Abstract
Many studies have demonstrated that microRNAs (miRNAs) may play vital roles in the development of breast cancer. The aim of this study was to examine the expression levels of miR-497 in human breast cancer and investigate whether its potential roles involved targeting Bcl-2. Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) was used to examine the expression levels of miR-497 in 48 breast cancer specimens and six breast cancer cell lines. MTT assay, colony formation assay, and flow cytometry were conducted to explore the potential functions of miR-497 in human MDA-MB-231 breast cancer cells. Correlation analysis and dual-luciferase reporter assay were performed to validate whether Bcl-2 was a direct target of miR-497. The effects of modulating miR-497 on endogenous levels of Bcl-2 were subsequently confirmed via qRT-PCR and western blot. MTT assay, colony formation assay and flow cytometry were used to indicate the roles of endogenous Bcl-2 in breast cancer cells. miR-497 expression levels were significantly decreased in human breast cancer specimens and cell lines (P<0.05). Overexpression of miR-497 in breast cancer cells suppressed cell proliferation and induced apoptosis. Correlation analysis indicated that miR-497 was highly inversely correlated with Bcl-2 protein expression in breast cancer specimens. Dual-luciferase reporter assays confirmed that Bcl-2 was a direct target of miR-497. qRT-PCR and western blot showed that miR-497 negatively regulated Bcl-2 protein expression but had no impact on mRNA expression of Bcl-2. Knockdown of Bcl-2 expression in MDA-MB-231 cells significantly suppressed cell proliferation and promoted apoptosis. Our study suggests that miR-497 may act as a breast cancer suppressor through negative regulation of Bcl-2 protein expression at the posttranscriptional levels. Therefore, targeting miR-497 may provide a novel strategy for the diagnosis and treatment of patients with this lethal disease.
Keywords: miR-497, Bcl-2, apoptosis, breast cancer
Introduction
Breast cancer is a major public and global health problem and accounts for 14% of female cancer deaths worldwide each year [1]. Therefore, it is essential to develop more effective methods forearly diagnosis and treatment.
MicroRNAs (miRNAs) are a class of endogenous short single strand non-coding RNA molecules [2], which are translated from exons of protein-coding and non-coding genes [2,3], and produce mature miRNA after a multistep process. The mature miRNA binds completely or partially to mRNA 3’ untranslated regions (UTRs) of specific protein-coding genes [4]. When the miRNA is perfectly complementary to its target gene, it can specifically cleave the target mRNA, and when it is partially complementary to its target, it can inhibit mRNA translation without influencing mRNA levels [5,6]. Many studies have demonstrated that one miRNA canregulate several hundreds of target mRNAs, conversely, one mRNA can be targeted by multiple miRNAs [7]. By targeting multiple transcripts, miRNAs play important roles in organ development, metabolism and other critical pathways, such assurvival, differentiation, proliferation and apoptosis [8]. Aberrant miRNA expression has been frequently observed in various types of human tumors, and several miRNAs have already been confirmed to play pivotal roles as oncogenes or tumor suppressor genes in breast cancer [9-11].
Previous studies indicated that miRNA-497 (miR-497) was downregulated in several kinds of tumors including human melanoma [12], gastric cancer [13], and adrenocortical carcinoma [14]. The expression levels of miR-497 was also significantly lower in breast cancer tissues and cell lines, and several targets of miR-497 have been previously identified, such as Raf-1, CCND1, Bcl-w, and CCNE1 [15-18].
B cell lymphoma 2 (Bcl-2) is a central player in the genetic program of eukaryotic cells favoring survival by inhibiting cell death [19]. Overexpression of Bcl-2 protein has been reported in many types of cancer, including leukemia [20], lymphomas [21,22], carcinomas [23] and breast cancer. Multiple studies have shown that Bcl-2 is a target of several miRNAs, such as miR-15 andmiR-16, that regulate Bcl-2 in chronic lymphocytic leukemia [24], and miR-34a that inhibits Bcl-2 in breast cancer [25]. Furthermore, miR-15b, miR-16 [26], miR-181b [27], miR-34a [28], miR-195, miR-24-2 and miR-365-2 [29] all modulate multidrug resistance by targeting Bcl-2.
Recent studies showed that miR-497 regulat esethanol-induced neuronal cell death through Bcl-2 protein [30], and miR-497 modulates multidrug resistance of human cancer celllines by targeting Bcl-2 in human gastric and lung cancer cell lines [13]. However, whether miRNA-497 targets Bcl-2 and modulates its apoptotic function in breast cancer is unknown.
In this study, we found that miR-497 was downregulated in human breast cancer clinical samples and cell lines. Ectopic expression of miR-497 inhibited proliferation and induced apoptosis by targeting Bcl-2 in breast cancer. Our findings showed that miR-497 negatively regulated Bcl-2 protein expression at the posttranscriptional levels.
Materials and methods
Ethics statement
Forty-eight pairs of breast cancer specimens and matched adjacent normal breast tissues were surgically collected from patients at the Department of Breast and Thyroid Surgery of the Shanghai Tenth People’s Hospital, China. Written informed consent was obtained from each patient and this study was approved by the Institutional Ethics Committee of Shanghai Tenth People’s Hospital (Ethics review approval document numbers: SHSY-IEC-pap-14-17).
Specimens
All samples were snap frozen in liquid nitrogen and confirmed as invasive, ductal breast cancer by trained pathologists. No patients received chemotherapy or radiotherapy prior to surgery.
Cell culture and transfection
Human MDA-MB-231, MCF-7, MDA-MB-435, T-74D, MDA-MB-468 and MDA-MB-453 breast cancer lines, MCF-10A non-malignant breast epithelial cells and HEK293T cells were purchased from Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (both from Gibco, Carlsbad, CA, USA), penicillin (100 U/ml) and streptomycin (100 μg/mL) (Enpromise, Hangzhou, China). Cells were incubated at 37°C in a humidified chamber supplemented with 5% CO2. Cells in the logarithmic growth phase (~90% confluence) were selected for the experiments.
miR-497 mimics, miR-497 inhibitor, non-specific negative control (NC) oligos, Bcl-2 siRNA, and siRNA negative control (siRNA NC) were purchased from GenePharma (Shanghai, China). For transfection, actively growing MDA-MB-231 cells (25×105 per well) were plated into six-well plates (BD Biosciences, Franklin Lakes, NJ, USA) and cultured with serum- and antibiotic-free DMEM. When cell density achieved 30-40% confluence, cells were transfected using Lipofectamine (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Complete media was changed 5 h after transfection.
Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
For detection of miR-497 expression, miRNAs were extracted using the miRcute miRNA isolation kit (Tiangen, Beijing, China) in accordance with the manufacturer’s instructions. miR-497 and U6 primers were purchased from GenePharma. The U6 primer used as an internal control was 5’-GTCCTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAATATGGAAC-3’ (stemloop primer), 5’-TGCGGGTGCTCGCTTCGCAGC-3’ (sense) and 5’-CCAGTGCAGGGTCCGAGGT-3’ (antisense). cDNA was generated by reverse transcription using the PrimeScript™ RT-PCR kit according to the manufacturer’s instructions (Takara, Tokyo, Japan). Real-time PCR was performed on a 7900HT fast RT-PCR instrument (Applied Biosystems, Singapore). qRT-PCR parameters for miRNA quantification were as follows: 2 min at 95°C, followed by 40 cycles of 30 s at 95°C and 45 s at 60°C. Each sample was tested in triplicate.
For Bcl-2 mRNA analysis, total RNAs was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions. cDNA was generated by reverse transcription using the PrimeScript RT-PCR kit (Takara) according to the manufacturer’s instructions. RT-PCR was performed on a 7900HT fast RT-PCR instrument using SYBR-Green. GAPDH mRNA levels were used for normalization. The primer (GenePharma) sequences were as follows: Bcl-2, 5’-GAACTGGGGGAGGATTGTGG-3’ (sense) and 5’-CCGGTTCAGGTACTCAGTCA-3’ (antisense); GAPDH, 5’-AAGGTCGGAGTCAACGGATT-3’ (sense) and 5’-CTGGAAGATGGTGATGGGATT-3’ (antisense). qRT-PCR parameters for relative quantification were as follows: 2 min at 95°C, followed by 40 cycles of 15 s at 95°C and 30 s at 60°C. Each sample was tested three times. The relative expression was calculated using the relative quantification equation (RQ) = 2-ΔΔCt [31].
Cell proliferation assay (MTT assay)
At 4-5 h after miR-497 mimics, miR-497 inhibitor, NC oligo, Bcl-2 siRNA or siRNA NC transfection, cells were trypsinized and counted. Cells were plated (1,000/well) in each well of 96-well plates (BD Biosciences) in 6 replicates and incubated at 37°C. Cell proliferation was assessed at 24, 48, 72, and 96 h post-transfection using the MTT assay. Briefly, 20 μL (5 mg/ml) MTT (Sigma, Santa Clara, CA, USA) solution was added to each well. After 4 h incubation at 37°C, the supernatant was discarded and 150 μL DMSO (Sigma) was added. After 10 min shaking (100 rpm), the absorbance at 490 nm of each sample was measured by a microplate spectrophotometer (Bio-Tek, USA). Each experiment was performed three times.
Colony formation assay
At 4 h after transfection, cells were trypsinized and counted, and 500 cells from each group were plated in a six-well plate in complete medium. The plates were shaken to disperse the cells equally. After incubation at 37°C with 5% CO2 for 7-10 days, or when the colonies were visible to the eyes, the culture was terminated. Complete medium was removed, and the plates were washed twice in phosphate buffered saline (PBS). The colonies were fixed by 95% ethanol for 10 min, dried and stained with 0.1% crystal violet solution for 10 min, and each plate was washed three times by water. The colonies containing >50 cells were counted, and images of the stained plates were obtained. The experiment was performed three times in triplicate.
Apoptosis assay
The Annexin V-FITC/PI kit was obtained from Beyotime Institute of Biotechnology (Jiangsu, China). After 48 h of transfection, as indicated in the experiment, cells were washed three times with ice-cold PBS and trypsinized. After adding 2.5 μL Annexin V-FITC reagent and 50 μL 1× binding buffer, the cells were incubated in the dark for 15 min at room temperature. Subsequently, 5 μL propidium iodide (PI) and 250 μL 1× binding buffer was added and the cells were incubated in the dark for 5 min at room temperature. Cells were gently resuspended in the Annexin V incubation reagent at a concentration of 1×105 to 106 cells per 100 μL. All samples were processed by flow cytometry (FACSCanto™ II, BD Biosciences). FACS analyses were performed in at least three replicates.
Dual-luciferase reporter assay
The dual-luciferase reporter vector and the mutant reporter vector were purchased from JOIN Genome (Hangzhou, China). The 3’-UTR of Bcl-2 containing the predicted miR-497 binding site was sub-cloned into the psiCHECK-2 reporter vector in the dual-luciferase reporter; the mutant constructs were generated by mutation. 293T cells were seeded in 12-well plates (BD Biosciences) in complete medium and incubated at 37°C with 5% CO2. The psiCHECK-2/Bcl-2 3’-UTR reporter plasmids (2 μg/well) were co-transfected with miR-497 mimics or miR-NC (75 nM) using Lipofectamine 2000 (Invitrogen) when 293T cells reached 80-90% confluence. psiCHECK-2/Bcl-2 3’-UTR mutant reporter plasmids (2 μg/well) were co-transfected with miR-497 mimics or miR-NC (75 nM). Complete media was changed 5 h after transfection. Thirty-six hours after transfection, luciferase activity was measured using the dual-luciferase reporter assay kit (Promega, Madison, WI, USA). Briefly, the cells were washed twice with PBS and lysed by incubation on ice for 30 min with passive lysis buffer (PLB, 250 μL/well). The supernatants were collected, and 20 μL of the aliquots were added to 96-well plates. The firefly luciferase (FL) reporter was measured by a microplate spectrophotometer immediately after adding 50 μL of Luciferase Assay Reagent II. Next, Stop & Glo® reagent (50 μL/well) was added to each well to initiate the Renilla luciferase (RL). RL activity was normalized to FL activity. All experiments were performed three times.
Western blot analysis
Cells were washed three times in ice-cold PBS and resuspended in RIPA lysis buffer (100 μL/well, Beyotime). After the cells were lysed on ice for 30 min, the cell lysate was centrifuged for 20 min at 12,000× g at 4°C (Eppendorf 5804R, Eppendorf Biotech, Germany). Supernatants were collected and the protein concentrations were quantified using a BCA protein assay kit (Beyotime). Protein samples were denatured with 5× SDS loading buffer (Beyotime) at 100°C for 15 min. Proteins of breast cancer specimens and matched adjacent normal breast tissues were obtained using the above methods. Protein samples (30 μg) were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, Beyotime) and transferred to a 0.45-μm nitrocellulose membrane (Beyotime). After 1 h blocking with 5% skim milk, the membrane was incubated overnight at 4°C with primary antibodies against Bcl-2 (1:1,000; Epitomics, USA) and β-actin (1:1,000; Santa Cruz Biotechnology). The membranes were incubated with secondary antibodies for 30 min at room temperature after three washes with PBST (Shanghai Engineering co). After three washes with PBST, immunereactive protein bands were detected with an Odyssey Scanning system (Li-Cor, Lincoln, NE, USA).
Statistical analysis
Semi-quantitative analysis was used for qRT-PCR analyses. Paired t-test was used to compare the expression levels of miR-497 between breast cancer specimens and matched adjacent normal breast tissues. Two independent sample t test was used to compare the statistical differences between miR-497 mimics and NC groups or inhibitor and NC groups. The relation between miR-497 and Bcl-2 protein was analyzed by correlation analysis. Data are presented as the means ± standard deviation (SD) from at least three independent experiments. Differences were considered significant for P-values <0.05. SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) statistical software were used in statistical analysis.
Results
MiR-497 expression is decreased in human breast cancer cell lines and clinical specimens
We measured the expression levels of miR-497 in several breast cancer cell lines, MDA-MB-231, MCF-7, MDA-MB-435, T-74D, MDA-MB-468, and MDA-MB-453 by qRT-PCR. Compared with MCF-10A cells, the above six breast cancer cell lines showed significantly decreased miR-497 expression (P<0.05 Figure 1A). The expression levels of miR-497 was the lowest in MDA-MB-231 cells, so this cell line was used in the subsequent experiments. The expression levels of miR-497 in 48 paired breast cancer specimens (0.14±0.01) was downregulated compared with matched adjacent normal breast tissues (P<0.05). These results indicated that the expression of miR-497 is significantly decreased in human breast cancer specimens and cell lines.
Figure 1.
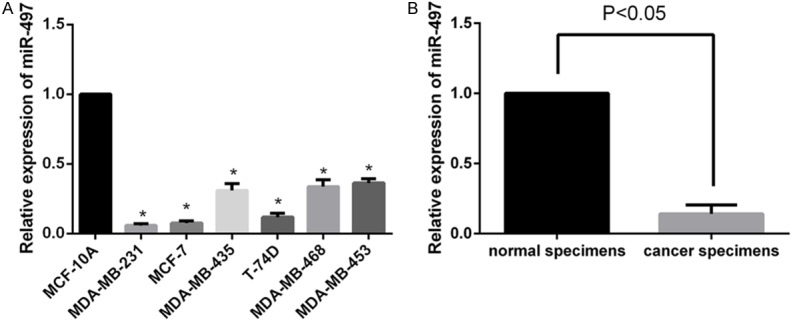
miR-497 is downregulated in human breast cancer cell lines and clinical specimens. A. Relative expression of miR-497 was downregulated in breast cancer cell lines compared to MCF-10A cells. B. Relative miR-497 expression in 48 paired breast cancer tissues was downregulated compared to matched adjacent normal breast tissues. Data represent the 2-ΔΔCt values ± SD,*P<0.05.
miR-497inhibits MDA-MB-231 cell proliferation
We next explored the roles of miR-497 in breast cancer cell proliferation by examining the effects of upregulated and downregulated miR-497 expression. First, we transfected MDA-MB-231 cells with miR-497 mimics, inhibitor or NC (100 nmol/l) for 48 h. We then measured miR-497 expression by qRT-PCR. Compared with the levels of the NC group, the miR-497 inhibitor group showed a significant decrease in miR-497 expression (P<0.05), and the miR-497 mimics group exhibited markedly higher miR-497 levels (P<0.05) (Figure 2A). This result showed transfection efficiency was high.
Figure 2.
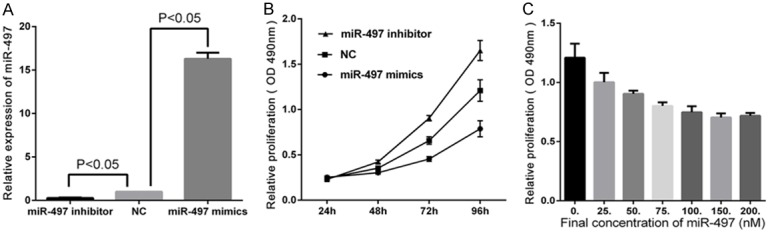
Overexpression of miR-497 repressed MDA-MB-231 cell proliferation. A. Relative miR-497 expression of miR-NC group were lower than miR-497 mimics group and higher than miR-497 inhibitor group. B. Upregulation of miR-497 significantly repressed the growth rate of MDA-MB-231 cells, while the inhibitor increased the growth rate (P<0.05). C. Dose-dependent effect of miR-497 mimics on cell growth. The optical density of each well was measured at the indicated time-points with a microplate reader at 490 nm. 100 nM was selected as the optimal concentration for subsequent analyses. The graph represents OD 490 nm ± SD, P<0.05.
Second, MTT assay showed that upregulation of miR-497 in MDA-MB-231 cells significantly repressed the growth rate at 48 h (14.4%), 72 h (30.8%) and 96 h (34.8%) compared with the NC group (P<0.05) (Figure 2B). Conversely, the miR-497 inhibitor increased the growth rate at 48 h (15.4%), 72 h (37.5%) and 96 h (36.6%) compared with the control cells (P<0.05). A dose-dependent effect of miR-497 mimics transfection at 72 h was also observed by MTT assay (Figure 2C), and a concentration of 100 nM was selected for use in the subsequent experiments. Colony formation assay results presented in Figure 3B showed fewer numbers of colonies in cells upregulated for miR-497 and higher numbers in cells transfected with miR-497 inhibitor (52.25±3.64 in miR-497 mimics group vs. 106.8±2.69 in miRNA NC group, P<0.05; 106.8±2.69 in miRNA NC group vs. 150.3±3.91 in miR-497 inhibitor group, P<0.05). These results suggest that miR-497 inhibits the growth of breast cancer cells and may induce cell apoptosis.
Figure 3.
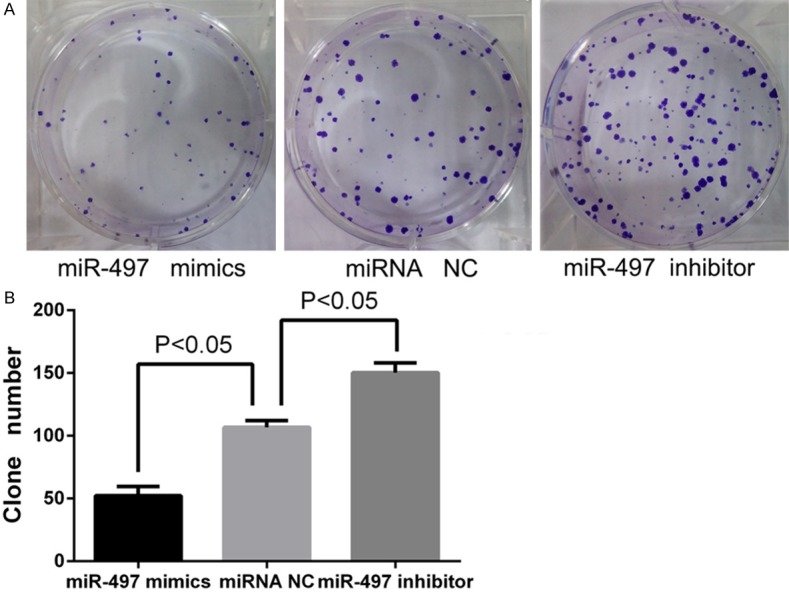
Overexpression of miR-497 inhibited colony formation ability of MDA-MB-231 cells. A. Crystal violet stained colonies from the colony formation assay for the miR-497 mimics group, miRNA NC group, and miR-497 inhibitor group. B. Cells transfected with miR-497 mimics exhibited fewer colonies than the NC group; cells transfected with miR-497 inhibitor exhibited higher numbers of colonies than the NC group (P<0.05).
Overexpression miR-497 induced breast cancer cell apoptosis
To investigate whether miR-497 can induce apoptosis of breast cancer cells, MDA-MB-231 cancer cells were transfected with 100 nmol/l of miR-497 mimics, inhibitor or NC for 48 h. Apoptotic cell death was evaluated using flow cytometric analysis of Annexin V-FITC/PI staining. As shown in Figure 4, the percentage of the early and late apoptotic cells was higher in the miR-497 mimic group (34.3±0.06%) compared with the miRNA NC group (3.5±0.04%) (P<0.05). Additionally, a small difference was observed between the miRNA NC and inhibitor group (3.5±0.04% vs. 2.3±0.04%, respectively; P<0.05). These results demonstrate that miR-497 can induce breast cancer cell apoptosis in vitro.
Figure 4.
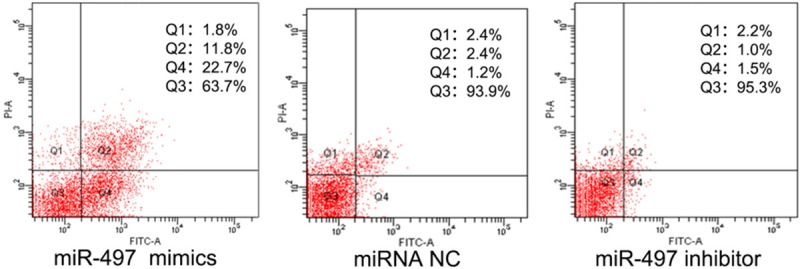
miR-497 mimics significantly induced early and late apoptosis of breast cancer cells. In contrast, cells transfected with negative control and inhibitor exhibited low levels of apoptosis (P<0.05).
The levels of miR-497are inversely correlated with Bcl-2 protein expression in breast cancer specimens
We used several online databases, including TargetScan, miRanda, miRBase and miRGen, to investigate potential targets of miR-497. Based on the results from these databases, we selected Bcl-2 as a possible candidate gene. To evaluate the putative relationship between miR-497 and Bcl-2, we first examined the correlation between the expression levels of miR-497 and Bcl-2 protein levels in breast cancer specimens and matched adjacent normal breast tissues by qRT-PCR and western blot analysis. In all breast cancer specimens, we found a highly inverse correlation betweenmiR-497 and Bcl-2 levels, with a coefficient of correlation of 95.43% (Figure 5).
Figure 5.

Bcl-2 protein expression was inversely correlated with miR-497 miRNA expression in breast cancer specimens. Normalized Bcl-2 protein expression (to β-actin) is shown compared to miR-497 levels by qRT-PCR. ACT: β-actin.
Bcl-2 is a direct target of miR-497
By analyzing the miR-497 and the Bcl-2 mRNA sequence, we found that seven nucleotides in miR-497 are complementary to the Bcl-2 cDNA (Figure 6A). Dual-luciferase reporter assay (Figure 6B) showed that the relative luciferase activity (FL/RL) of miR-497 mimics co-transfection of the vector containing the wild-type Bcl-2 3’-UTR sequence group significantly decreased compared with the negative control. However, this effect of miR-497 was abolished following co-transfection of the psiCHECK-2/Bcl-2 3’-UTR mutant. Together these data support the notion that Bcl-2 is a direct target of miR-497.
Figure 6.
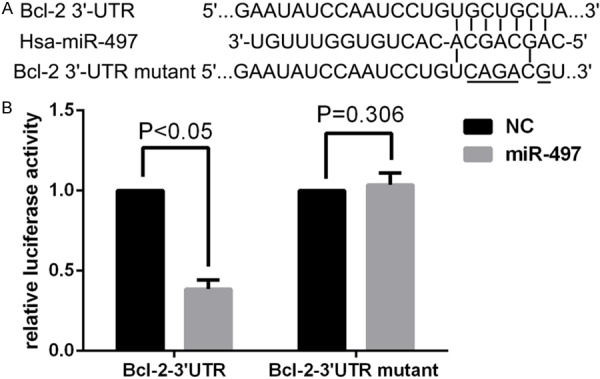
Bcl-2 is a direct target of miR-497. A. The miR-497 binding site in the 3’-UTR of Bcl-2 mRNA and the Bcl-2 3’-UTR mutant sequence. B. The relative luciferase activity (Firefly/Renilla) was measured in HEK293T cells after co-transfection of the Bcl-2 3’-UTR luciferase construct with miR-497 mimics or miR-NC and co-transfection of the Bcl-2 3’-UTR mutant luciferase construct with miR-497 mimics or miR-NC.
miR-497 negatively regulates Bcl-2 protein expression at the posttranscriptional level
miRNAs can downregulate a specific target by affecting mRNA translation or mRNA stability [7]. To determine whether miR-497 regulates endogenous Bcl-2 at the mRNA or protein level, miR-497 mimics, inhibitor, and NC were transfected into MDA-MB-231 cells, and the levels of Bcl-2 mRNA and protein were monitored 48 h after transfection by qRT-PCR and western blot. qRT-PCR analysis revealed no differences in the levels of Bcl-2 mRNA expression between any of the examined groups (Figure 7A). However, western blot showed that miR-497 mimics repressed the expression of endogenous Bcl-2 protein at 48 h after transfection compared with miRNA NC (Figure 7B). In contrast, miR-497 inhibitor significantly enhanced the expression of Bcl-2 compared with negative control inhibitor (Figure 7B).
Figure 7.
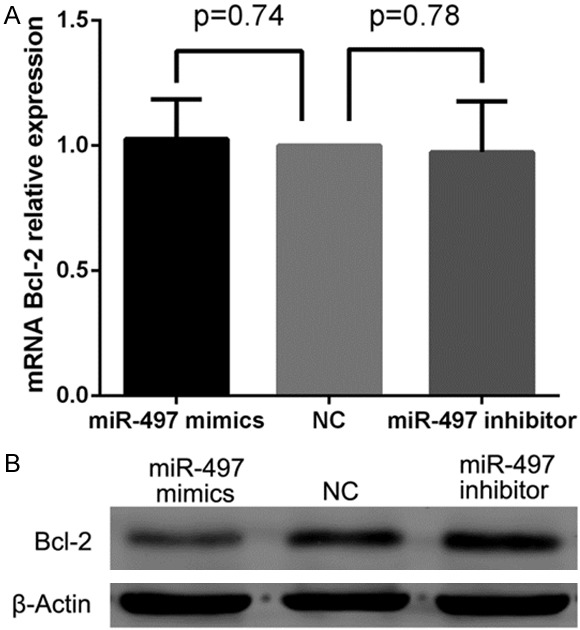
miR-497 negatively regulated Bcl-2 protein expression at the posttranscriptional level. A. No differences were detected in the levels of Bcl-2 mRNA between miR-497 mimics, inhibitor and negative control cells, P>0.05. B. Cells transfected with miR-497 mimic showed decreased endogenous Bcl-2 protein compared with NC. miR-497 inhibitor showed enhanced Bcl-2 protein expression compared with NC.
Taken together, these results suggest that miR-497 does not affect Bcl-2 mRNA stability and instead it negatively regulates Bcl-2 protein expression, at least in part, at the posttranscriptional level.
Effects of Bcl-2 on breast cancer cell proliferation and apoptosis
Because overexpression of miR-497 suppressed the proliferation and induced apoptosis of MDA-MB-231 breast cancer cells, and Bcl-2 was identified as a direct target of miR-497, we hypothesized that Bcl-2 was involved in proliferation and apoptosis mediated by miR-497. We next examined MDA-MB-231 cells knocked down for Bcl-2 expression by Bcl-2 siRNA. Western blot analysis confirmed that Bcl-2 protein was decreased in the siRNA group compared with siRNA NC group (Figure 8E). MTT analysis showed that the Bcl-2 siRNA group showed markedly inhibited cell proliferation, by 14.35%, 32.71% and 39.78% at 48, 72, and 96 h, respectively, compared with cells transfected with the control siRNA (P<0.05; Figure 8A). Furthermore, as shown in Figure 8B, the Bcl-2 siRNA group exhibited fewer colonies than the siRNA NC group (P<0.05), with colony numbers of 44.2±3.65 in the Bcl-2 siRNA group compared with 116.4±2.77 in the siRNA-NC group (P<0.05; Figure 8C). In addition, flow cytometry revealed a higher percentage of early and late apoptotic breast cancer cells in the Bcl-2 siRNA group (24.5±0.08%) compared with the siRNANC group (1.4±0.07%) (P<0.05; Figure 8D). These data indicated that downregulation of Bcl-2 expression inhibited cell proliferation and induced apoptosis, phenocopying the overexpression of miR-497 in MDA-MB-231 cells.
Figure 8.
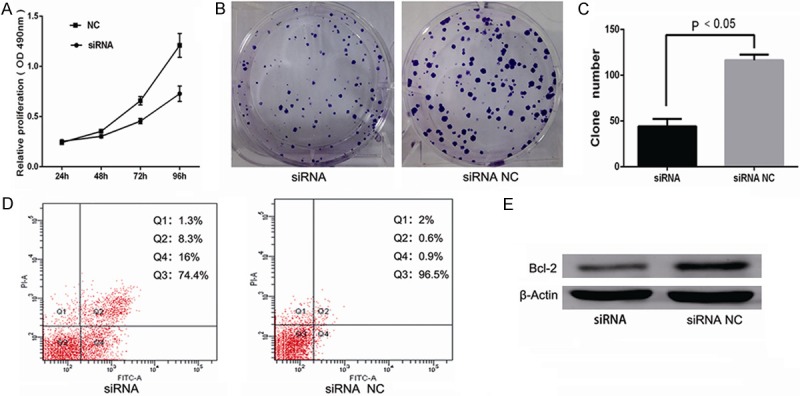
Effects of Bcl-2 in breast cancer cell proliferation and apoptosis. A. Cells transfected with Bcl-2 siRNA showed markedly inhibited cell proliferation compared with control siRNA (P<0.05). B, C. Cells transfected with Bcl-2 siRNA exhibited fewer colonies than the siRNA NC group (P<0.05). D. Inhibition of Bcl-2 promoted the ability of cells to undergo apoptosis (P<0.05). E. The level of Bcl-2 protein was decreased in the Bcl-2 siRNA group compared with siRNA NC group.
Discussion
Breast cancer is one of the most common malignant tumors in females. Surgery and chemotherapy are currently the main treatments, but some patients who undergo chemotherapy show early recurrence or metastasis, resulting in poor prognosis. The identification of new therapeutic targets for breast cancer treatment has been the subject of intensive worldwide research [32], and several molecularly targeted drugs such as raloxifene, letrozole and exemestane have been developed for the treatment of breast cancer. The discovery of the first miRNA, lin-4 in Caenorhabditis elegans, symbolized a new era of miRNA research. Over the last decades, the study on the roles of miRNA in the development of tumors has become a hot topic. Growing evidence indicate that miRNAs may contribute to cancer pathogenesis, and their biogenesis, functions and potential application have become active areas of research. Dysregulation of miRNAs is associated with the initiation and progression of breast cancer, since they may serve as oncogenes or tumor suppressors [11,33,34]. Thousands of miRNAs have been identified and may act as key regulators of carcinogenesis. A large number of studies suggest that more effective targets or targeted drugs for diagnosing and treating breast cancer may involve miRNAs.
Our qRT-PCR results showed that the expression levels of miR-497 in breast cancer specimens and six breast cancer cell lines were significantly lower than that in adjacent normal tissues and MCF-10A cells. Thus, we hypothesized that the expression of miR-497 is associated with breast cancer, and that miR-497 may function as a tumor suppressor. MTT assay, colony formation assay and apoptosis assay support the notion that miR-497 may function as a tumor suppressor in breast cancer.
Several targets of miR-497 have been identified in breast cancer. Shen L et al. found miR-497 induces apoptosis of breast cancer cells by targeting Bcl-w [17]; Luo Q et al. demonstrated miR-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer [18]. But other genes directly regulated by miR-497 may contribute to breast carcinogenesis. Database analysis identified the anti-apoptotic protein Bcl-2 as a potential target of miR-497. Furthermore, correlation analysis and dual-luciferase assays confirmed that Bcl-2 was a direct target of miR-497. Bcl-2 is an important anti-apoptotic protein which can regulate apoptosis, tumorigenesis and the cellular response to anticancer therapies. It can prohibit cytochrome crelease from the mitochondrial intermembrane space into the cytosol as a crucial regulator of the mitochondrial pathway of apoptosis [35].
Bcl-2 can regulate the mitochondrial pathway of apoptosis as an important anti-apoptotic protein through prohibiting release of cytochrome c from the mitochondrial intermembrane space into the cytosol [35]. Many studies have already shown that Bcl-2 is a direct target of miR-497 in neuronal cell, gastric and lung cancer cell lines [13,30]. However, in breast cancer models our study is the first to show the relevance of miR-497 modulation. Moreover, our results describe a novel mechanism of negative regulation of Bcl-2 at the posttranscriptional levels.
To investigate the molecular mechanism underlying the relationship between miR-497 and Bcl-2, we overexpressed miR-497 in MDA-MB-231 cells. Western blot showed that Bcl-2 protein levels were significantly decreased in miR-497 overexpressing cells, while qRT-PCR results showed no differences in Bcl-2 mRNA levels. These results indicated that miR-497 negatively regulates endogenous Bcl-2 protein expression at the posttranscriptional level and not at the mRNA levels.
Silencing Bcl-2 expression by siRNA in MDA-MB-231 cells led to inhibition of cellular proliferation and promoted apoptosis, consistent with the results of ectopic miR-497 expression in the same cells. These findings support the hypothesis that Bcl-2 is a new target of miR-497 in MDA-MB-231 cells.
In summary, our results showed that miR-497 was frequently downregulated not only in breast cancer cell lines, but also breast cancer specimens. Furthermore we observed a significant inverse correlation between the expression of Bcl-2 protein and miR-497 levels, and identified Bcl-2 as a direct physiological target of miR-497, providing a mechanism for the observed inverse correlation in breast cancer cells. Upregulation of miR-497 expression caused cellular growth inhibition and induced apoptosis by targeting Bcl-2. Together these data suggest that miR-497 may be a potential tumor suppressor that inhibits carcinogenesis in human breast cancer. Therefore, miR-497 may serve as a potential diagnostic and therapeutic target for breast cancer.
Acknowledgements
This research was supported by the National Natural Sciences Foundation of China for project 81272240. Furthermore, we give special thanks to all the teachers at the Central Laboratory of the Shanghai Tenth People’s hospital for their technical assistance in this research.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Griffiths-Jones S. Annotating noncoding RNA genes. Annu Rev Genomics Hum Genet. 2007;8:279–298. doi: 10.1146/annurev.genom.8.080706.092419. [DOI] [PubMed] [Google Scholar]
- 4.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 5.Vohradsky J, Panek J, Vomastek T. Numerical modelling of microRNA-mediated mRNA decay identifies novel mechanism of microRNA controlled mRNA downregulation. Nucleic Acids Res. 2010;38:4579–4585. doi: 10.1093/nar/gkq220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu C, Zhao Z. MicroRNA in the molecular mechanism of the circadian clock in mammals. Front Biosci (Landmark Ed) 2013;18:441–446. doi: 10.2741/4112. [DOI] [PubMed] [Google Scholar]
- 7.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi M, Guo N. MicroRNA expression and its implications for the diagnosis and therapeutic strategies of breast cancer. Cancer Treat Rev. 2009;35:328–334. doi: 10.1016/j.ctrv.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Le Quesne J, Caldas C. Micro-RNAs and breast cancer. Mol Oncol. 2010;4:230–241. doi: 10.1016/j.molonc.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iorio MV, Casalini P, Tagliabue E, Menard S, Croce CM. MicroRNA profiling as a tool to understand prognosis, therapy response and resistance in breast cancer. Eur J Cancer. 2008;44:2753–2759. doi: 10.1016/j.ejca.2008.09.037. [DOI] [PubMed] [Google Scholar]
- 12.Poell JB, van Haastert RJ, de Gunst T, Schultz IJ, Gommans WM, Verheul M, Cerisoli F, van Noort PI, Prevost GP, Schaapveld RQ, Cuppen E. A functional screen identifies specific microRNAs capable of inhibiting human melanoma cell viability. PLoS One. 2012;7:e43569. doi: 10.1371/journal.pone.0043569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu W, Zhu D, Lu S, Wang T, Wang J, Jiang B, Shu Y, Liu P. miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med Oncol. 2012;29:384–391. doi: 10.1007/s12032-010-9797-4. [DOI] [PubMed] [Google Scholar]
- 14.Ozata DM, Caramuta S, Velazquez-Fernandez D, Akcakaya P, Xie H, Hoog A, Zedenius J, Backdahl M, Larsson C, Lui WO. The role of microRNA deregulation in the pathogenesis of adrenocortical carcinoma. Endocr Relat Cancer. 2011;18:643–655. doi: 10.1530/ERC-11-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann U, Streichert T, Otto B, Albat C, Hasemeier B, Christgen H, Schipper E, Hille U, Kreipe HH, Langer F. Identification of differentially expressed microRNAs in human male breast cancer. BMC Cancer. 2010;10:109. doi: 10.1186/1471-2407-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang Y, Zou C, Zhang X, Liu S, Wang X, Zhao D, Sun Q, Zeng Z, Dress A, Lin MC, Kung HF, Rui H, Liu LZ, Mao F, Jiang BH, Lai L. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clin Cancer Res. 2011;17:1722–1730. doi: 10.1158/1078-0432.CCR-10-1800. [DOI] [PubMed] [Google Scholar]
- 17.Shen L, Li J, Xu L, Ma J, Li H, Xiao X, Zhao J, Fang L. miR-497 induces apoptosis of breast cancer cells by targeting Bcl-w. Exp Ther Med. 2012;3:475–480. doi: 10.3892/etm.2011.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Q, Li X, Gao Y, Long Y, Chen L, Huang Y, Fang L. MiRNA-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer. Cancer Cell Int. 2013;13:95. doi: 10.1186/1475-2867-13-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- 20.Majid A, Tsoulakis O, Walewska R, Gesk S, Siebert R, Kennedy DB, Dyer MJ. BCL2 expression in chronic lymphocytic leukemia: lack of association with the BCL2 938A>C promoter single nucleotide polymorphism. Blood. 2008;111:874–877. doi: 10.1182/blood-2007-07-098681. [DOI] [PubMed] [Google Scholar]
- 21.Iqbal J, Neppalli VT, Wright G, Dave BJ, Horsman DE, Rosenwald A, Lynch J, Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Campo E, Ott G, Muller-Hermelink HK, Delabie J, Jaffe ES, Grogan TM, Connors JM, Vose JM, Armitage JO, Staudt LM, Chan WC. BCL2 expression is a prognostic marker for the activated B-cell-like type of diffuse large B-cell lymphoma. J. Clin. Oncol. 2006;24:961–968. doi: 10.1200/JCO.2005.03.4264. [DOI] [PubMed] [Google Scholar]
- 22.Skirnisdottir I, Seidal T, Gerdin E, Sorbe B. The prognostic importance of p53, bcl-2, and bax in early stage epithelial ovarian carcinoma treated with adjuvant chemotherapy. Int J Gynecol Cancer. 2002;12:265–276. doi: 10.1046/j.1525-1438.2002.01121.x. [DOI] [PubMed] [Google Scholar]
- 23.Yanai S, Nakamura S, Takeshita M, Fujita K, Hirahashi M, Kawasaki K, Kurahara K, Sakai Y, Matsumoto T. Translocation t(14;18)/IGH-BCL2 in gastrointestinal follicular lymphoma: correlation with clinicopathologic features in 48 patients. Cancer. 2011;117:2467–2477. doi: 10.1002/cncr.25811. [DOI] [PubMed] [Google Scholar]
- 24.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med. 2013;13:109–117. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- 26.Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun S, Hong L, Liu J, Fan D. miR-15b and miR-16 modulate multidrug resistance by targeting BCL2 in human gastric cancer cells. Int J Cancer. 2008;123:372–379. doi: 10.1002/ijc.23501. [DOI] [PubMed] [Google Scholar]
- 27.Zhu W, Shan X, Wang T, Shu Y, Liu P. miR-181b modulates multidrug resistance by targeting BCL2 in human cancer cell lines. Int J Cancer. 2010;127:2520–2529. doi: 10.1002/ijc.25260. [DOI] [PubMed] [Google Scholar]
- 28.Kojima K, Fujita Y, Nozawa Y, Deguchi T, Ito M. MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate. 2010;70:1501–1512. doi: 10.1002/pros.21185. [DOI] [PubMed] [Google Scholar]
- 29.Singh R, Saini N. Downregulation of BCL2 by miRNAs augments drug-induced apoptosis--a combined computational and experimental approach. J Cell Sci. 2012;125:1568–1578. doi: 10.1242/jcs.095976. [DOI] [PubMed] [Google Scholar]
- 30.Yadav S, Pandey A, Shukla A, Talwelkar SS, Kumar A, Pant AB, Parmar D. miR-497 and miR-302b regulate ethanol-induced neuronal cell death through BCL2 protein and cyclin D2. J Biol Chem. 2011;286:37347–37357. doi: 10.1074/jbc.M111.235531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Ljungberg BJ, Jacobsen J, Rudolfsson SH, Lindh G, Grankvist K, Rasmuson T. Different vascular endothelial growth factor (VEGF), VEGF-receptor 1 and -2 mRNA expression profiles between clear cell and papillary renal cell carcinoma. BJU Int. 2006;98:661–667. doi: 10.1111/j.1464-410X.2006.06387.x. [DOI] [PubMed] [Google Scholar]
- 33.Luo Q, Li X, Li J, Kong X, Zhang J, Chen L, Huang Y, Fang L. MiR-15a is underexpressed and inhibits the cell cycle by targeting CCNE1 in breast cancer. Int J Oncol. 2013;43:1212–1218. doi: 10.3892/ijo.2013.2034. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Kong X, Zhang J, Luo Q, Li X, Fang L. MiRNA-26b inhibits proliferation by targeting PTGS2 in breast cancer. Cancer Cell Int. 2013;13:7. doi: 10.1186/1475-2867-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Liu X, Bhalla K, Kim CN, Ibrado AM, Cai J, Peng TI, Jones DP, Wang X. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]


