Abstract
Background: Undifferentiated pleomorphic sarcoma (UPS), previously known as malignant fibrous histiocytoma, comprises a series of high-grade soft tissue sarcomas, which fail to exhibit any specific line of differentiation by using currently available ancillary techniques. Studies on gene mutation screening occurring in UPS are rarely conducted. In this study, we described a case of UPS and analyzed its mutation changes. We detected 19 hotspot oncogenes in the case. To the best of our knowledge, this study is the first to use a high-throughput OncoCarta panel 1.0 and MassARRAY system to detect 238 known mutations in 19 hotspot oncogenes in UPS. In this study, our result revealed two missense mutations, namely, KRAS mutation (35G > A, G12D) and PIK3CA mutation (1636C > A, Q546K) in the case.
Keywords: Undifferentiated soft tissue sarcoma, mutations, MassARRAY
Introduction
Undifferentiated pleomorphic sarcoma (UPS), previously known as malignant fibrous histiocytoma (MFH), comprises a series of high-grade soft tissue sarcomas, which fail to exhibit any specific line of differentiation by using currently available ancillary techniques. In the 1960s, Ozzello first introduced MFH as a diagnosis for a group of malignant mesenchymal tumors composed primarily of pleomorphic and spindle-shaped cells with a predominantly storiform growth pattern. Moreover, these tumors did not fit into any of the recognized sarcoma categories [1]. By the end of the 1980s, pleomorphic MFH was the single largest category of sarcomas [2]. Nevertheless, by the advent of electron microscopy and immunohistochemical techniques, scientific evidence for a “fibrohistiocytic” line of differentiation in this group of tumors became clear [3]. Some scholars thought that a proportion of these tumors will likely prove to be of fibroblastic lineage. However, the recognized reproducible diagnostic criteria for these tumors remain challenging. Currently, World Health Organization has chosen to replace this term with the neutral and more accurate label of, i.e., UPS [4]. Moreover, the genetic aspects of UPS are difficult to evaluate because of the shifting diagnostic criteria used throughout the years. Therefore, we performed a gene mutation screening occurring in a case of UPS.
Case report
A 66-year-old woman presented with a tumor on the left thigh with non-specific cervical discomfort, no pain, and no compression symptoms was observed in this case. Laboratory tests indicated no significant abnormality. Computed tomography showed a mass with no clear boundary. Complete and extended surgical resection was performed. Macroscopic examination showed a tumor (16 cm × 10.8 cm × 7.9 cm) with a solid grayish cut surface and no clear envelope. Furthermore, the mass had storiform regional and polymorphic regional mixed distributions. The storiform area was constituted by blunt round spindle cells, and the polymorphic region contained abundant chromatin and nuclear irregular multinucleated giant cells. Compared with the storiform region, polymorphic region contained randomly arranged, obese or round fibroblast cells. However, those cells did not have the trend to grow around the vessels (Figure 1).
Figure 1.
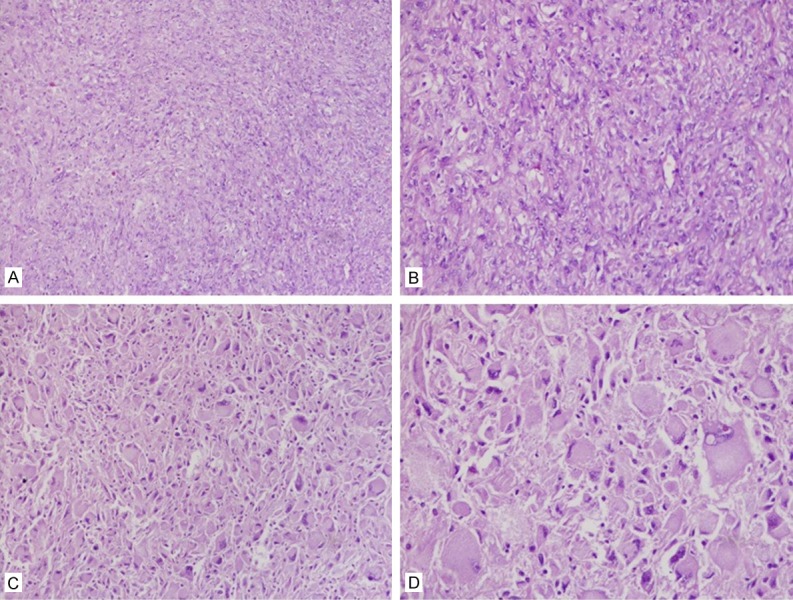
Major histologic feature of UPS. A, B. Numerous spindle tumor cells are set in a variably collagenized extracellular matrix. C, D. The polymorphic region contains abundant chromatin and nuclear irregular multinucleated giant cells.
The tumor cells were immunohistochemically determined to be diffusely positive for vimentin but negative for myogenin, desmin, MyoD1, EMA, smooth muscle actin, and CD34 (Figure 2). Furthermore, the tumor cells were the focal expression of CD68 (Figure 3), but negative for S-100, melan A, and HMB45.
Figure 2.
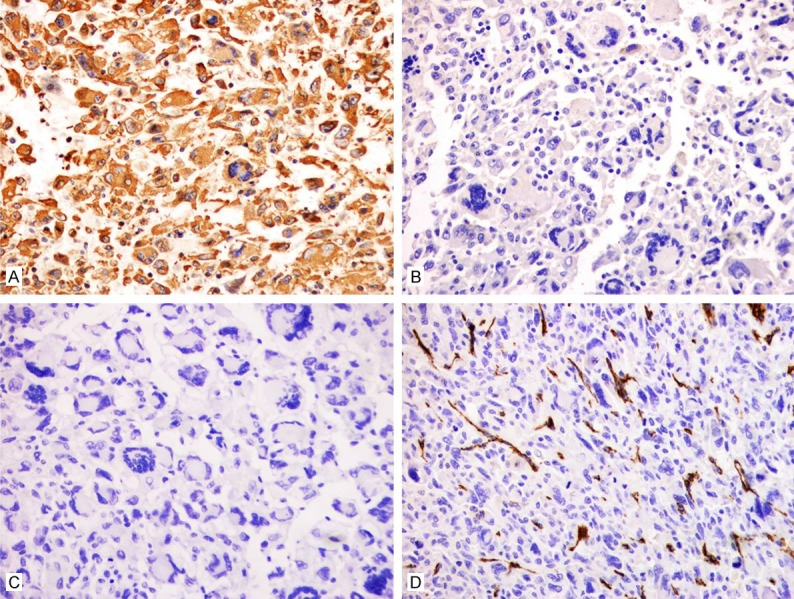
Immunohistochemical staining of differentially expressed proteins in UPS. A. Tumor cells showing diffused cytoplasmic immunoreactivity for vimentin. × 200. B. Tumor cells showing no immunoreactivity for EMA. × 200. C. Tumor cells showing no immunoreactivity for desmin. × 200. D. Tumor cells showing blood vessel immunoreactivity for CD34. × 200.
Figure 3.
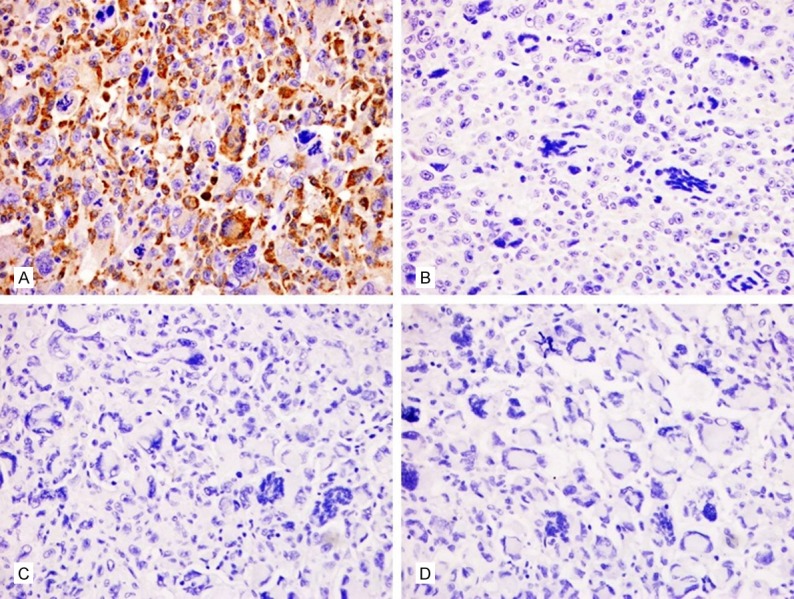
Immunohistochemical staining of differentially expressed proteins in UPS. A. Tumor cells showing cytoplasmic immunoreactivity for CD68. × 200. B. Tumor cells showing no immunoreactivity for S-100. × 200. C. Tumor cells showing no immunoreactivity for melan A. × 200. D. Tumor cells showing no immunoreactivity for HMB-45. × 200.
A MassARRAY system (Sequenom Inc., San Diego, CA, USA) based on matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and OncoCarta Panel v1.0 (Sequenom) detected 238 mutations in 19 genes (ABL1, JAK-2, CDK4, KRAS, HRAS, NRAS, AKT1, AKT2, KIT, EGFR, MET, ERBB2, PDGFA, BRAF, FGFR1, FGFR3, PIK3CA, RET, and FLT3). This system followed the manufacturer’s recommendations with minor modifications. The detailed procedure and conditions of a previous study were followed [5]. Mutation screening revealed the presence of KRAS and PIK3CA mutations in the present case. These mutations resulted in a G-A substitution at position 35, which led to the substitution of Gly by Asp, and a C-A substitution at position 1636, led to the substitution of Gln by substitute Lys (Figure 4).
Figure 4.
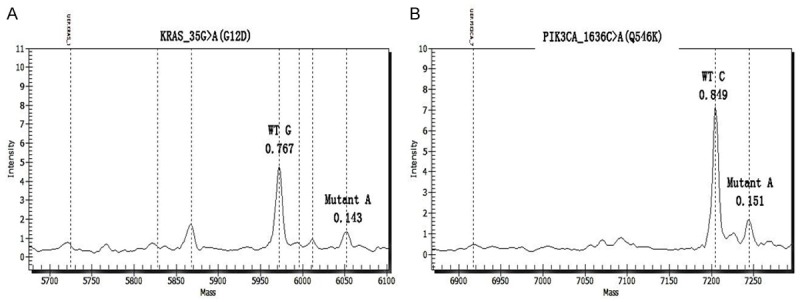
Compound KRAS and PIK3CA Mutations in UPS. Mass spectrogram of the (A) KRAS G12D and (B) PIK3CA Q546K mutations in the sample, indicating the substitution of GGU (Gly) by GAU (Asp) at codon 12 and CAA (Gln) by AAA (Lys) at codon 546.
The patient died because of tumor recurrence and metastases 12 months later in the case.
Discussion
UPS accounts for 5% to 10% of sarcomas in adults older than 40 years of age [4]. These tumors generally ascend in the deep soft tissues of older patients (in the sixth decade and older). The most common sites of origin are the proximal extremities, particularly the thigh and buttock. Histologically, tumors are composed of a haphazard or storiform arrangement of highly pleomorphic and spindle-shaped cells, and these cells have a variable amount of eosinophilic or amphophilic cytoplasm and numerous typical and atypical mitoses. Stroma is usually collagenous with an inflammatory infiltrate. UPS shows neither reproducible immunophenotype nor any pattern of protein expression that will enable more specific subclassification. Only vimentin is convincingly positive in most cases.
The genetic aspects of undifferentiated pleomorphic sarcoma are difficult to evaluate because of the shifting diagnostic criteria used throughout the years. The current study found that gene mutations contribute to tumor diagnosis and molecular targeted therapy. However, previous research that focused on a gene mutation associated with cancer provides only clues regarding the origin and development process of tumors. Mutations in KRAS, NRAS, HRAS, BRAF, PIK3CA, CTNNB1, PTPN11, and FGFR4 have been reported in rhabdomyosarcoma cases [6]. The presence of PIK3CA mutation has been associated with a significant increase in cancer-specific mortality [7].
To our knowledge, this study is the first to apply a high-throughput OncoCarta panel 1.0 and MassARRAY system to detect 238 known mutations in 19 hotspot oncogenes. This study also found the compound KRAS and PIK3CA mutations in UPS.
KRAS is a proto-oncogene that is located at 12p12.1. KRAS is the frequently altered gene with mutations occurring in 17% to 25% of all cancers. KRAS mutation is an important companion in a diagnostic test to guide the use of anti-EGFR antibody treatment of metastatic colorectal cancer. The U.S. Food and Drug Administration and the European Medicines Evaluation Agency require that KRAS mutation status should be determined prior to anti-EGFR treatment [8].
Mutations in the PIK3CA gene have been identified in many human solid tumors, including breast, colon, ovarian, liver, and lung cancers [9]. The majority of activating PIK3CA mutations has been identified in the helical (exon 9) and kinase domains (exon 20). Liao et al. sequenced PIK3CA through pyrosequencing in 1170 rectal and colon cancers in two prospective cohort studies and found 189 (16%) PIK3CA-mutated tumors. Patients with concomitant PIK3CA mutations in exons 9 and 20 significantly experienced the worst cancer-specific survival, and PIK3CA mutation is associated with poor prognosis of colorectal cancer patients [10]. Miron et al. showed that the frequency of PIK3CA mutations was essentially the same (approximately 30%) in ductal carcinoma in situ (DCIS), DCIS adjacent to invasive carcinoma, and invasive ductal breast carcinomas. Therefore, the mutation of PIK3CA was suggested to be an early event in breast cancer that is more expected to initiate breast tumor than in invasive progression [11]. Our mutation screening revealed the Q546K missense mutations, which result in a C-A substitution at position 1636, which led to the substitution of Gln by Lys at codon 546.
Limited data on UPS indicate an aggressive clinical course and high incidence of recurrence and metastasis. In the present case, the patient died because of tumor recurrence and metastases 12 months later. In future studies, a large sample size of UPS cases may aid in the investigation of molecular markers, which may improve the UPS treatment and diagnosis.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 81160322 and 81460404).
Disclosure of conflict of interest
None.
References
- 1.Ozzello L, Stout AP, Murray MR. Cultural characteristics of malignant histiocytomas and fibrous xanthomas. Cancer. 1963;16:331–344. doi: 10.1002/1097-0142(196303)16:3<331::aid-cncr2820160307>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 2.Weiss SW. Malignant fibrous histiocytoma. A reaffirmation. Am J Surg Pathol. 1982;6:773–784. doi: 10.1097/00000478-198212000-00009. [DOI] [PubMed] [Google Scholar]
- 3.Akerman M. Malignant fibrous histiocytoma--the commonest soft tissue sarcoma or a nonexistent entity? Acta Orthop Scand Suppl. 1997;273:41–46. [PubMed] [Google Scholar]
- 4.Fletcher CD. The evolving classification of soft tissue tumours - an update based on the new 2013 WHO classification. Histopathology. 2014;64:2–11. doi: 10.1111/his.12267. [DOI] [PubMed] [Google Scholar]
- 5.Liu CX, Li XY, Li CF, Chen YZ, Cui XB, Hu JM, Li F. Compound HRAS/PIK3CA mutations in Chinese patients with alveolar rhabdomyosarcomas. Asian Pac J Cancer Prev. 2014;15:1771–1774. doi: 10.7314/apjcp.2014.15.4.1771. [DOI] [PubMed] [Google Scholar]
- 6.Shukla N, Ameur N, Yilmaz I, Nafa K, Lau CY, Marchetti A, Borsu L, Barr FG, Ladanyi M. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18:748–757. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, Kure S, Chan AT, Engelman JA, Kraft P, Cantley LC, Giovannucci EL, Fuchs CS. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J. Clin. Oncol. 2009;27:1477–1484. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolton L, Reiman A, Lucas K, Timms J, Cree IA. KRAS mutation analysis by PCR: a comparison of two methods. PLoS One. 2015;10:e0115672. doi: 10.1371/journal.pone.0115672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 10.Liao X, Morikawa T, Lochhead P, Imamura Y, Kuchiba A, Yamauchi M, Nosho K, Qian ZR, Nishihara R, Meyerhardt JA, Fuchs CS, Ogino S. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18:2257–2268. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miron A, Varadi M, Carrasco D, Li H, Luongo L, Kim HJ, Park SY, Cho EY, Lewis G, Kehoe S, Iglehart JD, Dillon D, Allred DC, Macconaill L, Gelman R, Polyak K. PIK3CA mutations in in situ and invasive breast carcinomas. Cancer Res. 2010;70:5674–5678. doi: 10.1158/0008-5472.CAN-08-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]


