Abstract
What is life and how could it originate? This question lies at the core of understanding the cell as the smallest living unit. Although we are witnessing a golden era of the life sciences, we are ironically still far from giving a convincing answer to this question. In this short article, I argue why synthetic biology in conjunction with the quantitative sciences may provide us with new concepts and tools to address it.
“What is cell biology?” asks the Journal of Cell Biology on the occasion of its 60th anniversary. Raising this simple, yet fundamental question at a time when new data on cells are being collected by the minute is an excellent idea. Information is a necessary, but unfortunately by no means sufficient, requirement for understanding, and the vast amount of data we are now producing may help understand the details but obscure our vision of the cell as a whole. Living systems are inherently complex; this is one of their most distinctive features after billions of years on earth. Complexity is key for their adaptability and resilience, and is both the playground, and the result, of evolution. Unfortunately, the tolerable level of complexity in a connection of thoughts that our brain accepts as an “understanding” is usually rather low, and the most powerful scientific insights, derived by abstraction, have been formulated on the basis of only a few parameters. So either we give up on a systems-level understanding of a cell, and leave it to computers to compile, or we try a theoretical and experimental abstraction of the living cell from its manifold of actual representations. I would like to argue that the latter is possible and will help further our quest to understand the origins of life itself.
What is a cell and where does it come from?
Most of us would be able to instantly recognize a cell under a microscope. But when asked for its general distinctive features, we would start a complicated enumeration, doomed to be incomplete. Consequently, there is to date not a single agreed-on definition of cellular life. Instead, many contemporary researchers, being generally more humble than the pioneers of their disciplines, consider this question ill-posed and prefer not to address it. This is understandable as intellectual honesty, but it is rather unsatisfactory in a more general perspective. Leaving the central object of study undefined neglects its fundamental relevance and does not express sufficiently what biology is all about.
If we do not want to adhere to the persistent metaphysical notion of a “spirit” that fundamentally distinguishes living from nonliving systems, we will have to deliver an answer to the question of when and how a transition between these two regimes actually occurs in order arrive at a basic definition of a cell. We may remember the famous Wöhler experiment, demonstrating that a molecule like urea, previously considered to belong to the “sacred” sphere of animate matter, could be synthesized in a flask (Wöhler, 1828). Thus, there certainly isn’t any fundamental distinction to be made on the level of molecules. But regarding cells, we still do not have a strategy to escape the circular dictum of 19th century cell theory—attributed to Rudolf Virchow—that every cell derives from a cell (“omnis cellula e cellula”). Presumably, there wasn’t a cell right after the Big Bang, so where did the first one really come from? What did the molecules on earth (or anywhere else in the universe) look like before life made its first appearance? How did they self-assemble and self-organize into the first cell-like entity?
But do we even have to know this? Instead of taking a tedious historical approach to the origin of life, aiming to reconstruct the actual conditions and components of an early Earth, we may instead try and derive its preconditions and “driving forces” from physical and chemical laws (Pross, 2012). Erwin Schrödinger, the eminent physicist, made a very intriguing step in this direction (Schrödinger, 1944) by characterizing life as the generation of ordered structures far from equilibrium, using the continuous flow of (light or chemical) energy through them. Shortly after, Alan Turing formulated the mathematical conditions for biological self-organization and morphogenesis, involving remarkably few molecular species and parameters (Turing, 1952). So even if molecules and conditions have dramatically changed over the last billions of years, the same physical laws apply today, and may allow us to reconstitute another origin of life under laboratory conditions. In other words, a synthetic “Wöhler-like” approach, not to biological molecules but to the cell as a whole. This, if at all possible, will clearly require a huge cross-disciplinary effort, as major insights will have to come from chemistry as well as physics. But what can biology contribute?
Top-down and bottom-up: Entering the realm of synthetic biology
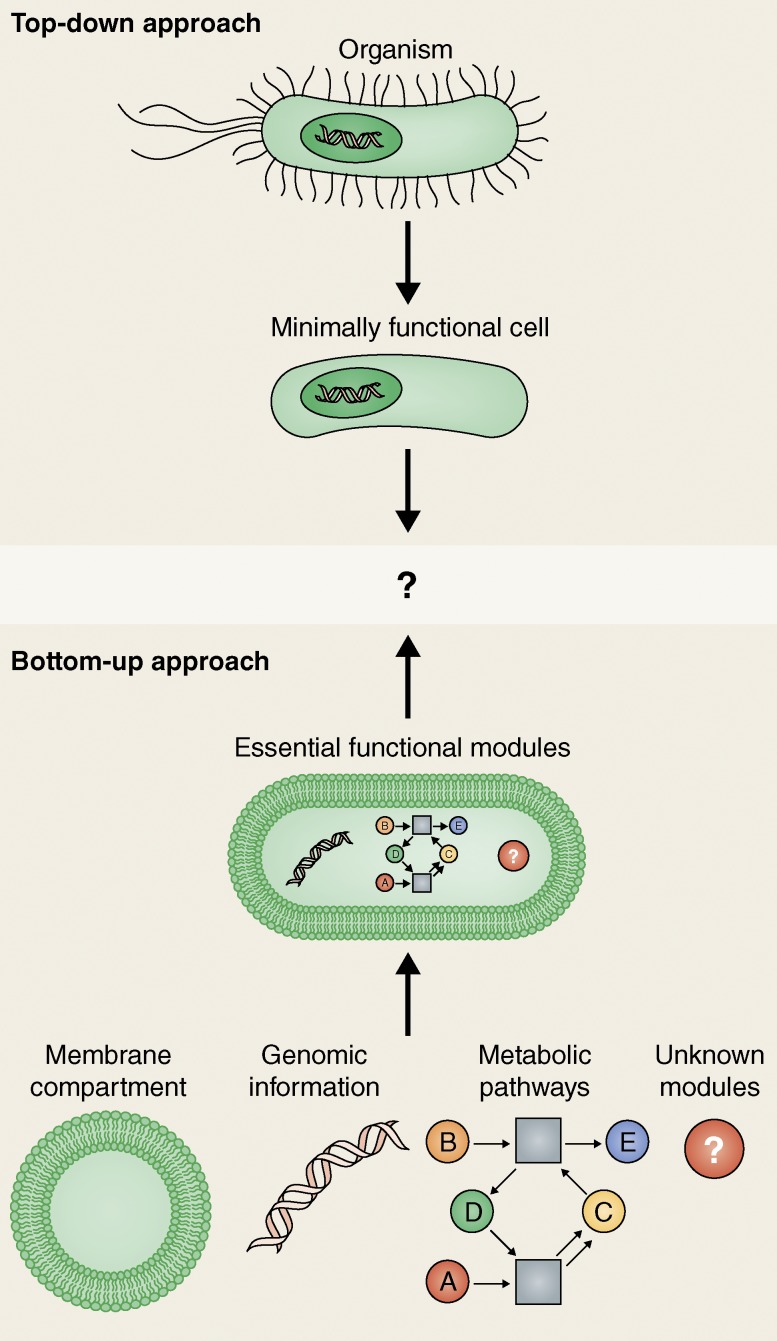
There is one modern branch of biology that raises some hopes that the old fundamental questions may be formulated in a new and more pragmatic way, interfacing better with the quantitative as well as technological disciplines. This is the concept of synthetic biology, with its distinctive idea of dissecting biological systems into modular functional units, or “bio-bricks,” that can ideally be recombined to assemble existing, but also wholly new biological systems (Fig. 1). Functional units, and in particular minimal functional units, may be much better defined than unknown ancient molecules whose exact composition could only be deduced from a postulated abundance of certain elements. In such a modular approach, the question would no longer be what the first cell and its environment actually looked like, but rather what elementary set of functional modules would have to be combined for the system to start showing features of life.
Figure 1.
Top-down versus bottom-up: strategies to arrive at the potential minimal living unit. While the top-down approach eliminates redundant genes from already living organisms, the bottom-up approach aims to jump-start cellular life from a fundamental system of functional modules, e.g., the basic ingredients of Ganti’s “chemoton.”
In fact, synthetic biology follows at least two different approaches toward such a minimal living unit. The more prominent one, often referred to as the “minimal genome” approach, is motivated by the biotechnological quest for an ultimately efficient production organism. Compared with known microorganisms, this idealized cell would contain the absolute minimum number of genes required for metabolism and replication in an optimal environment (Fig. 1). The underlying idea is that the genomes of even the smallest known organisms contain a substantial degree of redundancy, or other features imposed on them by coevolution with competitors, that could in principle be eliminated by genome engineering without compromising their principle viability. The minimal genome has become a prominent entry in synthetic biology literature, after the Venter group first formulated what the minimal genome of a free-living organism would have to look like (although much smaller genomes of symbionts have been identified) and termed it Mycoplasma laboratorium. They then fully synthesized and transplanted another small (but not quite minimal) Mycoplasma mycoides genome into another Mycoplasma species that actually thrived and replicated (Gibson et al., 2010). Recently, a similar transplantation was accomplished with a full yeast chromosome (Annaluru et al., 2014).
The minimal genome approach is arguably the best testable way to explore what functional modules can be taken away from an existing system for it to still remain alive. However, it is more than questionable whether, using this strategy, one will ever find out what the smallest possible configuration of a living system looks like. This may be illustrated by taking a highly evolved technical unit—say, a car—and removing a module of recent innovation, e.g., the car’s electrical system. The car may immediately stop its operation, even though it still contains the part essential for the working of a car, i.e., a combustion engine.
Thus, a more conceptually rewarding, but also obviously more risky, synthetic biology approach would be to try and assemble a living system from scratch, combining its necessary and sufficient modules from the bottom-up (Fig. 1; Szostak et al., 2001; Schwille, 2011). If successful, this strategy would indeed result in the very transition that supposedly occurred for the first time ∼3.5 billion years ago: the transition from nonliving to living. However, in contrast to the minimal genome concept, which relies on systems that nature has already successfully established, this strategy can’t rely on living or fossil examples, but instead requires a detailed master plan, or at least a convincing hypothesis about the minimal set of functional modules to “jump-start” life—a true intellectual act of synthesis. Is it possible to create such a master plan?
Revisiting Gánti’s chemoton theory: The three pillars of life
Although this short article is not the place for a detailed discussion of the definitions of life, we first need to determine what particular aspects and features need to be addressed first in a bottom-up reconstitution approach. Where to start? Out of the many more or less sophisticated theories and models, I would like to introduce a particularly elegant one to be revisited in interdisciplinary enterprises, taking the modern physical, chemical, and biological insights into account. This is the “chemoton” theory of the Hungarian biochemist Tibor Gánti (Gánti, 2003). In short, Ganti described the cell as a chemical automaton characterized by three main features: (1) a self-reproducing chemical motor (e.g., metabolism), (2) a chemical information system (e.g., DNA/RNA), and (3) a chemical boundary system (e.g., membrane).
The first two elementary features of metabolism and information replication, and their interdependence, have been subjects of heated chicken-or-egg discussions on the origin of life in the genomic era. The third pillar of Gánti’s chemoton, the boundary system, however, has long been neglected. Apart from physical chemists and soft matter physicists, who have always been fascinated by the rich phenomenology of bilayers composed of amphiphiles in aqueous environments, the biological interest in membranes, the paradigmatic cell boundaries, arose mainly from their role of harboring channels, receptors, and energy converting machinery. It is only quite recently, under the notion of a “protocell,” that the replicating membrane vesicle has entered the focus of fundamental considerations of how to emulate the necessary self-replication of boundaries, and how to couple this to information replication and evolution (Chen et al., 2004). In fact, simple fatty acid vesicles can be easily grown and transformed, and even spontaneously divide as a result of surface or volume growth, although their suitability as reliable containers for genetic material is limited. Regarding the more biologically suitable phospholipid membranes, it has primarily been the technical advances of the last decade in creating useful models, such as giant unilamellar vesicles (GUVs)—cell-sized, free-standing membranes perfectly adapted to observation by light microscopy—that moved the fundamental task of compartment generation and replication forward on the agenda of bottom-up synthetic biology (Rasmussen et al., 2008).
Toward regulated compartment division: Lessons from Escherichia coli
As outlined in the previous sections, defining a minimal compartment to be divided and replicated along with the genetic material contained within seems to be a relatively well-defined but crucial task toward the synthesis of a cell. In this last section, I will shortly illustrate my own recent work aiming for exactly that: a minimal model system to reconstitute controlled division of a cell-like compartment from the bottom-up. True to the synthetic biology concept of identifying modules of biological functionality, we have been searching for relatively simple but well-described mechanisms that could be mimicked in a first approach, and then reduced and conceptually further simplified after recognition of their most fundamental features. We chose the cell division system of E. coli, an arguably simple but by no means minimal organism, that, however, supports the molecular biology and biochemistry for reconstitution experiments. E. coli is rod-shaped, grows mainly along its long axis, and divides symmetrically by assembling a contractile protein ring at its central plane. The positioning of this ring is accomplished by a peculiar oscillation of regulatory proteins MinCDE between the cell poles (Raskin and de Boer, 1999).
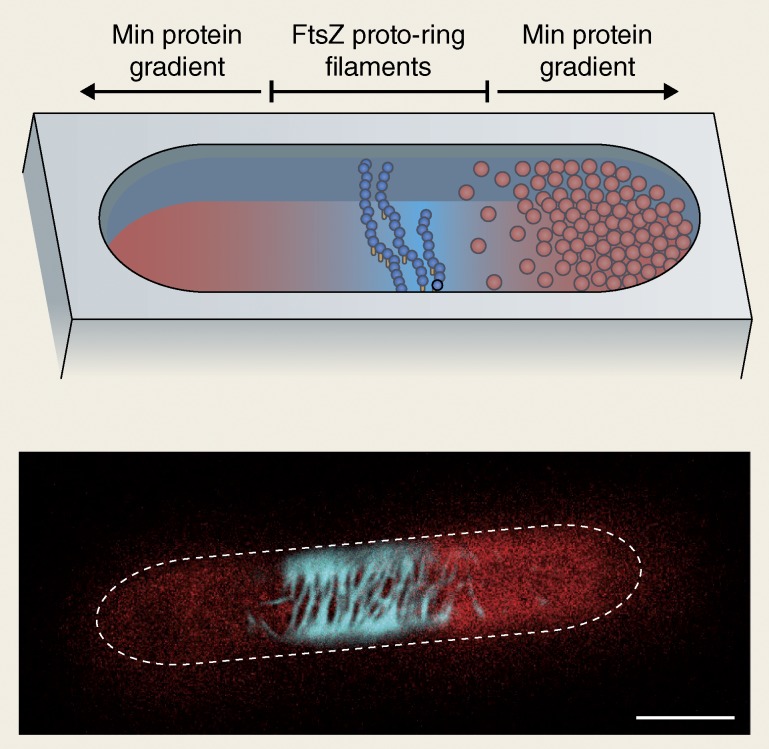
Most remarkably, this Min protein system can be demonstrated to represent the smallest possible set of biochemical agents for initiating self-organization and pattern formation (Loose et al., 2008), requiring two proteins, ATP as an energy source, and a membrane as a template to yield wave-like dynamic protein gradients on flat extended membranes. Introduced into cell-shaped compartments, the cellular oscillations could be reconstituted (Zieske and Schwille, 2013). Moreover, the bottom-up experiments clearly demonstrated how time-averaged gradients act as spatial cues to direct downstream processes (Zieske and Schwille, 2014). Specifically, proto-ring filaments of FtsZ, the primary constituent of the contractile Z ring, could be exclusively positioned in the middle of the synthetic compartment (Fig. 2). Although this is only the first, and by no means the crucial step toward reconstituting division of a membrane compartment based on protein self-assembly and self-organization, this shows the advantage of the approach of dissecting a phenomenon into simple functional modules, which can in principle be derived from any organism or even designed from scratch.
Figure 2.
Toward cell division in vitro. Shown is a model of reconstitution of protein self-organization and gradient formation in artificial cell-shaped containers by minimal functional elements of the E. coli cell division machinery. Reaction/diffusion–induced oscillations of MinCDE proteins position FtsZ protofilaments to the center of the compartment, mimicking the first step in Z ring assembly. Adapted from Zieske and Schwille (2014).
Outlook: Engineering with biological modules
Modern cell biology has, despite its huge advances in the past 50 years, not yet arrived at a fundamental understanding of our main topic of research: the cell. Specifically, we are still unable to pinpoint the line of division between the living and the nonliving. Even less are we able to reproduce the transition between these domains, which are thus still considered separate. However, we know so much more than any generation before us about the exact features and functions of living systems. Speaking and thinking in terms of functional modules, as synthetic biology has lately taught us to do, the possibility of reconstituting and bringing together known and biologically established functionalities allows us to build with a cellular “Lego” toolkit with unprecedented efficiency. Thus, the time may be just right to once again consider the possibility of constructing not only with and within biological systems, but to extend the synthetic approach to the very construction of biological systems from nonbiological ones, as accomplished at least once by nature, billions of years ago. It goes without saying that this enterprise is not a genuinely biological one, because biology is only what results from it. But it will require the specific expertise and insights of biologists, and particularly cell biologists, into the many essential functional elements that are at the core of life as we know it.
Acknowledgements
The author's ongoing work on bottom-up synthetic biology is supported by the Federal Ministry of Education and Research and the Max Planck Society in the framework of the research network MaxSynBio. Illustrations were provided by Neil Smith (www.neilsmithillustration.co.uk).
The author declares no competing financial interests.
References
- Annaluru N., Muller H., Mitchell L.A., Ramalingam S., Stracquadanio G., Richardson S.M., Dymond J.S., Kuang Z., Scheifele L.Z., Cooper E.M., et al. 2014. Total synthesis of a functional designer eukaryotic chromosome. Science. 344:55–58. 10.1126/science.1249252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.A., Roberts R.W., and Szostak J.W.. 2004. The emergence of competition between model protocells. Science. 305:1474–1476. 10.1126/science.1100757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gánti T. 2003. The Principles of Life. Oxford University Press, Oxford, England. 220 pp. [Google Scholar]
- Gibson D.G., Glass J.I., Lartigue C., Noskov V.N., Chuang R.Y., Algire M.A., Benders G.A., Montague M.G., Ma L., Moodie M.M., et al. 2010. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 329:52–56. 10.1126/science.1190719 [DOI] [PubMed] [Google Scholar]
- Loose M., Fischer-Friedrich E., Ries J., Kruse K., and Schwille P.. 2008. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 320:789–792. 10.1126/science.1154413 [DOI] [PubMed] [Google Scholar]
- Pross A. 2012. What is Life? How Chemistry Becomes Biology. Oxford University Press, Oxford, England. 200 pp. [Google Scholar]
- Raskin D.M., and de Boer P.A.J.. 1999. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc. Natl. Acad. Sci. USA. 96:4971–4976. 10.1073/pnas.96.9.4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S., Bedau M.A., Chen L., Deamer D., Krakauer D.C., Packard N.H., and Stadler P.F., editors. 2008. Protocells: Bridging Nonliving and Living Matter. MIT Press, Cambridge, MA. 712 pp. [Google Scholar]
- Schrödinger E. 1944. What is Life? The Physical Aspects of the Living Cell. Cambridge University Press, Cambridge, England. 194 pp. [Google Scholar]
- Schwille P. 2011. Bottom-up synthetic biology: engineering in a tinkerer’s world. Science. 333:1252–1254. 10.1126/science.1211701 [DOI] [PubMed] [Google Scholar]
- Szostak J.W., Bartel D.P., and Luisi P.L.. 2001. Synthesizing life. Nature. 409:387–390. 10.1038/35053176 [DOI] [PubMed] [Google Scholar]
- Turing A.M. 1952. The chemical basis of morphogenesis. Philos. Trans. R. Soc. Lond. 237:37–72. 10.1098/rstb.1952.0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wöhler F. 1828. Ueber kuenstliche Bildung des Harnstoffs. Ann. Phys. 88:253–256. 10.1002/andp.18280880206 [DOI] [Google Scholar]
- Zieske K., and Schwille P.. 2013. Reconstitution of pole-to-pole oscillations of min proteins in microengineered polydimethylsiloxane compartments. Angew. Chem. Int. Ed. Engl. 52:459–462. 10.1002/anie.201207078 [DOI] [PubMed] [Google Scholar]
- Zieske K., and Schwille P.. 2014. Reconstitution of self-organizing protein gradients as spatial cues in cell-free systems. eLife. 3 10.7554/eLife.03949 [DOI] [PMC free article] [PubMed] [Google Scholar]