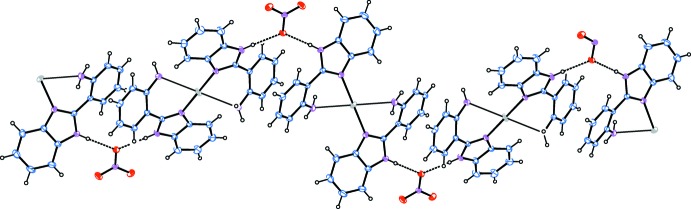
The geometry around silver(I) metal atom in the title complex is distorted square planar with two normal Ag—N bonds and two long Ag—N bonds.
Keywords: crystal structure, silver(I) complex, distorted square-planar geometry, benzimidazole, N—H⋯O hydrogen bonds
Abstract
In the cation of the title salt, [Ag(C13H11N3)2]NO3, the AgI atom lies on a crystallographic inversion center and is coordinated by four N atoms from two bidentate 2-(1H-benzimidazol-2-yl)aniline ligands in a distorted square-planar geometry. The Ag—N(aniline) bond [2.729 (2) Å] is significantly longer than the Ag—N(imidazole) bond [2.165 (1) Å]. In the ligand, the aniline ring is twisted by 37.87 (6)° from the mean plane of the benzimidazole ring system. The nitrate anion lies on a crystallographic twofold rotation axis which passes through the N atom and one of the O atoms. In the crystal, N—H⋯O hydrogen bonds link the components, forming a layer parallel to the bc plane.
Chemical context
Azole and benzazole derivatives have been of interest in an important group in biological systems (Esparza-Ruiz et al., 2011 ▸; Hock et al., 2013 ▸). Benzimidazoles have shown antiviral and antitumor activity (Wang et al., 2007 ▸; Ramla et al., 2007 ▸). Some transition metal complexes with benzimidazole derivatives are important biological molecules (Sánchez-Guadarrama et al., 2009 ▸; Gökçe et al., 2005 ▸). The complexes of silver(I) with a series of benzimidazole-based N-heterocyclic carbenes have shown in vitro antibacterial potential against E. coli and B. subtilis bacteria (Haque et al., 2015 ▸). Recently, we reported on the synthesis and structural features of a zinc complex with a benzimidazole derivative (Kim & Kang, 2015 ▸). In a continuation of our research in this area, the title complex has been synthesized and characterized by single crystal diffraction.
Structural commentary
The cationic AgI complex adopts a distorted square-planar geometry with four N atoms of two bidentate 2-(1H-benzimidazol-2-yl)aniline ligands (Fig. 1 ▸). The AgI atom lies on a crystallographic inversion center. The smaller N2—Ag1—N17 angle is 74.8 (1)° and the other is 105.2 (1)°. The benzimidazole ring system (N2–C10) is almost planar with an r.m.s. deviation of 0.015 (2) Å from the corresponding least-squares plane defined by the nine constituent atoms. The dihedral angle between the benzimidazole ring system and the aniline ring is 37.87 (6)°. This twisting is a driving force in the formation of the weak Ag1—N17 bonding in the Ag complex. The Ag1—N17 bond length of 2.729 (2) Å is much longer than the Ag1—N2 bond length of 2.165 (1) Å. Typical Ag—N bond lengths are within the range 2.1–2.4 Å (Gulbransen & Fitchett, 2012 ▸; Pettinari et al., 2013 ▸; Sun et al., 2006 ▸). However, the bond length of 2.729 (2) Å is shorter than the sum of the van der Waals radii of N and Ag atoms (1.55 and 1.70 Å, respectively; Bondi, 1964 ▸). In the heterocyclic imidazole ring, the N2—C10 bond [1.331 (2) Å] is shorter than the other N—C bonds [N2—C3 1.388 (2), C8—N9 1.380 (2), N9—C10 1.352 Å], which means the N2—C10 bond has double-bond character. In the nitrate counter-anion, atoms N18 and O20 lie on a crystallographic twofold rotation axis.
Figure 1.
Molecular structure of the title compound, showing the atom-numbering scheme and 30% probability ellipsoids. The N—H⋯O hydrogen bond is indicated by a dashed line. [Symmetry codes: (i) −x, −y, −z + 1; (ii) −x, y, −z +  .]
.]
Supramolecular features
In the crystal, the N—H group of the 2-(1H-benzimidazol-2-yl)aniline ligand interacts strongly with the counter-anion, giving rise to a nearly linear hydrogen bond (Table 1 ▸), which forms a zigzag chain along the c axis (Fig. 2 ▸). Another weak N—H⋯O hydrogen bond between the NH2 group and the anion (Table 1 ▸) links the chains into a layer parallel to the bc plane.
Table 1. Hydrogen-bond geometry (, ).
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| N9H9O20 | 0.81(3) | 2.05(3) | 2.8588(18) | 178(3) |
| N17H17BO19i | 0.89(3) | 2.35(3) | 3.214(2) | 164(2) |
Symmetry code: (i)  .
.
Figure 2.
Part of the crystal structure of the title compound, showing molecules linked by intermolecular N—H⋯O hydrogen bonds (dashed lines).
Database survey
A search of the Cambridge Structural Database (Version 5.36 with one update; Groom & Allen, 2014 ▸) returned 2993 entries for crystal structures of benzimidazoles. Most of them are crystal structures of metal complexes. However, there are only four entries with the ligand 2-(1H-benzimidazol-2-yl)aniline or 2-(2-aminophenyl)-1H-benzimidazole bonded to a transition metal: a Zn complex (Eltayeb et al., 2011 ▸), an Ni (Esparza-Ruiz et al., 2011 ▸), an Re (Machura et al., 2011 ▸) and an Ru (Malecki, 2012 ▸).
Synthesis and crystallization
To a stirred solution of Ag(NO3) (0.085 g, 0.5 mmol) in acetonitrile (5 ml) was added a solution of 2-(1H-benzimidazol-2-yl)aniline (0.211 g, 1.0 mmol) in acetonitrile (10 ml) at 333 K. After 24 h of stirring, the solution turned ivory in color. Single crystals of the title complex were obtained by slow evaporation of the solvent at room temperature within three weeks.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. H atoms of the NH and NH2 groups were located in a difference Fourier map and refined freely [refined distances; N—H = 0.81 (3)–0.89 (3) Å]. Other H atoms were positioned geometrically and refined using a riding model, with C—H = 0.93 Å, and with U iso(H) = 1.2U eq(C).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | [Ag(C13H11N3)2]NO3 |
| M r | 588.37 |
| Crystal system, space group | Orthorhombic, P b c n |
| Temperature (K) | 296 |
| a, b, c () | 11.9903(2), 10.1377(2), 20.1115(5) |
| V (3) | 2444.63(9) |
| Z | 4 |
| Radiation type | Mo K |
| (mm1) | 0.87 |
| Crystal size (mm) | 0.18 0.16 0.15 |
| Data collection | |
| Diffractometer | Bruker SMART CCD area detector |
| Absorption correction | Multi-scan (SADABS; Bruker, 2002 ▸) |
| T min, T max | 0.846, 0.872 |
| No. of measured, independent and observed [I > 2(I)] reflections | 62591, 3043, 2446 |
| R int | 0.034 |
| (sin /)max (1) | 0.667 |
| Refinement | |
| R[F 2 > 2(F 2)], wR(F 2), S | 0.030, 0.080, 1.06 |
| No. of reflections | 3043 |
| No. of parameters | 182 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| max, min (e 3) | 0.44, 0.48 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015015315/is5411sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015015315/is5411Isup2.hkl
CCDC reference: 1419095
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
This work was supported by the research fund of Chungnam National University.
supplementary crystallographic information
Crystal data
| [Ag(C13H11N3)2](NO3) | Dx = 1.599 Mg m−3 |
| Mr = 588.37 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbcn | Cell parameters from 9923 reflections |
| a = 11.9903 (2) Å | θ = 2.6–28.3° |
| b = 10.1377 (2) Å | µ = 0.87 mm−1 |
| c = 20.1115 (5) Å | T = 296 K |
| V = 2444.63 (9) Å3 | Block, colourless |
| Z = 4 | 0.18 × 0.16 × 0.15 mm |
| F(000) = 1192 |
Data collection
| Bruker SMART CCD area-detector diffractometer | 2446 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.034 |
| φ and ω scans | θmax = 28.3°, θmin = 2.0° |
| Absorption correction: multi-scan (SADABS; Bruker, 2002) | h = −15→15 |
| Tmin = 0.846, Tmax = 0.872 | k = −13→13 |
| 62591 measured reflections | l = −26→26 |
| 3043 independent reflections |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.030 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.080 | w = 1/[σ2(Fo2) + (0.0371P)2 + 1.1132P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max < 0.001 |
| 3043 reflections | Δρmax = 0.44 e Å−3 |
| 182 parameters | Δρmin = −0.48 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Ag1 | 0.0000 | 0.0000 | 0.5000 | 0.04858 (10) | |
| N2 | −0.04065 (13) | 0.18355 (14) | 0.45056 (7) | 0.0336 (3) | |
| C3 | −0.09293 (15) | 0.29248 (18) | 0.47845 (9) | 0.0340 (4) | |
| C4 | −0.13679 (17) | 0.3119 (2) | 0.54221 (10) | 0.0436 (4) | |
| H4 | −0.1356 | 0.2450 | 0.5739 | 0.052* | |
| C5 | −0.18161 (19) | 0.4335 (2) | 0.55614 (11) | 0.0527 (5) | |
| H5 | −0.2119 | 0.4487 | 0.5980 | 0.063* | |
| C6 | −0.1829 (2) | 0.5349 (3) | 0.50918 (12) | 0.0588 (6) | |
| H6 | −0.2133 | 0.6161 | 0.5208 | 0.071* | |
| C7 | −0.14013 (19) | 0.5180 (2) | 0.44611 (12) | 0.0512 (5) | |
| H7 | −0.1417 | 0.5852 | 0.4146 | 0.061* | |
| C8 | −0.09453 (15) | 0.39484 (18) | 0.43215 (9) | 0.0365 (4) | |
| N9 | −0.04206 (14) | 0.34549 (15) | 0.37641 (8) | 0.0355 (3) | |
| H9 | −0.030 (2) | 0.381 (3) | 0.3412 (13) | 0.056 (7)* | |
| C10 | −0.01033 (14) | 0.22026 (16) | 0.38965 (8) | 0.0300 (3) | |
| C11 | 0.05731 (14) | 0.14278 (16) | 0.34296 (8) | 0.0295 (3) | |
| C12 | 0.13818 (14) | 0.20841 (18) | 0.30566 (9) | 0.0364 (4) | |
| H12 | 0.1448 | 0.2995 | 0.3095 | 0.044* | |
| C13 | 0.20848 (15) | 0.1415 (2) | 0.26327 (9) | 0.0405 (4) | |
| H13 | 0.2605 | 0.1871 | 0.2378 | 0.049* | |
| C14 | 0.20058 (17) | 0.00563 (19) | 0.25906 (9) | 0.0407 (4) | |
| H14 | 0.2497 | −0.0409 | 0.2320 | 0.049* | |
| C15 | 0.12026 (16) | −0.06123 (18) | 0.29469 (9) | 0.0386 (4) | |
| H15 | 0.1155 | −0.1525 | 0.2911 | 0.046* | |
| C16 | 0.04616 (16) | 0.00555 (16) | 0.33594 (9) | 0.0320 (4) | |
| N17 | −0.03508 (17) | −0.06622 (18) | 0.37016 (8) | 0.0410 (4) | |
| H17A | −0.092 (2) | −0.021 (2) | 0.3769 (11) | 0.042 (6)* | |
| H17B | −0.048 (2) | −0.145 (3) | 0.3528 (13) | 0.068 (8)* | |
| N18 | 0.0000 | 0.5893 (2) | 0.2500 | 0.0314 (4) | |
| O19 | 0.02905 (14) | 0.64820 (15) | 0.19918 (8) | 0.0534 (4) | |
| O20 | 0.0000 | 0.4645 (2) | 0.2500 | 0.0446 (5) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ag1 | 0.07365 (18) | 0.03391 (13) | 0.03818 (13) | 0.00637 (10) | 0.00441 (10) | 0.01569 (8) |
| N2 | 0.0448 (8) | 0.0272 (7) | 0.0287 (7) | −0.0002 (6) | 0.0025 (6) | 0.0041 (6) |
| C3 | 0.0372 (9) | 0.0314 (9) | 0.0332 (8) | −0.0026 (7) | 0.0043 (7) | 0.0015 (7) |
| C4 | 0.0487 (10) | 0.0466 (11) | 0.0355 (9) | −0.0064 (9) | 0.0113 (8) | 0.0006 (8) |
| C5 | 0.0548 (12) | 0.0550 (13) | 0.0483 (12) | −0.0016 (10) | 0.0205 (10) | −0.0094 (10) |
| C6 | 0.0637 (15) | 0.0460 (12) | 0.0666 (15) | 0.0147 (11) | 0.0202 (11) | −0.0103 (11) |
| C7 | 0.0612 (13) | 0.0357 (11) | 0.0565 (13) | 0.0134 (9) | 0.0141 (11) | 0.0052 (9) |
| C8 | 0.0413 (10) | 0.0312 (9) | 0.0369 (9) | 0.0029 (7) | 0.0066 (7) | 0.0026 (7) |
| N9 | 0.0483 (8) | 0.0268 (8) | 0.0315 (8) | 0.0070 (6) | 0.0069 (7) | 0.0071 (6) |
| C10 | 0.0359 (8) | 0.0250 (8) | 0.0291 (8) | −0.0005 (6) | 0.0001 (6) | 0.0034 (6) |
| C11 | 0.0344 (8) | 0.0267 (8) | 0.0273 (8) | 0.0027 (7) | −0.0026 (6) | 0.0022 (6) |
| C12 | 0.0386 (9) | 0.0290 (8) | 0.0415 (10) | −0.0005 (7) | 0.0013 (7) | 0.0020 (7) |
| C13 | 0.0335 (9) | 0.0455 (11) | 0.0427 (10) | −0.0002 (8) | 0.0043 (7) | −0.0007 (8) |
| C14 | 0.0381 (9) | 0.0439 (11) | 0.0402 (11) | 0.0089 (8) | −0.0035 (7) | −0.0098 (8) |
| C15 | 0.0467 (10) | 0.0280 (9) | 0.0411 (10) | 0.0051 (8) | −0.0075 (8) | −0.0072 (7) |
| C16 | 0.0386 (8) | 0.0298 (9) | 0.0278 (8) | −0.0011 (7) | −0.0074 (7) | 0.0013 (6) |
| N17 | 0.0558 (10) | 0.0306 (9) | 0.0366 (9) | −0.0083 (8) | 0.0003 (8) | 0.0004 (7) |
| N18 | 0.0370 (10) | 0.0232 (10) | 0.0341 (10) | 0.000 | −0.0049 (9) | 0.000 |
| O19 | 0.0731 (10) | 0.0381 (8) | 0.0491 (8) | −0.0022 (7) | 0.0064 (7) | 0.0155 (7) |
| O20 | 0.0759 (14) | 0.0209 (8) | 0.0368 (10) | 0.000 | 0.0055 (9) | 0.000 |
Geometric parameters (Å, º)
| Ag1—N2i | 2.1653 (14) | C10—C11 | 1.468 (2) |
| Ag1—N2 | 2.1653 (14) | C11—C12 | 1.395 (2) |
| Ag1—N17 | 2.7288 (17) | C11—C16 | 1.405 (2) |
| N2—C10 | 1.331 (2) | C12—C13 | 1.377 (3) |
| N2—C3 | 1.388 (2) | C12—H12 | 0.9300 |
| C3—C8 | 1.394 (2) | C13—C14 | 1.383 (3) |
| C3—C4 | 1.400 (2) | C13—H13 | 0.9300 |
| C4—C5 | 1.374 (3) | C14—C15 | 1.379 (3) |
| C4—H4 | 0.9300 | C14—H14 | 0.9300 |
| C5—C6 | 1.396 (3) | C15—C16 | 1.391 (3) |
| C5—H5 | 0.9300 | C15—H15 | 0.9300 |
| C6—C7 | 1.379 (3) | C16—N17 | 1.397 (3) |
| C6—H6 | 0.9300 | N17—H17A | 0.83 (2) |
| C7—C8 | 1.392 (3) | N17—H17B | 0.89 (3) |
| C7—H7 | 0.9300 | N18—O19ii | 1.2340 (17) |
| C8—N9 | 1.380 (2) | N18—O19 | 1.2340 (17) |
| N9—C10 | 1.352 (2) | N18—O20 | 1.265 (3) |
| N9—H9 | 0.81 (3) | ||
| N2i—Ag1—N2 | 180.0 | N9—C10—C11 | 122.12 (15) |
| N2i—Ag1—N17 | 105.23 (6) | C12—C11—C16 | 118.98 (16) |
| N2—Ag1—N17 | 74.77 (5) | C12—C11—C10 | 118.21 (15) |
| C10—N2—C3 | 105.83 (14) | C16—C11—C10 | 122.78 (16) |
| C10—N2—Ag1 | 126.97 (12) | C13—C12—C11 | 121.57 (17) |
| C3—N2—Ag1 | 126.86 (11) | C13—C12—H12 | 119.2 |
| N2—C3—C8 | 109.18 (15) | C11—C12—H12 | 119.2 |
| N2—C3—C4 | 130.65 (17) | C12—C13—C14 | 119.08 (18) |
| C8—C3—C4 | 120.14 (17) | C12—C13—H13 | 120.5 |
| C5—C4—C3 | 117.37 (19) | C14—C13—H13 | 120.5 |
| C5—C4—H4 | 121.3 | C15—C14—C13 | 120.38 (18) |
| C3—C4—H4 | 121.3 | C15—C14—H14 | 119.8 |
| C4—C5—C6 | 121.8 (2) | C13—C14—H14 | 119.8 |
| C4—C5—H5 | 119.1 | C14—C15—C16 | 121.11 (17) |
| C6—C5—H5 | 119.1 | C14—C15—H15 | 119.4 |
| C7—C6—C5 | 121.8 (2) | C16—C15—H15 | 119.4 |
| C7—C6—H6 | 119.1 | C15—C16—N17 | 119.05 (17) |
| C5—C6—H6 | 119.1 | C15—C16—C11 | 118.75 (17) |
| C6—C7—C8 | 116.3 (2) | N17—C16—C11 | 122.18 (17) |
| C6—C7—H7 | 121.8 | C16—N17—Ag1 | 103.64 (11) |
| C8—C7—H7 | 121.8 | C16—N17—H17A | 111.8 (15) |
| N9—C8—C7 | 131.96 (17) | Ag1—N17—H17A | 80.7 (16) |
| N9—C8—C3 | 105.46 (15) | C16—N17—H17B | 113.6 (17) |
| C7—C8—C3 | 122.57 (18) | Ag1—N17—H17B | 128.7 (17) |
| C10—N9—C8 | 107.99 (15) | H17A—N17—H17B | 114 (2) |
| C10—N9—H9 | 122.6 (19) | O19ii—N18—O19 | 122.1 (2) |
| C8—N9—H9 | 129.4 (19) | O19ii—N18—O20 | 118.96 (11) |
| N2—C10—N9 | 111.53 (15) | O19—N18—O20 | 118.96 (11) |
| N2—C10—C11 | 126.15 (15) |
Symmetry codes: (i) −x, −y, −z+1; (ii) −x, y, −z+1/2.
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N9—H9···O20 | 0.81 (3) | 2.05 (3) | 2.8588 (18) | 178 (3) |
| N17—H17B···O19iii | 0.89 (3) | 2.35 (3) | 3.214 (2) | 164 (2) |
Symmetry code: (iii) −x, y−1, −z+1/2.
References
- Bondi, A. (1964). J. Phys. Chem. 68, 441–451.
- Bruker (2002). SADABS, SAINT and SMART. Bruker AXS Inc., Madison, Wisconsin, USA.
- Eltayeb, N. E., Teoh, S. G., Chantrapromma, S. & Fun, H.-K. (2011). Acta Cryst. E67, m1062–m1063. [DOI] [PMC free article] [PubMed]
- Esparza-Ruiz, A., Peña-Hueso, A., Mijangos, E., Osorio-Monreal, G., Nöth, H., Flores-Parra, A., Contreras, R. & Barba-Behrens, N. (2011). Polyhedron, 30, 2090–2098.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Gökçe, M., Utku, S., Gür, S., Özkul, A. & Gümüş, F. (2005). Eur. J. Med. Chem. 40, 135–141. [DOI] [PubMed]
- Groom, C. R. & Allen, F. H. (2014). Angew. Chem. Int. Ed. 53, 662–671. [DOI] [PubMed]
- Gulbransen, J. L. & Fitchett, C. M. (2012). CrystEngComm, 14, 5394–5397.
- Haque, R. A., Choo, S. Y., Budagumpi, S., Iqbal, M. A. & Al-Ashraf Abdullah, A. (2015). Eur. J. Med. Chem. 90, 82–92. [DOI] [PubMed]
- Hock, S. J., Schaper, L., Herrmann, W. A. & Kühn, F. E. (2013). Chem. Soc. Rev. 42, 5073–5089. [DOI] [PubMed]
- Kim, Y. & Kang, S. K. (2015). Acta Cryst. E71, m85–m86. [DOI] [PMC free article] [PubMed]
- Machura, B., Wolff, M., Gryca, I., Palion, A. & Michalik, K. (2011). Polyhedron, 30, 2275–2285.
- Małecki, J. G. (2012). Struct. Chem. 23, 461–472.
- Pettinari, C., Marchetti, F., Orbisaglia, S., Pettinari, R., Ngoune, J., Gómez, M., Santos, C. & Álvarez, E. (2013). CrystEngComm, 15, 3892–3907.
- Ramla, M. M., Omar, M. A., Tokuda, H. & El-Diwani, H. I. (2007). Bioorg. Med. Chem. 15, 6489–6496. [DOI] [PubMed]
- Sánchez-Guadarrama, O., López-Sandoval, H., Sánchez-Bartéz, F., Gracia-Mora, I., Höpfl, H. & Barba-Behrens, N. (2009). J. Inorg. Biochem. 103, 1204–1213. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Sun, Q., Bai, Y., He, G., Duan, C., Lin, Z. & Meng, Q. (2006). Chem. Commun. pp. 2777–2779. [DOI] [PubMed]
- Wang, X. A., Cianci, C. W., Yu, K.-L., Combrink, K. D., Thuring, J. W., Zhang, Y., Civiello, R. L., Kadow, K. F., Roach, J., Li, Z., Langley, D. R., Krystal, M. & Meanwell, N. A. (2007). Bioorg. Med. Chem. Lett. 17, 4592–4598. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989015015315/is5411sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015015315/is5411Isup2.hkl
CCDC reference: 1419095
Additional supporting information: crystallographic information; 3D view; checkCIF report