Abstract
MicroRNAs (miRNAs) are small, non-coding RNAs of endogenous origin. Accumulating studies have shown aberrant miRNA expression plays an important role in many tumor types. However, the mechanisms by which miRNAs regulate esophageal squamous cell carcinoma (ESCC) development remain poorly understood. In the present study, we assayed expression level of miR-192 in ESCC tissues and cell lines by real-time PCR, and defined the target gene and biological function by luciferase reporter assay, Western blot and apoptosis assay. We first verified that the expression level of miR-192 was significantly increased in ESCC tissues and cancer cells. Moreover, miR-192 over-expression inhibited cells apoptosis and promoted ESCC cells proliferation. We further demonstrated that miR-192 directly targeted 3’-UTR of Bim gene, and inhibited its protein expression. Importantly, Bim could reduce ESCC cells apoptosis ability induced by miR-192. These data suggest an important role of miR-192 in the molecular etiology of ESCC and implicate the potential application of miR-192 in ESCC therapy.
Keywords: miR-192, ESCC, Bim, apoptosis
Introduction
Esophageal cancer is the sixth most common malignancy worldwide [1,2]. Esophageal squamous cell carcinoma (ESCC) accounts for > 90% of cases of esophageal cancer in most Asian countries, including China [3-5]. Although treatment and perioperative management have evolved in recent years with dramatic advances in diagnosis, operative methods, and combined chemoradiotherapy, the prognosis of patients with ESCC is not ideal. Only a small subset of patients (20%-30%) exhibits a 5-year survival rate after surgery [6,7]. Therefore, there is a requirement for understanding the mechanisms involved in ESCC progression.
MicroRNAs (miRNAs) are conserved, endogenous, small, noncoding RNAs which negatively regulate gene expression either by translational repression, or target mRNA degradation through binding to the 3’UTR of target gene [8]. It is now well established that miRNAs may have causal roles in many normal/tumor cellular processes, such as development, differentiation, proliferation and apoptosis, and increasing sensitivity or resistance to chemotherapy [9,10]. Aberrant expressions of some miRNAs in cancer have been reported.
MiR-192 was first cloned by Lagos-Quintana et al. [11] and later confirmed by Lim et al. [12]. The miR-192 gene is located on human chromosome 11 and is transcribed as a cluster with miR-194 [13]. MiR-192 was reported to be up-regulated in multiple cancer types including gastric cancer, hepatocellular carcinoma, neuroblastoma, and pancreatic ductal adenocarcinoma [14-18]. The biological effects of miR-192 in these cancers have been partially identified, miR-192 could enhance cell proliferation and migration, reduce cell apoptosis and promotes cell cycle progression from the G0/G1 to the S phase by regulating of key factors in these progress such as smad-interacting protein 1 (SIP1) and Dicer [15,16]. However, miR-192 was also found down-regulated in some cancer types, such as colon cancer, colorectal cancer and lung cancer [19-21]. It may also function as a tumor suppressor. Up to now, little is known about the role of miR-192 in ESCC progression.
In our study, we found that miR-192 was over-expressed in ESCC cell lines and tumor tissues compared to normal squamous epithelial cell line and adjacent corresponding tissues respectively, suggesting miR-192 might act as a tumor oncogene in ESCC. miR-192 expression was associated with TNM stage and lymph node metastasis of ESCC, which indicated miR-192 might be involved in the pathogenesis of ESCC. Besides, we identified that apoptosis regulator Bim was one of direct target genes of miR-192. MiR-192 is able to regulate apoptosis of ESCC cells through paralyzing the function of Bim.
Materials and methods
Samples
Fresh samples from ESCC and corresponding normal adjacent tissue were obtained from 50 patients at Second Hospital Affiliated to Hebei Medical University between January 2008 and November 2010. The samples immediately snap frozen in liquid nitrogen, and stored at -80°C until RNA extraction. The tumors were classified according to World Health Organization classification. The study was approved by hospital ethical committee, and every patient had written informed consent. Clinicopathological information of the patients about age, sex, stage and lymph node metastasis was obtained from patient records, which were summarized in Table 1.
Table 1.
Relationship between miR-192 expression and clinicopathological factors in 50 ESCC patients
| Mir-192 Expression | |||||
|---|---|---|---|---|---|
|
|
|||||
| Factor | Low | High | Z Value | P Value | |
| Age | |||||
| < 60 | 10 | 15 | |||
| = 60 | 15 | 10 | 2.000 | 0.157 | |
| Gender | |||||
| Man | 16 | 21 | |||
| Woman | 9 | 4 | 2.599 | 0.107 | |
| Stage | |||||
| I+II | 20 | 9 | |||
| III+IV | 5 | 16 | 9.934 | 0.002 | |
| Lymph node status | |||||
| Nagative | 14 | 7 | |||
| Positive | 11 | 18 | 4.023 | 0.045 | |
Cell culture and transfection
The human esophageal carcinoma cell lines KYSE-150, KYSE-510, EC-9706 and immortalized human esophageal epithelial cell SHEE were kindly provided by Dr. Zhang Xun (Tianjin Chest Hospital). TE13 was purchased from The American Type Culture Collection (Manassas, VA). The cells were cultured in RPMI-1640 (Invitrogen, Carlsbad, CA) medium containing 10% fetal bovine serum (FBS, GIBCO), 100 IU/ml penicillin and 100 mg/ml streptomycin maintained at 37°C in humidified air containing 5% CO2. For transfection, cells were cultured to 80% confluence and transfected with recombinant eukaryotic vector and empty vector using Lipofectamine 2000 (Invitrogen, CA, USA) according to the manufacturer’s recommendation.
Quantitative real-time PCR
Quantitative RT-PCR was performed to validate the miRNA expression level. qRT-PCR was carried out using SYBR®Premix Ex TaqTM (Takara, Japan). PCR were carried out in triplicate and analyzed using the ABI Prism 7900HT fast real-time PCR system (Applied Biosystems, Life technologies, USA). The relative quantification values for each miRNA were calculated by the 2-ΔΔCt method using U6 as an internal reference. All primers were shown in Table S1.
Plasmid constructions
Genomic sequence of human miR-192, including, 200 bp flanking sequence, was amplified from human genome, then inserted into the BamHI/EcoRI site of the pcDNA3.1 vector (Invitrogen), named as pcDNA3.1-miR-192. The full-length 3’untranslated region (3’UTR) of Bim was amplified from human genomic DNA, and was cloned into the downstream of the firefly luciferase coding region of pMIR-GLOTM Luciferase vector (Promega, USA). The recombined vector was named as pMIR-Bim. Mutations of miR-192 binding sites were introduced by site-directed mutagenesis and the resulted vector was named pMIR-Bim-Mut. Primers used for the constructions were listed in Table S1. All the constructions were confirmed by sequencing.
Proliferation assay
MTT assay was used to analyze cell proliferation. EC9706 and KYSE-150 cells were transfected with either pcDNA3.1-miR-192 (miR-192) or empty vector (NC). After 24 h transfection, cells were seeded into 96-well plate at 5.0 × 103 cells/ml and continue cultured for 24, 48, 72, and 96 h, respectively. At each time point, 10 μl MTT reagent (5 mg/ml, Sigma) was added to each well, successive incubated for 4 h at 37°C. The supernatant was removed and 200 μl DMSO (Invitrogen) was added to dissolve the formazan crystals for 30 min. Spectrometric absorbance at a wavelength of 570 nm was measured on microplate reader (Spectra Max M5, MD, USA). Each sample was tested in triplicate and all experiments were performed three times.
Apoptosis analysis
Cells were transfected as above, 48 h after transfection, cells were harvested and washed with ice-cold phosphate-buffered saline (PBS), and then subjected to FITC Annexin V Apoptosis Detection kitI (BD Pharmingen, USA) for staining, which was followed by flow cytometric analysis using a FACScan instrument (Beckman Coulter, USA) and the test were repeated for three times with triplicate per experiment.
Dual-luciferase reporter assay
Cells were seeded into 24-well plates and cotransfected with 200 ng of pMIR-Bim or pMIR-Bim-Mut vector and 100 ng of pcDNA3.1-miR-192 vector or pcDNA 3.1 empty vectors, and the pRL-TK plasmid (Promega, Madison, WI) which was used as internal normalization. After 48 h, cells were lysed using the lysis buffer (Promega). Luciferase reporter gene assay was implemented using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. All experiments were performed at least three times.
Western blotting
Cells were transfected with either pcDNA3.1-miR-192, pCMV-Tag-2b-Bim or small interfere RNA target Bim (siBim). Total cell extracts prepared from cells using RIPA buffer (Beyotime, China), were resolved on 10% gradient SDS-polacrylamide gel and transferred to NC membranes. Membranes were blocked for 1 hour in 5% skim milk in TBST and incubated with primary antibody overnight at 4°C, followed by the incubation with appropriate HRP-conjugated secondary antibody at optimized concentration. The primary antibodies used in this study were as follows: anti-Bim antibody (1:1000, CST), anti-caspase3 antibody (1:1000, Santa Cruz), anti-caspase9 antibody (1:1000, Santa Cruz) and anti-β-actin antibody (1:5000, CST). The densitometry of Western blot results was measured using ImageJ software.
Statistical analysis
The data were presented as mean ± standard deviation (SD). MiR-192 expression in 50 pairs of primary ESCC tissues and corresponding adjacent tissues was compared by Wilcoxon signed-rank test. A chi-square test was used to analyze the relationship between miR-192 expression levels and clinicopathologic characters. T-test was used to determine the significant differences between control and treatment groups. Statistical analysis was performed using SPSS16.0 software, and P < 0.05 was considered to be a statistically significant difference.
Results
miR-192 is up-regulated in ESCC cells and tissue specimens
Firstly, we analyzed the expression level of miR-192 by qRT-PCR in human esophageal epithelial cell SHEE and four human esophageal carcinoma cell lines including KYSE150, KYSE510, EC9706 and TE13. The expression of miR-192 was significantly increased in esophageal carcinoma cell lines compared with SHEE (Figure 1A). Thus, we thought miR-192 may be an oncogene in human ESCC cells. Further, we detected miR-192 expression in both ESCC tissues and corresponding adjacent tissues by qRT-PCR. Significantly, we found that miR-192 expression was increased in ESCC tissues (n = 50, Figure 1B), which indicated that miR-192 was acted as an oncogene. After analyzing the clinical information of patients, we found that the aberrant expression level of miR-192 was associated with the pathological stage and lymph node metastasis of patients (Table 1, P < 0.05), indicating that miR-192 may play an important role the pathogenesis of ESCC.
Figure 1.
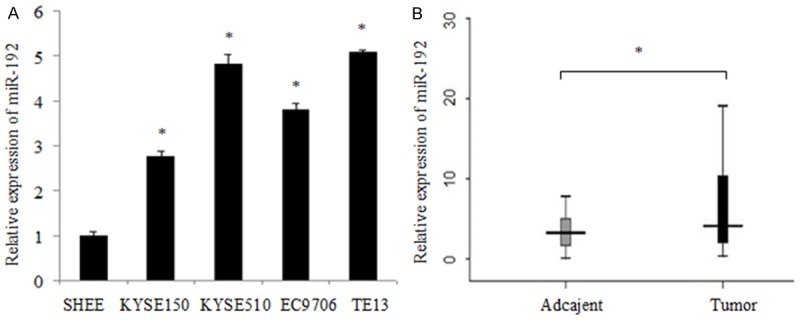
MiR-192 shows high expression in ESCC cells and tissue specimens. A. The relative mRNA levels of miR-192 were detected by qRT-PCR and normalized against an endogenous control (U6 RNA). Data are reported as mean ± SD for three independent experiments. B. qRT-PCR analysis of miR-192 expression in 50 pairs of primary ESCC tissues and their corresponding adjacent tissues. MiR-192 expression in those two types of tissues was compared by way of Wilcoxon signed-rank test. *indicates significant difference (P < 0.05).
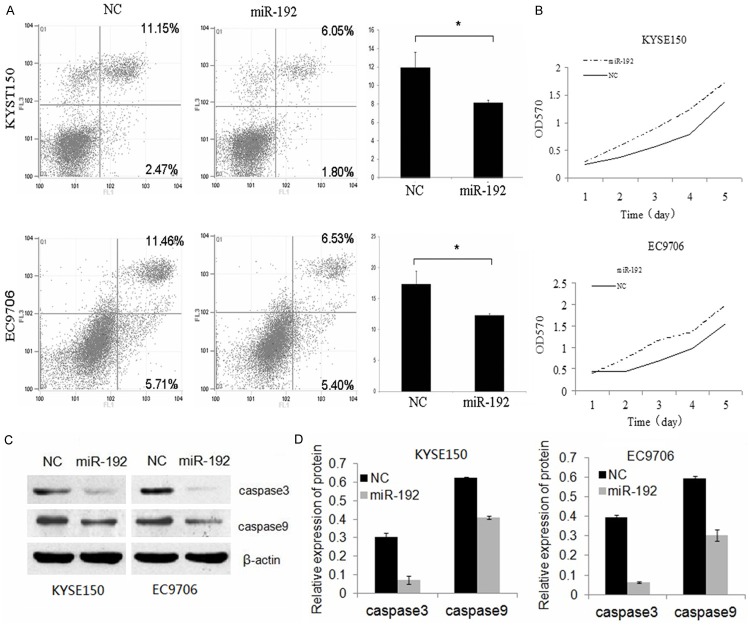
MiR-192 inhibited cell apoptosis while promote cell proliferation of ESCC cells
To detect the functional roles of miR-192, we then examined the effect of miR-192 on cell apoptosis and cell proliferation. Flow cytometry analysis showed that upregulation of miR-192 in both KYSE150 and EC9706 cells inhibited cell apoptosis, compared to the control group (Figure 2A). Additionally, we used MTT assay to investigate the effect of miR-192 on cell proliferation. As shown in Figure 2B, when transfected with pcDNA3.1-miR-192 plasmids, the proliferation ability of KYSE150 and EC9706 cells was upregulated when compared with the control. Furthermore, in order to detect the inhibition affection of cell apoptosis, we detected the viability of Caspase 3 and 9 in cells transfected with pcDNA3.1-miR-192 plasmids and control to explore the correlation between miR-192 and apoptosis. The results indicated that both the cleaved Caspase 3 and 9 was decreased in cells transfected with pcDNA3.1-miR-192 plasmids comparing with the cells transfected with control (Figure 2C, 2D). This result suggested that the aberrant increasing level of miR-192 may have the ability of suppressing cell apoptosis in ESCC cell lines.
Figure 2.
MiR-192 inhibited cell apoptosis and promote cell proliferation of ESCC cells. A. Significantly decreased number of apoptosis was observed in KYSE150 and EC9706 cells after transfected miR-192. B. The KYSE-150 and EC-9706 cells proliferation ability were increased after transfected miR-192. C. The protein of the cleaved Caspase3 and 9 were examined by western blot in cells treated with miR-16 and control. D. shows the relative gray values of each band (normalized to β-actin). Protein bands from three independent Western blot assays were quantified using Image J software. Data are reported as mean ± SD (*P < 0.05, Student’s t- test).
MiR-192 directly targets Bim
To gain further insight into the molecular mechanism by which miR-192 suppresses apoptosis of esophageal carcinoma cells, we searched for genes targeted by miR-192 using biological target prediction software TargestScan, miRanda, TargetScan, and PicTar. A gene named BCL2L11 (BCL2-like 11), also named Bim, had drawn our attention. Bim, a pro-apoptotic protein that is involved in the intrinsic apoptosis pathway, was found to have a putative miR-192 binding site within its 3’UTR (Figure 3A). To detect whether Bim is regulated by miR-192, the wild type or mutant 3’UTR sequence of Bim was cloned into pMIR reporter vector, respectively, as shown in Figure 3B. The luciferase activity of pMIR-Bim-3’UTR-wt construct was significantly decreased upon the over-expression of miR-192 in KYSE150 cells, whereas its mutant counterpart was not (Figure 3C). In addition, the protein level of Bim in KYSE150 and EC9706 cells was dramatically reduced by miR-192, but mRNA level was not changed (Figure 3D-F). Taken together, these data indicated that Bim was the direct target of miR-192 at least in esophageal cancer.
Figure 3.
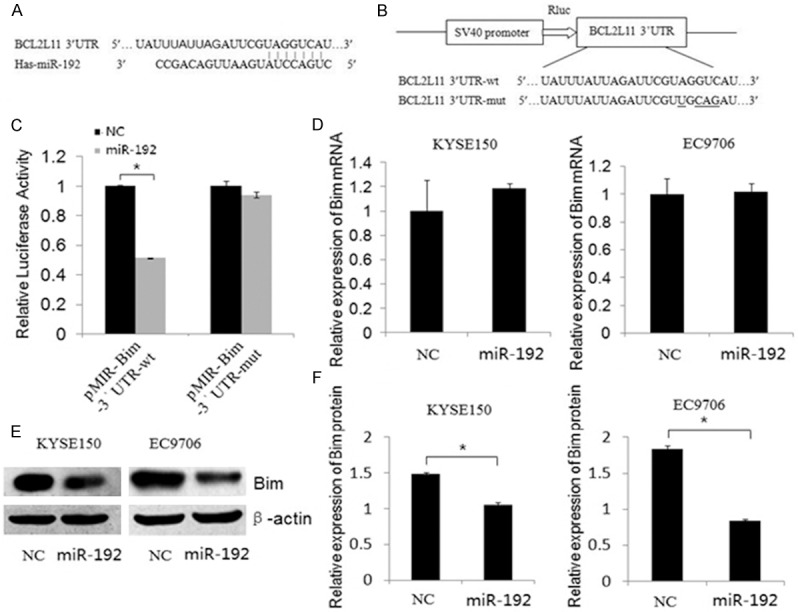
MiR-192 directly inhibits the expression of Bim through its 3’UTR of ESCC cells. A. The miR-192 binding site predicted in the 3’UTR of Bim mRNA. B. Mutant was generated at the seed region of Bim 3’UTR as indicated by the underline. A 3’UTR fragment of Bim mRNA containing wild-type or mutant of the miR-192 binding sequence was cloned into the downstream of the luciferase gene in pMIR vector. C. KYSE-150 cells was transfected with pMIR reporter vectors containing either wild-type or mutant Bim 3’UTR (indicated as pMIR-Bim-3’UTR-wt and pMIR- Bim-3’UTR-mut) with either pcDNA3.1 (indicated as NC) or pcDNA3.1-miR-192 vector (indicated as miR-192). Luciferase activity was determined 48 h after transfection. D. Bim mRNA was detected by qRT-PCR in cell lines transfected with pcDNA3.1 (indicated as NC) or pcDNA3.1-miR-192 vector (indicated as miR-192). E, F. The protein levels of Bim was examined by Western blot in cells transfected with different plasmids. Figure F shows the relative gray values of each band (normalized to β-actin). Data are reported as mean ± SD (*P < 0.01, Student’s t- test).
Bim contributes to miR-192 suppressed apoptosis of ESCC cells
Having demonstrated Bim as a direct target of miR-192, we next examined the importance of Bim in miR-192 mediated cell apoptosis. Ectopic expression of Bim in EC9706 cells significantly enhanced cell apoptosis (Figure 4A), however, silencing Bim by siRNAs in EC9706 cells decreased apoptosis ability of the cells (Figure 4B), revealing its positive roles in the contribution of ESCC cells apoptosis. Meanwhile, the transfecting or silencing efficiency of Bim in the cells was detected by Western blot (Figure 4A, 4B lower). Then, we accessed whether the functional effect of miR-192 on ESCC cells was dependent on Bim. As shown in Figure 4C, ectopic expression of Bim abrogated miR-192 effects on cell apoptosis (Figure 4C). In parallel, the protein level of Bim was confirmed by Western blot (Figure 4C, lower). Collectively, these results suggested that Bim functions as a target of miR-192, responsible for miR-192 mediated regulation of the apoptosis of ESCC cells.
Figure 4.
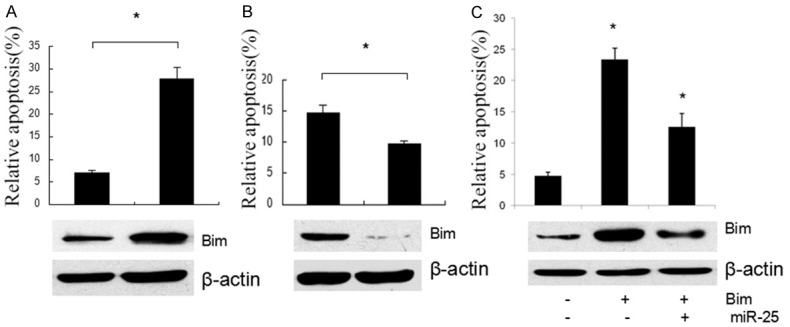
Effects of Bim in miR-192 Suppressed apoptosis of ESCC Cells. A. Significantly increased number of apoptosis was observed in the EC9706 cells after Bim transfection. B. Silencing Bim by siRNAs in EC9706 cells decreased apoptosis ability of the cells. C. Significantly decreased number of apoptosis was observed in the EC9706 cells after Bim transfection with or without miR-25 mimics compared to control. The protein expression level of Bim was assessed by Western blotting in each group.
Discussion
The increasing evidence has shown miRNAs may be a novel class of oncogenes/tumor suppressors and the correlation between miRNAs and cancers has become a focus for the diagnosis and therapy of cancer. MiR-192 is over-expressed in several human tumors, such as HCC, gastric cancer, neuroblastoma and pancreatic cancer [14-18]. Furthermore, miR-192 expression was associated with chemotherapy resistancer [23], or tumor metastasis [15,16]. However, miR-192 expression was reduced in colon cancer, breast cancer, colorectal cancer and lung cancer [19-22]. Suggesting a tumor suppressor gene-like function. In our study, we found that miR-192 was up-regulated in ESCC tissues and cell lines. Thus, we supposed that miR-192 may be a tumor oncogene in ESCC. However, the potential role of miR-192 in the progression of ESCC still need to be further investigated.
Next, we investigated the function of miR-192 in ESCC cells. Our data showed that miR-192 highly reduced the cell apoptosis and promoted the cell proliferation of ESCC cells in vitro. Furthermore, we found miR-192 could affect the apoptosis related protein caspase3 and caspase9 expression. Therefore, our data suggested that the increased expression of miR-192 may contribute to the apoptosis of cancer cells and consequently facilitate the advanced development of human cancers like ESCC.
A single miRNA can coordinate a large number of target genes [24]. Several miR-192 targets have been identified in different cells and organs. RB1 was found to be a target of miR-192, which mediates an effect on cell apoptosis through the caspase pathway [20]. Galina et al [15] indicated that miR-192 increase cell proliferation and migration ability of NB cells by directly target Dicer1. miR-192 inhibits nucleotide excision repair by targeting ERCC3 and ERCC4 in HepG2.2.15 cells [25]. miR-192 directly target SIP1 mediates the growth-promoting effects of miR-192 in PDAC [16]. Our present data suggest Bim as a functional target of miR-192 by luciferase reporter gene assays, RT-PCR and Western blot analysis method, respectively. Bim is a BH3-only protein (BOP), a pro-apoptotic member of the Bcl-2 protein family and its expression is regulated both transcriptionally and posttranslationally [26,27]. The important role of pro-apoptotic protein Bim has been strongly underlined in numerous papers, due to its function in inducing apoptosis and facilitating the proliferation of cancer cells [28-30].
In summary, we investigated the role of miR-192 in ESCC development. Our founding suggests that miR-192 may be a tumor oncogene in ESCC. The aberrant expression level of miR-192 in ESCC tissues was high associated with TNM stage and lymph node metastasis. According to our results, over-expressed level of miR-192 induced the inhibition of cell apoptosis, thus, we assumed that the loss of cell apoptosis could result in the proliferation of tumor cell. In conclusion, miR-192 may affect the proliferation and apoptosis of ESCC cells through targeting pro-apoptosis regulator Bim. Additionally, miR-192 may serve as a potential therapeutic candidate in the treatment of ESCC.
Acknowledgements
This research is supported by Hebei Natural Science Foundation (No. H2013078091) and Key project of Hebei Health Bureu (No. 20110068).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Lin SW, Abnet CC, Freedman ND, Murphy G, Risques R, Prunkard D, Rabinovitch P, Pan QJ, Roth MJ, Wang GQ, Wei WQ, Lu N, Taylor PR, Qiao YL, Dawsey SM. Measuring telomere length for the early detection of precursor lesions of esophageal squamous cell carcinoma. BMC Cancer. 2013;13:578. doi: 10.1186/1471-2407-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu C, Li M, Hu C, Duan H. Prognostic role of microRNA polymorphisms in patients with advanced esophageal squamous cell carcinoma receiving platinum based chemotherapy. Cancer Chemother Pharmacol. 2014;73:335–341. doi: 10.1007/s00280-013-2364-x. [DOI] [PubMed] [Google Scholar]
- 3.Kim D, Cho J, Kim K, Shim YM. Chyle leakage patterns and management after oncologic esophagectomy: A retrospective cohort study. Thoracic Cancer. 2014;5:391–397. doi: 10.1111/1759-7714.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Migita K, Sho M, Shimada K, Yasuda S, Yamato I, Takayama T, Matsumoto S, Wakatsuki K, Hotta K, Tanaka T, Ito M, Konishi N, Nakajima Y. Significant involvement of herpesvirus entry mediator in human esophageal squamous cell carcinoma. Cancer. 2014;120:808–817. doi: 10.1002/cncr.28491. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, Zhang W, Liu X, Zhang X, He J, Feng Q, Zhou Z, Wang L, Yin W, Xiao Z. Prognosis of esophageal squamous cell carcinoma patients with preoperative radiotherapy: Comparison of different cancer staging systems. Thoracic Cancer. 2014;5:204–210. doi: 10.1111/1759-7714.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morita M, Yoshida R, Ikeda K, Egashira A, Oki E, Sadanaga N, Kakeji Y, Yamanaka T, Maehara Y. Advances in esophageal cancer surgery in Japan: an analysis of 1000 consecutive patients treated at a single institute. Surgery. 2008;143:499–508. doi: 10.1016/j.surg.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Sugimachi K, Matsuoka H, Ohno S, Mori M, Kuwano H. Multivariate approach for assessing the prognosis of clinical oesophageal carcinoma. Br J Surg. 1988;75:1115–8. doi: 10.1002/bjs.1800751122. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 10.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNA swith a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 11.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 13.Hino K, Tsuchiya K, Fukao T, Kiga K, Okamoto R, Kanai T, Watanabe M. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA. 2008;14:1433–1442. doi: 10.1261/rna.810208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X, Zhou X, Gan J. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One. 2014;9:e107986. doi: 10.1371/journal.pone.0107986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feinberg-Gorenshtein G, Guedj A, Shichrur K, Jeison M, Luria D, Kodman Y, Ash S, Feinmesser M, Edry L, Shomron N, Weizman A, Yaniv I, Avigad S. MiR-192 directly binds and regulates Dicer1 expression in neuroblastoma. PLoS One. 2013;8:e78713. doi: 10.1371/journal.pone.0078713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao C, Zhang J, Zhang S, Yu D, Chen Y, Liu Q, Shi M, Ni C, Zhu M. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncol Rep. 2013;30:276–84. doi: 10.3892/or.2013.2420. [DOI] [PubMed] [Google Scholar]
- 17.Chen Q, Ge X, Zhang Y, Xia H, Yuan D, Tang Q, Chen L, Pang X, Leng W, Bi F. Plasma miR-122 and miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncol Rep. 2014;31:1863–70. doi: 10.3892/or.2014.3004. [DOI] [PubMed] [Google Scholar]
- 18.Gui J, Tian Y, Wen X, Zhang W, Zhang P, Gao J, Run W, Tian L, Jia X, Gao Y. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci. 2011;120:183–93. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang Y, Song Y, Wang Z, Liu Z, Gao P, Liang J, Zhu J, Xing C, Xu H. microRNA-192, -194 and -215 are frequently downregulated in colorectal cancer. Exp Ther Med. 2012;3:560–566. doi: 10.3892/etm.2011.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng S, Cong S, Zhang X, Bao X, Wang W, Li H, Wang Z, Wang G, Xu J, Du B, Qu D, Xiong W, Yin M, Ren X, Wang F, He J, Zhang B. MicroRNA-192 targeting retinoblastoma 1 inhibits cell proliferation and induces cell apoptosis in lung cancer cells. Nucleic Acids Res. 2011;39:6669–78. doi: 10.1093/nar/gkr232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karaayvaz M, Pal T, Song B, Zhang C, Georgakopoulos P, Mehmood S, Burke S, Shroyer K, Ju J. Prognostic significance of miR-215 in colon cancer. Clin Colorectal Cancer. 2011;10:340–7. doi: 10.1016/j.clcc.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu F, Meng X, Tong Q, Liang L, Xiang R, Zhu T, Yang S. BMP-6 inhibits cell proliferation by targeting microRNA-192 in breast cancer. Biochim Biophys Acta. 2013;1832:2379–90. doi: 10.1016/j.bbadis.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 23.Hummel R, Sie C, Watson DI, Wang T, Ansar A, Michael MZ, Van der Hoek M, Haier J, Hussey DJ. MicroRNA signatures in chemotherapy resistant esophageal cancer cell lines. World J Gastroenterol. 2014;20:14904–12. doi: 10.3748/wjg.v20.i40.14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J, Zhou J, Wu J, Shao C. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32:1183–1189. doi: 10.1093/carcin/bgr105. [DOI] [PubMed] [Google Scholar]
- 26.Gillings AS, Balmanno K, Wiggins CM, Johnson M, Cook SJ. Apoptosis and autophagy: BIM as a mediator of tumour cell death in response to oncogene-targeted therapeutics. FEBS J. 2009;276:6050–6062. doi: 10.1111/j.1742-4658.2009.07329.x. [DOI] [PubMed] [Google Scholar]
- 27.Akiyama T, Dass CR, Choong PF. Bim-targeted cancer therapy: a link between drug action and underlying molecular changes. Mol Cancer Ther. 2009;8:3173–3180. doi: 10.1158/1535-7163.MCT-09-0685. [DOI] [PubMed] [Google Scholar]
- 28.Muthalagu N, Junttila MR, Wiese KE, Wolf E, Morton J, Bauer B, Evan GI, Eilers M, Murphy DJ. BIM is the primary mediator of MYC-induced apoptosis in multiple solid tissues. Cell Rep. 2014;8:1347–53. doi: 10.1016/j.celrep.2014.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang ZC, Li YY, Wang HY, Fu S, Wang XP, Zeng MS, Zeng YX, Shao JY. Knockdown of miR-214 promotes apoptosis and inhibits cell proliferation in nasopharyngeal carcinoma. PLoS One. 2014;9:e86149. doi: 10.1371/journal.pone.0086149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braicu C, Pileczki V, Irimie A, Berindan-Neagoe I. p53siRNA therapy reduces cell proliferation, migration and induces apoptosis in triple negative breast cancer cells. Mol Cell Biochem. 2013;381:61–8. doi: 10.1007/s11010-013-1688-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.