Abstract
Tim-3 (T cell immunoglobulin and mucin domain 3), belonging to the member of the novel Tim family, has been confirmed that it plays a critical negative role in regulating the immune responses against viral infection and carcinoma. Recently, it has also been reported that the over-expression of Tim-3 is associated with poor prognosis in solid tumors. However, the role of Tim-3 in colorectal cancer remains largely unknown. In the current study, we aim to investigate the expression of Tim-3 in colorectal carcinoma and discuss the relationship between Tim-3 expression and colon cancer prognosis, thus speculating the possible role of Tim-3 in colon cancer progression. Colon cancer tissues and paired normal tissue were obtained from 201 patients with colon cancer for preparation of tissue microarray. Tim-3 expression was evaluated by immunohistochemical staining. The Tim-3 expression level was evaluated by q-RT-PCR, western blot and immunocytochemistry in four colon cancer cell lines (HT-29, HCT116, LoVo, SW620). Tim-3 was expressed in 92.5% tumor tissue samples and 86.5% corresponding normal tissue samples. Expression of Tim-3 was significantly higher in tumor tissues than in normal tissues (P < 0.0001). Tim-3 expression in colon cancer tissues is in correlation with colon cancer lymphatic metastasis and TNM (P < 0.0001). Multivariate analysis demonstrated that Tim-3 expression could be a potential independent prognostic factor for colon cancer patients (P < 0.0001). Kaplan-Meier survival analysis result showed that patients with higher Tim-3 expression had a significantly shorter survival time than those with lower Tim-3 expression patients. Our results indicated that Tim-3 might participate in the tumorgenesis of colon cancer and Tim-3 expression might be a potential independent prognostic factor for patients with colorectal cancer.
Keywords: Tim-3, colon cancer, prognosis, overall survival
Introduction
Colorectal cancer (CRC) is the third leading cancer and the fourth cause of cancer deaths worldwide, with 1.2 million estimated new cases and 609,000 estimated deaths in 2008 [1]. In China, CRC is the fourth most commonly diagnosed tumor and the fifth most frequently cause of mortality among tumor suffers. There were about 153760 new diagnosed of CRC patients and 78700 people taken away their lives [2]. The high mortality rate primarily result from the difficulty in diagnosing CRC at early stages and the deficiency of more effective therapy method. Certainly, the barren manner to judge prognosis accurately at the beginning of the diagnosed colon cancer patients, thus drawing less attention on tumor and selecting fewer effective therapies, is also a significant factor. As health stander is increasingly improved greatly, more effective treatment method and prognosis indicator for CRC are necessary urgently. Consequently, novel more effective therapy method and prognosis effectors should be explored as soon as possible.
T-cell immunoglobulin-and mucin domain-3-containing molecule 3 (Tim-3) was originally identified as a membrane protein of CD4+ Th1 cell rather than Th2 cell [3-5]. Parallel to other co-inhibitory check-point inhibitors-PD, CTLA-4, Tim-3 plays a negative role in host, especially to immune system. Engagement of Tim-3 with its ligand galectin-9 negatively regulates interferon γ secretion T cells [6] and promoting the death of interferon (IFN)-gamma-inducing Th-1 cells [7,8]. Emerging data also suggest that Tim-3 may take center stage in T cell exhaustion-a state that T cells fail to proliferate and exert effective functions such as cytotoxicity and cytokine secretion in response to antigen stimulation. Primarily, evidence have identified that Tim-3 was expressed on exhausted T cells in both human cancer [9,10] and in preclinical models of cancer [11,12]. Further, to melanoma patients, CD8+ T cell dysfunction has an intimate relationship with the check-point inhibitors-Tim-3 [13]. Moreover, evidence suggested that Tim-3 suppresses CD4+ T cell activation through the interleukin-6-STAT3 pathway [14]. Inspiringly, studies results had demonstrated that blocking Tim-3 can restore T cell proliferation and enhance cytokine production to HIV and HCV patients [7,15]. Given these observations, it appears that Tim-3 may be an effective target in reversing T cell exhaustion, thus efficient treating neoplasm patients.
Recently, Tim-3 was reported to be expressed in melanoma cells, contributing to low adhesion of tumor cells and favoring of the survival of melanoma cells [16]. Evidence also suggested Tim-3 was identified expressed on many carcinoma cells such as cervical cancer cell, Gastric Cancer cells, prostate cancer cell, Clear cell renal carcinoma cell [17-20] and Tim-3 directly plays a significant role in the occurrence and development of tumors [19-21]. The role of Tim-3 expressed in tumor cells may promote tumor genesis, proliferation and invasion capacity directly or via suppressing immunity. In addition, researches indicated that the proliferation and invasion capacity of tumor cells whose Tim-3 gene were knocked down were diminished [17,19,20]. Consequently, Tim-3 maybe a new target to treat cancer effectively. Now, investigating the role of Tim-3 in tumor has been becoming a hotspot. However, whether Tim-3 is also expressed on CRC and the role of Tim-3 in colon cancer remain unknown.
In this study, we investigated the expression of Tim-3 in colorectal cancer cell lines and in colon cancer tissues and revealed the relationship between Tim-3 expression and colon cancer prognosis further.
Materials and methods
Cell culture
Human colon cancer cell lines HT-29, SW-620, HCT116, and LoVo, were received from Department of Enze Medical Research Centre, Taizhou Hospital of Wenzhou Medical University. Cells were cultured in the medium according to ATCC mixed with 10% heat-inactivated fetal bovine serum (FBS) in an incubator (contain 5% CO2) at 37°C with a humidified atmosphere. Culture medium was replaced every 2-3 days. When cells were in logarithmic growth phase, collected them for further experiments.
Western blot
HT-29, SW-620, HCT116, and LoVo CRC cell lines to detect Tim-3 were lysed in a lysis buffer purchased from Beyotime Institute of Biotechnolog (p0013c, Beyotime, China) and centrifuged at 12000 rpm for 30 min. Then mixed with 5 × loading buffer and boiled the mixture for 5 minutes in boiling water. Equal amounts of protein extracts (80 µg) for electrophoresis loaded in SDS polyacrylamide gel. Then transfered the protein to a polyvinylidene fluoride membrane. The membrane was blocked for 2 hours at room temperature with buffer containing 20 mmol/L Tris-Hcl (pH7.5), 500 mmol/l NaCl, and 5% nonfat milk. Then incubated the membrane with contain Tim-3 antibody (1:1000, Abcam, US) buffer overnight at 4°C overnight. Then washed the membrane with PBS buffer contain 0.1% Tween20 and incubated with a horseradish peroxidase-labeled secondary antibody rabbit anti-mouse IgG (1:10000) for 2 hours at room temperature. Finally, the blots were developed using an enhanced chemiluminescence detection system (Amersham Life Science) and analyzed. We utilized Hela and 293T cell line-s predominantly expressed Tim-3 protein as positive control, while THP-1 cell line rarely expresses Tim-3 protein as negative control. β-actin was used as internal control to normalize the blots.
qRT-PCR
Total cellular RNA was isolated from cell by Trizol reagent (Invitrogen, Shanghai, China). Then RNA was reverse transcribed to generate cDNA using Thermo Scientific RevertAid First Strand cDNA Synthesis Kit (Thermo, NY, USA) according to the manufacturer’s protocol. Then use primers of Tim-3 5sense primer (5’-TACTGCTGCCGGATCCTAAT-3’) and a 3 antisense primer (5’-ACCTTGGCTGGTTTGATGAC-3’) to amplify Tim-3 and use 5 sense primer (5’-CTCACGAAACTGGAATAAGC-3’) and a 3 antisense primer (5’-AAGCCACACGTACTAAAGGT-3’) to amplify a 200-bp β-actin as control, Quantitative real-time PCR (qRT-PCR) was performed with SYBR Green Master Mix (Roche, Shanghai, China) on a LightCycler 480II system (Roche). Then checked the quantity of the PCR product in a 2% agarose gel. Make the solution to become lukewarm and added 0.1 mg/ml ethidium bromide. Poured the gel on a gel-casting tray and lead it to solidify. Placed the gel in an electrophoresis tank with 1 × TAE buffer. Mixed The PCR product samples with 6 × DNA loading dye and loaded them on the gel. Lastly, electrophoresed the gel at 2 volts/cm and took image in a gel documentation system (Bio Rad Gel Doc).
Patients
This study was approved by the ethics committee of Taizhou Hospital, Wenzhou Medical University for analysis of human tissues. All patients gave informed written consent for analysis of their tissue for research purposes. CRC patients underwent initial resection at the Taizhou Hospital Wenzhou Medical University from 2005 to 2010 were enrolled. The patients’ age ranged from 26 to 90 years (median age, 65 years). All of the selected colon cancer tissues met the following inclusion criteria: (1) the patient underwent curative resection, (2) the patient had a regular follow-up, (3) there were adequate specimens. Exclusion criteria were (1) history of previous malignant disease or a second primary tumor, (2) familial adenomatous polyposis patients, (3) preoperative chemotherapy and/or radiation. All patients were diagnosed colon cancer according to World Health Organization criteria by two specialists of the hospital. Following clinical parameters were collected in this study: age at the time of surgery, gender, cancer embolus, differentiation, depth of invasion, lymph metastasis, distant metastasis, American Joint Committee on Cancer (AJCC) stage (TNM) and survival time.
Follow-up
Basically, patients were evaluated at the hospital or contacted by telephone or e-mails, every 3 months for the first year, every 6 months for the second year, and annually three after. The primary end point of this study was overall survival (OS) defined as the time from one month later after surgery to death from any cause. The secondary end point was the end of follow-up.
Immunohistochemical staining
Briefly, the tissue microarray (TMA) were roasted overnight at 60°C, then deparaffinized and rehydrated through graded alcohols before being exposed to the antigen retrieval system (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) in high-pressure boiler for 30 second. Endogenous peroxidase was blocked with 3% hydrogen peroxide for 15 minutes at room temperature. Then rinsed the slides in 0.01 mol/L phosphate-buffered saline solution (PBS) for (3 × 5 min). Following incubation with the primary mouse polyclonal antibody (1:200, Abcam, US) 1 hour at room temperature. After washing the slides with PBS (3 × 5 min), incubated the slides with rabbit antimouse avidin-biotin peroxidase complex kit (1; 50, Gene Tech, China). Then stained the slides with DAB peroxidase substrate kit (Gene Tech, CA). Hematoxylin and eosin staining was performed on the microarry for histopathological evaluation. Known immunostaining-positive slides were used as positive controls. Negative controls with normal serum or isotope IgG were performed in every procedures. Cell lines (HT-29, SW620, HT-629, LoVo) were routinely cultured, embedded in paraffin and processed as tissue slides.
Evaluation of the IHC variable
All microarries were independently analyzed by two experienced pathologists with no prior knowledge of clinicopathological parameters under a light microscope and the images were recorded by digital camera. To evaluate Tim-3 density, the stained sections were screened under low power (× 100-fold magnification) to identify representative fields. Tim-3 positive cells were then counted under high power (HP; × 400-fold magnification) in 5 fields of vision using the Leica DMI 4000B inverted research microscope (Leica Microsystems, Wetzlar, Germany). Through comparing to the immunoreactivity of positive controls that were included in each process, pathologist could estimate percentages of Tim-3-positive CRC cells and the immunostaining intensity. The stain intensity was divided into 4 categories: no staining = 0, weak staining = 1, moderate staining = 2 and strong staining = 3 and the percentage of the stain cells were judged at same time. Then the staining results were revealed with semi-quantitative analysis (HSCORE system = stain intensity × the percentage of the stain cells). The interobserver agreement for the Tim-3 count was 88%. Disagreements were reevaluated until a consensus was reached.
Statistical analysis
The SPSS statistical 18.0 software was used to analyze the research data. Paired t-test was used to analyze Tim-3 expression between tumor tissues and normal tissues. The Kaplan-Meier method was used to evaluate the overall survival rate as a function of time. The Cox proportional hazard model was used in univariate and multivariate analysis of prognostic factor. Value of P < 0.05 was considered to be statistically significantly.
Results
Tim-3 is expressed in CRC cell lines
The above referenced cell lines of CRC were used to detect Tim-3 mRNA and protein through qRT-PCR and western blot. The results were shown in Figure 1A and 1B. There was no difference of expression of Tim-3 in the level of mRNA, whereas difference of Tim-3 protein expression is apparent. Interestingly, STAT3 were expressed in all of the four cell lines and pSTAT3 of HCT116 is activated (Figure 1A).
Figure 1.
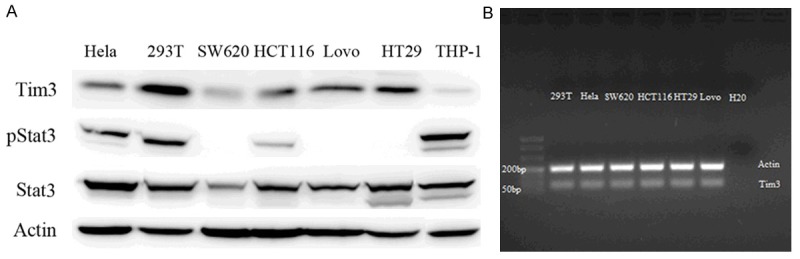
Expression of Tim-3 in colorectal cancer cell lines. Tim-3 protein was verified by western blot in SW620, HCT116, HT29, LoVo cell lines (A). Hela and 293T cell lines as positive control and THP-1 as negative control. The Tim-3 expression was also detected in the mRNA level (B).
Tim-3 expression in colon cancer tissues
According to recent research reports Tim-3 is expressed not only in various immunocytes but also in several kinds of carcinoma cells [18-21]. The phenomena attract us to explore whether Tim-3 is also expressed in colorectal carcinoma tissues. From the (Figure 2), the brown-stained part of the immunohistochemical analysis image is Tim-3 Protein (Figure 2A-F). As shown in Table 1, Tim-3 positive colon carcinoma cells could be detected in 92.5% (186/201) of colon cancer specimens, of which 58.7% (118/201) showed Tim-3 expression HSCORE over 200 (Figure 2A), 33.8% (68/201) showed Tim-3 expression HSCORE less than 200 (Figure 2C) and 7.4% (15/201) negative (-) (Figure 2E) Tim-3 expression, and Tim-3 can be detected in 86.6% (174/201) of normal colon specimens, in which 9.5% (19/201) showed Tim-3 expression HSCORE over 200 (Figure 2B), 77.1% (155/201) showed Tim-3 expression HSCORE less than 200 (Figure 2D), 13.4% (27/201) negative (-) (Figure 2F) Tim-3 expression. Tim-3 expression in cancer lesions was significantly higher than in compared normal tissues (P < 0.0001, not shown). In addition, cell lines HT-29 and SW620 showed cytoplasmic and nuclear positivity of Tim-3 staining and HCT116, LoVo showed cytoplasmic Tim-3 staining (Figure 3A-D).
Figure 2.
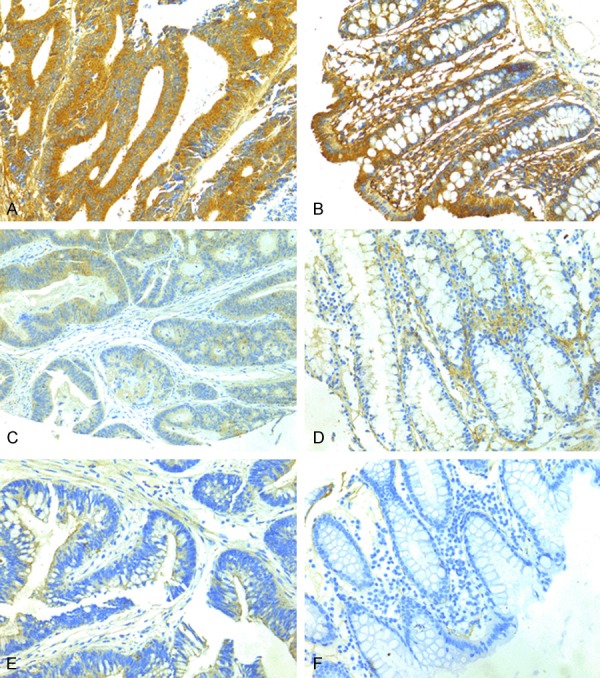
Immunohistochemical staining of Tim-3. (A) Tim-3 expression strong stain, (C) weak stain, (E) negative stain in tumor tissues and the Tim-3 expression (B) strong stain, (D) weak stain, and (F) negative stain corresponding normal tissues.
Table 1.
Correlation between Tim-3 expression and clinicopathologic parameters in patients with colon cancer (n = 201)
| Clinicopathologic parameters | Patients (n) | Tim-3 expression | P Value | |
|---|---|---|---|---|
|
| ||||
| < 200 | ≥ 200 | |||
| Age | 0.599 | |||
| ≤ Median (65 y) | 103 | 46 | 57 | |
| > Median | 98 | 37 | 61 | |
| Gender | 0.634 | |||
| Male | 116 | 49 | 67 | |
| Female | 85 | 34 | 51 | |
| Cancer embolus | 0.075 | |||
| Yes | 42 | 10 | 32 | |
| No | 159 | 73 | 86 | |
| Differentiation | 0.242 | |||
| Well | 28 | 13 | 15 | |
| Middle | 162 | 63 | 99 | |
| Poor | 11 | 7 | 4 | |
| Depth of invasion | 0.135 | |||
| T1 | 3 | 0 | 3 | |
| T2 | 16 | 5 | 11 | |
| T3 | 46 | 20 | 26 | |
| T4 | 136 | 58 | 78 | |
| Lymph metastasis | < 0.0001 | |||
| N0 | 114 | 60 | 54 | |
| N1 | 56 | 18 | 38 | |
| N2 | 31 | 5 | 26 | |
| Distant metastasis | 0.845 | |||
| Negative | 192 | 79 | 113 | |
| Positive | 9 | 4 | 5 | |
| TNM | < 0.0001 | |||
| I | 15 | 4 | 11 | |
| II | 92 | 55 | 37 | |
| III | 82 | 20 | 62 | |
| IV | 12 | 4 | 8 | |
Figure 3.
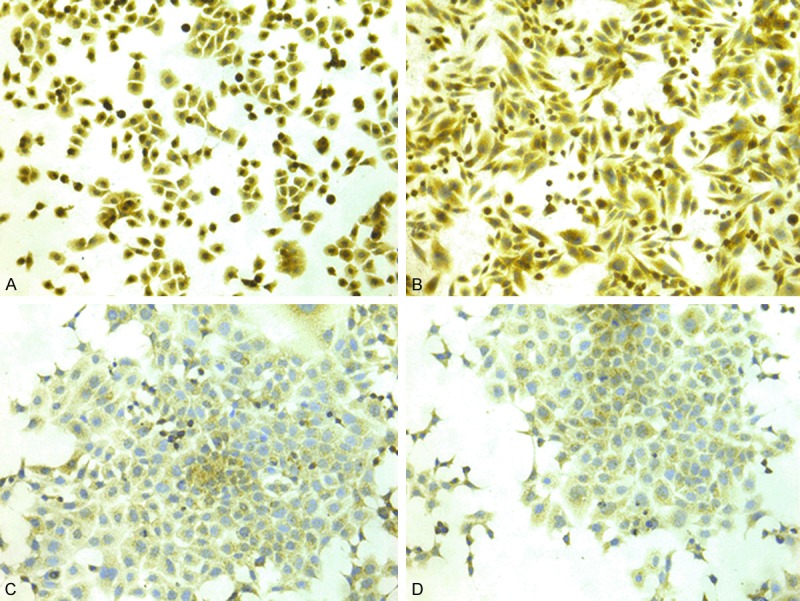
Immunocytochemical staining of Tim-3 in colorectal cancer cell lines. Tim-3 expression in cell lines of HT-29 (A), SW620 (B), HCT116 (C) and LoVo (D).
Correlation of Tim-3 expression with clinicopathologic parameters
Correlation between Tim-3 expression and various clinicopathologic parameters was implemented in the 201 colon tumor samples as shown in Table 1, Tim-3 expression has positively significant correlation with lymph metastasis, TNM (all values of P < 0.0001) in CRC tissues. However, as shown in Table 1, the Tim-3 expression has no significant correlation with age, gender, cancer embolus, differentiation, depth of invasion, and distant metastasis (P > 0.05). Notably, the frequency of higher Tim-3 expression was higher in patients with differentiation middle/poor and in T3/T4 depth of invasion stages than those with well differentiation and in T1/T2 depth of invasion stages (Table 1).
Tim-3 expression and overall survival
Thirty days after postoperative complications period, all the 201 patients were conducted follow-up. The post period of follow-up last 120 months and the sheathed stage of it ranged from 2 to 103 months (median 61 months). HSCORE of Tim-3 expression was classified into two kinds (HSCORE < 200 and HSCORE ≥ 200). Firstly, the expression of Tim-3 in colon tissues had a statistically significant correlation with a shorter of the survival probability (P < 0.05, data not shown). During the follow-up, there were 54 (26.9%) patients died. Among these deaths, the ten-year survival rate of patients with Tim-3 expression HSCORE ≥ 200 was significantly lower than those HSCORE < 200 (P < 0.0001, log-rank test) (Figure 4). In addition, the survival time difference was directly shown in Kaplan-Meier survival curve, which was plotted according to the HSCORE assortment above (Figure 4). As shown in Table 2, the results suggested that TNM (P = 0.003), distant metastasis (P = 0.011), cancer embolus (P = 0.002) and Tim-3 expression (P < 0.0001) had significance relationship with survival rate (P < 0.05). All these analysis suggested that Tim-3 expression could be an independent prognostic factor for colon cancer patients (P < 0.0001).
Figure 4.
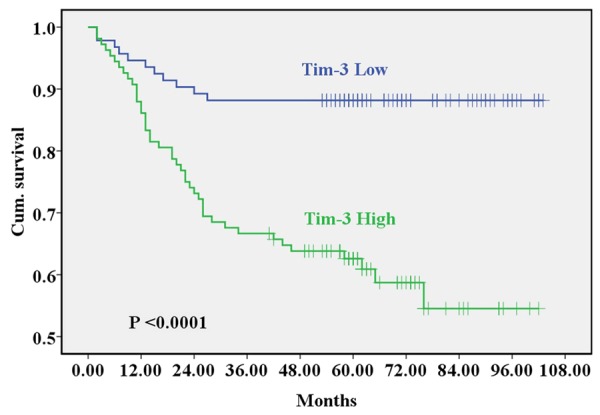
The Kaplan-Meier survival analysis comparative curves between negative/weak Tim-3 expression and strong Tim-3 expression in colon cancer patients (P < 0.05).
Table 2.
Correlation of clinicopathologic parameters and Tim-3 expression with the survival rate of colon cancer patients (n = 201)
| Clinicopathologic Parameters | Patients (n) | 10 years survival Tim-3 expression | Overall survival rate (P Value) | |
|---|---|---|---|---|
|
| ||||
| < 200 | ≥ 200 | |||
| Age | 0.181 | |||
| ≤ Median (65 y) | 103 | 46 | 31 | |
| Median | 98 | 36 | 34 | |
| Gender | 0.673 | |||
| Male | 116 | 48 | 37 | |
| Female | 85 | 34 | 28 | |
| Cancer embolus | 0.002 | |||
| Yes | 42 | 10 | 8 | |
| No | 159 | 72 | 57 | |
| Differentiation | 0.346 | |||
| Well | 28 | 12 | 6 | |
| Middle | 162 | 63 | 58 | |
| Poor | 11 | 7 | 1 | |
| Depth of invasion | 0.204 | |||
| T1 | 3 | 0 | 3 | |
| T2 | 16 | 5 | 8 | |
| T3 | 46 | 20 | 16 | |
| T4 | 136 | 57 | 38 | |
| Lymph metastasis | 0.108 | |||
| N0 | 114 | 60 | 36 | |
| N1 | 56 | 18 | 20 | |
| N2 | 31 | 4 | 9 | |
| Distant metastasis | 0.011 | |||
| Negative | 192 | 78 | 62 | |
| Positive | 9 | 4 | 3 | |
| TNM | 0.003 | |||
| I | 15 | 4 | 9 | |
| II | 92 | 55 | 26 | |
| III | 82 | 19 | 29 | |
| IV | 12 | 4 | 1 | |
| Tim-3 expression | < 0.0001 | |||
| < 200 | 83 | 82 | - | |
| ≥ 200 | 118 | - | 65 | |
Discussion
Tim-3 was originally identified as a surface protein of Th1 cell rather than Th2 cell and Tim-3 could induce Th1 cell apoptosis through interact with its ligand [8]. In addition, Tim-3 has also been found on many other immune cell lines and tumor cell lines [16,22,23]. What’s more, whatever Tim-3 acts as an independent predictor or bear the direct effect to promote tumor evolution, Tim-3 play an all-important role. Here, we conformed Tim-3 was expressed in colon cancer tissues and corresponding compared normal tissues from clinical patients. In addition, we found Tim-3 protein in tumor tissues had a significant correlation with the clinicopathologic parameters according to the statistics. Remarkably, we also found that the high Tim-3 expression patients had a statistic trend of lower survival rate than those with weak/negative Tim-3 expression. The univariate and multivariate analyses also suggested that Tim-3 expression state in tumor tissues could be a potential independent prognostic predictor for colorectal cancer patients.
Though Tim-3 makes significant contribution to tumorigenesis and development, the mechanisms of regulation Tim-3 expression in tumor are not fully understood. New evidence suggested that microenvironment could transform cellular Tim-3 expression status. Huang X et al. [14] found that Tim-3 is preferentially expressed in lymphoma-derived ECs rather than ECs from reactive lymph nodes in the level of mRNA and protein. Similarly, for gastric cancer and cervical cancer Tim-3 expression in tumor tissues was significantly higher than compared normal tissues [17,18]. In line with our study, we also demonstrated that Tim-3 expression in tumor tissues was significantly higher than compared normal tissues. Therefore, we hypothesize that there are factors to induce variation of Tim-3 expression in malignant tumor tissues. Wiener et al. [16] verified that Tim-3 is expressed in melanoma cells and could be up regulation through TGF-β1. Moreover, TGF-β1 plays an important role in tumor development [24,25]. Therefore, tumor environment may affect the expression of TGF-β1, thus regulating Tim-3 expression of tumor cells.
Tim-3 expressed by immune cells plays important roles in regulating innate and acquired immunity. In antiviral and antitumor immunity, evidence has demonstrated that Tim-3 expressed by immune cells not only suppressed innate immunity but also induced CD4+ T cells and CD8+ T cells exhausted [26]. In our study, we have demonstrated that colon cancer cells expressed Tim-3 and Tim-3 expression could be used as an independent prognostic factor for colon cancer patients. As a result, we conjecture that Tim-3 may play an important role in onset, growth, and dissemination of colon carcinoma. However, the function of Tim-3 in colon cancer has not yet been demonstrated explicitly. Geng et al found that Tim-3 could be a form of secreted and bind to T cells by interfering with its ligand(s), thus facilitating tumor growth through impaired immune response [27]. The Tim-3 expression in colon carcinoma cells may work in similar mechanisms of suppressing immune system thus assisting tumor cells escape immune surveillance and tumor growth. However, can Tim-3 play a role in colon cancer cell directly? According to published studies, Tim-3 could promote tumor growth directly, which could be identified by when these tumor cell lines were exposure to Tim-3 siRNA resulted in significant inhibition of cell proliferation and invasion [17,19,28]. Therefore, Tim-3 may also promote tumor growth directly in some ways. Cao et al. have defined Tim-3 can activate the IL-6-STAT3-pSTAT3 pathway [14]. In addition, evidence suggested that the IL-6-STAT3 pathway plays an important role in tumor growth and metastasis [29,30]. In our studies, we demonstrated that all of the four colon cell lines could express STAT3. Fascinatingly, pSTAT3 of HCT116 is activated. Based on these previous findings and our results from this study, we hypothesize that Tim-3 might directly facilitate tumor growth through the IL-6-STAT3 pathway or IL-6-STAT3-pSTAT3 pathway. Further study on the correlation among Tim-3 and colon cancer cells could be conducted to support the hypothesis.
In conclusion, we demonstrated colon carcinoma cells expressed Tim-3 for the first time. In addition, we confirmed that Tim-3 expression could as an independent prognostic factor for patients with colon cancer patients. We also demonstrated that colon cancer cells could express STAT3 or STAT3-pSTAT3-potential mechanism for Tim-3 promoting cancer growth. Therefore, Tim-3 may be a good target for further study to clarify colon carcinogenic mechanism.
Disclosure of conflict of interest
None.
References
- 1.Karsa LV, Lignini TA, Patnick J, Lambert R, Sauvaget C. The dimensions of the CRC problem. Best Pract Res Clin Gastroenterol. 2010;24:381–396. doi: 10.1016/j.bpg.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 2.Huang Y, Li W, Chu D, Zheng J, Ji G, Li M, Zhang H, Wang W, Du J, Li J. Overexpression of matrix metalloproteinase-21 is associated with poor overall survival of patients with colorectal cancer. J Gastrointest Surg. 2011;15:1188–1194. doi: 10.1007/s11605-011-1519-5. [DOI] [PubMed] [Google Scholar]
- 3.Olson TS, Ley K. Chemokines and chemokine receptors in leukocyte trafficking. Am J Physiol Regul Integr Comp Physiol. 2002;283:R7–28. doi: 10.1152/ajpregu.00738.2001. [DOI] [PubMed] [Google Scholar]
- 4.Sallusto F, Mackay CR, Lanzavecchia A. The role of chemokine receptors in primary, effector, and memory immune responses. Annu Rev Immunol. 2000;18:593–620. doi: 10.1146/annurev.immunol.18.1.593. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida R, Imai T, Hieshima K, Kusuda J, Baba M, Kitaura M, Nishimura M, Kakizaki M, Nomiyama H, Yoshie O. Molecular cloning of a novel human CC chemokine EBI1-ligand chemokine that is a specific functional ligand for EBI1, CCR7. J Biol Chem. 1997;272:13803–13809. doi: 10.1074/jbc.272.21.13803. [DOI] [PubMed] [Google Scholar]
- 6.Oikawa T, Kamimura Y, Akiba H, Yagita H, Okumura K, Takahashi H, Zeniya M, Tajiri H, Azuma M. Preferential involvement of Tim-3 in the regulation of hepatic CD8+ T cells in murine acute graft-versus-host disease. J Immunol. 2006;177:4281–4287. doi: 10.4049/jimmunol.177.7.4281. [DOI] [PubMed] [Google Scholar]
- 7.Jones RB, Ndhlovu LC, Barbour JD, Sheth PM, Jha AR, Long BR, Wong JC, Satkunarajah M, Schweneker M, Chapman JM, Gyenes G, Vali B, Hyrcza MD, Yue FY, Kovacs C, Sassi A, Loutfy M, Halpenny R, Persad D, Spotts G, Hecht FM, Chun TW, McCune JM, Kaul R, Rini JM, Nixon DF, Ostrowski MA. Tim-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. J Exp Med. 2008;205:2763–2779. doi: 10.1084/jem.20081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu C, Anderson AC, Schubart A, Xiong H, Imitola J, Khoury SJ, Zheng XX, Strom TB, Kuchroo VK. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol. 2005;6:1245–1252. doi: 10.1038/ni1271. [DOI] [PubMed] [Google Scholar]
- 9.Arai Y, Saito H, Ikeguchi M. Upregulation of TIM-3 and PD-1 on CD4+ and CD8+ T Cells Associated with Dysfunction of Cell-Mediated Immunity after Colorectal Cancer Operation. Yonago Acta Med. 2012;55:1–9. [PMC free article] [PubMed] [Google Scholar]
- 10.Baitsch L, Baumgaertner P, Devevre E, Raghav SK, Legat A, Barba L, Wieckowski S, Bouzourene H, Deplancke B, Romero P, Rufer N, Speiser DE. Exhaustion of tumor-specific CD8(+) T cells in metastases from melanoma patients. J Clin Invest. 2011;121:2350–2360. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207:2187–2194. doi: 10.1084/jem.20100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Q, Munger ME, Veenstra RG, Weigel BJ, Hirashima M, Munn DH, Murphy WJ, Azuma M, Anderson AC, Kuchroo VK, Blazar BR. Coexpression of Tim-3 and PD-1 identifies a CD8+ T-cell exhaustion phenotype in mice with disseminated acute myelogenous leukemia. Blood. 2011;117:4501–4510. doi: 10.1182/blood-2010-10-310425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fourcade J, Sun Z, Benallaoua M, Guillaume P, Luescher IF, Sander C, Kirkwood JM, Kuchroo V, Zarour HM. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J Exp Med. 2010;207:2175–2186. doi: 10.1084/jem.20100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang X, Bai X, Cao Y, Wu J, Huang M, Tang D, Tao S, Zhu T, Liu Y, Yang Y, Zhou X, Zhao Y, Wu M, Wei J, Wang D, Xu G, Wang S, Ma D, Zhou J. Lymphoma endothelium preferentially expresses Tim-3 and facilitates the progression of lymphoma by mediating immune evasion. J Exp Med. 2010;207:505–520. doi: 10.1084/jem.20090397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golden-Mason L, Palmer BE, Kassam N, Townshend-Bulson L, Livingston S, McMahon BJ, Castelblanco N, Kuchroo V, Gretch DR, Rosen HR. Negative immune regulator Tim-3 is overexpressed on T cells in hepatitis C virus infection and its blockade rescues dysfunctional CD4+ and CD8+ T cells. J Virol. 2009;83:9122–9130. doi: 10.1128/JVI.00639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiener Z, Kohalmi B, Pocza P, Jeager J, Tolgyesi G, Toth S, Gorbe E, Papp Z, Falus A. TIM-3 is expressed in melanoma cells and is upregulated in TGF-beta stimulated mast cells. J Invest Dermatol. 2007;127:906–914. doi: 10.1038/sj.jid.5700616. [DOI] [PubMed] [Google Scholar]
- 17.Cao Y, Zhou X, Huang X, Li Q, Gao L, Jiang L, Huang M, Zhou J. Tim-3 expression in cervical cancer promotes tumor metastasis. PLoS One. 2013;8:e53834. doi: 10.1371/journal.pone.0053834. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Jiang J, Jin MS, Kong F, Cao D, Ma HX, Jia Z, Wang YP, Suo J, Cao X. Decreased galectin-9 and increased Tim-3 expression are related to poor prognosis in gastric cancer. PLoS One. 2013;8:e81799. doi: 10.1371/journal.pone.0081799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piao YR, Piao LZ, Zhu LH, Jin ZH, Dong XZ. Prognostic value of T cell immunoglobulin mucin-3 in prostate cancer. Asian Pac J Cancer Prev. 2013;14:3897–3901. doi: 10.7314/apjcp.2013.14.6.3897. [DOI] [PubMed] [Google Scholar]
- 20.Yuan J, Jiang B, Zhao H, Huang Q. Prognostic implication of TIM-3 in clear cell renal cell carcinoma. Neoplasma. 2014;61:35–40. [PubMed] [Google Scholar]
- 21.Wu J, Liu C, Qian S, Hou H. The expression of Tim-3 in peripheral blood of ovarian cancer. DNA Cell Biol. 2013;32:648–653. doi: 10.1089/dna.2013.2116. [DOI] [PubMed] [Google Scholar]
- 22.Zhuang X, Zhang X, Xia X, Zhang C, Liang X, Gao L, Zhang X, Ma C. Ectopic expression of TIM-3 in lung cancers: a potential independent prognostic factor for patients with NSCLC. Am J Clin Pathol. 2012;137:978–985. doi: 10.1309/AJCP9Q6OVLVSHTMY. [DOI] [PubMed] [Google Scholar]
- 23.Yan J, Zhang Y, Zhang JP, Liang J, Li L, Zheng L. Tim-3 expression defines regulatory T cells in human tumors. PLoS One. 2013;8:e58006. doi: 10.1371/journal.pone.0058006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baritaki S, Sifakis S, Huerta-Yepez S, Neonakis IK, Soufla G, Bonavida B, Spandidos DA. Overexpression of VEGF and TGF-beta1 mRNA in Pap smears correlates with progression of cervical intraepithelial neoplasia to cancer: implication of YY1 in cervical tumorigenesis and HPV infection. Int J Oncol. 2007;31:69–79. [PubMed] [Google Scholar]
- 25.Hasegawa Y, Takanashi S, Kanehira Y, Tsushima T, Imai T, Okumura K. Transforming growth factor-beta1 level correlates with angiogenesis, tumor progression, and prognosis in patients with nonsmall cell lung carcinoma. Cancer. 2001;91:964–971. [PubMed] [Google Scholar]
- 26.Sanchez-Fueyo A, Tian J, Picarella D, Domenig C, Zheng XX, Sabatos CA, Manlongat N, Bender O, Kamradt T, Kuchroo VK, Gutierrez-Ramos JC, Coyle AJ, Strom TB. Tim-3 inhibits T helper type 1-mediated auto- and alloimmune responses and promotes immunological tolerance. Nat Immunol. 2003;4:1093–1101. doi: 10.1038/ni987. [DOI] [PubMed] [Google Scholar]
- 27.Geng H, Zhang GM, Li D, Zhang H, Yuan Y, Zhu HG, Xiao H, Han LF, Feng ZH. Soluble form of T cell Ig mucin 3 is an inhibitory molecule in T cell-mediated immune response. J Immunol. 2006;176:1411–1420. doi: 10.4049/jimmunol.176.3.1411. [DOI] [PubMed] [Google Scholar]
- 28.Yuan J, Jiang B, Zhao H, Huang Q. Prognostic implication of TIM-3 in clear cell renal cell carcinoma. Neoplasma. 2013 [Epub ahead of print] [PubMed] [Google Scholar]
- 29.Tu B, Du L, Fan QM, Tang Z, Tang TT. STAT3 activation by IL-6 from mesenchymal stem cells promotes the proliferation and metastasis of osteosarcoma. Cancer Lett. 2012;325:80–88. doi: 10.1016/j.canlet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Yang X, Liang L, Zhang XF, Jia HL, Qin Y, Zhu XC, Gao XM, Qiao P, Zheng Y, Sheng YY, Wei JW, Zhou HJ, Ren N, Ye QH, Dong QZ, Qin LX. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58:158–170. doi: 10.1002/hep.26305. [DOI] [PubMed] [Google Scholar]


