | 0.087* | |
| C14 | 0.7641 (4) | 0.1488 (14) | 0.0423 (11) | 0.058 (3) | |
| H14A | 0.7795 | 0.1633 | 0.1366 | 0.088* | |
| H14B | 0.7588 | 0.0373 | 0.0246 | 0.088* | |
| H14C | 0.7871 | 0.1917 | −0.0306 | 0.088* | |
| P1 | 0.58082 (9) | 0.6220 (4) | 0.6714 (2) | 0.0514 (7) | |
| F1 | 0.5510 (2) | 0.7847 (7) | 0.6576 (6) | 0.0737 (19) | |
| F2 | 0.5482 (3) | 0.5900 (10) | 0.8133 (7) | 0.097 (3) | |
| F3 | 0.6235 (3) | 0.7031 (11) | 0.7626 (8) | 0.117 (3) | |
| F4 | 0.6122 (2) | 0.6531 (8) | 0.5300 (6) | 0.0635 (17) | |
| F5 | 0.5377 (3) | 0.5381 (11) | 0.5818 (8) | 0.094 (3) | |
| F6 | 0.6097 (3) | 0.4592 (7) | 0.6862 (8) | 0.094 (2) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Fe1 | 0.0415 (6) | 0.0379 (6) | 0.0388 (6) | 0.0011 (6) | −0.0025 (7) | −0.0031 (8) |
| Cl1 | 0.0505 (14) | 0.0455 (15) | 0.0452 (12) | −0.0004 (14) | −0.0107 (12) | −0.0074 (12) |
| Cl2 | 0.0602 (13) | 0.0356 (11) | 0.0522 (13) | 0.0070 (10) | −0.0035 (13) | 0.0016 (13) |
| N1 | 0.053 (4) | 0.054 (5) | 0.036 (5) | 0.008 (4) | 0.005 (4) | 0.003 (4) |
| N2 | 0.048 (5) | 0.045 (5) | 0.037 (4) | 0.002 (4) | −0.002 (4) | 0.003 (4) |
| N3 | 0.039 (5) | 0.045 (5) | 0.042 (4) | −0.006 (4) | −0.009 (4) | −0.004 (4) |
| N4 | 0.041 (5) | 0.048 (6) | 0.047 (4) | −0.001 (5) | −0.002 (4) | −0.009 (4) |
| C1 | 0.066 (8) | 0.055 (9) | 0.052 (6) | −0.002 (7) | 0.001 (5) | −0.006 (6) |
| C2 | 0.054 (7) | 0.059 (8) | 0.058 (7) | 0.005 (6) | 0.024 (5) | 0.011 (6) |
| C3 | 0.061 (8) | 0.056 (8) | 0.049 (6) | −0.022 (6) | 0.012 (5) | −0.002 (5) |
| C4 | 0.067 (7) | 0.052 (7) | 0.058 (6) | −0.012 (6) | −0.004 (6) | −0.002 (5) |
| C5 | 0.059 (7) | 0.055 (8) | 0.051 (6) | −0.007 (6) | 0.002 (6) | −0.008 (5) |
| C6 | 0.072 (8) | 0.067 (8) | 0.040 (5) | −0.008 (6) | −0.009 (5) | 0.000 (5) |
| C7 | 0.086 (8) | 0.070 (8) | 0.046 (6) | −0.011 (6) | 0.007 (6) | 0.004 (6) |
| C8 | 0.076 (8) | 0.044 (7) | 0.058 (6) | 0.003 (6) | 0.007 (5) | 0.006 (5) |
| C9 | 0.086 (9) | 0.047 (7) | 0.086 (8) | 0.002 (7) | 0.032 (7) | 0.002 (6) |
| C10 | 0.092 (8) | 0.030 (5) | 0.079 (9) | −0.005 (5) | 0.005 (8) | 0.007 (6) |
| C11 | 0.067 (9) | 0.052 (9) | 0.073 (7) | 0.025 (7) | −0.008 (6) | 0.003 (6) |
| C12 | 0.068 (7) | 0.053 (8) | 0.059 (6) | −0.004 (6) | −0.022 (6) | 0.004 (6) |
| C13 | 0.071 (8) | 0.068 (8) | 0.035 (5) | −0.021 (6) | 0.007 (5) | 0.014 (5) |
| C14 | 0.053 (7) | 0.059 (8) | 0.064 (6) | 0.010 (6) | 0.013 (5) | −0.003 (6) |
| P1 | 0.0442 (14) | 0.0654 (18) | 0.0445 (16) | 0.0126 (13) | 0.0024 (11) | 0.0086 (13) |
| F1 | 0.069 (4) | 0.063 (4) | 0.090 (5) | 0.024 (3) | 0.029 (3) | 0.022 (3) |
| F2 | 0.123 (7) | 0.094 (6) | 0.075 (4) | 0.050 (5) | 0.058 (4) | 0.038 (4) |
| F3 | 0.073 (5) | 0.171 (9) | 0.107 (5) | 0.025 (5) | −0.027 (4) | −0.090 (5) |
| F4 | 0.060 (4) | 0.077 (5) | 0.054 (3) | 0.004 (3) | 0.012 (3) | 0.000 (3) |
| F5 | 0.054 (4) | 0.131 (8) | 0.097 (5) | −0.021 (5) | 0.017 (4) | −0.032 (5) |
| F6 | 0.123 (5) | 0.081 (5) | 0.079 (4) | 0.046 (4) | 0.038 (5) | 0.019 (4) |
Geometric parameters (Å, º)
| Fe1—N4 | 2.188 (8) | C6—C7 | 1.549 (14) |
| Fe1—N1 | 2.197 (7) | C6—H6A | 0.9900 |
| Fe1—N3 | 2.223 (7) | C6—H6B | 0.9900 |
| Fe1—N2 | 2.229 (8) | C7—H7A | 0.9900 |
| Fe1—Cl2 | 2.278 (3) | C7—H7B | 0.9900 |
| Fe1—Cl1 | 2.288 (3) | C8—C9 | 1.534 (15) |
| N1—C11 | 1.437 (13) | C8—H8A | 0.9900 |
| N1—C10 | 1.461 (11) | C8—H8B | 0.9900 |
| N1—C1 | 1.559 (12) | C9—C10 | 1.493 (15) |
| N2—C13 | 1.473 (12) | C9—H9A | 0.9900 |
| N2—C3 | 1.480 (14) | C9—H9B | 0.9900 |
| N2—C2 | 1.506 (13) | C10—H10A | 0.9900 |
| N3—C12 | 1.453 (13) | C10—H10B | 0.9900 |
| N3—C5 | 1.490 (13) | C11—C12 | 1.528 (15) |
| N3—C6 | 1.501 (12) | C11—H11A | 0.9900 |
| N4—C14 | 1.458 (12) | C11—H11B | 0.9900 |
| N4—C7 | 1.482 (13) | C12—H12A | 0.9900 |
| N4—C8 | 1.538 (15) | C12—H12B | 0.9900 |
| C1—C2 | 1.469 (16) | C13—H13A | 0.9800 |
| C1—H1A | 0.9900 | C13—H13B | 0.9800 |
| C1—H1B | 0.9900 | C13—H13C | 0.9800 |
| C2—H2A | 0.9900 | C14—H14A | 0.9800 |
| C2—H2B | 0.9900 | C14—H14B | 0.9800 |
| C3—C4 | 1.516 (13) | C14—H14C | 0.9800 |
| C3—H3A | 0.9900 | P1—F3 | 1.564 (7) |
| C3—H3B | 0.9900 | P1—F5 | 1.575 (8) |
| C4—C5 | 1.467 (14) | P1—F4 | 1.580 (6) |
| C4—H4A | 0.9900 | P1—F6 | 1.591 (6) |
| C4—H4B | 0.9900 | P1—F1 | 1.601 (6) |
| C5—H5A | 0.9900 | P1—F2 | 1.602 (7) |
| C5—H5B | 0.9900 | ||
| N4—Fe1—N1 | 88.9 (3) | N3—C6—C7 | 110.3 (9) |
| N4—Fe1—N3 | 81.8 (3) | N3—C6—H6A | 109.6 |
| N1—Fe1—N3 | 79.8 (3) | C7—C6—H6A | 109.6 |
| N4—Fe1—N2 | 166.8 (3) | N3—C6—H6B | 109.6 |
| N1—Fe1—N2 | 81.5 (3) | C7—C6—H6B | 109.6 |
| N3—Fe1—N2 | 87.6 (3) | H6A—C6—H6B | 108.1 |
| N4—Fe1—Cl2 | 96.7 (3) | N4—C7—C6 | 108.8 (8) |
| N1—Fe1—Cl2 | 168.3 (2) | N4—C7—H7A | 109.9 |
| N3—Fe1—Cl2 | 90.9 (2) | C6—C7—H7A | 109.9 |
| N2—Fe1—Cl2 | 91.2 (2) | N4—C7—H7B | 109.9 |
| N4—Fe1—Cl1 | 92.4 (2) | C6—C7—H7B | 109.9 |
| N1—Fe1—Cl1 | 93.2 (2) | H7A—C7—H7B | 108.3 |
| N3—Fe1—Cl1 | 171.0 (2) | C9—C8—N4 | 116.3 (9) |
| N2—Fe1—Cl1 | 97.2 (2) | C9—C8—H8A | 108.2 |
| Cl2—Fe1—Cl1 | 96.70 (11) | N4—C8—H8A | 108.2 |
| C11—N1—C10 | 108.8 (9) | C9—C8—H8B | 108.2 |
| C11—N1—C1 | 111.8 (7) | N4—C8—H8B | 108.2 |
| C10—N1—C1 | 106.8 (8) | H8A—C8—H8B | 107.4 |
| C11—N1—Fe1 | 113.3 (6) | C10—C9—C8 | 117.9 (10) |
| C10—N1—Fe1 | 113.6 (6) | C10—C9—H9A | 107.8 |
| C1—N1—Fe1 | 102.3 (6) | C8—C9—H9A | 107.8 |
| C13—N2—C3 | 106.7 (8) | C10—C9—H9B | 107.8 |
| C13—N2—C2 | 110.6 (8) | C8—C9—H9B | 107.8 |
| C3—N2—C2 | 109.7 (8) | H9A—C9—H9B | 107.2 |
| C13—N2—Fe1 | 108.4 (6) | N1—C10—C9 | 115.5 (9) |
| C3—N2—Fe1 | 113.3 (6) | N1—C10—H10A | 108.4 |
| C2—N2—Fe1 | 108.2 (6) | C9—C10—H10A | 108.4 |
| C12—N3—C5 | 109.2 (8) | N1—C10—H10B | 108.4 |
| C12—N3—C6 | 112.3 (8) | C9—C10—H10B | 108.4 |
| C5—N3—C6 | 107.7 (8) | H10A—C10—H10B | 107.5 |
| C12—N3—Fe1 | 111.4 (6) | N1—C11—C12 | 115.2 (9) |
| C5—N3—Fe1 | 113.6 (6) | N1—C11—H11A | 108.5 |
| C6—N3—Fe1 | 102.5 (5) | C12—C11—H11A | 108.5 |
| C14—N4—C7 | 111.3 (8) | N1—C11—H11B | 108.5 |
| C14—N4—C8 | 101.4 (8) | C12—C11—H11B | 108.5 |
| C7—N4—C8 | 109.3 (8) | H11A—C11—H11B | 107.5 |
| C14—N4—Fe1 | 112.0 (6) | N3—C12—C11 | 116.5 (9) |
| C7—N4—Fe1 | 109.7 (6) | N3—C12—H12A | 108.2 |
| C8—N4—Fe1 | 113.0 (6) | C11—C12—H12A | 108.2 |
| C2—C1—N1 | 110.6 (9) | N3—C12—H12B | 108.2 |
| C2—C1—H1A | 109.5 | C11—C12—H12B | 108.2 |
| N1—C1—H1A | 109.5 | H12A—C12—H12B | 107.3 |
| C2—C1—H1B | 109.5 | N2—C13—H13A | 109.5 |
| N1—C1—H1B | 109.5 | N2—C13—H13B | 109.5 |
| H1A—C1—H1B | 108.1 | H13A—C13—H13B | 109.5 |
| C1—C2—N2 | 109.6 (9) | N2—C13—H13C | 109.5 |
| C1—C2—H2A | 109.8 | H13A—C13—H13C | 109.5 |
| N2—C2—H2A | 109.8 | H13B—C13—H13C | 109.5 |
| C1—C2—H2B | 109.8 | N4—C14—H14A | 109.5 |
| N2—C2—H2B | 109.8 | N4—C14—H14B | 109.5 |
| H2A—C2—H2B | 108.2 | H14A—C14—H14B | 109.5 |
| N2—C3—C4 | 119.6 (9) | N4—C14—H14C | 109.5 |
| N2—C3—H3A | 107.4 | H14A—C14—H14C | 109.5 |
| C4—C3—H3A | 107.4 | H14B—C14—H14C | 109.5 |
| N2—C3—H3B | 107.4 | F3—P1—F5 | 179.0 (5) |
| C4—C3—H3B | 107.4 | F3—P1—F4 | 91.0 (4) |
| H3A—C3—H3B | 107.0 | F5—P1—F4 | 89.8 (3) |
| C5—C4—C3 | 116.1 (9) | F3—P1—F6 | 90.5 (5) |
| C5—C4—H4A | 108.3 | F5—P1—F6 | 88.9 (5) |
| C3—C4—H4A | 108.3 | F4—P1—F6 | 88.7 (4) |
| C5—C4—H4B | 108.3 | F3—P1—F1 | 90.0 (4) |
| C3—C4—H4B | 108.3 | F5—P1—F1 | 90.6 (4) |
| H4A—C4—H4B | 107.4 | F4—P1—F1 | 92.0 (3) |
| C4—C5—N3 | 117.6 (9) | F6—P1—F1 | 179.2 (4) |
| C4—C5—H5A | 107.9 | F3—P1—F2 | 89.8 (5) |
| N3—C5—H5A | 107.9 | F5—P1—F2 | 89.4 (4) |
| C4—C5—H5B | 107.9 | F4—P1—F2 | 179.2 (4) |
| N3—C5—H5B | 107.9 | F6—P1—F2 | 91.5 (4) |
| H5A—C5—H5B | 107.2 | F1—P1—F2 | 87.8 (3) |
| C11—N1—C1—C2 | 69.7 (11) | C8—N4—C7—C6 | −156.2 (9) |
| C10—N1—C1—C2 | −171.5 (9) | Fe1—N4—C7—C6 | −31.9 (11) |
| Fe1—N1—C1—C2 | −51.9 (10) | N3—C6—C7—N4 | 57.8 (11) |
| N1—C1—C2—N2 | 58.7 (11) | C14—N4—C8—C9 | −176.9 (9) |
| C13—N2—C2—C1 | 85.8 (10) | C7—N4—C8—C9 | 65.5 (11) |
| C3—N2—C2—C1 | −156.8 (8) | Fe1—N4—C8—C9 | −56.9 (10) |
| Fe1—N2—C2—C1 | −32.7 (9) | N4—C8—C9—C10 | 60.6 (14) |
| C13—N2—C3—C4 | −177.2 (10) | C11—N1—C10—C9 | −63.7 (11) |
| C2—N2—C3—C4 | 63.0 (12) | C1—N1—C10—C9 | 175.6 (9) |
| Fe1—N2—C3—C4 | −58.0 (12) | Fe1—N1—C10—C9 | 63.5 (11) |
| N2—C3—C4—C5 | 61.0 (15) | C8—C9—C10—N1 | −64.2 (14) |
| C3—C4—C5—N3 | −62.3 (14) | C10—N1—C11—C12 | 145.2 (10) |
| C12—N3—C5—C4 | −62.7 (11) | C1—N1—C11—C12 | −97.1 (11) |
| C6—N3—C5—C4 | 175.1 (9) | Fe1—N1—C11—C12 | 17.9 (12) |
| Fe1—N3—C5—C4 | 62.3 (11) | C5—N3—C12—C11 | 142.4 (10) |
| C12—N3—C6—C7 | 69.2 (10) | C6—N3—C12—C11 | −98.1 (11) |
| C5—N3—C6—C7 | −170.6 (8) | Fe1—N3—C12—C11 | 16.2 (12) |
| Fe1—N3—C6—C7 | −50.5 (9) | N1—C11—C12—N3 | −23.2 (15) |
| C14—N4—C7—C6 | 92.6 (11) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1A···F4 | 0.99 | 2.59 | 3.244 (13) | 124 |
| C2—H2A···F1i | 0.99 | 2.36 | 3.249 (12) | 148 |
| C3—H3B···Cl2 | 0.99 | 2.73 | 3.304 (11) | 117 |
| C5—H5B···Cl2 | 0.99 | 2.67 | 3.277 (11) | 120 |
| C6—H6B···Cl2 | 0.99 | 2.91 | 3.453 (10) | 115 |
| C8—H8A···Cl1 | 0.99 | 2.78 | 3.337 (10) | 116 |
| C8—H8B···Cl2ii | 0.99 | 2.78 | 3.726 (11) | 159 |
| C9—H9A···Cl2iii | 0.99 | 2.76 | 3.571 (12) | 140 |
| C10—H10A···Cl1 | 0.99 | 2.69 | 3.311 (11) | 121 |
| C11—H11A···F2iv | 0.99 | 2.51 | 3.097 (13) | 118 |
| C11—H11B···F5i | 0.99 | 2.41 | 3.360 (15) | 161 |
| C13—H13A···Cl1 | 0.98 | 2.63 | 3.198 (10) | 117 |
| C13—H13B···F1v | 0.98 | 2.59 | 3.323 (11) | 131 |
| C14—H14A···Cl1 | 0.98 | 2.97 | 3.554 (10) | 119 |
| C14—H14B···Cl1vi | 0.98 | 2.89 | 3.772 (11) | 150 |
| C14—H14B···Cl2 | 0.98 | 2.76 | 3.265 (11) | 113 |
Symmetry codes: (i) −x+1, −y+1, z−1/2; (ii) −x+3/2, y+1/2, z−1/2; (iii) x, y+1, z; (iv) x, y, z−1; (v) x, y−1, z; (vi) −x+3/2, y−1/2, z−1/2.
References
- Boyington, J. C., Gaffney, B. J. & Amzel, L. M. (1993). Science, 260, 1482–1486. [DOI] [PubMed]
- England, J., Prakash, J., Cranswick, M. A., Mandal, D., Guo, Y., Münck, E., Shaik, S. & Que, L. Jr (2015). Inorg. Chem. 54, 7828–7839. [DOI] [PubMed]
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Feng, Y., England, J. & Que, L. Jr (2011). ACS Catal. 1, 1035–1042.
- Hubin, T. J. (2003). Coord. Chem. Rev. 241, 27–46.
- Hubin, T. J., McCormick, J. M., Alcock, N. W. & Busch, D. H. (2001). Inorg. Chem. 40, 435–444. [DOI] [PubMed]
- Hubin, T. J., McCormick, J. M., Collinson, S. R., Alcock, N. W., Clase, H. J. & Busch, D. H. (2003). Inorg. Chim. Acta, 346, 76–86.
- Hubin, T. J., McCormick, J. M., Collinson, S. R., Buchalova, M., Perkins, C. M., Alcock, N. W., Kahol, P. K., Raghunathan, A. & Busch, D. H. (2000). J. Am. Chem. Soc. 122, 2512–2522.
- Jang, H. G., Cox, D. D. & Que, L. Jr (1991). J. Am. Chem. Soc. 113, 9200–9204.
- Jones, D. G., Wilson, K. R., Cannon-Smith, D. J., Shircliff, A. D., Zhang, Z., Chen, Z., Prior, T. J., Yin, G. & Hubin, T. J. (2015). Inorg. Chem. 54, 2221–2234. [DOI] [PubMed]
- McClain II, J. M., Maples, D. L., Maples, R. D., Matz, D. L., Harris, S. M., Nelson, A. D. L., Silversides, J. D., Archibald, S. J. & Hubin, T. J. (2006). Acta Cryst. C62, m553–m555. [DOI] [PubMed]
- Murray, K. S. (1974). Coord. Chem. Rev. 12, 1–35.
- Ortiz de Montellano, P. R. (1986). In Cytochrome P450: Structure, Mechanism, and Biochemistry. New York: Plenum Press.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Springborg, J. (2003). Dalton Trans. pp. 1653–1665.
- Stoe & Cie (2002). X-AREA. Stoe & Cie, Darmstadt, Germany.
- Wallar, B. J. & Lipscomb, J. D. (1996). Chem. Rev. 96, 2625–2658. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Wong, E. H., Weisman, G. R., Hill, D. C., Reed, D. P., Rogers, M. E., Condon, J. S., Fagan, M. A., Calabrese, J. C., Lam, K.-C., Guzei, I. A. & Rheingold, A. L. (2000). J. Am. Chem. Soc. 122, 10561–10572.
- Yin, G., Buchalova, M., Danby, A. M., Perkins, C. M., Kitko, D., Carter, J. D., Scheper, W. M. & Busch, D. H. (2006). Inorg. Chem. 45, 3467–3474. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015015340/zl2640sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015015340/zl2640Isup2.hkl
CCDC reference: 1419250
Additional supporting information: crystallographic information; 3D view; checkCIF report