A microtubule-independent membranous “spindle envelope” confines spindle assembly and accounts for faithful chromosome segregation.
Abstract
The mitotic spindle is a microtubular assembly required for chromosome segregation during mitosis. Additionally, a spindle matrix has long been proposed to assist this process, but its nature has remained elusive. By combining live-cell imaging with laser microsurgery, fluorescence recovery after photobleaching, and fluorescence correlation spectroscopy in Drosophila melanogaster S2 cells, we uncovered a microtubule-independent mechanism that underlies the accumulation of molecules in the spindle region. This mechanism relies on a membranous system surrounding the mitotic spindle that defines an organelle-exclusion zone that is conserved in human cells. Supported by mathematical modeling, we demonstrate that organelle exclusion by a membrane system causes spatio-temporal differences in molecular crowding states that are sufficient to drive accumulation of mitotic regulators, such as Mad2 and Megator/Tpr, as well as soluble tubulin, in the spindle region. This membranous “spindle envelope” confined spindle assembly, and its mechanical disruption compromised faithful chromosome segregation. Thus, cytoplasmic compartmentalization persists during early mitosis to promote spindle assembly and function.
Introduction
During mitosis, chromosomes must be accurately segregated to opposite sides of the cell by a microtubule (MT)-based structure known as the mitotic spindle. Additionally, an MT-independent structure, the so-called “spindle matrix,” has been proposed to assist mitosis. Originally envisioned as a static scaffold that could act as a tensile element that mediates chromosome motion (Pickett-Heaps et al., 1984) and/or determines spindle length (Mitchison et al., 2005), more recent findings suggest that the spindle matrix is rather a dynamic assembly of molecules confined to the vicinity of the spindle (Lince-Faria et al., 2009; Schweizer et al., 2014). However, the mechanism that underlies the MT-independent accumulation of proteins in the spindle region remains unknown.
Membranes derived from the nuclear envelope (NE) and ER surround the mitotic spindle (Hepler and Wolniak, 1984), but their role in mitosis remains poorly understood. A membranous “lamin B matrix” was proposed to work as a scaffold for the recruitment of proteins required for mitotic spindle assembly and function (Tsai et al., 2006). Importantly, the formation of a lamin B matrix relies on cytoplasmic Dynein (Ma et al., 2009), suggesting a dependence on MTs, and has been implicated in force balance with Kinesin-like motors to promote efficient spindle assembly (Civelekoglu-Scholey et al., 2010; Goodman et al., 2010). Supported by mathematical modeling, these findings led to the proposal that lamin B and possibly other membranous components surrounding the spindle play a structural role in the orchestration of cell division, even in systems undergoing open mitosis (Liu and Zheng, 2009; Civelekoglu-Scholey et al., 2010; Poirier et al., 2010; Zheng, 2010; Capalbo et al., 2011).
Here, we used Drosophila melanogaster S2 and human HeLa cells to dissect the mechanism that drives the MT-independent accumulation of proteins in the spindle region. We show that this process is a consequence of spatial confinement by a membranous, organelle-exclusion spindle envelope, which is required for normal spindle architecture and faithful chromosome segregation.
Results and discussion
Megator and soluble tubulin accumulate in the spindle region independently of MTs
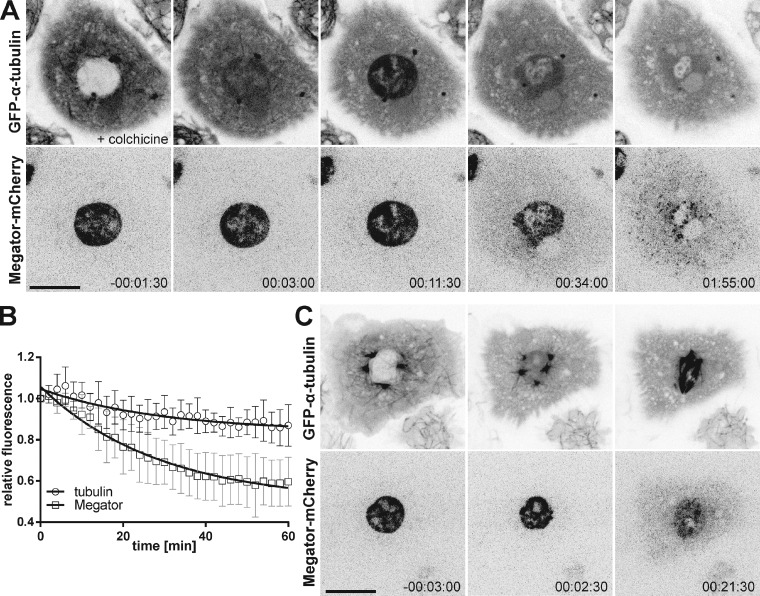
A pool of the conserved nucleoporin Megator persists in the spindle region of Drosophila S2 cells after MT depolymerization (Lince-Faria et al., 2009). To elucidate the MT-independent mechanism behind the enrichment of Megator in the spindle region, we monitored GFP–α-tubulin and Megator-mCherry in Drosophila S2 cells by live-cell imaging, after depolymerizing MTs with colchicine before NEB (Fig. 1, A and B). We observed that after NEB (monitored by influx of soluble tubulin), Megator remained confined to the spherical “nuclear” space for ∼10 min before decreasing exponentially and spreading into the cytoplasm (Fig. 1, A and B; and Video 1). As reported previously in Caenorhabditis elegans and Drosophila embryos (Hayashi et al., 2012; Yao et al., 2012), soluble tubulin also accumulated in the nuclear region after NEB, reaching the highest level (∼1.5-fold the levels in the surrounding cytoplasm) within a few minutes, upon which it decreased exponentially (Fig. 1, A and B). Notably, soluble tubulin and Megator were excluded from chromosomes (Fig. 1, A and B) and were also enriched in the “nuclear” region soon after NEB during an unperturbed mitosis (Fig. 1 C). Thus, soluble tubulin and Megator accumulate in the spindle region during early mitosis in an MT-independent manner.
Figure 1.
Megator and soluble tubulin accumulate in the spindle region independently of MTs. (A) S2 cell expressing Megator-mCherry and GFP–α-tubulin upon MT depolymerization with colchicine before NEB. (B) Normalized relative fluorescence (nuclear/cytoplasm) of GFP–α-tubulin and Megator-mCherry. The relative fluorescence was set to 1 at time 0 (maximal accumulation of soluble tubulin in the nuclear region). Data points represent means obtained from seven cells, to which an exponential curve was fit. Error bars represent standard deviations. (C) Accumulation of GFP–α-tubulin and Megator-mCherry in the nuclear region after NEB in an unperturbed mitosis. Time is given in hours:minutes:seconds, relative to NEB. Bars, 10 µm.
Soluble tubulin passively diffuses into the nuclear space at NEB
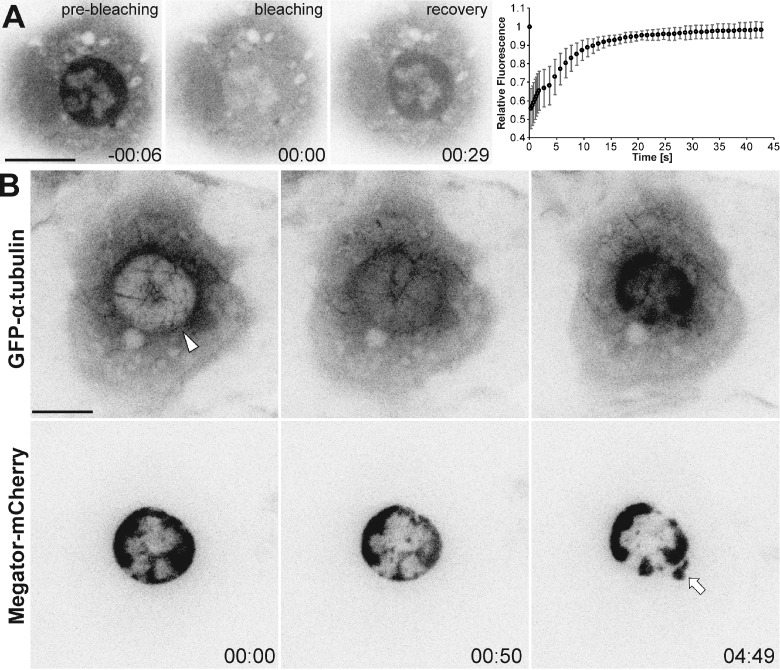
To analyze the dynamics of soluble tubulin in the nuclear region during early mitosis, we performed FRAP of GFP–α-tubulin in S2 cells, after MT depolymerization with colchicine at NEB. We found that after photobleaching the entire nuclear region, GFP–α-tubulin reaccumulated with a half-life of ∼4 s, with a percentage of recovery (nuclear/cytoplasmic ratio) close to 100% (Fig. 2 A). Thus, most if not all soluble tubulin in the nuclear region is mobile, in line with previous reports in sea urchin embryos (Salmon et al., 1984), but in contrast to a recently reported RanGTP-dependent immobile fraction in C. elegans (Hayashi et al., 2012).
Figure 2.
Soluble tubulin passively diffuses into the nuclear space at NEB. (A) FRAP of GFP–α-tubulin in the nuclear region of mitotic S2 cells after MT depolymerization with colchicine. Bleaching was conducted at time 00:00 (minutes:seconds). The relative fluorescence (nuclear/cytoplasm) was set to 1 before bleaching. Data points represent means obtained from 10 cells, error bars represent standard deviations. Soluble tubulin is highly mobile and exchanges between compartments (t1/2 = 4.495 s; SD = 1.552 s; recovery = 98.5%; SD = 4.3%; n = 10 cells). (B) Laser microsurgery in an interphase S2 cell expressing Megator-mCherry and GFP–α-tubulin after MT depolymerization with colchicine. The arrowhead indicates the cut region. Membrane blebbing was occasionally observed in the cut region (arrow). Time is given in minutes:seconds, relative to laser microsurgery. Bars, 10 µm.
To test whether tubulin passively enters/accumulates in the nuclear region, we performed laser microsurgery on the NE in interphase cells after MT depolymerization with colchicine. This caused immediate influx and accumulation of soluble tubulin in the nucleus, while Megator remained confined in this region (Fig. 2 B and Video 2). Occasionally, Megator and tubulin showed some leakage in the perforated region, possibly due to the formation of a membrane bleb (Tinevez et al., 2009). Thus, the influx/accumulation of soluble tubulin in the nuclear space is normally prevented until NEB, upon which tubulin passively diffuses between compartments.
A membranous spindle envelope confines Megator and soluble tubulin during early mitosis
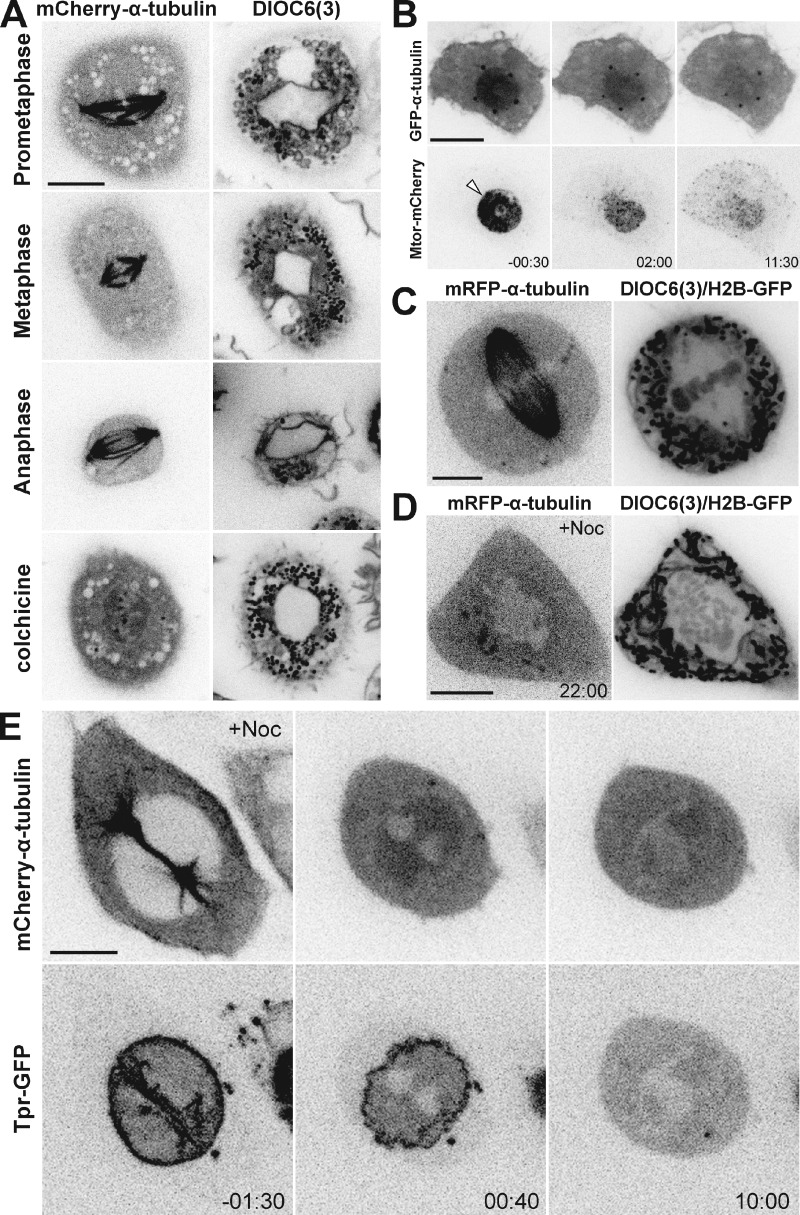
Drosophila embryos and cells undergo a “semi-open” mitosis within a membranous spindle envelope (Stafstrom and Staehelin, 1984; Harel et al., 1989; Debec and Marcaillou, 1997). To investigate the distribution of membranes and membranous organelles during mitosis in living cells, we generated an S2 cell line stably expressing mCherry–α-tubulin and added DiOC6(3), a lipophilic green-fluorescent vital dye. In parallel, we transiently expressed a membrane-tagged GFP by fusing it with the human T cell receptor CD8 (D’Avino et al., 2006) in S2 cells stably expressing mRFP–α-tubulin and histone H2B-GFP. We observed that the spindle region was surrounded by membranes that persisted until anaphase (Fig. 3 A, Fig. S1 A, and Video 3). These membranes did not form a diffusion barrier for small molecules as they were open near centrosomes, but deformed while the spindle changed shape and appeared to keep the spindle region clear of large membranous organelles (e.g., mitochondria). This organelle-exclusion zone was independent of MTs and accumulated soluble tubulin after NEB in colchicine-treated cells (Fig. 3 A). Importantly, the apparent integrity of the membranous spindle envelope gradually decreased from prophase to anaphase, similar to Lamin B (Fig. S1, A and B; and Video 3), which might explain the decrease of Megator and soluble tubulin in the nuclear region after NEB.
Figure 3.
A membranous spindle envelope confines Megator and soluble tubulin during early mitosis. (A) Live S2 cells expressing mCherry–α-tubulin treated with DiO6C(3). (B) Live S2 cell expressing Megator-mCherry and GFP–α-tubulin in which the spindle envelope has been disrupted by laser microsurgery (arrowhead) after MT depolymerization with colchicine. (C and D) Live HeLa cells expressing mRFP–α-tubulin and histone H2B-GFP treated with DiOC6(3), with/without nocodazole as indicated. (E) Live HeLa cell expressing Tpr-GFP and mCherry–α-tubulin after nocodazole-induced MT depolymerization before NEB. Time is in minutes:seconds, relative to laser microsurgery (B) or NEB (D and E). Bars, 10 µm.
To directly test whether membranes are necessary to confine Megator and soluble tubulin in the nuclear region during early mitosis, we performed laser microsurgery to severely disrupt the spindle envelope in mitotic cells (160 consecutive pulses compared with a single pulse in interphase) stably expressing Megator-mCherry and GFP–α-tubulin, after MT depolymerization with colchicine. We observed an immediate decrease in the confinement of Megator and soluble tubulin that was faster than in unperturbed cells at NEB (Fig. 3 B; Fig. S2, A and B; and Video 4). Thus, a membranous system supports elevated concentrations of soluble tubulin and Megator in the spindle region during early mitosis in S2 cells.
An MT-independent organelle-exclusion system surrounds the spindle region in human cells
The spindle region in human cells works as an organelle-exclusion zone (McIntosh and Landis, 1971). Because mitosis in human cells is considered to be open, it implies that the mitotic spindle itself prevents large organelles from invading the region occupied by chromosomes. However, some studies suggest that organelle exclusion is independent of MTs (Wheatley, 1990). One possibility is that ER membranes that remain as an interconnected network around the mitotic spindle (Paweletz, 1981; Hepler and Wolniak, 1984; Ellenberg et al., 1997) exclude large organelles from this region in human cells. To test this, we analyzed membrane distribution in live HeLa cells stably expressing mRFP–α-tubulin and histone H2B-GFP, after addition of DiOC6(3). We observed that, similar to S2 cells, the spindle region of HeLa cells is surrounded by membranes and free of membranous organelles (Fig. 3 C). Importantly, this organelle-exclusion zone persisted after MT depolymerization with nocodazole (Fig. 3 D). These data from living cells support previous ultrastructural studies (Moll and Paweletz, 1980; Wheatley, 1990; McCullough and Lucocq, 2005) and indicate that a membranous system, not MTs, prevents the invasion of the spindle region by large organelles in human cells.
To investigate whether this membranous system accounts for protein accumulation in the spindle region in human cells, we analyzed mitotic HeLa cells expressing mouse Tpr, the mammalian orthologue of Megator (Lince-Faria et al., 2009), fused to GFP and mCherry–α-tubulin. To rule out the contribution from MTs, they were depolymerized with nocodazole before NEB. We observed that soluble tubulin enriched 1.16-fold in the nuclear region (SD = 0.08; n = 10 cells) shortly after NEB (Fig. 3 E and Video 5). Tpr also accumulated in the nuclear region, while remaining associated with the disassembling NE (Fig. 3 E and Video 5). However, the enrichment of soluble tubulin and Tpr in the nuclear region rapidly decreased, indicating that, despite the presence of an organelle-exclusion system, the MT-independent accumulation of proteins in the spindle region is limited to early mitosis in human cells.
Protein accumulation in the nuclear/spindle region is a consequence of different molecular crowding states
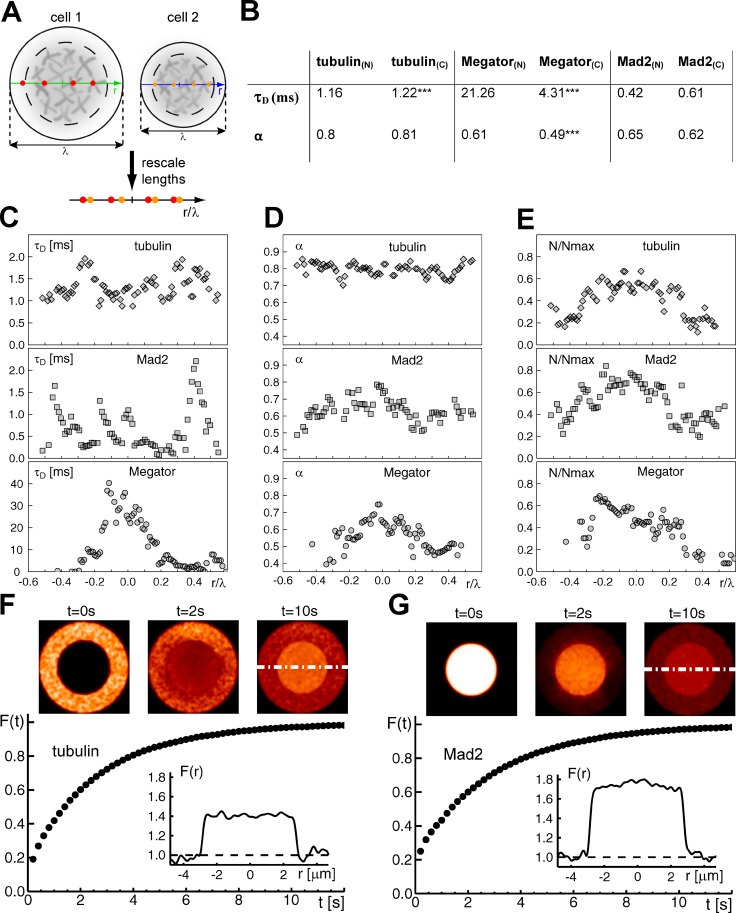
To investigate how a fenestrated organelle-exclusion envelope drives protein accumulation in the nuclear/spindle region during early mitosis, we characterized the diffusion properties of GFP–α-tubulin and Megator-mCherry at a single-molecule level using fluorescence correlation spectroscopy (FCS) in S2 cells. To ensure the independence from MTs, they were depolymerized with colchicine. While soluble tubulin showed similar anomalous diffusion inside and outside the nuclear region, Megator diffusion was about five times slower in the nuclear region when compared with the rest of the cytoplasm (Fig. 4, A–E), where Megator diffused more anomalously (i.e., more obstructed). These differences might reflect interactions with at least two other nuclear-derived proteins: Chromator and EAST (Qi et al., 2004, 2005).
Figure 4.
Protein accumulation in the nuclear/spindle region is a consequence of different molecular crowding states. (A) Cells with different diameters (λ) and different spatial positions of the FCS measurements were made comparable by expressing measurement positions relative to the cell size (red and orange bullets). Dashed circles indicate the nuclear space; gray-shaded objects symbolize condensed chromatin. (B) FCS analysis of GFP–α-tubulin, Megator-mCherry, and mRFP-Mad2 during mitosis after MT depolymerization with colchicine. Depicted are the mean or median values of the dwell time τD and the anomaly degree (α) of the diffusional motion within the focus in both the nuclear region (N) and cytoplasm (C). ***, P < 0.001. (C–E) FCS measurements were performed at 10 loci per cell (tubulin, 11 cells; Megator, 15 cells; Mad2, 15 cells). Position data were rescaled (nuclear region: λ = −0.2, . . . , +0.2). The number of fluorescent molecules (N) within the focus was relativized to the maximum number of fluorescent molecules (Nmax) detected in each cell to account for varying expression levels. (F and G) Modeling of protein diffusion into/out of the nuclear region. Diffusion of soluble tubulin (F) into and Mad2 (G) out of the nuclear region progresses within few seconds to a steady state, as indicated by the apparent fluorescence in the nuclear region Fn(t) and the cytoplasm Fc(t). Representative fluorescence images of the initial condition, an intermediate state, and the steady state are shown in the top panels. Dashed white lines indicate the respective line scans used for quantification of fluorescence (inset graphs).
In line with the observed increase of fluorescence in the nuclear space, the number of mobile tubulin and Megator molecules was increased ∼1.4-fold in this compartment when compared with the surrounding cytoplasm (Fig. 4 E), suggesting that the entire pool of soluble tubulin and Megator is mobile within the nuclear region. Importantly, GFP and mCherry signals did not cross-correlate, indicating that soluble tubulin and Megator are not bound to each other.
Because the spindle-assembly checkpoint protein Mad2 is also confined to the spindle region in an MT-independent manner (Lince-Faria et al., 2009), next we used the same approach in cells expressing mRFP-Mad2 and α-tubulin–GFP. We found that Mad2 diffused similarly within the nuclear region and the cytoplasm, with mobile Mad2 enriched ∼1.8-fold in the nuclear region when compared with the cytoplasm (Fig. 4, B–E). Notably, the dwell time of Mad2 within the focus fluctuated significantly, possibly reflecting the existence of different soluble pools (Fig. 4 C). Mad2 and tubulin signals also did not cross-correlate. Thus, confinement of soluble tubulin, Megator, and Mad2 to the spindle region is not caused by binding of these proteins to a common stationary substrate.
To test whether a fenestrated organelle-exclusion envelope could account for protein accumulation in the spindle region during early mitosis, we simulated the influx of soluble tubulin/efflux of Mad2 into/out of the nuclear space at NEB, using the FCS-measured diffusion properties and considering different amounts of inaccessible volumes in the cytoplasm due to the presence of large membranous organelles that cannot penetrate the spindle envelope. To account for this structure, a fenestrated barrier around the nuclear region was assigned, allowing protein influx/efflux at only 25% of its sites. By blocking 33% and 36% of the cytoplasm for soluble tubulin and Mad2, respectively (see Materials and methods), simulations yielded the experimentally observed fluorescence ratio between the nuclear region and the cytoplasm within few seconds (Fig. 4, F and G). These findings suggest that a size-exclusion spindle envelope is sufficient to drive accumulation of soluble tubulin and Mad2 in the spindle region, as a consequence of distinct molecular crowding states.
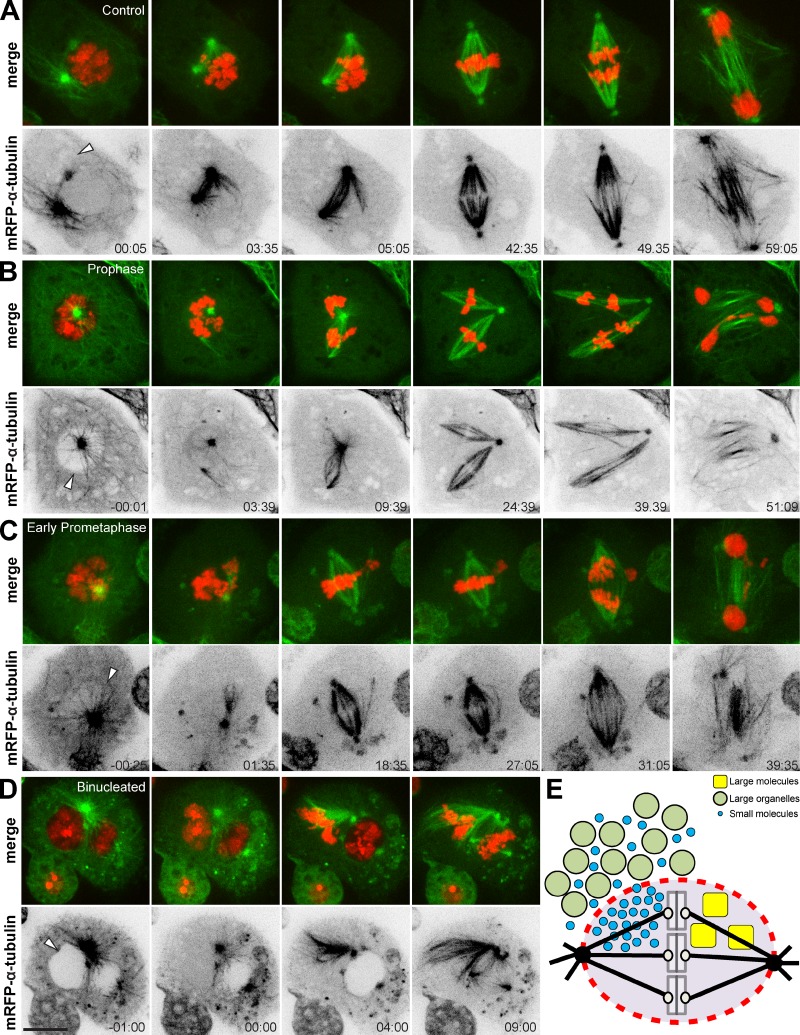
Artificial disruption of the nuclear/spindle envelope in mitosis causes aberrant spindle formation and chromosome missegregation
Previous works, supported by mathematical modeling, proposed that a membranous spindle envelope promotes bipolar spindle assembly and function (Liu and Zheng, 2009; Civelekoglu-Scholey et al., 2010; Poirier et al., 2010; Zheng, 2010; Capalbo et al., 2011). Here we tested the role of a membranous spindle envelope for spindle assembly by means of laser-induced disruption (160 consecutive pulses) of the nuclear/spindle envelope during early mitosis in live S2 cells expressing histone H2B-GFP and mRFP–α-tubulin. As a control, we performed laser microsurgery at a random site in the cytoplasm shortly after NEB and found that 10 out of 11 cells progressed normally through mitosis (Fig. 5 A). In contrast, we observed that immediately after cutting the nuclear/spindle envelope, acentrosomal MTs nucleated in the proximity of the cut region and grew into the cytoplasm and into the nucleus/nuclear region (Fig. 5, B–D; and Videos 6 and 7). In line with these findings, membrane resealing after laser microsurgery could only be observed in 2 out of 6 cells that transiently expressed CD8-GFP (Fig. S3). Notably, acentrosomal MT growth occasionally led to MT-mediated transport of chromosomes outside the nuclear region (5 out of 21 cells; Fig. 5, B and C) and/or resulted in the formation of multipolar spindles (6 out of 21 cells; Fig. 5, B and C), while markedly increasing the frequency of lagging chromosomes/chromatin bridges during anaphase/telophase (13 out of 21 cells; Fig. 5, B and C). Finally, in another independent experiment, we disrupted the NE of one nucleus in a binucleated cell. We observed that, immediately after laser microsurgery, soluble tubulin entered that nucleus, in which chromosomes instantly condensed (Fig. 5 D and Video 8). Spindle assembly also started immediately, extending beyond the nuclear space (Fig. 5 D and Video 8). NEB of the unperturbed nucleus started only several minutes later and spindle assembly was confined to the nuclear region (Fig. 5 D and Video 8). These data support the finding that the spindle envelope confines spindle assembly by constraining MT nucleation and growth to ensure proper chromosome segregation.
Figure 5.
Artificial disruption of the nuclear/spindle envelope in mitosis causes aberrant spindle formation and chromosome missegregation. (A–D) S2 cells expressing mRFP–α-tubulin and histone H2B-GFP. The nuclear/spindle envelope was consecutively disrupted by laser microsurgery (arrowheads). (A) Control cell progressing through mitosis. Laser microsurgery was performed at a random site in the cytoplasm shortly after NEB (arrowhead). (B) Disruption of the NE in prophase or the spindle envelope in early prometaphase (C) caused aberrant spindle formation and chromosome missegregation. (D) Chromosomes of the severed nucleus in a binucleated prophase cell condensed prematurely, and centrosome-nucleated MTs grew in the cytoplasm toward the cut region. NEB of the unsevered nucleus initiated only several minutes later, and MTs grew predominantly into the nuclear space. Time is given in minutes:seconds, relative to laser microsurgery. Bars, 10 µm. (E) Model: a membranous spindle envelope preserves different molecular crowding states between the cytoplasm and the spindle region during mitosis. Large assemblies such as membranous organelles are excluded from the spindle region. Small molecules like soluble tubulin or Mad2 equilibrate in all available spaces after NEB and accumulate in the less crowded spindle region. Large nuclear assemblies, such as Tpr, are retained in the spindle region.
The role of an organelle-exclusion envelope during mitosis
The cytoplasm is more crowded than the nucleoplasm, meaning that there is more “free” space in the nucleus for nano-sized particles (Guigas et al., 2007). When proteins like Megator, soluble tubulin, or Mad2, which are restricted to the nucleus or the cytoplasm during interphase, diffusively equilibrate in all available spaces within the cell after NEB, this consequently leads to an enrichment of these proteins in the less crowded spindle region (Fig. 5 D). While other roles cannot be excluded, the spindle envelope might act as a selective barrier to impede the invasion of the spindle region by large organelles, while retaining Megator complexes (Fig. 5 E) that might show gel-like properties (Yao et al., 2012). Importantly, cyclin B/Cdk1 and many other mitotic regulators, such as Polo/Plk1, Mad2, and Fizzy/Cdc20, as well as soluble tubulin and chromosomes, accumulate in the spindle region during mitosis (Moutinho-Santos et al., 1999; Raff et al., 2002; Lince-Faria et al., 2009; Hayashi et al., 2012). This might facilitate interactions between chromosomes and MTs during spindle assembly, not only by providing a physical constraint for MT growth and chromosome distribution, but also by creating a biochemical milieu that favors spindle assembly (Fig. 5 E). Importantly, an MT-independent organelle-exclusion system is conserved in human cells and might therefore represent a common strategy to control mitosis in space and time.
Materials and methods
Cell culture
Drosophila S2 cells were cultured in Schneider’s insect medium (Gibco) supplemented with 10% FBS (Invitrogen) in a 25°C incubator. Human HeLa cells were cultured in DMEM supplemented with 10% FBS (all from Invitrogen) in a 37°C incubator with 5% CO2.
Transgenic cell lines
The S2 cell line stably expressing mCherry–α-tubulin was generated by transfecting cells with a pAc-mCherry–α-tubulin plasmid (provided by G. Goshima, Nagoya University, Nagoya, Japan) together with a pCo-Blast vector (Invitrogen) using Cellfectin (Invitrogen). Cells were selected with blasticidin (Invitrogen) 3 d after transfection. S2 cells stably expressing mRFP–α-tubulin and histone H2B-GFP (provided by G. Goshima, University of Cambridge,Cambridge, England, UK) were transiently transfected with a pMT-CD8-GFP plasmid (provided by P.P. D`Avino) using X-tremeGene (Invitrogen). HeLa cells stably expressing Tpr-GFP (MitoCheck) were transiently transfected with an mCherry–α-tubulin plasmid using X-tremeGENE (Invitrogen). All other cell lines have been described previously (Lince-Faria et al., 2009; Logarinho et al., 2012).
Drug treatments
Expression from a pMT promoter was induced with 500 µM CuSO4 for a minimum of 18 h (Megator-mCherry) or 700 µM CuSO4 for 5–12 h (CD8-GFP). MTs were depolymerized with 100 µM colchicine or 3.3 µM nocodazole (both from Sigma-Aldrich). Membranes were stained with 1 µM DiOC6(3) (Enzo Life Sciences).
Live-cell imaging
S2 cells were imaged in concanavalin A–coated (0.25 mg/ml; EMD Millipore) glass-bottom dishes (Ibidi) at 25°C in Schneider’s insect medium (Gibco) + 10% FBS (Invitrogen) using a 100× 1.4 NA Plan-Apochromatic differential interference contrast (DIC) objective lens (Carl Zeiss) mounted on an inverted microscope (TE2000U; Nikon) equipped with a CSU-X1 spinning-disk confocal head (Yokogawa Electric Corporation) and with two laser lines (488 nm and 561 nm). Images were detected with an iXonEM+ EM-CCD camera (Andor Technology), using the NIS-Elements software (Nikon). HeLa cells were imaged in the same system at 37°C in L15 medium + 10% FBS (Invitrogen). Images were analyzed in ImageJ, processed (contrast adjustments) in Photoshop CS4 (Adobe), and represent maximum intensity projections of multiple z stacks (step size: 0.5–1 µm).
Immunofluorescence microscopy
S2 cells transiently expressing CD8-GFP were grown on concanavalin A–coated (0.5 mg/ml; EMD Millipore) glass coverslips at 25°C in Schneider’s insect medium (Gibco) + 10% FBS (Invitrogen) and fixed with 4% paraformaldehyde for 10 min and subsequently extracted with 0.1% Triton X-100 for 10 min. After short washes in PBS and blocking with 10% FBS, cells were incubated with mouse anti-lamin Dm0 (clone ADL67, 1:50; Developmental Studies Hybridoma Bank) and rat anti–α-tubulin (YOL1/34, 1:100; Serotec), followed by short washes in PBS and incubation with Alexa Fluor 568 and 647 (1:1,000; Invitrogen). DNA was counterstained with DAPI (1 µg/ml; Sigma-Aldrich) before coverslips were mounted in 90% glycerol + 10% Tris, pH 8.5, + 0.5% N-propylgallate on glass slides. Images were acquired on an AxioImager Z1 (100×, Plan Apochromatic oil DIC objective lens, 1.4 NA; all from Carl Zeiss) equipped with a charge-coupled device (CCD) camera (ORCA-R2; Hamamatsu Photonics) using the Zen software (Carl Zeiss) and blind deconvolved using Autoquant X (Media Cybernetics). Images were processed (contrast adjustments) in Adobe Photoshop CS4 (Adobe) and represent either maximum intensity projections of deconvolved z stacks (step size: 0.22 µm; DAPI; α-tubulin) or a single slice (CD8-GFP; lamin Dm0).
FRAP
FRAP of soluble GFP–α-tubulin was performed on an inverted microscope (TE2000U; Nikon; 100× plan-apochromatic DIC objective lens; 1.4 NA; Carl Zeiss) equipped with an iXonEM+ EM-CCD camera (Andor Technology). Bleaching was conducted for 400 ms after acquisition of one prebleach image. Images were acquired every ∼200 ms and analyzed in ImageJ. The fluorescence intensity of the bleached area was normalized using the intensity of an area in the cytoplasm. GraphPad Prism 5 (GraphPad Software) was used for curve fitting.
FCS
FCS was performed with a confocal laser scanning microscope (SP5-TCSPC; Leica) equipped with a water immersion objective lens (HCX Plan Apochromat 63× 1.2 W CORR) and an FCS unit (Picoquant). Samples were illuminated using the 488-nm line of an Argon laser; fluorescence was detected using a band-pass filter (500–530 nm). The pinhole was set to one Airy unit. FCS data at each position were acquired for 40 s and correlation curves were fitted with a previously developed expression (Weiss et al., 2004):
Here, S denotes the elongation of the confocal volume along the optical axis and τD is the mean dwell time of fluorescent particles in the confocal volume. The prefactor A includes the inverse number of fluorescent particles in the confocal volume and also the photophysics of the fluorophore. In each cell, FCS data were acquired at 10 positions across the cell diameter. To make data from different cells comparable, the distance r of each measurement position from the cell center was rescaled with the cell diameter, λ.
Simulations
Diffusion of tubulin and Mad2 into/out of the nuclear region after NEB was modeled by Monte Carlo simulations of particles that move on a square lattice (lattice constant Δx = 30 nm). In accordance with experimental observations, we assumed cells and nuclei to be concentric circular regions with the nucleus occupying ∼1/3 of the cell’s area. Therefore, lattice sites outside the cell’s radius (Rc = 5 µm) were made unavailable, and the nuclear radius was set to . We considered different amounts of occupied volumes that are inaccessible for tubulin (33%) or Mad2 (36%) in the cytoplasm, and in the nuclear space all sites remained accessible. The difference in occupied volumes reflects the different effective sizes of tubulin and Mad2, and the chosen values led to the experimentally observed fluorescence ratio between the nuclear region and the cytoplasm. With these choices, inaccessible volumes in the nuclear region were assumed to be on length scales well below 30 nm whereas cytoplasmic obstacles (composed of 2 × 2 lattice sites) also took into account somewhat larger entities like organelles. To account for the spindle envelope, an annulus around the nuclear region (thickness: two lattice sites) was assigned to ∼75% of inaccessible sites to hinder influx/efflux of proteins. Indeed, this choice was necessary to reach experimentally observed time scales for equilibration of tubulin and Mad2, as extrapolating the diffusional data obtained with FCS on small spatial scales predicted a roughly tenfold faster equilibration. For the simulations, a total number of n = 5 × 105 particles was initially distributed randomly on available lattice sites in the cytoplasm (tubulin) or in the nucleus (Mad2). Then, particles were allowed to move according to a continuous time random walk: particles were assigned individual waiting times drawn from a power-law distribution, p(T)∼1/Tb, which enforces a subdiffusive movement of particles with a tuneable anomaly α = b − 1 (α = 0.8 for tubulin; α = 0.65 for Mad2). After its waiting time had elapsed, each particle was allowed to move to one of the four next-neighbor lattice sites (randomly chosen) unless this site was unavailable (‘blind ant algorithm’). Then, the particle was again assigned a waiting time. The time step of the simulation was determined by demanding the particle’s mean square displacement in the simulations to agree with FCS experiments on tubulin or Mad2 (i.e., not only α = 0.8 or 0.65 but also τD = 1.1 ms or 0.5 ms needed to be reproduced, respectively). To allow for a comparison of simulation results with real microscopy images, we visualized the temporally changing concentration profiles by blurring the particle occupancy on the lattice with a Gaussian filter (radius 250 nm, similar to the diffraction-limited optics used in experiments).
Laser microsurgery
Laser microsurgery was performed with a doubled-frequency laser (FQ-500-532; Elforlight) on an inverted microscope (TE2000U; Nikon), using a 100× 1.4 NA plan-apochromatic DIC objective lens (Nikon) equipped with an iXonEM+ EM-CCD camera (Andor Technology). Disruption of the nuclear/spindle envelope was performed by 1 pulse (interphase) or 160 consecutive pulses (200 Hz repetition rate) (mitosis). The pulse width was 8–10 ns and the pulse energy was 1.5–2 μJ. The laser microsurgery setup has been described previously (Pereira et al., 2009).
Statistical analysis
Normality tests (Shapiro-Wilk) and two-sample t tests were performed in Origin Pro 8 (OriginLab).
Online supplemental material
Fig. S1 demonstrates that spindle envelope integrity decreases over time. Fig. S2 shows quantifications regarding the effect of laser microsurgery on protein confinement. Fig. S3 shows membrane dynamics upon laser microsurgery. Video 1 shows that soluble tubulin and Megator are spatially confined during mitosis. Video 2 shows that soluble tubulin can accumulate in the interphase nucleus. Video 3 demonstrates that spindle envelope integrity decreases over time. Video 4 shows that spindle envelope disruption causes loss of protein confinement. Video 5 shows that MT-independent accumulation of proteins in the spindle region is conserved in human cells. Videos 6, 7, and 8 demonstrate that artificial disruption of the nuclear/spindle envelope causes aberrant spindle formation and chromosome missegregation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201506107/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201506107.dv.
Supplementary Material
Acknowledgments
The authors would like to thank the support of the Human Frontier Science Program throughout this project. We also thank G. Goshima and P. P. D`Avino for the kind gift of reagents and A. J. Pereira for help with laser microsurgery.
N. Schweizer held a doctoral fellowship (SFRH/BD/69198/2010) from Fundação para a Ciência e a Tecnologia (FCT) of Portugal. Work in the H. Maiato laboratory is funded by the seventh framework program grant PRECISE from the European Research Council and FLAD Life Science 2020.
The authors declare no competing financial interests.
References
- Capalbo L., D’Avino P.P., Archambault V., and Glover D.M.. 2011. Rab5 GTPase controls chromosome alignment through Lamin disassembly and relocation of the NuMA-like protein Mud to the poles during mitosis. Proc. Natl. Acad. Sci. USA. 108:17343–17348. 10.1073/pnas.1103720108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G., Tao L., Brust-Mascher I., Wollman R., and Scholey J.M.. 2010. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-B envelope. J. Cell Biol. 188:49–68. 10.1083/jcb.200908150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino P.P., Savoian M.S., Capalbo L., and Glover D.M.. 2006. RacGAP50C is sufficient to signal cleavage furrow formation during cytokinesis. J. Cell Sci. 119:4402–4408. 10.1242/jcs.03210 [DOI] [PubMed] [Google Scholar]
- Debec A., and Marcaillou C.. 1997. Structural alterations of the mitotic apparatus induced by the heat shock response in Drosophila cells. Biol. Cell. 89:67–78. 10.1016/S0248-4900(99)80082-3 [DOI] [PubMed] [Google Scholar]
- Ellenberg J., Siggia E.D., Moreira J.E., Smith C.L., Presley J.F., Worman H.J., and Lippincott-Schwartz J.. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138:1193–1206. 10.1083/jcb.138.6.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman B., Channels W., Qiu M., Iglesias P., Yang G., and Zheng Y.. 2010. Lamin B counteracts the kinesin Eg5 to restrain spindle pole separation during spindle assembly. J. Biol. Chem. 285:35238–35244. 10.1074/jbc.M110.140749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigas G., Kalla C., and Weiss M.. 2007. The degree of macromolecular crowding in the cytoplasm and nucleoplasm of mammalian cells is conserved. FEBS Lett. 581:5094–5098. 10.1016/j.febslet.2007.09.054 [DOI] [PubMed] [Google Scholar]
- Harel A., Zlotkin E., Nainudel-Epszteyn S., Feinstein N., Fisher P.A., and Gruenbaum Y.. 1989. Persistence of major nuclear envelope antigens in an envelope-like structure during mitosis in Drosophila melanogaster embryos. J. Cell Sci. 94:463–470. [DOI] [PubMed] [Google Scholar]
- Hayashi H., Kimura K., and Kimura A.. 2012. Localized accumulation of tubulin during semi-open mitosis in the Caenorhabditis elegans embryo. Mol. Biol. Cell. 23:1688–1699. 10.1091/mbc.E11-09-0815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler P.K., and Wolniak S.M.. 1984. Membranes in the mitotic apparatus: their structure and function. Int. Rev. Cytol. 90:169–238. 10.1016/S0074-7696(08)61490-4 [DOI] [PubMed] [Google Scholar]
- Lince-Faria M., Maffini S., Orr B., Ding Y., Cláudia Florindo C.E., Sunkel A., Tavares J., Johansen K.M., Johansen, and Maiato H.. 2009. Spatiotemporal control of mitosis by the conserved spindle matrix protein Megator. J. Cell Biol. 184:647–657. 10.1083/jcb.200811012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., and Zheng Y.. 2009. A requirement for epsin in mitotic membrane and spindle organization. J. Cell Biol. 186:473–480. 10.1083/jcb.200902071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho E., Maffini S., Barisic M., Marques A., Toso A., Meraldi P., and Maiato H.. 2012. CLASPs prevent irreversible multipolarity by ensuring spindle-pole resistance to traction forces during chromosome alignment. Nat. Cell Biol. 14:295–303. 10.1038/ncb2423 [DOI] [PubMed] [Google Scholar]
- Ma L., Tsai M.Y., Wang S., Lu B., Chen R., Iii J.R., Zhu X., and Zheng Y.. 2009. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat. Cell Biol. 11:247–256. 10.1038/ncb1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough S., and Lucocq J.. 2005. Endoplasmic reticulum positioning and partitioning in mitotic HeLa cells. J. Anat. 206:415–425. 10.1111/j.1469-7580.2005.00407.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R., and Landis S.C.. 1971. The distribution of spindle microtubules during mitosis in cultured human cells. J. Cell Biol. 49:468–497. 10.1083/jcb.49.2.468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T.J., Maddox P., Gaetz J., Groen A., Shirasu M., Desai A., Salmon E.D., and Kapoor T.M.. 2005. Roles of polymerization dynamics, opposed motors, and a tensile element in governing the length of Xenopus extract meiotic spindles. Mol. Biol. Cell. 16:3064–3076. 10.1091/mbc.E05-02-0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll E., and Paweletz N.. 1980. Membranes of the mitotic apparatus of mammalian cells. Eur. J. Cell Biol. 21:280–287. [PubMed] [Google Scholar]
- Moutinho-Santos T., Sampaio P., Amorim I., Costa M., and Sunkel C.E.. 1999. In vivo localisation of the mitotic POLO kinase shows a highly dynamic association with the mitotic apparatus during early embryogenesis in Drosophila. Biol. Cell. 91:585–596. 10.1111/j.1768-322X.1999.tb01104.x [DOI] [PubMed] [Google Scholar]
- Paweletz N. 1981. Membranes in the mitotic apparatus. Cell Biol. Int. Rep. 5:323–336. 10.1016/0309-1651(81)90001-1 [DOI] [PubMed] [Google Scholar]
- Pereira A.J., Matos I., Lince-Faria M., and Maiato H.. 2009. Dissecting mitosis with laser microsurgery and RNAi in Drosophila cells. Methods Mol. Biol. 545:145–164. 10.1007/978-1-60327-993-2_9 [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps J., Spurck T., and Tippit D.. 1984. Chromosome motion and the spindle matrix. J. Cell Biol. 99:137s–143s. 10.1083/jcb.99.1.137s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier C.C., Zheng Y., and Iglesias P.A.. 2010. Mitotic membrane helps to focus and stabilize the mitotic spindle. Biophys. J. 99:3182–3190. 10.1016/j.bpj.2010.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Rath U., Wang D., Xu Y.Z., Ding Y., Zhang W., Blacketer M.J., Paddy M.R., Girton J., Johansen J., and Johansen K.M.. 2004. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila. Mol. Biol. Cell. 15:4854–4865. 10.1091/mbc.E04-07-0579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Rath U., Ding Y., Ji Y., Blacketer M.J., Girton J., Johansen J., and Johansen K.M.. 2005. EAST interacts with Megator and localizes to the putative spindle matrix during mitosis in Drosophila. J. Cell. Biochem. 95:1284–1291. 10.1002/jcb.20495 [DOI] [PubMed] [Google Scholar]
- Raff J.W., Jeffers K., and Huang J.Y.. 2002. The roles of Fzy/Cdc20 and Fzr/Cdh1 in regulating the destruction of cyclin B in space and time. J. Cell Biol. 157:1139–1149. 10.1083/jcb.200203035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon E.D., Leslie R.J., Saxton W.M., Karow M.L., and McIntosh J.R.. 1984. Spindle microtubule dynamics in sea urchin embryos: analysis using a fluorescein-labeled tubulin and measurements of fluorescence redistribution after laser photobleaching. J. Cell Biol. 99:2165–2174. 10.1083/jcb.99.6.2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer N., Weiss M., and Maiato H.. 2014. The dynamic spindle matrix. Curr. Opin. Cell Biol. 28:1–7. 10.1016/j.ceb.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Stafstrom J.P., and Staehelin L.A.. 1984. Dynamics of the nuclear envelope and of nuclear pore complexes during mitosis in the Drosophila embryo. Eur. J. Cell Biol. 34:179–189. [PubMed] [Google Scholar]
- Tinevez J.Y., Schulze U., Salbreux G., Roensch J., Joanny J.F., and Paluch E.. 2009. Role of cortical tension in bleb growth. Proc. Natl. Acad. Sci. USA. 106:18581–18586. 10.1073/pnas.0903353106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.Y., Wang S., Heidinger J.M., Shumaker D.K., Adam S.A., Goldman R.D., and Zheng Y.. 2006. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 311:1887–1893. 10.1126/science.1122771 [DOI] [PubMed] [Google Scholar]
- Weiss M., Elsner M., Kartberg F., and Nilsson T.. 2004. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys. J. 87:3518–3524. 10.1529/biophysj.104.044263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley D.N. 1990. Mitosis and protein synthesis. 3. Organelle relocation during normal and colcemid-arrested M-phase in HeLa S-3 cells. Cytobios. 63:109–130. [PubMed] [Google Scholar]
- Yao C., Rath U., Maiato H., Sharp D., Girton J., Johansen K.M., and Johansen J.. 2012. A nuclear-derived proteinaceous matrix embeds the microtubule spindle apparatus during mitosis. Mol. Biol. Cell. 23:3532–3541. 10.1091/mbc.E12-06-0429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. 2010. A membranous spindle matrix orchestrates cell division. Nat. Rev. Mol. Cell Biol. 11:529–535. 10.1038/nrm2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.