Abstract
We aimed to assess protein expressions of p16 and pRB in breast cancer and explore the clinicopathologic implications. Tissue microarray (TMA) was constructed with 406 cases of breast cancer. The cases were subgrouped into luminal A, luminal B, HER-2, and triple negative breast cancer (TNBC) based on the results of immunohistochemical stains for ER, PR, HER-2, and Ki-67 and fluorescent in situ hybridization (FISH) for HER-2. One hundred and sixty-eight cases were allocated to the subgroup luminal A; 87 cases to the luminal B; 32 cases to the HER-2; and 119 cases to the TNBC. The TNBC group showed the highest negative rate for p16, and the luminal B and HER-2 groups showed the highest positive rate for p16 (P < 0.001). Alteration of p16 was the highest in the luminal B and HER-2 groups, and pRB expression rate was the highest in the HER-2 group and lowest in the luminal A group. In addition, p16(+)/pRB(+) type was the most common in the luminal B group, p16(+)/pRB(-) in the luminal A group, and p16(-)/pRB(+) in the TNBC group (P < 0.001). Altered p16/pRB(+) and non-altered p16/pRB(+) type was the most common in the luminal B, and altered p16/pRB(-) and non-altered p16/pRB(+) type was the most common in the luminal A (P < 0.001). Alteration of p16 was correlated with higher Ki67 labeling index (LI) (P = 0.013), and p16 negativity was correlated with ER negativity (P = 0.002), PR negativity (P = 0.004), and higher Ki67 LI (P < 0.001). pRB positivity was correlated with PR negativity (P = 0.009), HER-2 positivity (P = 0.001), and higher Ki-67 LI (P < 0.001). In luminal group A, p16 alteration was correlated with shorter DFS in univariate analysis (P = 0.024). In conclusion, Expression rates of p16 and pRB differ according to the molecular subgroups of breast cancer and they subsequently correlate with clnicopathologic factors.
Keywords: Breast cancer, p16, RB
Introduction
Unrestricted cellular proliferation due to deregulation of cell cycle is the key feature of malignancy, and an alteration in the cell cycle check points is one of the most common abnormalities encountered in the molecular basis of human cancer. Among these cell cycle check points are p16 and pRB, the former a cyclin dependent kinase 2 (CDKN2) gene products [1] serving as a negative regulator of cell cycle and the latter an important regulator of cell cycle repressing progression from G1 phase to S phase [2]. Phosphorylation by cyclin D makes pRB inactive, thus igniting the start of cell cycle. On the other hand, p16 suppresses cyclin D activity by bonding with cyclin-dependent kinase 4 and 6, slowing down the cell replication rate by allowing pRB to stay in its dephosphorylated, active status [3]. Because p16 and pRB have such an essential role in cell proliferation, altered expressions of p16 and pRB and their correlation with clinicopathologic factors have been reported in various types of human cancer [4-8]. However, reports on their altered expressions in breast cancer and their subsequent clinicopathologic implications are limited in number [9-14]. We investigated their differential expression patterns in breast cancer according to the molecular subtypes and explored the clinicopathologic significance.
Materials and methods
Patient selection and histologic evaluation
Patients who underwent surgical excision of invasive ductal carcinoma, NOS, from January 2000 to December 2006 in Yonsei University Severance Hospital were subject to the study. Patients who received preoperative chemotherapy or hormonal therapy were excluded. This study was approved by the Institutional Review Board of Yonsei University Severance Hospital, and was exempt from informed consent of the patients. Hematoxylin and Eosin (H&E) stained slides of all cases were reviewed by a breast pathologist (Koo, JS). Histological grade was assessed using the Nottingham grading system [15]. Clinicopathologic parameters evaluated in each case included patient age at initial diagnosis, lymph node metastasis, tumor recurrence, distant metastasis, and patient survival.
Tissue microarray
On H&E-stained slides of tumors, a representative area was selected and a corresponding spot was marked on the surface of the paraffin block. Using a biopsy needle, the selected area was punched out and a 3-mm tissue core was placed into a 6 × 5 recipient block. Tissue of invasive tumor was extracted. More than 2 tissue cores were extracted to minimize extraction bias. Each tissue core was assigned with a unique tissue microarray location number that was linked to a database containing other clinicopathologic data.
Immunohistochemistry
All immunohistochemistry was performed with formalin-fixed, paraffin-embedded tissue sections. Briefly, 5-μm-thick sections were obtained with a microtome, transferred onto adhesive slides, and dried at 62°C for 30 minutes. After incubation with primary antibodies, immunodetection was performed with biotinylated anti-mouse immunoglobulin, followed by peroxidase-labeled streptavidin using a labeled streptavidin biotin kit with 3,3’-diaminobenzidine chromogen as the substrate. The primary antibody incubation step was omitted in the negative control. Positive control tissue was used as per the manufacturer’s recommendation. Slides were counterstained with Harris hematoxylin.
Interpretation of immunohistochemical staining
All immunohistochemical markers were accessed by light microscopy. A cut-off value of 1% or more positively stained nuclei was used to define ER and PR positivity [16]. HER-2 staining was analyzed according to the American Society of Clinical Oncology (ASCO)/College of American Pathologists (CAP) guidelines using the following categories: 0 = no immunostaining; 1+ = weak incomplete membranous staining, less than 10% of tumor cells; 2+ = complete membranous staining, either uniform or weak in at least 10% of tumor cells; and 3+ = uniform intense membranous staining in at least 30% of tumor cells [17]. HER-2 immunostaining was considered positive when strong (3+) membranous staining was observed, whereas cases with 0 to 1+ were regarded as negative. Cases showing 2+ HER-2 expression were evaluated for HER-2 amplification by fluorescent in situ hybridization (FISH).
Immunohistochemical stains for p16 and pRB were assessed semi-quantitatively under the light microscopy according to the previously reported method [18]. Staining results in tumor and stromal cells were assessed as 0: negative or immunostaining in < 10% of the tumor, 1: expression in 11-25% of the tumor, 2: positive in 26%-50% of the tumor, 3: positive in 51%-75% of the tumor, and 4: positive in 76%-100% of the tumor. For p16, grade 1 and 2 were considered low positive, 3 and 4 high positive, and grade 1 and 4 were considered as altered expression [19]. For pRB, grade 1 or higher was considered positive.
Tumor phenotype classification
In this study, we classified breast cancer phenotypes according to the immunohistochemistry results for ER, PR, HER-2, Ki-67 and FISH results for HER-2 as follows [20]: luminal A type, ER or/and PR positive, HER-2 negative and Ki-67 LI < 14%; Luminal B type, (HER-2 negative) ER or/and PR positive, HER-2 negative and Ki-67 LI ≥ 14%; (HER-2 positive) ER or/and PR positive and HER-2 overexpressed or/and amplified; HER-2 overexpression type, ER and PR negative and HER-2 overexpressed or/and amplified; TNBC type: ER, PR, and HER-2 negative.
Statistical analysis
Data were analyzed using SPSS for Windows, Version 12.0 (SPSS Inc., Chicago, IL, USA). For determination of statistical significance, Student’s t and Fisher’s exact tests were used for continuous and categorical variables, respectively. In the case of analyzing data with multiple comparisons, a corrected p-value with the application of the Bonferroni multiple comparison procedure was used. Statistical significance was set to P < 0.05. Kaplan-Meier survival curves and log-rank statistics were employed to evaluate time to tumor recurrence and overall survival. Multivariate regression analysis was performed using the Cox proportional hazards model.
Results
Basal characteristics of breast cancer
The clinicopathologic features of breast cancer subject to this study are shown in Table 1. Among the total number of 406 cases, 168 were luminal A, 87 were luminal B, 32 were HER-2, and 119 were TNBC. These molecular subtypes showed difference in histologic grade (P < 0.001), tumor stage (P = 0.022), and Ki67 LI (P < 0.001): luminal A showed lower histologic grade, less advanced tumor stage, and lower Ki67 LI; HER-2 and TNBC showed higher histologic grade, advanced tumor stage, and higher Ki67 LI (Table 2).
Table 1.
Source, clone, and dilution of used antibodies
| Antibody | Clone | Dilution | Company |
|---|---|---|---|
| Molecular subtype related | |||
| ER | SP1 | 1:100 | Thermo Scientific, CA, USA |
| PR | PgR | 1:50 | DAKO, Denmark |
| HER-2 | Polyclonal | 1:1500 | DAKO, Denmark |
| Ki-67 | MIB-1 | 1:1000 | Abcam, Cambridge, UK |
| Cell cycle related | |||
| p16 | 2D9A12 | 1:200 | Abcam, Cambridge, UK |
| Phospho pRB (S780) | Polyclonal | 1:100 | Abcam, Cambridge, UK |
Table 2.
Clinicopathologic characteristics of patients according to breast cancer phenotype
| Parameter | Total (n = 406) (%) | Luminal A (n = 168) (%) | Luminal B (n = 87) (%) | HER-2 (n = 32) (%) | TNBC (n = 119) (%) | P-value |
|---|---|---|---|---|---|---|
| Age (years) | 0.284 | |||||
| ≤ 50 | 229 (56.4) | 93 (55.4) | 55 (63.2) | 14 (43.8) | 67 (56.3) | |
| > 50 | 177 (43.6) | 75 (44.6) | 32 (36.8) | 18 (56.2) | 52 (43.6) | |
| Histologic grade | < 0.001 | |||||
| I/II | 265 (65.3) | 151 (89.9) | 56 (64.4) | 15 (46.9) | 43 (36.1) | |
| III | 141 (34.7) | 17 (10.1) | 31 (35.6) | 17 (53.1) | 76 (63.9) | |
| Tumor stage | 0.022 | |||||
| T1 | 203 (50.0) | 98 (58.3) | 43 (49.4) | 14 (43.8) | 48 (40.3) | |
| T2/T3 | 203 (50.0) | 70 (41.7) | 44 (50.6) | 18 (56.2) | 71 (59.7) | |
| Nodal metastasis | 0.426 | |||||
| Absent | 250 (61.6) | 101 (60.1) | 49 (56.3) | 20 (62.5) | 80 (67.2) | |
| Present | 156 (38.4) | 67 (39.9) | 38 (43.7) | 12 (37.5) | 39 (32.8) | |
| Estrogen receptor status | < 0.001 | |||||
| Negative | 156 (38.4) | 2 (1.2) | 3 (3.4) | 32 (100.0) | 119 (100.0) | |
| Positive | 250 (61.6) | 166 (98.8) | 84 (96.6) | 0 (0.0) | 0 (0.0) | |
| Progesterone receptor status | < 0.001 | |||||
| Negative | 201 (49.5) | 22 (13.1) | 28 (32.2) | 32 (100.0) | 119 (100.0) | |
| Positive | 205 (50.5) | 146 (86.9) | 59 (67.8) | 0 (0.0) | 0 (0.0) | |
| HER-2 status | < 0.001 | |||||
| Negative | 331 (81.5) | 168 (100.0) | 44 (50.6) | 0 (0.0) | 119 (100.0) | |
| Positive | 75 (18.5) | 0 (0.0) | 43 (49.4) | 32 (100.0) | 0 (0.0) | |
| Ki-67 LI (%) | < 0.001 | |||||
| ≤ 14 | 229 (56.4) | 168 (100.0) | 26 (29.9) | 16 (50.0) | 229 (56.4) | |
| > 14 | 177 (43.6) | 0 (0.0) | 61 (70.1) | 16 (50.0) | 177 (43.6) | |
| Tumor recurrence | 41 (10.1) | 10 (6.0) | 8 (9.2) | 6 (18.8) | 17 (14.3) | 0.042 |
| No. of patient deaths | 43 (10.6) | 10 (6.0) | 9 (10.3) | 7 (21.9) | 17 (14.3) | 0.020 |
TNBC, triple negative breast cancer.
Expression of p16 and pRB according to the molecular subtypes
For each molecular subtype of breast cancer, p16 expression (P < 0.001), p16 alteration status (P = 0.018), and pRB expression (P < 0.001) were different. P16 negativity rate was higher in TNBC and high rate of p16 expression was more frequently observed in luminal B and HER-2. P16 alteration was higher in luminal B and HER-2 groups, and pRB expression was higher in HER-2 group and lower in luminal B group (Table 3 and Figure 1).
Table 3.
Expression of metabolism-related proteins according to breast cancer subtype
| Parameter | Total (n = 406) (%) | Luminal A (n = 168) (%) | Luminal B (n = 87) (%) | HER-2 (n = 32) (%) | TNBC (n = 119) (%) | P-value |
|---|---|---|---|---|---|---|
| p16 | < 0.001 | |||||
| Negative | 31 (7.6) | 3 (1.8) | 9 (10.3) | 1 (3.1) | 18 (15.1) | |
| Low positive | 212 (52.2) | 102 (60.7) | 37 (42.5) | 18 (56.2) | 55 (46.2) | |
| High positive | 163 (40.1) | 63 (37.5) | 41 (47.1) | 13 (40.6) | 46 (38.7) | |
| Alteration of p16 | 0.018 | |||||
| No | 212 (52.2) | 102 (60.7) | 37 (42.5) | 18 (56.2) | 55 (46.2) | |
| Yes | 194 (47.8) | 66 (39.3) | 50 (57.5) | 14 (43.8) | 64 (53.8) | |
| pRB | < 0.001 | |||||
| Negative | 339 (83.5) | 156 (92.9) | 62 (71.3) | 21 (65.6) | 100 (84.0) | |
| Positive | 67 (16.5) | 12 (7.1) | 25 (28.7) | 11 (34.4) | 19 (16.0) |
TNBC, triple negative breast cancer.
Figure 1.
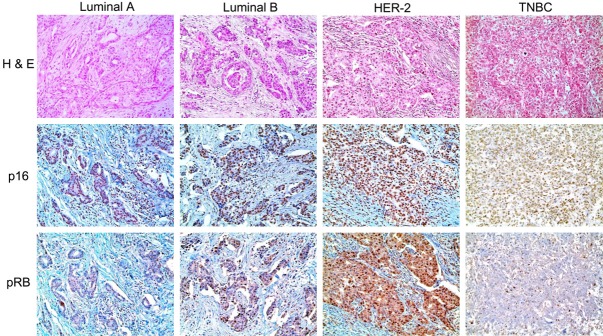
Immunohistochemical expression of p16 and pRB according to the molecular subtype. p16 negativity rate is higher in TNBC and high rate of p16 expression is observed in luminal B and HER-2. p16 alteration is higher in luminal B and HER-2 groups, and pRB expression is higher in HER-2 group and lower in luminal B group.
The combined status of p16 and pRB expressions was different according to the molecular subtypes (P < 0.001). P16(+)/pRB(+) was higher in luminal B, p16(+)/pRB(-) was higher in luminal A, p16(-)/pRB(+) and p16(-)/pRB(-) were higher in TNBC. The combined status of p16 alteration and pRB expression was also different according to the molecular subtypes (P < 0.001). Altered p16/pRB(+) and non-altered p16/pRB(+) types were higher in luminal B and altered p16/pRB(-) and non-altered p16/pRB(-) types were higher in luminal A (Table 4).
Table 4.
Expression of metabolism-related proteins according to breast cancer subtype
| Parameter | Total (n = 406) (%) | Luminal A (n = 168) (%) | Luminal B (n = 87) (%) | HER-2 (n = 32) (%) | TNBC (n = 119) (%) | P-value |
|---|---|---|---|---|---|---|
| p16/pRB status | < 0.001 | |||||
| p16(+)/pRB(+) | 64 (100.0) | 12 (18.8) | 24 (37.5) | 11 (17.2) | 17 (26.6) | |
| p16(+)/pRB(-) | 311 (100.0) | 153 (49.2) | 54 (17.4) | 20 (6.4) | 84 (27.0) | |
| p16(-)/pRB(+) | 3 (100.0) | 0 (0.0) | 1 (33.3) | 0 (0.0) | 2 (66.7) | |
| p16(-)/pRB(-) | 28 (100.0) | 3 (10.7) | 8 (28.6) | 1 (3.6) | 16 (57.1) | |
| Altered p16/pRB status | < 0.001 | |||||
| Altered p16/pRB(+) | 31 (100.0) | 4 (12.9) | 12 (38.7) | 5 (16.1) | 10 (32.3) | |
| Altered p16/pRB(-) | 163 (100.0) | 62 (38.0) | 38 (23.3) | 9 (5.5) | 54 (33.1) | |
| Non-altered p16/pRB(+) | 36 (100.0) | 8 (22.2) | 13 (36.1) | 6 (16.7) | 9 (25.0) | |
| Non-altered p16/pRB(-) | 176 (100.0) | 94 (53.4) | 24 (13.6) | 12 (6.8) | 46 (26.1) |
TNBC, triple negative breast cancer.
Correlation between p16 and pRB expressions and clinicopathologic factors
Assessment of correlation between p16 and pRB expressions and clinicopathologic factors revealed correlation of p16 alteration with higher Ki67 LI (P = 0.013), and p16 negativity with ER negativity (P = 0.002), PR negativity (P = 0.004), and higher Ki67 LI (P < 0.001). pRB positivity was correlated with PR negativity (P = 0.009), HER-2 positivity (P = 0.001), and higher Ki67 LI (P < 0.001) (Table 5).
Table 5.
Correlation between clinicopathologic factors and p16/pRB status
| Parameters | p16 alteration | p16 status | pRB status | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| No n = 212 (%) | Yes n = 194 (%) | p-value | Negative n = 31 (%) | Positive n = 375 (%) | p-value | Negative n = 339 (%) | Positive n = 67 (%) | p-value | |
| Age (years) | 0.359 | 0.576 | 0.452 | ||||||
| ≤ 50 | 115 (54.2) | 114 (58.8) | 16 (51.6) | 213 (56.8) | 194 (57.2) | 35 (52.2) | |||
| > 50 | 97 (45.8) | 80 (41.2) | 15 (48.4) | 162 (43.2) | 145 (42.8) | 32 (47.8) | |||
| Histologic grade | 0.449 | 0.204 | 0.108 | ||||||
| I/II | 142 (67.0) | 123 (63.4) | 17 (54.8) | 248 (66.1) | 227 (67.0) | 38 (56.7) | |||
| III | 70 (33.0) | 71 (36.6) | 14 (45.2) | 127 (33.9) | 112 (33.0) | 29 (43.3) | |||
| Tumor stage | 0.427 | 0.093 | 0.894 | ||||||
| T1 | 110 (51.9) | 93 (47.9) | 11 (35.5) | 192 (51.2) | 170 (50.1) | 33 (49.3) | |||
| T2/T3 | 102 (48.1) | 101 (52.1) | 20 (64.5) | 183 (48.8) | 169 (49.9) | 34 (50.7) | |||
| Nodal metastasis | 0.925 | 0.422 | 0.944 | ||||||
| Absent | 131 (61.8) | 119 (61.3) | 17 (54.8) | 233 (62.1) | 209 (61.7) | 41 (61.2) | |||
| Present | 81 (38.2) | 75 (38.7) | 14 (45.2) | 142 (37.9) | 130 (38.3) | 26 (38.8) | |||
| ER status | 0.187 | 0.002 | 0.242 | ||||||
| Negative | 75 (35.4) | 81 (41.8) | 20 (64.5) | 136 (36.3) | 126 (37.2) | 30 (44.8) | |||
| Positive | 137 (64.6) | 113 (58.2) | 11 (35.5) | 239 (63.7) | 213 (62.8) | 37 (55.2) | |||
| PR status | 0.237 | 0.004 | 0.009 | ||||||
| Negative | 99 (46.7) | 102 (52.6) | 23 (74.2) | 178 (47.5) | 158 (46.6) | 43 (64.2) | |||
| Positive | 113 (53.3) | 92 (47.4) | 8 (25.8) | 197 (52.5) | 181 (53.4) | 24 (35.8) | |||
| HER-2 status | 0.418 | 0.895 | 0.001 | ||||||
| Negative | 176 (83.0) | 155 (79.9) | 25 (80.6) | 306 (81.6) | 286 (84.4) | 45 (67.2) | |||
| Positive | 36 (17.0) | 39 (20.1) | 6 (19.4) | 69 (18.4) | 53 (15.6) | 22 (32.8) | |||
| Ki-67 LI (%) | 0.013 | < 0.001 | < 0.001 | ||||||
| ≤ 14 | 132 (62.3) | 97 (50.0) | 8 (25.8) | 221 (58.9) | 207 (61.1) | 22 (32.8) | |||
| > 14 | 80 (37.7) | 97 (50.0) | 23 (74.2) | 154 (41.1) | 132 (38.9) | 45 (67.2) | |||
When the combined status of p16 and pRB expressions was correlated with clinicopathologic factors, p16/pRB type showed correlation with histologic grade (P = 0.029), ER status (P = 0.001), HER-2 status (P = 0.003), and Ki67 LI (P < 0.001). On the other hand, p16(+)/pRB(-) type showed lower histologic grade, ER positivity, PR positivity, HER-2 negativity, and lower Ki67 LI. P16(+)/pRB(+) type and p16(-)/pRB(-) type showed higher histologic grade, ER negativity, PR negativity, HER-2 positivity, and higher Ki67 LI. The combined status of p16 alteration and pRB expression was correlated with HER-2 status (P = 0.007) and Ki67 LI (P < 0.001); altered p16/pRB(+) type and non-altered p16/pRB(+) type showed higher HER-2 positivity and higher Ki67 LI; altered p16/pRB(-) and non-altered p16/pRB(-) type was higher in HER-2 negativity and lower Ki67 LI (Figure 2).
Figure 2.
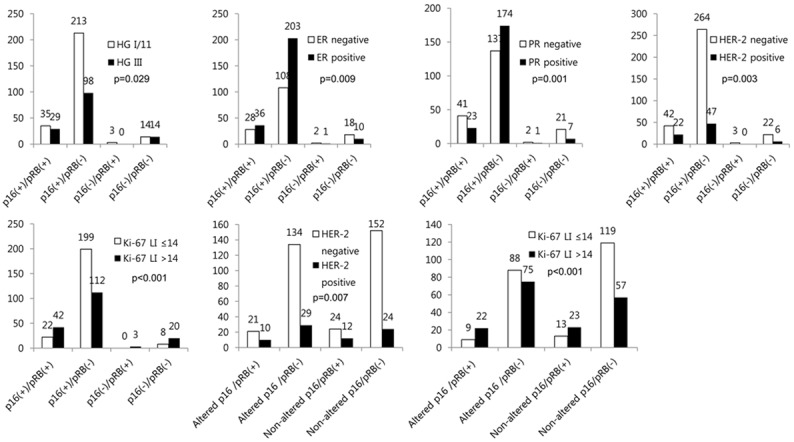
Correlation between combined p16/pRB status and clinicopathologic factors.
Impact of p16 and pRB status on breast cancer prognosis
Expressions of p16 and pRB did not have statistically significant impact on the breast cancer prognosis in univariate analysis (Table 6). However, in the molecular subtype analysis, p16 alteration correlated with shorter DFS in the luminal A type (P = 0.024, Figure 3).
Table 6.
Univariate analysis of the impact of p16 and pRB status in breast cancers on prognosis by the log-rank test
| Parameter | Number of patients/recurrence/death | Disease-free survival | Overall survival | ||
|---|---|---|---|---|---|
|
|
|
||||
| Mean survival (95% CI) months | P-value | Mean survival (95% CI) months | P-value | ||
| p16 | 0.279 | 0.369 | |||
| Negative | 31/5/5 | 95 (83-106) | 117 (102-132) | ||
| Positive | 375/36/38 | 126 (122-130) | 128 (124-131) | ||
| Alteration of p16 | 0.108 | 0.922 | |||
| No | 212/17/23 | 128 (123-133) | 128 (123-132) | ||
| Yes | 194/24/20 | 119 (113-125) | 125 (119-131) | ||
| pRB | 0.900 | 0.706 | |||
| Negative | 339/34/35 | 125 (121-130) | 127 (123-131) | ||
| Positive | 67/7/8 | 123 (115-131) | 123 (115-131) | ||
| p16/pRB status | n/a | 0.538 | |||
| p16(+)/pRB(+) | 64/7/7 | n/a | 124 (116-132) | ||
| p16(+)/pRB(-) | 311/29/31 | n/a | 127 (123-132) | ||
| p16(-)/pRB(+) | 3/0/1 | n/a | 94 (36-151) | ||
| p16(-)/pRB(-) | 28/5/4 | n/a | 119 (104-134) | ||
| Altered p16/ pRB status | 0.458 | 0.974 | |||
| Altered p16/pRB(+) | 31/4/4 | 120 (106-133) | 121 (108-133) | ||
| Altered p16/pRB(-) | 163/20/16 | 114 (108-120) | 125 (119-132) | ||
| Non-altered p16/pRB(+) | 36/3/4 | 111 (102-119) | 124 (114-135) | ||
| Non-altered p16/pRB(-) | 176/14 /19 | 128 (123-134) | 128 (122-133) | ||
Figure 3.
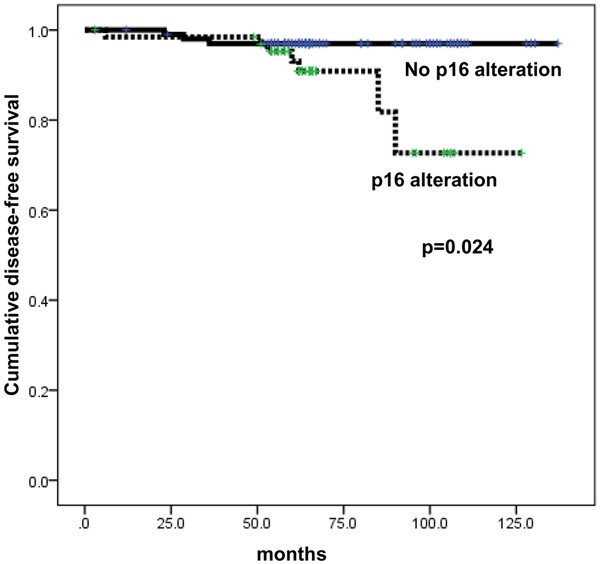
Disease-free survival according to p16 alteration status in luminal A breast cancer.
Discussion
Our assessment of p16 and pRB expressions in breast cancer showed correlation between pRB positivity and PR negativity (P = 0.009), HER-2 positivity (P = 0.001), and higher Ki67 LI (P < 0.001), in contrast to the previous study showing pRB negativity correlating with ER negativity and higher histologic grade [10]. The latter used antibody for non-phosphorylated form of pRB, but we used antibody for phosphorylated form of pRB. Since non-phosphorylated form is an active form of pRB and phosphorylated form is an inactive form, the previous study and our current study identified active and inactive forms of pRB, respectively. Therefore, it is only natural that the previous study showed pRB negativity correlating with ER negativity and higher histologic grade, both related with aggressive behavior of breast cancer, and our study showed pRB positivity correlating with PR negativity, HER-2 positivity, and higher Ki67 LI, all three related with aggressive behavior of breast cancer. In other words, pRB positivity represents pRB alteration in this study. P16 negativity was correlated with ER negativity (P = 0.002), PR negativity (P = 0.004), and higher Ki67 LI (P < 0.001). p16 normally abdicates cell replication, hence p16 negativity showing correlation with ER negativity, PR negativity, and higher Ki67 LI that are related with breast cancer aggressiveness can be explained.
The previous study has reported that p16 does not have any impact on clinicopathologic factors of breast cancer [10], but this discrepancy can be explained by the type of antibody used and the difference in criteria for interpretation. Moreover, as altered p16/pRB(+) type is expected to be most aggressive in the aspect of tumor cell proliferation, our results demonstrating higher HER-2 positivity and higher Ki67 LI in the subtype correspond to the expectation.
P16 and pRB were differentially expressed according to the molecular subtypes; p16 negativity was highest in the TNBC and high p16 expression was observed in luminal B and HER-2 types. P16 alteration was higher in luminal B and HER-2 types, and pRB expression was high in HER-2 type and low in luminal A type. These results cannot be compared with the results of the previous study since the latter did not include assessment of molecular subtypes. However, cases of altered expression of p16 and/or pRB showed correlation with aggressive parameters, and such results seem to concord with the results of the previous study [10,11,13].
In conclusion, the status of p16 and pRB alteration is different in each of the molecular subtypes of breast cancer, and the rate of abnormal expression is higher in HER-2 type.
Acknowledgements
This study was supported by a grant from the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (1420080). This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2015R1A1A1A05001209).
Disclosure of conflict of interest
None.
References
- 1.Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day RS 3rd, Johnson BE, Skolnick MH. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 2.Sellers WR, Kaelin WG Jr. Role of the retinoblastoma protein in the pathogenesis of human cancer. J. Clin. Oncol. 1997;15:3301–3312. doi: 10.1200/JCO.1997.15.11.3301. [DOI] [PubMed] [Google Scholar]
- 3.Rocco JW, Sidransky D. p16 (MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 2001;264:42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- 4.Caponio MA, Addati T, Popescu O, Petroni S, Rubini V, Centrone M, Trojano G, Simone G. P16 (INK4a) protein expression in endocervical, endometrial and metastatic adenocarcinomas of extra-uterine origin: diagnostic and clinical considerations. Cancer Biomark. 2014;14:169–175. doi: 10.3233/CBM-130326. [DOI] [PubMed] [Google Scholar]
- 5.Macha MA, Rachagani S, Pai P, Gupta S, Lydiatt WM, Smith RB, Johansson SL, Lele SM, Kakar SS, Lee JH, Meza J, Ganti AK, Jain M, Batra SK. MUC4 regulates cellular senescence in head and neck squamous cell carcinoma through p16/Rb pathway. Oncogene. 2015;34:1698–708. doi: 10.1038/onc.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller LB, Meurer L, Lopes AB, Antunes LC, Vanazzi S, Fagundes RB. Stepwise expression of CDKN2A and RB1 proteins in esophageal mucosa from patients at high risk for squamous cell carcinoma. Appl Immunohistochem Mol Morphol. 2014;22:669–673. doi: 10.1097/PAI.0000000000000011. [DOI] [PubMed] [Google Scholar]
- 7.Srivastava V, Patel B, Kumar M, Shukla M, Pandey M. Cyclin D1, retinoblastoma and p16 protein expression in carcinoma of the gallbladder. Asian Pac J Cancer Prev. 2013;14:2711–2715. doi: 10.7314/apjcp.2013.14.5.2711. [DOI] [PubMed] [Google Scholar]
- 8.Tarakji B, Alenzi F, Al-Khuraif AA. Assessment of inverse correlation of p16 and pRb expression in carcinoma ex pleomorphic adenoma. Pol J Pathol. 2013;64:144–148. doi: 10.5114/pjp.2013.36015. [DOI] [PubMed] [Google Scholar]
- 9.Bazarov AV, Lee WJ, Bazarov I, Bosire M, Hines WC, Stankovich B, Chicas A, Lowe SW, Yaswen P. The specific role of pRb in p16 (INK4A) -mediated arrest of normal and malignant human breast cells. Cell Cycle. 2012;11:1008–1013. doi: 10.4161/cc.11.5.19492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dublin EA, Patel NK, Gillett CE, Smith P, Peters G, Barnes DM. Retinoblastoma and p16 proteins in mammary carcinoma: their relationship to cyclin D1 and histopathological parameters. Int J Cancer. 1998;79:71–75. doi: 10.1002/(sici)1097-0215(19980220)79:1<71::aid-ijc14>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 11.Geradts J, Wilson PA. High frequency of aberrant p16 (INK4A) expression in human breast cancer. Am J Pathol. 1996;149:15–20. [PMC free article] [PubMed] [Google Scholar]
- 12.Gorgoulis VG, Koutroumbi EN, Kotsinas A, Zacharatos P, Markopoulos C, Giannikos L, Kyriakou V, Voulgaris Z, Gogas I, Kittas C. Alterations of p16-pRb pathway and chromosome locus 9p21-22 in sporadic invasive breast carcinomas. Mol Med. 1998;4:807–822. [PMC free article] [PubMed] [Google Scholar]
- 13.Grupka NL, Bloom C, Singh M. Expression of retinoblastoma protein in breast cancer metastases to sentinel nodes: evaluation of its role as a marker for the presence of metastases in non-sentinel axillary nodes, and comparison to p16INK4a. Appl Immunohistochem Mol Morphol. 2006;14:63–70. doi: 10.1097/01.pai.0000161486.72621.4a. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen NH, Emdin SO, Cajander J, Landberg G. Deregulation of cyclin E and D1 in breast cancer is associated with inactivation of the retinoblastoma protein. Oncogene. 1997;14:295–304. doi: 10.1038/sj.onc.1200833. [DOI] [PubMed] [Google Scholar]
- 15.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 16.Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, Hicks DG, Lester S, Love R, Mangu PB, McShane L, Miller K, Osborne CK, Paik S, Perlmutter J, Rhodes A, Sasano H, Schwartz JN, Sweep FC, Taube S, Torlakovic EE, Valenstein P, Viale G, Visscher D, Wheeler T, Williams RB, Wittliff JL, Wolff AC. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 18.Lee K, Jung ES, Choi YJ, Lee KY, Lee A. Expression of pRb, p53, p16 and cyclin D1 and their clinical implications in urothelial carcinoma. J Korean Med Sci. 2010;25:1449–1455. doi: 10.3346/jkms.2010.25.10.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shariat SF, Tokunaga H, Zhou J, Kim J, Ayala GE, Benedict WF, Lerner SP. p53, p21, pRB, and p16 expression predict clinical outcome in cystectomy with bladder cancer. J. Clin. Oncol. 2004;22:1014–1024. doi: 10.1200/JCO.2004.03.118. [DOI] [PubMed] [Google Scholar]
- 20.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]


