Abstract
As a representative fluoroquinolone antibacterial, ciprofloxacin is frequently used to treat infections caused by bacteria such as E. coli. It is much meaningful to explore ciprofloxacin susceptibility and investigate a possible mechanism of drug susceptibility changes in E. coli ATCC25922 exposed to the environmental stress of simulated microgravity. The subculture of E. coli lasted for 7 days under simulated microgravity conditions (SMG) and normal microgravity (NG) conditions. On the 8th day, the cultures were divided into three groups: (1) NG group (continuous NG cultures); (2) SMG group (continuous SMG cultures); (3) SMCNG group (simulated microgravity change into normal gravity cultures). Ciprofloxacin (a final concentration of 0.125 μg/ml) sensitivity and expression of acrAB-tolC genes were detected in E. coli cells. The count and percentage of viable cells in the SMG cultures bacteria exposed to ciprofloxacin were higher than that in NG cultures and reduced to the levels of NG group when they were subcultivated from SMG to NG. The expressions of efflux pump genes (acrA, acrB and tolC) were upregulated in SMG culture and downregulated to the levels of NG group when they were subcultivated from SMG to NG. Susceptibility to ciprofloxacin and expression of acrAB-tolC genes in E. coli could be reversibly affected by SMG conditions. Over expression of efflux pump genes acrAB-tolC perhaps played an important role in decreased CIP susceptibility under SMG.
Keywords: Simulated microgravity, E. coli, ciprofloxacin, drug susceptibility, acrAB-tolC genes
Introduction
The weightless environment in space flight is a special stressful status. Microgravity induces a vast array of changes in organisms ranging from bacteria to humans. Studies have showed that the microgravity has a series of adverse impacts on the microorganisms [1,2]. If, during extended space travel or residence, someone picks up some sort of bacterial infection, it is very important to know if the medicine being used will be effective.
Several types of E. coli exist as part of the normal flora of the human gut and have many beneficial functions. E. coli is often considered an opportunistic pathogen because it could cause infection whenever it has the opportunity. When it shifts from intestine to other organs and tissues-it often causes a variety of human infections in human body, such as peritonitis, cholecystitis and cystitis [3]. The E. coli cell itself could be affected by various stressful environmental factors including the weightless environment [4-6]. The changes in biological features are particularly complicating and astronauts’ immune systems are not as strong in space. They may increase the risk of E. coli infection and reduce the efficacy of antibiotic treatment in eradicating bacteria [7]. As a representative fluoroquinolone antibacterial, ciprofloxacin is also frequently used to treat infections caused by bacteria such as E. coli. This study may lead to a greater understanding of antibiotic efficacy under microgravity.
Microgravity is a condition where the physical force of gravity is reduced. The rotary cell culture system (RCCS) manufactured by Synthecon (Houston, TX) was used to generate a simulated microgravity environment in ground-based studies. This apparatus consists of a moto, a cylindrical high-aspect ratio vessel (HARV) bioreactor and a platform on which the vessel is rotated. Aeration was achieved through a semipermeable membrane at the back of the vessel. The mixture of bacteria, inoculums and medium were used to fill HARVs. Air bubbles are removed to eliminate turbulence and ensure a low shear environment. It is then attached to the platform. If the axis of rotation is parallel to gravity, normal gravity (NG) conditions are achieved. If the axis of rotation is perpendicular to gravity, the vessel randomizes the gravitational vectors felt on the cultured cells, allowing the cells to experience “free-fall” and maintaining the cells in a continuous suspended orbit (Figure 1) [8].
Figure 1.
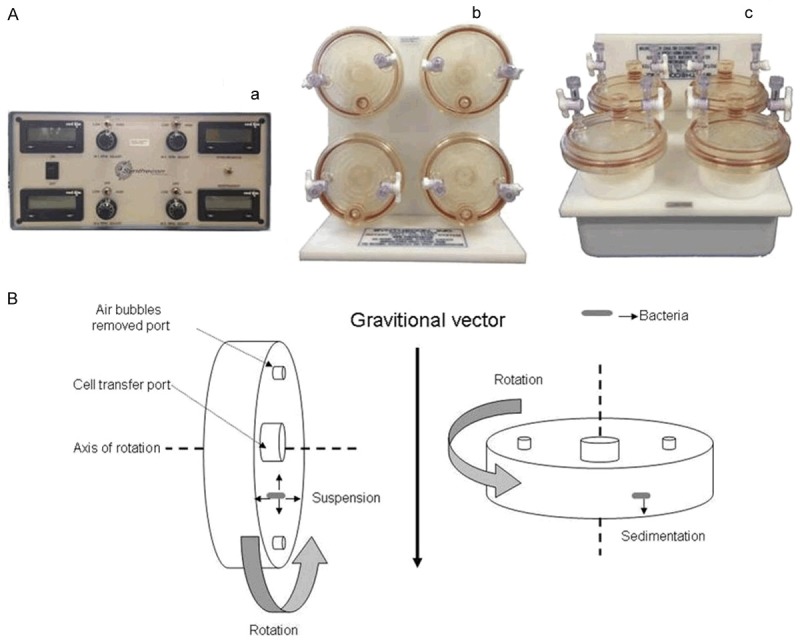
A. RCCS system used to generate a simulated microgravity environment in ground-based investigations (Synthecon Inc, Houston, Texas). a. moto; b. HARVs and a platform (The axis of rotation is perpendicular to gravity); c. HARVs and a platform (The axis of rotation is parallel to gravity). B. The high aspect ratio vessel (HARV) is used to simulate microgravity conditions.
Materials and methods
Bacterial strains, medium and growth conditions
E. coli ATCC25922 was obtained from the China General Microbiological Culture Collection Center, which was susceptible to more antimicrobial categories. Cells were grown at 220 rpm and 37°C for 12-16 h in Luria-Bertani (LB) medium. Overnight cultures were diluted 1:1000000 into fresh medium. The fresh cultures were added respectively into the HARVs of normal gravity group (NG) and simulated microgravity group (SMG) and air bubbles are removed. The cultured solution was taken and diluted 1:1000000 into fresh medium every 24 h, and the remaining was discarded. New culture medium was added into HARVs for continuous rotating culture, and the subculture lasted. On the 8th days of culture, the cultured solution was taken and diluted 1:10 into fresh medium and new culture medium was added into HARV for 3 h rotating culture. Bacterial cells were collected from the HARVs representing mid-late log phase and quickly frozen on dry ice and stored at -80°C for RNA extraction. Normal gravity group continued to horizontally rotate. Simulated microgravity group was divided into simulated microgravity group which continued to perpendicularly rotate and normal gravity group (SMCNG) which changed perpendicular rotation to horizontal rotation. SMCNG group experienced gravitational change from simulated microgravity to normal gravity. Three replicate HARVs were used for each condition and the rotation speed was 25 rpm.
Drug susceptibility testing under simulated microgravity
On the 8th day, after 3 h the cultures (optical density at 600 nm of 0.50-0.6) were used for the measurement of antibiotic susceptibility. The optical density of the cultures was adjusted to 0.100. Ciprofloxacin was added to a final concentration of 0.125 μg/ml. The fresh CIP-containing cultures were added respectively into the HARVs of normal gravity group (NG) and simulated microgravity group (SMG) and air bubbles are removed for 3 h rotating culture. Normal gravity group continued to horizontally rotate. Simulated microgravity group was divided into simulated microgravity group which continued to horizontally rotate and normal gravity group (SMCNG) which changed perpendicular rotation to horizontal rotation. Three replicate HARVs were used for each condition and the rotation speed was 25 rpm.
MTT assay
100 µl E. coli culture dilutions (1:10) were placed in 96-well microtiter plate followed by addition of 10 µl of 5 mg/ml methyl thiazolyl tetrazolium (MTT, Sigma-Aldrich, United States) to each well. The plate was incubated in the dark environment at 37°C for 2 to 5 h. The supernatant was removed from the wells and the formazan was dissolved in 100 µl of DMSO for 15 min. The absorbance was measured at 570 nm for MTT assay using an ELISA reader (Bio-Rad Model 680 microplate reader, Bio-Rad laboratories, Unite States) [9].
Flow cytometry assay
Dead and live bacteria were stained using LIVE/DEAD BacLight Bacteria Viability Kit™ (Invitrogen Molecular probes Inc., Oslo, Norway). This kit utilizes fluorescent dyes that stain live and dead bacteria fluorescent green and fluorescent red, respectively. Samples were washed twice and diluted in filtered sterile normal saline. The average cell concentration was kept constant at 7.5×106 cfu/ml. We prepared 100% proportions of live and dead cells using 70% alcohol killed E. coli cells as control samples (Figure 2). Dual staining was also performed for all the samples. All the samples were then incubated in the dark for 20 min and placed on ice before flow cytometry analysis (FACS Calibur, Becton and Dickinson Company, Unite States). Flow cytometric detectors and compensation settings for the different quadrants were performed using control samples.
Figure 2.
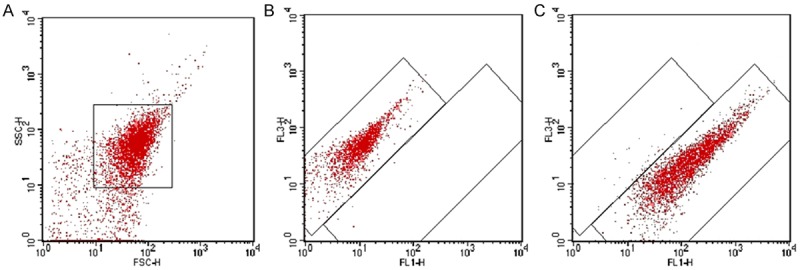
FSC/SSC scatter diagram was used for gating to set target bacterial cells, and FL-1/FL-3 scatter diagram was used to analyze the ratio of inactivated and viable bacterial cells. A. Bacteria cells; B. 100% dead cells; C. 100% viable cells.
RNA extraction
1 ml of cultures was centrifuged at 13000 rpm for 10 min. After centrifugation, the supernatant was discarded, 200 μl of lysis solution (a mixture of TE [Tris-ethylene diamine tetra-acetic acid] and lysozyme) was added and the samples were incubated for 1 h at 37°C. Total RNA was isolated with the RNeasy mini purification kit (Qiagen, Valencia, CA, United States), including an additional step of oncolumn DNase digestion with RNase-free DNase (Qiagen, Valencia, CA, United States). RNA concentration and quality was determined by using an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). RNA quality was considered good after appearance of two intact peaks in the electropherogram, the presence of two distinct bands representing 23S and 16S rRNA, absence of high molecular weight bands during capillary gel electrophoresis, and a ratio of A260/A280 close to 1.9.
Reverse transcriptase (RT)
RT reactions were conducted using the AMV first strand cDNA synthesis kit (New England Biolabs, New England). 5 μl of total RNA, 1 μl of random hexamers (0.2 g/l) and 5 μl Rnase-free ddH2O were added to a 200 μl PCR tube. Prior to cDNA synthesis, denaturation was performed at 70°C for 5 min. After this step, 4.0 μl reaction buffer (5×), 2.0 μl dNTPs (10 mmol/l), 2 μl of AMV reverse transcriptase (10 U/μl) and 1.0 μl RNase inhibitor (20 U/μl) were added to each reaction to a final volume of 20 μl. The reaction conditions were 5 min at 37°C, 60 min at 42°C and 10 min at 70°C.
Real-time quantitative PCR (qRT-PCR)
Real-time PCR was performed in a StepOneTM Real-Time PCR System (Applied Biosystems, Unite States) using a SybrGreen PCR Master Mix (Applied Biosystems, Unite States) and the Universal Thermal Cycling conditions, which included 2 min at 95°C (enzyme activation), followed by 40 cycles of 95°C for 15 s (denaturation) and 60°C (anneal/extension) for 40 s. At the end of each PCR reaction, a melting curve was also performed. Primers (Table 1) were used at a concentration of 10 μM and the cDNA template was diluted 1/8 in the mix reaction. Relative gene expression was evaluated using the 2-ΔΔCT method to calculate fold induction of expression of the target gene. Normalization of the transcriptional levels was done comparing expression of the 16S gene as an endogenous control.
Table 1.
Sequences of primers used for RT-PCR reactions, designed with the Primer ExpressTM 5.0 software
| Primes | Primers sequences 5’→3’ | Amplicon size (bp) and Melting temperature (°C) |
|---|---|---|
| acrA-F | ATCGCAGAAGTTCGTCCTCA | 136, 58 |
| acrA-R | CCTTTCGCACTGTCGTATGTC | |
| acrB-F | TGAAGAGTTCGGCAAAATCC | 182, 57.8 |
| acrB-R | GATGTCGTAGTTCTCACCACCC | |
| tolC-F | TGCTCCCCATTCTTATCGG | 142, 58.3 |
| tolC-R | TTCAAAGGCAGCATCACGA |
Statistical analysis
All data were analyzed by SPSS 17.0 software and were expressed as mean ± SE. For the comparison of multiple samples, the t-test assesses whether the means of two groups are statistically different from each other. A value of P<0.05 was considered to be statistically significant.
Results
Susceptibility to CIP of E. coli under simulated gravity
On the 8th day, the relative number and the percentage of viable cells in the NG, SMG and SMCNG group were assayed by MTT (Figure 3) and the flow cytometer (Figure 4) respectively. Compared with the NG group, a statistically significant increase of E. coli cells’ viability was detected in SMG group. Compared with SMG group, a statistically significant decrease of E. coli cells’ viability was detected in the SMCNG group.
Figure 3.
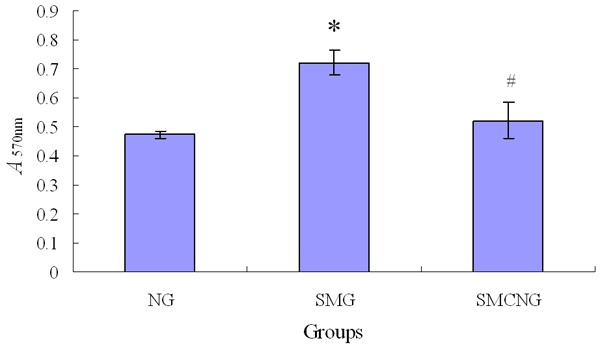
The relative number of viable cells exposed to CIP under simulated microgravity (Compared with NG group, *P<0.05; Compared with SMG group, #P<0.05).
Figure 4.
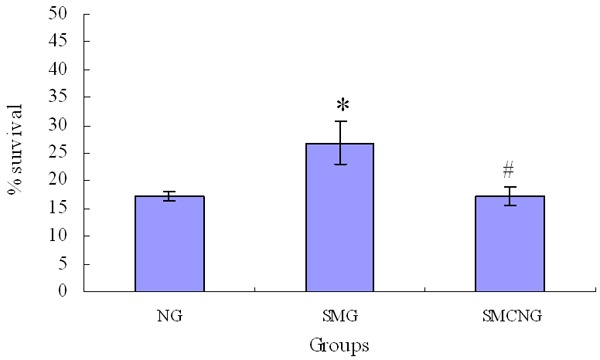
The percentages of viable cells exposed to CIP under simulated microgravity (Compared with NG group, *P<0.05; Compared with SMG group, #P<0.05).
Changes in gene expression of E. coli under simulated gravity
On the 8th day of culture in RCCS, gene expression of E. coli in the NG group, SMG and SMCNG group were assayed respectively by real-time PCR (Figure 5). Up-regulated genes acrA, acrB and tolC have 3.4, 3.4, 3.8-fold change expression levels respectively under SMG conditions compared with NG. Altered expression of these genes can recover when E. coli cultured under SMG are brought to culture under NG.
Figure 5.
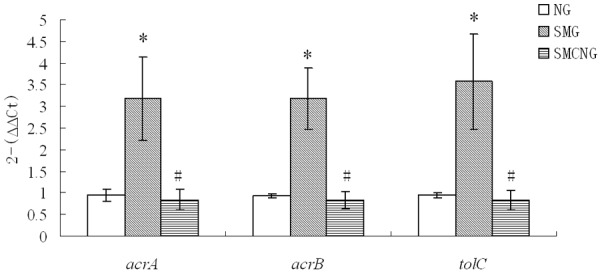
qRT-PCR analysis to determine LSMMG-induced alterations of gene expression related to fluoroquinolone drug sensitivity in E. Coli (Compared with NG group, *P<0.05; Compared with SMG group, #P<0.05).
Discussion
Spaceflight conditions may alter antimicrobial susceptibility. The changes in different antibiotic sensitivity observed in different microbes under microgravity or simulated microgravity conditions are not consistent. Minimum inhibitory concentrations (MICs) of the E. coli isolated against both colistin and kanamycin increased significantly (from 4 mg/l to >16 mg/l) when the bacteria were grown and tested aboard the flight module compared with control experiments conducted on the ground. There were smaller increases in MICs of the S. aureus isolate against oxacillin, erythromycin and chloramphenicol, although very large increases in the thickness of the staphylococcal cell wall were noted following in-flight growth in the absence of antibiotic [10]. At 24 h, the E. coli biofilms were thicker under low sheer modeled microgravity (LSMMG) than their normal-gravity counterparts and exhibited increased resistance to two antibiotics (penicillin and chloramphenicol). Biofilms of a mutant of E. coli, deficient in σs, were impaired in developing LSMMG conferred resistance to the general stressors but not to the antibiotics, indicating other pathways of LSMMG-conferred resistance [11]. Haloferax mediterranei were grown in simulated microgravity (SMG) with the rotary cell culture system. It’s resistance to the antibiotics bacitracin, erythromycin, and rifampicin increased markedly [12]. Simulated microgravity increased Candida albicans’ susceptibility to fluconazole and the susceptibility take on a rising trend with prolonged culture time [13]. Acinetobacter baumannii cells were incubated for 4-5 h under normal earth gravity or LSMMG conditions and no changes in resistance patterns were detected by E-test. Antimicrobials included in the study were amikacin, ampicillin/sulbactam, azithromycin, ceftazidime, gatifloxacin, gentamicin, imipenem, meropenem, minocycline, ticarcillin/clavulanic acid, tigecycline, and trimethoprim/sulfamethoxazole [14].
However, due to limitations of the experimental implementation in space, many researchers began to study the bacterial susceptibility to antibiotics by ground-based facilities for simulation of microgravity. The behavior of bacteria cells grown in a HARV simulates the low-shear microgravity conditions encountered in spaceflight. But many laboratory methodologies for bacterial antimicrobial susceptibility testing can be very difficult to accomplish in HARV, such as the American Journal of Clinical Standards Committee (NCCLS) recommended agar diffusion method and E-test. Despite bacteria growth under microgravity or simulated microgravity conditions, some antimicrobial susceptibility test is not conducted under microgravity or simulated microgravity condition but under normal gravity condition [13,14]. Changes in antibiotic sensitivity caused by simulated microgravity are not constant. Bacterial samples collected on the crew during flight in the Apollo-Soyouz Test Project Mission presented higher antibiotic resistance than controls. When spaceflight bacteria subculture was done in the ground environment, their antibiotic sensitivity recovered to preflight status [15].
E. coli cells grow in continued subculture in HARV under NG and SMG for 7days. In order to avoid the effects of gravity on the experimental results during susceptibility testing process, on the 8th day, CIP and cells cultured under NG and SMG were added into HARVs and continued to be subcultured under NG or SMG conditions respectively. CIP and cells cultured under SMG were added into HARVs and changed to be subcultured under NG. By observing the count and percentage of viable cells in culture, we assessed the susceptibility of E. coli to ciprofloxacin. A statistically significant increase of cells’ viability was detected in SMG group compared with the NG group. A statistically significant decrease of cells’ viability was detected in the SMCNG group compared with SMG group. Therefore, under the experimental conditions, we believe that simulate microgravity reduces the sensitivity of E. coli to ciprofloxacin. Reduced sensitivity is recoverable when E. coli returns to NG conditions.
The bacterial responses to antibiotic drug treatments have proven to be quite complex, involving multiple genetic pathways [16]. Fluoroquinolones are a group of antimicrobial agents with a very good activity for Gram-negative. Molecular mechanisms of fluoroquinolone drug sensitivity in bacterial cells may be mutation or changes in expression levels of drug sensitivity related genes [17]. Generally, fluoroquinolone resistance has been attributed to point mutations in the fluoroquinolone resistance-determining regions of the target genes, such as gyrA [18]. But, we speculate that changes in drug sensitivity under simulated gravity may be related to the latter because decreased CIP sensitivity in E. coli caused by microgravity can recovery under normal gravity. Gene expression analysis of E. coli indicated that the expressions of hundred genes were significantly altered in simulated microgravity conditions compared to that of normal gravity conditions [19,20].
Bacterial efflux pumps are important in resistance to many classes of antimicrobial agents, including fluoroquinolones [21]. The major antibiotic efflux pump of E. coli is AcrAB-TolC. AcrAB-TolC imparts a strong intrinsic resistance phenotype to many clinically significant molecules in E. coli. This complex is composed of a pump, AcrB encoded by acrB, and a periplasmic protein, AcrA encoded by acrA, that exports substrates through a common outer membrane porin, TolC encoded by tolC [22]. The increased level of resistance to fluoroquinolones, such as ciprofloxacin which causes multiple resistance phenotypes is attributed to over activation of multidrug efflux pumps, mainly AcrAB-TolC pump in E. coli [23]. Compared with normal gravity, microgravity caused 3-4 fold increased expression of acrA, arcB and tolC. Over expression of these genes recovered when E. coli cultured under microgravity were brought to culture under normal gravity.
In short, microgravity has been shown to correlate with distinctive changes in CIP susceptibility and gene expression in E. coli. Ciprofloxacin Susceptibility decreased in E. coli under simulated microgravity and reverted to normal values when in-microgravity cultures were subcultured under normal gravity. Changes in CIP sensitivity and gene expression affected by simulated microgravity were consistent and were closely related to gravity condition. Over expression of efflux pump genes acrAB-tolC perhaps played important role in decreased CIP susceptibility under simulated microgravity.
Acknowledgements
This work was supported by the Major Programs of the Military Medical Science and Technique Foundation during 12th Five-Year Plan Period (BWS11J051).
Disclosure of conflict of interest
None.
References
- 1.Nickerson CA, Ott CM, Wilson JW, Ramamurthy R, Pierson DL. Microbial responses to microgravity and other low-shear environments. Microbiol Mol Biol Rev. 2004;68:345–361. doi: 10.1128/MMBR.68.2.345-361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenzweig JA, Abogunde O, Thomas K, Lawal A, Nguyen YU, Sodipe A, Jejelowo O. Spaceflight and modeled microgravity effects on microbial growth and virulence. Appl Microbiol Biotechnol. 2010;85:885–891. doi: 10.1007/s00253-009-2237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hua Y, An X, Pei G, Li S, Wang W, Xu X, Fan H, Huang Y, Zhang Z, Mi Z, Chen J, Li J, Zhang F, Tong Y. Characterization of the morphology and genome of an Escherichia coli podovirus. Arch Virol. 2014;159:3249–3256. doi: 10.1007/s00705-014-2189-x. [DOI] [PubMed] [Google Scholar]
- 4.Baker PW, Meyer ML, Leff LG. Escherichia coli growth under modeled reduced gravity. Microgravity Sci Technol. 2004;15:39–44. doi: 10.1007/BF02870967. [DOI] [PubMed] [Google Scholar]
- 5.Kim HW, Matin A, Rhee MS. Microgravity alters the physiological characteristics of Escherichia coli O157:H7 ATCC 35150, ATCC 43889, and ATCC 43895 under different nutrient conditions. Appl Environ Microbiol. 2014;80:2270–2278. doi: 10.1128/AEM.04037-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang A, Pierson DL, Koenig DW, Mishra SK, Demain AL. Effect of simulated microgravity and shear stress on microcin B17 production by Escherichia coli and on its excretion into the medium. Appl Environ Microbiol. 1997;63:4090–4092. doi: 10.1128/aem.63.10.4090-4092.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tixador R, Gasset G, Moatti N, Lapchine L, Woldringh C, Toorop P, Moatti JP, Delmotte F, Tap G. Behavior of bacteria and antibiotics under space conditions. Aviat Space Environ Med. 1994;65:551–556. [PubMed] [Google Scholar]
- 8.Fang A, Pierson DL, Mishra SK, Koenig DW, Demain AL. Gramicidin S production by Bacillus brevis in simulated microgravity. Curr Microbiol. 1997;34:199–204. doi: 10.1007/s002849900168. [DOI] [PubMed] [Google Scholar]
- 9.Banzi EC, Costa AR, Puppin-Rontani RM, Babu J, Garca-Godoy F. Inhibitory effects of a cured antibacterial bonding system on viability and metabolic activity of oral bacteria. Dent Mater. 2014;30:e238–244. doi: 10.1016/j.dental.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Tixador R, Richoilley G, Gasset G, Templier J, Bes JC, Moatti N, Lapchine L. Study of minimal inhibitory concentration of antibiotics on bacteria cultured in vitro in space (Cytos 2 experiment) Aviat Space Environ Med. 1985;56:748–751. [PubMed] [Google Scholar]
- 11.Lynch SV, Mukundakrishnan K, Benoit MR, Ayyaswamy PS, Matin A. Escherichia coli biofilms formed under low-shear modeled microgravity in a ground-based system. Appl Environ Microbiol. 2006;72:7701–7710. doi: 10.1128/AEM.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dornmayr-Pfaffenhuemer M, Legat A, Schwimbersky K, Fendrihan S, Stan-Lotter H. Responses of haloarchaea to simulated microgravity. Astrobiology. 2011;11:199–205. doi: 10.1089/ast.2010.0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang W, Xu B, Yi Y, Huang Y, Li XO, Jiang F, Zhou J, Zhang J, Cui Y. Effects of simulated microgravity by RCCS on the biological features of Candida albicans. Int J Clin Exp Pathol. 2014;7:3781–3790. [PMC free article] [PubMed] [Google Scholar]
- 14.Hensley DM. Maintenance of antimicrobial susceptibility of Acinetobacter baumannii in modeled microgravity. Clin Lab Sci. 2010;23:84–88. [PubMed] [Google Scholar]
- 15.Lapchine L, Moatti N, Gasset G, Richoilley G, Templier J, Tixador R. Antibiotic activity in space. Drugs Exp Clin Res. 1986;12:933–938. [PubMed] [Google Scholar]
- 16.Hawkey PM. Mechanisms of quinolone action and microbial response. J Antimicrob Chemother. 2003;51(Suppl 1):29–35. doi: 10.1093/jac/dkg207. [DOI] [PubMed] [Google Scholar]
- 17.Becker S, Soukup JM, Gilmour MI, Devlin RB. Stimulation of human and rat alveolar macrophages by urban air particulates: effects on oxidant radical generation and cytokine production. Toxicol Appl Pharmacol. 1996;141:637–48. doi: 10.1006/taap.1996.0330. [DOI] [PubMed] [Google Scholar]
- 18.Goldsmith CA, Frevert C, Imrich A, Sioutas C, Kobzik L. Alveolar macrophage interaction with air pollution particulates. Environ Health Perspect. 1997;105:1191–5. doi: 10.1289/ehp.97105s51191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li N, Wang M, Oberley TD, Sempf JM, Nel AE. Comparison of the pro-oxidative and proinflammatory effects of organic diesel exhaust particle chemicals in bronchial epithelial cells and macrophages. J Immunol. 2002;169:4531–4541. doi: 10.4049/jimmunol.169.8.4531. [DOI] [PubMed] [Google Scholar]
- 20.Raja V, Eric M, Laura L. Changes in Gene Expression of E. coli under Conditions of Modeled Reduced Gravity. Microgravity Sci Technol. 2008;20:41–57. [Google Scholar]
- 21.Grkovic S, Brown MH, Skurray RA. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev. 2002;66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


