Abstract
With the emergence of individualized medicine and the increasing amount and complexity of available medical data, a growing need exists for the development of clinical decision-support systems based on prediction models of treatment outcome. In radiation oncology, these models combine both predictive and prognostic data factors from clinical, imaging, molecular and other sources to achieve the highest accuracy to predict tumour response and follow-up event rates. In this Review, we provide an overview of the factors that are correlated with outcome—including survival, recurrence patterns and toxicity—in radiation oncology and discuss the methodology behind the development of prediction models, which is a multistage process. Even after initial development and clinical introduction, a truly useful predictive model will be continuously re-evaluated on different patient datasets from different regions to ensure its population-specific strength. In the future, validated decision-support systems will be fully integrated in the clinic, with data and knowledge being shared in a standardized, instant and global manner.
Introduction
Over the past decade, we have witnessed advances in cancer care, with many new diagnostic methods and treatment modalities becoming available,1 including advances in radiation oncology.2 The abundance of new options and the progress in individualized medicine has, however, created new challenges. For example, achieving level I evidence is increasingly difficult given the numerous disease and patient parameters that have been discovered, resulting in an ever-diminishing number of ‘homogeneous’ patients.3 This reality contrasts to a certain extent with classic evidence-based medicine, whereby randomized trials are designed for large populations of patients. Thus, new strategies are needed to find evidence for subpopulations on the basis of patient and disease characteristics.4
For each patient, the clinician needs to consider state-of-the-art imaging, blood tests, new drugs, improved modalities for radiotherapy planning and, in the near future, genomic data. Medical decisions must also consider quality of life, patient preferences and, in many health-care systems, cost efficiency. This combination of factors renders clinical decision making a dauntingly complex, and perhaps inhuman, task because human cognitive capacity is limited to approximately five factors per decision.3 Furthermore, dramatic genetic,5 transcriptomic,6 histological7 and microenvironmental8 heterogeneity exists within individual tumours, and even greater heterogeneity exists between patients.9 Despite these complexities, individualized cancer treatment is inevitable. Indeed, intratumoural and intertumoural variability might be leveraged advantageously to maximize the therapeutic index by increasing the effects of radiotherapy on the tumour and decreasing those effects on normal tissues.10–12
The central challenge, however, is how to integrate diverse, multimodal information (clinical, imaging and molecular data) in a quantitative manner to provide specific clinical predictions that accurately and robustly estimate patient outcomes as a function of the possible decisions. Currently, many prediction models are being published that consider factors related to disease and treatment, but without standardized assessments of their robustness, reproducibility or clinical utility.13 Consequently, these prediction models might not be suitable for clinical decision-support systems for routine care.
In this Review, we highlight prognostic and predictive models in radiation oncology, with a focus on the methodological aspects of prediction model development. Some characteristic prognostic and predictive factors and their challenges are discussed in relation to clinical, treatment, imaging and molecular factors. We also enumerate the steps that will be required to present these models to clinical professionals and to integrate them into clinical decision-support systems (CDSSs).
Methodological aspects
Factors for prediction
The overall aim of developing a prediction model for a CDSS is to find a combination of factors that accurately anticipate an individual patient’s outcome.14 These factors include, but are not limited to, patient demographics as well the results of imaging, pathology, proteomic and genomic testing, the presence of key biomarkers and, crucially, the treatment undertaken. ‘Outcome’ can be defined as tumour response to radio-therapy, toxicity evolution during follow up, rates of local recurrence, evolution to metastatic disease, survival or a combination of these end points. Although predictive factors (that is, factors that influence the response to a specific treatment) are necessary for decision support, prognostic factors (that is, factors that influence response in the absence of treatment)15 are equally important in revealing the complex relationship with outcome. Herein, we refer to both of these terms generically as ‘features’ because, for a predictive model, correlation with outcome must be demonstrable.
Model development stages
The procedure for finding a combination of features correlated with outcome is analogous to the development of biomarker assays.16 In that framework, we can distinguish qualification and validation. Qualification demonstrates that the data are indicative or predictive of an end point, whereas validation is a formalized process used to demonstrate that a combination of features is both reliable and suitable for the intended purpose. That is, we need to identify features, test whether they are predictive in independent datasets and then determine whether treatment decisions made using these features improve outcome. The complete cycle of model development entails several stages (Figure 1).
Figure 1.
Schematic overview of methodological processes in clinical decision-support system development, describing model development, assessment of clinical usefulness and what ideally to publish. The coloured, parallel lines represent heterogeneous data, which have been split early for independent validation (but without internal cross-validation).
In the hypothesis-generation stage, one must consider the end point to predict, the timing of the treatment decision and the available data at these time points. In the data-selection step, a review of potential features is first conducted, ideally by an expert panel. A practical inventory of the available data and sample-size calculations are recommended, especially for the validation phase.17,18 Data from both clinical trials (high quality, low quantity, controlled, biased selection) and clinical practice (low quality, high quantity, unbiased selection) are useful, but selection biases must be identified in both cases and the inclusion criteria should be equivalent. For all features, including the characteristics of the treatment decision, data heterogeneity is a requirement to identify predictive features and to have the freedom to tailor treatment.
Next, performance measures for models are determined, and these measures include the area under the receiver operating characteristic curve (AUC), accuracy, sensitivity, specificity and c-index of censored data.19 AUC, which has values between 0 and 1 (with 1 denoting the best model and 0.5 randomness), is the most commonly used performance measure. However, for time-to-event models, the c-index and hazard ratio are more appropriate because both can handle censored data.
The preprocessing stage deals with missing data (imputation strategies; that is, replacing missing values by calculated estimates),20 identifying incorrectly measured or entered data21 as well as discretizing (if applicable) and normalizing data to avoid sensitivity for different orders of data scales.22 If an external, independent dataset is not available for validation, the available data must be split (in a separate stage) into a model-training dataset and a validation set, the latter of which is used subsequently in the validation step. In the feature-selection stage, the ratio of the number of evaluated features to number of outcome events must be kept as low as possible to avoid overfitting. When a model is overfitted, it is specifically and exclusively trained for the training data (including its data noise) and, as a result, performs poorly on new data. Data-driven preselection of features is, therefore, recommended.23 Univariate analyses are commonly used to prioritize the features–that is, testing each feature individually and ranking them on their strength of correlation with outcome.
Predicting outcomes
In the next stage, the input data are fed into a model that can classify all possible patient outcomes. Traditional statistical24 and machine-learning models25 can be considered. For two or more classes (for example, response versus no response), one might consider logistic regression, support vector machines, decision trees, Bayesian networks or Naive Bayes algorithms.26,27 For time-to-event outcomes, whether censored or not, Cox proportional hazards models28 or the Fine and Gray model28 of competing risks are most common. The choice of model depends on the type of outcome (for example, logistic regression for two or more outcomes, or Cox regression for survival-type data) and the type of input data (for example, Bayesian networks require categorized data, whereas support-vector machines can deal with continuous data). In general, several models with similar properties can be tested to find the optimal model for the available data. A simple model is, however, preferred because it is expected to be robust to a wider range of data than a more complex model.
Performance on the training dataset is upwards-biased because the features were selected. Thus, external validation data must be used, which can be derived from a separate institute or independent trial. When data are limited, internal validation can be considered using random split, temporal split or k-fold cross-validation techniques.29 The developed model should have a benefit over standard decision making, and must be assessed prospectively in the clinic in the penultimate stage of development. Models must be compared against predictions by clinicians30,31 and to standard prognostic and predictive factors.32 Critically, to demonstrate the improvement of patient outcome, quality of life and/or reduced toxicity,33 clinical trials must be conducted whereby the random assignment of patients is based on the prediction model output. Fulfilling this requirement will generate the final evidence that the model is improving health care by comparing, in a controlled way, the tailored treatments with standard treatments in the clinic.
Finally, the prediction models and data can be published, enabling the wider oncological community to evaluate them. Full transparency on the data and methodology is the key towards global implementation of the model into CDSSs. This suggestion is similar to clinical ‘omics’ publications for which the raw data, the code used to derive the results from the raw data, evidence for data provenance (the process that led to a piece of data) and a written description of nonscriptable analysis steps are routinely made available.34
In practice, this cycle of development usually begins by identifying clinical parameters, because these are widely and instantly available in patient information systems and clinical trials. These clinical variables also form the basis for extending prediction models with imaging or molecular data.
Clinical features
Decision making in radiotherapy is mainly based on clinical features, such as the patient performance status, organ function and grade and extent of the tumour (for example, as defined by the TNM system). In almost all studies, such features have been found to be prognostic for survival and development of toxicity.35–37 Consequently, these features should be evaluated in building robust and clinically acceptable radiotherapy prognostic and predictive models. Moreover, measurement of some clinical variables, such as performance status, can be captured with minimal effort.
Even the simplest questionnaire, however, should be validated as is the case for laboratory measurements of organ function or parameters measured from blood.38,39 Furthermore, a standardized protocol should be available to ensure that comparisons are possible between centres and questionnaires over time.40 Moreover, why specific features were chosen for measurement should be clearly explained. For example, if haemoglobin measurements were only taken in patients with fatigue, the resulting bias would demand caution when including and interpreting the measurements. Only when clinical parameters are recorded prospectively with the same scrutiny as laboratory measurements will observational studies become as reliable as randomized trials.41,42
Toxicity measurements and scoring should also build on validated scoring systems, such as the Common Terminology Criteria for Adverse Events (CTCAE), which can be scored by the physician or patient.43,44 Indeed, a meta-analysis showed that high-quality toxicity assessments from observational trials are similar to those of randomized trials.45,46 However, a prospective protocol must clarify which scoring system was used and how changes in toxicity score were dealt with over time with respect to treatment.
Finally, to ensure a standardized interpretation, the reporting of clinical and toxicity data and their analyses should be performed in line with the STROBE (Strengthening the Reporting of Observational Studies in Epidemiology) statement for observational studies and genetic-association studies, which is represented as checklists of items that should be addressed in reports to facilitate the critical appraisal and interpretation of these type of studies.47,48
Treatment features
Currently, image-guided radiotherapy (IGRT) is a highly accurate cancer treatment modality in delivering its agent (radiation) to the tumour.49 Furthermore, very accurate knowledge of the effects of radiation on normal tissue has been obtained.50 With modern radio-therapy techniques—such as intensity-modulated radiotherapy, volumetric arc therapy or particle-beam therapy—the treatment dose can be sculpted around the target volume with dosimetric accuracy of a few percentage points. IGRT ensures millimetre precision to spare the organs at risk as much as possible.51
For prediction modelling, recording features that are derived from planned spatial and temporal distribution of the radiotherapy dose is crucial. Additionally, features must be recorded that describe the efforts undertaken during treatment to ensure that the dose is delivered as planned (that is, in vivo dosimetry); a delicate balance exists between tumour control and treatment-related toxicity.52 Additional therapies, such as (concurrent) chemotherapy, targeted agents and surgery, and their features must also be recorded because these have various effects on outcome.32,53 An example is the difference between concurrent versus sequential chemoradiation, which has a major influence on the occurrence of acute oesophagitis that induces dysphagia.54
With respect to the spatial dimension of radiotherapy, how to combine information about the spatially variable dose distribution for every subvolume of the target tumour (or organ) with the global effect to the tumour or adjacent normal tissue remains indeterminate. Dose-response relationships for tumour tissues are often reported in terms of mean (biologically equivalent) dose, although voxel-based measures have also been reported.55 Mean doses or doses to a prescription point inside the tumour are easily determined and reported and can suffice for many applications. However, spatial characteristics might be more relevant in personalized approaches to ensure radioresistant areas of the tumour receive higher doses.55 For normal tissue toxicity, dose features—including the mean and maximum dosage, as well as the volume of the normal tissue receiving a certain dose—are important. For example, V20 <35% is a common threshold to prevent lung toxicity.56
Clinical dose–volume histogram analysis for pneumonitis after the 3D treatment for non-small-cell lung cancer was first described in 1991.57 In 2010, a series of detailed reviews of all frequently irradiated organs (the QUANTEC project) was described,50 showing that, as for the tumour, care must be taken when assessing dose at the organ level. For example, in some organs, the volume receiving a certain dose is important (such as the oesophagus or lung) because of their proximity to other vital structures, whereas the maximum dose to a small region of other organs might be most important (such as for the spinal cord) because preserving its post-treatment function is crucial. Predicting complications to normal tissue is an active research area in ongoing, large, prospective multicentre projects, including ALLEGRO58 and others.59–61
Although important, in general, one must be careful about relying completely on planned-radiotherapy dose-based predictions because patients display wide variability in toxicity development. The reasons for this variability include many known clinical and molecular-based features as well as the quality of the treatment execution. The focus on the planned radio-therapy dose distribution as the prime determinant of outcome is perhaps the most common pitfall in prediction models because deviations from the original plan during the time of treatment frequently occur.62 The accuracy of prediction models is expected to increase when measured dose is used, as this measure reflects the effect of radiotherapy most accurately. Figure 2 shows an example of these variations in a patient with prostate cancer. Dose reconstructions (2D and 3D), Gamma-Index calculations and dose–volume histograms during treatment can help in identifying increasingly accurate dose-related features,63,64 such as radiation pneumonitis65 and oesophagitis.66
Figure 2.
The importance of considering measured dose for outcome prediction for a patient with prostate cancer. a | Original planning CT scan that includes contours of the prostate (red), bladder (yellow), exterior wall of the rectum (blue) and seminal vesicles (green). b | Contoured CT scan after 16 fractions of radiotherapy. c | Reconstructed 3D dose after 16 fractions of radiotherapy. d | Calculated dose differences (expressed as a 3D Gamma Index) after 16 fractions of radiotherapy. e | Dose–volume histograms at fractions 1, 6, 11, 16, 21 and 26 (dashed lines) as well as pretreatment histograms (solid lines). Clear deviations are visible from the planned dose–volume histogram for the rectum and bladder.
The temporal aspect of fractionated radiotherapy is also an active area of research. The fact that higher radiation doses are required to control a tumour when treatment is prolonged is well-known, and increasing evidence suggests that accelerated regimens giving the same physical dose can improve outcome.67,68 A multi-centre analysis of patients with head-and-neck cancer treated with radiotherapy alone showed that the potential doubling time of the tumour before treatment was not a predictor for local control.69 Alongside the classic explanation of accelerated repopulation,70 changes in cell loss, hypoxia and selection of radioresistant stem cells have each been suggested as underlying causes of this observation, the possible implications of which include shorter overall treatment times with higher doses per fraction and the avoidance of breaks during treatment.71,72 Overall, treatment time is an accessible feature that is correlated with local failure in several tumour sites.73,74
Ideally, the spatial and temporal dimensions of radio-therapy would be exploited by showing a fractional dose distribution in a tumour radioresistance (and normal- tissue radiosensitivity) map that is continuously updated during treatment. However, such an image of radio-resistance does not yet exist. If it did, CDSSs would guide the planning and modification of the spatial and temporal distribution of radiation in such a way as to maintain or improve the balance between tumour control and the probability of normal tissue complications continuously during treatment, instead of the current approach that delivers radiation as planned with an identical dose to the tumour as a whole.
Imaging features
Medical imaging has a fundamental role in radiation oncology, particularly for treatment planning and response monitoring.75,76 Technological advances in noninvasive imaging—including improved temporal and spatial resolution, faster scanners and protocol standardization—have enabled the field to move towards the identification of quantitative noninvasive imaging biomarkers.77–79
Metrics based on tumour size and volume are the most commonly used image-based predictors of tumour response to therapy and survival,80–87 and rely on CT and MRI technology for 3D measurement.88–90 Although used in clinical practice, tumour size and volume measurements are subject to interobserver variability that can be attributed to differences in tumour delineations.85–87, 91,92 Moreover, the optimal measurement technique and definitions of appropriate response criteria, in terms of changes in tumour size, are unclear.93 Additionally, tumour motion and image artefacts are additional sources of variability.94,95 To overcome these issues, automated tumour delineation methods have been introduced96–99 on the basis of, for example, the selection of ranges of Hounsfield units (which represent the linear attenuation coefficient of the X-ray beam by the tissue) on CT that define a certain tissue type, or calculation of the gradient of an image (mathematical filter) to reveal the borders between tissue types. Extensive evaluation, however, is needed before these methods can be used routinely in the clinic.100–102
A commonly used probe for the metabolic uptake of the tumour is 18F-fluorodeoxyglucose (FDG) for PET imaging.103,104 The pretreatment maximum standardized uptake value (SUV, which is the normalized FDG uptake for an injected dose according to the patient’s body weight) is strongly associated with overall survival and tumour recurrence in a range of tumour sites, including the lung, head and neck, rectum, oesophagus and cervix.105–111 Furthermore, several studies have shown that changes in SUV during and after treatment are early predictors of tumour recurrence.112–115 FDG–PET measurements, however, are dependent on a number of factors, including injected dose, baseline glucose concentration, FDG clearance, image reconstruction methods used and partial-volume effects.116,117 Standardization of these factors across institutions is, therefore, fundamental to enable comparisons and validation of data from FDG–PET imaging.118,119
Multiple studies have shown that diffusion-weighted MRI parameters, such as the apparent diffusion coefficient (ADC), which is a measure of water mobility in tissues, can accurately predict response and survival in multiple tumour sites.120–124 However, lack of reproducibility of ADC measurements—due to lack of standardization of instruments between vendors and of internationally accepted calibration protocols—remains a bottleneck in these types of studies.125 Evaluations of different time points in dynamic contrast-enhanced MRI have also been used to describe tumour perfusion.90,126–128 Indeed, hypothesis-driven preclinical129 and xenograft studies130,131 support these clinical studies. For example, assessment of the correlation of features from imaging (such as lactate level and the extent of reoxygenation) with tumour control is possible.130,131
Increasingly advanced image-based features are currently being investigated. For example, routine clinical imaging can capture both tumour heterogeneity and post-treatment changes, which can be analysed to identify functional biomarkers (Figure 3). Changes in Hounsfield units in contrast-enhanced CT are directly proportional to the quantity of contrast agent present in the tissue and have been used as a surrogate for tumour perfusion.132,133 Indeed, reductions of Hounsfield units following treatment have been used to evaluate treatment response in rectal, hepatic and pulmonary cancers.134,135
Figure 3.
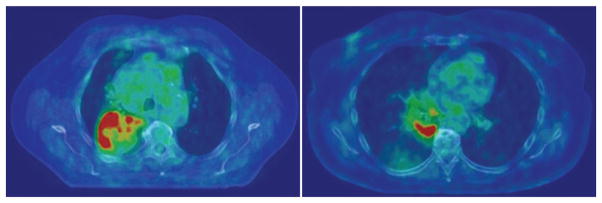
Axial FDG–PET and CT images of two different patients with NSCLC. Tumour imaging biomarkers describing, for example, textural heterogeneity, FDG uptake and tumour size can be assessed noninvasively before, during and after radiotherapy and associated with treatment outcome. Abbreviations: FDG, 18F-fluorodeoxyglucose; NSCLC, non-small-cell lung cancer.
Standardizing the extraction and quantification of a large number of traits derived from diagnostic imaging are now being considered in new imaging marker approaches.79 Through advanced image-analysis methods, we can quantify descriptors of tumour heterogeneity (such as variance or entropy of the voxel values) and the relationship of the tumour with adjacent tissues.136–138 These analytical methods enable high-throughput evaluation of imaging parameters that can be correlated with treatment outcome and, potentially, with biological data. Indeed, qualitative imaging parameters on CT and MRI scans have been used to predict mRNA abundance variation in hepatocellular carcinomas and brain tumours.139–141 Furthermore, a combination of anatomical, functional and metabolic imaging techniques might be used to capture pathophysiological and morphological tumour characteristics in a noninvasive manner, including apparent intratumoural heterogeneity.142
Molecular features
Biological markers are also valuable clinical decision-support features; these include prognostic and predictive factors for outcomes, such as tumour response and normal-tissue tolerance. Despite these strengths, trials of molecular biomarkers are prone to experimental variability; for this reason standardizing assay criteria, trial design and analysis are imperative if multiple molecular markers are to be used in predictive modelling.16
Tumour response
Next to tumour size, tumour control after radiotherapy is largely determined by three criteria: intrinsic radio-sensitivity, cell proliferation and the extent of hypoxia.143 In addition, large tumours intuitively require higher doses of radiation than small tumours because there are simply more cells to kill—this requirement is true even if intrinsic radiosensitivity, hypoxia and repopulation rates are equal. Several approaches have been developed to measure these additional three parameters to predict tumour response to radiotherapy.
Intrinsic radiosensitivity
Malignant tumours display wide variation in intrinsic radiosensitivity, even between tumours of similar origin and histological type.144 Attempts to assess the radio-sensitivity of human tumours have relied on determining the ex vivo tumour survival fraction.145 Those studies and others have shown that tumour cell radiosensitivity is a significant prognostic factor for radiotherapy outcome in both cervical146 and head-and-neck147 carcinomas. However, these colony assays suffer from technical disadvantages that include a low success rate (<70%) for human tumours and the time needed to produce data, which can be up to several weeks.
Other studies have included assessments of chromosome damage, DNA damage, glutathione levels and apoptosis.148 Indeed, some clinical studies using such assays have shown correlations with radiotherapy outcome, whereas others have not.149 However, these cell-based functional assays only have limited clinical utility as predictive assays, despite being useful in confirming a mechanism that underlies differences in the response of tumours to radiotherapy. For example, some studies have provided encouraging data showing that immunohistochemical staining for γ-histone H2AX, a marker of DNA damage, might be a useful way to assess intrinsic radiosensitivity very early after the start of treatment.150,151 Double-stranded breaks are generated when cells are exposed to ionizing radiation or DNA-damaging chemotherapeutic agents, which rapidly results in the phosphorylation of γ-histone H2AX. γ-Histone H2AX is the most sensitive marker that can be used to examine the DNA damage and its subsequent repair, and it can be detected by immunoblotting and immunostaining using microscopic or flow cytometric detection. Clinically, two biopsies (one before and one after treatment) are needed to assess the γ-histone H2AX status, which is not always easy to implement in practice.
Hypoxia
Tumour hypoxia is the key factor involved in determining resistance to treatment and malignant progression; it is a negative prognostic factor after treatment with radio-therapy, chemotherapy and surgery.152,153 Indeed, some data show that hypoxia promotes both angiogenesis and metastasis and, therefore, has a key role in tumour progression.154 Although a good correlation has been demonstrated between pimonidazole (a chemical probe of hypoxia) staining and outcome after radiotherapy in head-and-neck cancer,155 the same relationship has not been found in cervical cancer.156 In light of these contrasting results, one of the hypotheses put forward to explain this is that hypoxia tolerance is more important than hypoxia itself.157
The use of fluorinated derivatives of such chemical probes also enables their detection by noninvasive PET.158–160 Although this approach requires administration of a drug, it does benefit from sampling the whole tumour and not just a small part of it. Another possible surrogate marker of hypoxia is tumour vasculature; the prognostic significance of tumour vascularity has been measured as both intercapillary distance (thought to reflect tumour oxygenation) and microvessel density (the ‘hotspot’ method that provides a histological assessment of tumour angiogenesis). Some studies have found positive correlations with outcome—mainly using micro-vessel density in cervical cancer—whereas others have shown negative correlations.161 Some concerns have been raised about the extent to which biopsies taken randomly truly represent the usually large, heterogeneous tumours.
Proliferation
If the overall radiotherapy treatment time is prolonged, for example, for technical reasons (breakdown of a linear particle accelerator) or because of poor tolerance by the patient to the treatment, higher doses of radiation are required for tumour control—clearly indicating that the influence of tumour proliferation is important.162 Although proliferation during fractionated radiotherapy is clearly an important factor in determining outcome, reliable measurement methods are not yet available. To understand why radiation leads to an accelerated repopulation response in some tumours and not in others, a greater understanding of the response at both the cellular and molecular level is required.
Normal-tissue tolerance
Inherent differences in cellular radiosensitivity among patients dominate normal-tissue reactions more than other contributing factors.164 That is, the radiation doses given to most patients might in actuality be too low for an optimal cure because 5% of patients are very sensitive; these 5% of patients are so sensitive that they skew what is ‘optimal’ radiotherapy to the lower end of the spectrum, to the detriment of the majority of patients who are not as sensitive. Future CDSSs should be able to distinguish such overly sensitive patients and classify them separately so they receive different treatments to the less-sensitive patients.
Several small165 and large166 in vitro studies found a correlation between radiosensitivity and severity of late effects, namely radiation-induced fibrosis of the breast, but these findings were not consistent because no standardized quality assurance exists for radio-therapy in vivo.167,168 Similar discrepancies were later found using rapid assays that measure chromosomal damage,169 DNA damage170 and clonogenic cell survival.171 For example, the lymphocyte apoptosis assay has been used in a prospective trial as a stratification factor to assess late toxicity using letrozole as radio-sensitizer in patients with breast cancer.172 Cytokines such as TGF-β, which influences fibroblast proliferation and differentiation, are known to have a central role in fibrosis and senescence.173,174 Currently, the relationships between the lymphocyte predictive assay, TGF-β and late complications are purely correlative and a clear molecular explanation is lacking. Genome-wide association studies (GWAS) and the analysis of single nucleotide polymorphisms (SNPs) in candidate genes have also shown promise in identifying normal-tissue tolerance,175,176 although these do not often validate results from independent studies.177 In general, the problem with all these studies has been the wide experimental variability rather than interindividual differences in radiosensitivity. Normal-tissue tolerance is the dose-limiting factor for the administration of radiotherapy, therefore, any CDSS should be based on predictors of tumour control and the probability of complications.
Representation of predictions
Although the decisions made in the process of developing predictive models will determine the characteristics of a multivariate model (for example, which features are selected and the overall prediction accuracy), the success of the model depends on other factors, such as its availability and interactivity, which increases the acceptability. Even models based on large patient populations, with proper external validation, can fail to be accepted within the health-care community if the model and its output are not easily interpretable, if there is a lack of opportunity to apply the model or if the clinical usefulness is not proven or reported.178
Although some models, such as decision trees, implicitly have a visual representation that is somewhat interpretable, most models do not. One highly interpretable representation of a set of features is the nomogram.179 The nomogram was originally used in the early 20th century to make approximate graphical computations of mathematical equations. In medicine, nomograms have experienced a revival, reflected by the increasing number of studies reporting them.180–184 Figure 4 shows an example of a published clinical nomogram of local control in larynx cancer in which values for the selected features directly relate to a prediction score. The sum of these scores corresponds to a probability of local control within 2 or 5 years.181
Figure 4.
A published nomogram for local control in patients with cancer of the larynx treated with radiotherapy. Clinical and treatment variables are associated with local control status at follow-up durations of 2 and 5 years. The predictors are age of the patient (in years), haemoglobin level (in mmol/l), clinical tumour stage (T-stage), clinical nodal stage (N-stage), patient’s sex and equivalent dose (in Gy). A probability for local control can be calculated by drawing a vertical line from each predictor value to the score scale at the top—’points’. After manually summing up the scores, the ‘total points’ correspond to the probability of local control, which are estimated by drawing a vertical line from this value to the bottom scales to estimate local control.181
Another idea for increasing acceptability of computer- assisted personalized medicine is to make prediction models available on the internet. If interactive, peer-reviewed models are provided with sufficient background information, clinicians can test them using their own patient data. Such a system would provide retrospective validation of the multiple features by the wider community, as well as provide an indication on the clinical usefulness of the methodology. The best-known website with interactive clinical prediction tools is Adjuvant! Online.185 This website provides decision support for adjuvant therapy (for example, chemotherapy and hormone therapy) after surgery for patients with early-stage cancer. Many researchers have evaluated the models available on this prediction website, thereby refining them with additional predictors and updated external validations.186,187 A prediction website that focuses on decision support for radiotherapy was recently established.188 The aim of this website is to let users work with and validate the interactive models developed for patients with cancer treated with radio-therapy, which contributes to CDSS development in general by demonstrating the potential of these predictions and raising the awareness of their existence and limitations.
Future prospects
The major focus of this Review, thus far, has been model development, validation and presentation (including the features from different domains that might be considered as predictive and prognostic). Although an accurate outcome prediction model forms the basis of a CDSS, additional considerations must be made before a new CDSS can be used in daily radiation oncology practice.
First, any decision a patient or physician makes is based on a balance between its benefits (survival, local control and quality of life) and harms (toxic effects, complications, quality of life and financial cost). For example, an increased radiation dose usually results in both a higher probability of tumour control, but a concomitant higher probability of normal-tissue complications. Identifying the right balance between harm and benefit is a deeply personal choice that can vary substantially among patients. Thus, a CDSS should simultaneously predict local control, survival, treatment toxicity, quality of life and cost. The system should represent these predictions and the balance between them in a way that is not only clear to the physician, but also to the patient, to achieve shared decision making.
Additionally, any prediction using a CDSS should be accompanied by a confidence interval. Accurately evaluating the confidence interval is an active and challenging area of research because uncertainties in the input features, missing features, size and quality of the training set and the intrinsic uncertainty of cancer must be incorporated to specify the uncertainty in the prediction for an individual patient. Without knowing if two possible decisions have a statistically significant and clinically meaningful difference in outcome, clinical decision support is difficult. Always sharing the data on which the model was based is a crucial prerequisite for this effort.
Current prediction models for decision support can only assist in distinguishing very high-level decisions—such as palliative versus curative treatment, sequential versus concurrent chemoradiation, surgery versus a watch-and-wait approach. The radiation oncology community, however, is probably more interested in decisions such as intensity-modulated radiotherapy versus 3D-conformal radiotherapy or accelerated versus non-accelerated treatment. The current prediction models are simply not trained on datasets with these detailed subgroups and are not, therefore, accurate enough to support these decisions. Whether learning from increasingly diverse patient groups and adding other features will sufficiently improve the current models is unclear. As a result, tightly controlled studies using evidence-based medicine approaches are still crucial to guide clinical practice.
Finally, CDSSs should be seen as medical devices that require stringent acceptance, commissioning and quality assurance by the local institute. The key part of the commissioning and subsequent quality assurance is to validate the accuracy of the prediction model in the local patient population. Indeed, local patient data should be collected and the predicted outcomes compared with actual outcomes to convince local physicians that the support system works in their local setting. This ‘local validation’ should be done at the commissioning stage, but should be repeated to ensure the decision support remains valid, despite changes in local practice. Validation studies need to indicate what will be the required commission frequency.
This required quality assurance also enables the improvement of the system as more patient data becomes available. Using routine patient data to extract knowledge and apply that knowledge immediately is called ‘rapid learning’.3,189 Rapid learning via continuously updated CDSSs offers a way to quickly learn from retrospective data and include new data sets (such as randomized controlled trial results) to adapt treatment protocols and deliver personalized decision support.
As a data-driven discipline with well-established standards, such as DICOM–RT (digital imaging and communications in medicine in radiotherapy), radio-therapy offers an excellent starting point for adopting these rapid-learning principles (Figure 5). Aside from the importance of local data capture, which is still often lacking for (patient-reported) outcome and toxicity in particular, the quantity and heterogeneity of data that is necessary for rapid learning requires the pooling of data in a multi-institutional, international fashion.190,191 One method of pooling data is to replicate routine clinical data sources in a distributed de-identified data warehouse, such as what is done in an international Computer-Aided Theragnostics network.192 Examples of initiatives that create large centralized data and tissue infrastructures for routine radiation oncology patients are GENEPI,193 the Radiogenomics Consortium,194 ALLEGRO58 and ULICE.195 These initiatives also facilitate studies for external validation, reproducibility and hypothesis generation.190
Figure 5.
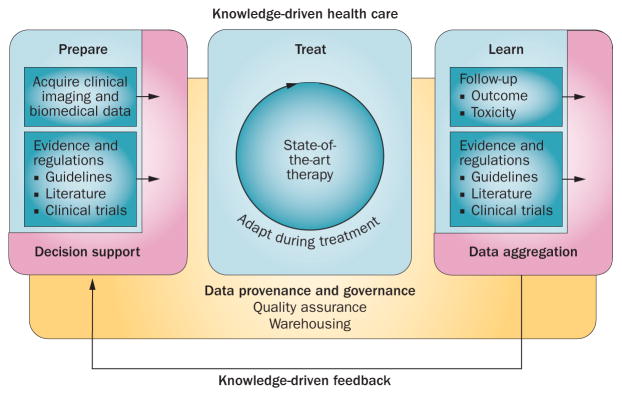
Knowledge-driven health-care principles using a clinical decision-support system in conjunction with standard evidence and regulations to choose the optimal treatment. In learning from follow-up data, knowledge is fed back to improve the clinical decision-support system and adapt regulations.
As datasets become larger (both in number of patients and in number of features per patient) high-throughput methods, both molecular196–201 and imaging-based,79 can produce large numbers of features that correlate with outcome.68,70,202–204 A limited application of these techniques has already transformed our understanding of radiotherapy response. For example, GWAS have associated SNPs with radiation toxicity.205,206 Similarly, mRNA-abundance microarrays have been used to predict tumour response and normal-tissue toxicity in both patient and in-vitro studies,207–211 as well as to create markers that reflect biological phenotypes that are important for radiation response, such as hypoxia212,213 and proliferation.214 Both the data analysis and validation are important but challenging aspects of model development.196,215 For example, the studies described above207–214 suffer from the substantial multiple-testing problem (that is, a large number of measured features compared with the sample number), which renders their results preliminary. Human input and large, robust validation studies are, therefore, needed before features from high-throughput techniques can be included in CDSSs.216–218
Although studies on a single feature can be informative, only its combination into multimodal, multivariate models can be expected to provide a more holistic view of the response to radiation. By combining events at different levels using systems-biology-like approaches, creating tumour-specific and patient-specific models of the effects and implications of radiation therapy should become possible (Figure 6). Indeed, future studies will not only need to identify the individual components related to radiation response, but will also need to establish the interactions and relations amongst them.219 Although this approach has not yet been applied to model radiotherapy responses, at least one study has demonstrated that combining multiple high-throughput data types can be used to map molecular cancer characteristics.220 Combining models at different levels (societal, patient, whole tumour or organ, local tumour or organ, and cellular) is expected to lead to an increasingly holistic and accurate CDSS for the individual patient. Evidence that longitudinal data have added value to predicting outcome in, for example, repeated PET-imaging221 and tumour-perfusion222 studies is growing, implying that this data need to be taken into account as candidates for future CDSSs.
Figure 6.
A simplified schematic representation of systems biology applied to radiotherapy. a | On the basis of in-vitro, in-vivo and patient data, modules representing the three biological categories (gene expression, immunohistochemical data and mutation data) important for radiotherapy response can be created. b | For an individual patient, appropriate molecular data will be accumulated. c | Combining the individual patient data with the modules will provide knowledge on specific module alterations (such as a deletion [X], upregulation [red] or downregulation [blue]), which can be translated to information on relative radioresistance and the molecular ‘weak’ spots of the tumour. This information will subsequently indicate whether dose escalation is necessary and which targeted drug is most effective for the patient. Part b used with permission from the National Academy of Sciences © Duboisa, L. J. Proc. Natl Acad. Sci. USA 108, 14620–14625 (2011).
Despite the challenges that remain, the vision of predictive models leading to CDSSs that are continuously updated via rapid learning on large datasets is clear, and numerous steps have already been taken. These include universal data-quality assurance programmes and semantic interoperability issues.223 However, we believe that this truly innovative journey will lead to necessary improvement of healthcare effectiveness and efficiency. Indeed, investments are being made in research and innovation for health-informatics systems, with an emphasis on interoperability and standards for secured data transfer, which shows that ‘eHealth’ will be among the largest health-care innovations of the coming decade.223,224
Conclusions
Accurate, externally validated prediction models are being rapidly developed, whereby multiple features related to the patient’s disease are combined into an integrated prediction. The key, however, is standardization—mainly in data acquisition across all areas, including molecular-based and imaging-based assays, patient preferences and possible treatments. Standardization requires harmonized clinical guidelines, regulated image acquisition and analysis parameters, validated biomarker assay criteria and data-sharing methods that use identical ontologies. Assessing the clinical usefulness of any CDSS is just as important as standardizing the development of externally validated accurate prediction models with high-quality data, preferably by standardizing the design of clinical trials. These crucial steps are the basis of validating a CDSS, which, in turn, will stimulate developments in rapid-learning health-care and will enable the next major advances in shared decision making.
Review criteria.
The PubMed and MEDLINE databases were searched for articles published in English (not restricted by date of publication) using a range of key phrases, including but not limited to: “PET imaging”, “heterogeneity in imaging”, “tumour response in radiotherapy”.
Key points.
Many prediction models that consider factors related to disease and treatment are available, but lack standardized assessments of their robustness, reproducibility or clinical utility
The complete cycle of model development for decision making in radiotherapy involves several stages, including selection of data, performance measure, classification and external validation
Clinical decision-support systems (CDSSs) based on validated predictors will be crucial to implement personalized radiation oncology
Tolerance of normal tissue is the dose-limiting factor for the administration of radiotherapy, therefore, any CDSS should be based on predictors of tumour control and the probability of complications
Rapid-learning healthcare will enable the increasingly rapid validation of CDSSs, which, in turn, will enable the next major advances in shared decision making
Acknowledgments
We acknowledge financial support from the Center for Translational Molecular Medicine framework (AIR FORCE), European Union sixth and seventh framework programme (ARTFORCE and METOXIA), INTERREG (www.eurocat.info), QuIC-ConCePT (funded by the Innovative Medicine Initiative Joint Undertaking) and the Dutch Cancer Society (KWF UM 2011-5020 and KWF UM 2009-4454).
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
P. Lambin and R. G. P. M. van Stiphout contributed equally to this manuscript, leading the research efforts for the data, discussion of the article content and writing of the manuscript. M. H. W. Starmans, E. Rios-Velazquez, E. Roelofs, W. van Elmpt, A. C. Begg, and D. De Ruysscher contributed to the discussion of the article’s content and helped to write the manuscript. G. Nalbantov and H. J. W. L. Aerts helped to write the manuscript. P. C. Boutros and A. Dekker researched the data, contributed to the discussion of the article’s content and helped to write the manuscript. P. Granone and V. Valentini contributed to the discussion of the article’s content. All authors edited the manuscript before submission.
Contributor Information
Philippe Lambin, Department of Radiation Oncology (MAASTRO), GROW School for Oncology and Developmental Biology, Maastricht University Medical Centre+, Maastricht, 6229 ET, The Netherlands.
Ruud G. P. M. van Stiphout, Department of Radiation Oncology (MAASTRO), GROW School for Oncology and Developmental Biology, Maastricht University Medical Centre+, Maastricht, 6229 ET, The Netherlands
Maud H. W. Starmans, Department of Radiation Oncology (MAASTRO), GROW School for Oncology and Developmental Biology, Maastricht University Medical Centre+, Maastricht, 6229 ET, The Netherlands
Emmanuel Rios-Velazquez, Department of Radiation Oncology (MAASTRO), GROW School for Oncology and Developmental Biology, Maastricht University Medical Centre+, Maastricht, 6229 ET, The Netherlands.
Georgi Nalbantov, Department of Radiation Oncology (MAASTRO), GROW School for Oncology and Developmental Biology, Maastricht University Medical Centre+, Maastricht, 6229 ET, The Netherlands.
Hugo J. W. L. Aerts, Departments of Radiation Oncology and Biostatistics and Computational Biology, Dana–Farber Cancer Institute, Harvard University, Boston, MA 02215, USA
Erik Roelofs, Department of Radiation Oncology (MAASTRO), GROW School for Oncology and Developmental Biology, Maastricht University Medical Centre+, Maastricht, 6229 ET, The Netherlands.
Wouter van Elmpt, Department of Radiation Oncology (MAASTRO), GROW School for Oncology and Developmental Biology, Maastricht University Medical Centre+, Maastricht, 6229 ET, The Netherlands.
Paul C. Boutros, Informatics and Biocomputing Platform, Ontario Institute for Cancer Research, Toronto, ON M5G 0A3, Canada
Pierluigi Granone, Department of Surgery, Catholic University of the Sacred Heart, Policlinico Universitario Agostino Gemelli, Rome 00168, Italy.
Vincenzo Valentini, Department of Radiotherapy, Catholic University of the Sacred Heart, Policlinico Universitario Agostino Gemelli, Rome 00168, Italy.
Adrian C. Begg, Division of Biological Stress Response, H3, The Netherlands Cancer Institute, Amsterdam, 1066 CX, The Netherlands
Dirk De Ruysscher, Department of Radiation Oncology (MAASTRO), GROW School for Oncology and Developmental Biology, Maastricht University Medical Centre+, Maastricht, 6229 ET, The Netherlands.
Andre Dekker, Department of Radiation Oncology (MAASTRO), GROW School for Oncology and Developmental Biology, Maastricht University Medical Centre+, Maastricht, 6229 ET, The Netherlands.
References
- 1.Vogelzang NJ, et al. Clinical cancer advances 2011: annual report on progress against cancer from the American Society of Clinical Oncology. J Clin Oncol. 2012;30:88–109. doi: 10.1200/JCO.2011.40.1919. [DOI] [PubMed] [Google Scholar]
- 2.Fraass BA, Moran JM. Quality, technology and outcomes: evolution and evaluation of new treatments and/or new technology. Semin Radiat Oncol. 2012;22:3–10. doi: 10.1016/j.semradonc.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Abernethy AP, et al. Rapid-learning system for cancer care. J Clin Oncol. 2010;28:4268–4274. doi: 10.1200/JCO.2010.28.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maitland ML, Schilsky RL. Clinical trials in the era of personalized oncology. CA Cancer J Clin. 2011;61:365–381. doi: 10.3322/caac.20135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bachtiary B, et al. Gene expression profiling in cervical cancer: an exploration of intratumor heterogeneity. Clin Cancer Res. 2006;12:5632–5640. doi: 10.1158/1078-0432.CCR-06-0357. [DOI] [PubMed] [Google Scholar]
- 7.Boyd CA, Benarroch-Gampel J, Sheffield KM, Cooksley CD, Riall TS. 415 patients with adenosquamous carcinoma of the pancreas: a population-based analysis of prognosis and survival. J Surg Res. 2012;174:12–19. doi: 10.1016/j.jss.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milosevic MF, et al. Interstitial fluid pressure in cervical carcinoma: within tumor heterogeneity, and relation to oxygen tension. Cancer. 1998;82:2418–2426. doi: 10.1002/(sici)1097-0142(19980615)82:12<2418::aid-cncr16>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 9.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;468:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suit H, Skates S, Taghian A, Okunieff P, Efird JT. Clinical implications of heterogeneity of tumor response to radiation therapy. Radiother Oncol. 1992;25:251–260. doi: 10.1016/0167-8140(92)90244-o. [DOI] [PubMed] [Google Scholar]
- 11.Aerts HJ, et al. Identification of residual metabolic-active areas within NSCLC tumours using a pre-radiotherapy FDG-PET-CT scan: a prospective validation. Lung Cancer. 2012;75:73–76. doi: 10.1016/j.lungcan.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Aerts HJ, et al. Identification of residual metabolic-active areas within individual NSCLC tumours using a pre-radiotherapy (18) Fluorodeoxyglucose–PET–CT scan. Radiother Oncol. 2009;91:386–392. doi: 10.1016/j.radonc.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vickers AJ. Prediction models: revolutionary in principle, but do they do more good than harm? J Clin Oncol. 2011;29:2951–2952. doi: 10.1200/JCO.2011.36.1329. [DOI] [PubMed] [Google Scholar]
- 14.Bright TJ, et al. Effect of clinical decision-support systems: a systematic review. Ann Intern Med. 2012;157:29–43. doi: 10.7326/0003-4819-157-1-201207030-00450. [DOI] [PubMed] [Google Scholar]
- 15.Clark GM. Prognostic factors versus predictive factors: examples from a clinical trial of erlotinib. Mol Oncol. 2008;1:406–412. doi: 10.1016/j.molonc.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dancey JE, et al. Guidelines for the development and incorporation of biomarker studies in early clinical trials of novel agents. Clin Cancer Res. 2010;16:1745–1755. doi: 10.1158/1078-0432.CCR-09-2167. [DOI] [PubMed] [Google Scholar]
- 17.Peek N, Arts DG, Bosman RJ, van der Voort PH, de Keizer NF. External validation of prognostic models for critically ill patients required substantial sample sizes. J Clin Epidemiol. 2007;60:491–501. doi: 10.1016/j.jclinepi.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 18.Vergouwe Y, Steyerberg EW, Eijkemans MJ, Habbema JD. Substantial effective sample sizes were required for external validation studies of predictive logistic regression models. J Clin Epidemiol. 2005;58:475–483. doi: 10.1016/j.jclinepi.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 19.Steyerberg EW, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21 :128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aittokallio T. Dealing with missing values in large-scale studies: microarray data imputation and beyond. Brief Bioinform. 2010;11:253–264. doi: 10.1093/bib/bbp059. [DOI] [PubMed] [Google Scholar]
- 21.Ludbrook J. Outlying observations and missing values: how should they be handled? Clin Exp Pharmacol Physiol. 2008;35:670–678. doi: 10.1111/j.1440-1681.2007.04860.x. [DOI] [PubMed] [Google Scholar]
- 22.Jayalakshmi T, Santhakumaran A. Statistical normalization and back propagation for classification. Int J Comput Theory Eng. 2011;3:89–93. [Google Scholar]
- 23.Huan L, Motoda H. Feature Selection for Knowledge Discovery and Data Mining. Kluwer Academic Publishers; Norwell, MA: 1998. [Google Scholar]
- 24.Harrell FE. Regression Modeling Strategies. Springer; New York: 2001. [Google Scholar]
- 25.Bishop CM. Pattern Recognition and Machine Learning. Springer; New York: 2007. [Google Scholar]
- 26.Lee SM, Abbott PA. Bayesian networks for knowledge discovery in large datasets: basics for nurse researchers. J Biomed Inform. 2003;36:389–399. doi: 10.1016/j.jbi.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Cruz JA, Wishart DS. Applications of machine learning in cancer prediction and prognosis. Cancer Inform. 2007;2:59–77. [PMC free article] [PubMed] [Google Scholar]
- 28.Putter H, Fiocco M, Geskus RB. Tutorial in biostatistics: competing risks and multi-state models. Stat Med. 2007;26:2389–2430. doi: 10.1002/sim.2712. [DOI] [PubMed] [Google Scholar]
- 29.Moons KG, et al. Risk prediction models: II. External validation, model updating, and impact assessment. Heart. 2012;98:691–698. doi: 10.1136/heartjnl-2011-301247. [DOI] [PubMed] [Google Scholar]
- 30.Dehing-Oberije C, et al. Development, external validation and clinical usefulness of a practical prediction model for radiation-induced dysphagia in lung cancer patients. Radiother Oncol. 2010;97:455–461. doi: 10.1016/j.radonc.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 31.Specht MC, Kattan MW, Gonen M, Fey J, Van Zee KJ. Predicting nonsentinel node status after positive sentinel lymph biopsy for breast cancer: clinicians versus nomogram. Ann Surg Oncol. 2005;12:654–659. doi: 10.1245/ASO.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 32.Dehing-Oberije C, et al. Tumor volume combined with number of positive lymph node stations is a more important prognostic factor than TNM stage for survival of non-small-cell lung cancer patients treated with (chemo)radiotherapy. Int J Radiat Oncol Biol Phys. 2008;70:1039–1044. doi: 10.1016/j.ijrobp.2007.07.2323. [DOI] [PubMed] [Google Scholar]
- 33.Vickers AJ, Kramer BS, Baker SG. Selecting patients for randomized trials: a systematic approach based on risk group. Trials. 2006;7 :30. doi: 10.1186/1745-6215-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baggerly KA, Coombes KR. What information should be required to support clinical “omics” publications? Clin Chem. 2011;57:688–690. doi: 10.1373/clinchem.2010.158618. [DOI] [PubMed] [Google Scholar]
- 35.Klopp AH, Eifel PJ. Biological predictors of cervical cancer response to radiation therapy. Semin Radiat Oncol. 2012;22:143–150. doi: 10.1016/j.semradonc.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 36.Kristiansen G. Diagnostic and prognostic molecular biomarkers for prostate cancer. Histopathology. 2012;60:125–141. doi: 10.1111/j.1365-2559.2011.04083.x. [DOI] [PubMed] [Google Scholar]
- 37.Dehing-Oberije C, et al. Development and external validation of prognostic model for 2-year survival of non-small-cell lung cancer patients treated with chemoradiotherapy. Int J Radiat Oncol Biol Phys. 2009;74:355–362. doi: 10.1016/j.ijrobp.2008.08.052. [DOI] [PubMed] [Google Scholar]
- 38.Ang CS, Phung J, Nice EC. The discovery and validation of colorectal cancer biomarkers. Biomed Chromatogr. 2011;25:82–99. doi: 10.1002/bmc.1528. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt ME, Steindorf K. Statistical methods for the validation of questionnaires--discrepancy between theory and practice. Methods Inf Med. 2006;45:409–413. [PubMed] [Google Scholar]
- 40.Garrido-Laguna I, et al. Validation of the Royal Marsden Hospital prognostic score in patients treated in the Phase I Clinical Trials Program at the MD Anderson Cancer Center. Cancer. 2012;118:1422–1428. doi: 10.1002/cncr.26413. [DOI] [PubMed] [Google Scholar]
- 41.Shrier I, et al. Should meta-analyses of interventions include observational studies in addition to randomized controlled trials? A critical examination of underlying principles. Am J Epidemiol. 2007;166:1203–1209. doi: 10.1093/aje/kwm189. [DOI] [PubMed] [Google Scholar]
- 42.Tzoulaki I, Siontis KC, Ioannidis JP. Prognostic effect size of cardiovascular biomarkers in datasets from observational studies versus randomised trials: meta-epidemiology study. BMJ. 2011;343:d6829. doi: 10.1136/bmj.d6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trotti A, Colevas AD, Setser A, Basch E. Patient-reported outcomes and the evolution of adverse event reporting in oncology. J Clin Oncol. 2007;25:5121–5127. doi: 10.1200/JCO.2007.12.4784. [DOI] [PubMed] [Google Scholar]
- 44.Trotti A, et al. CTCAE v3.0: development of a comprehensive grading system for the adverse effects of cancer treatment. Semin Radiat Oncol. 2003;13:176–181. doi: 10.1016/S1053-4296(03)00031-6. [DOI] [PubMed] [Google Scholar]
- 45.Golder S, Loke YK, Bland M. Meta-analyses of adverse effects data derived from randomised controlled trials as compared to observational studies: methodological overview. PLoS Med. 2011;8:e1001026. doi: 10.1371/journal.pmed.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steg PG, et al. External validity of clinical trials in acute myocardial infarction. Arch Intern Med. 2007;167:68–73. doi: 10.1001/archinte.167.1.68. [DOI] [PubMed] [Google Scholar]
- 47.Little J, et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE statement. Ann Intern Med. 2009;150:206–215. doi: 10.7326/0003-4819-150-3-200902030-00011. [DOI] [PubMed] [Google Scholar]
- 48.von Elm E, et al. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet. 2007;370:1453–1457. doi: 10.1016/S0140-6736(07)61602-X. [DOI] [PubMed] [Google Scholar]
- 49.Dawson LA, Sharpe MB. Image-guided radiotherapy: rationale, benefits, and limitations. Lancet Oncol. 2006;7:848–858. doi: 10.1016/S1470-2045(06)70904-4. [DOI] [PubMed] [Google Scholar]
- 50.Bentzen SM, et al. Quantitative analyses of normal tissue effects in the clinic (QUANTEC): an introduction to the scientific issues. Int J Radiat Oncol Biol Phys. 2010;76 (Suppl 3):3–9. doi: 10.1016/j.ijrobp.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verellen D, et al. Innovations in image-guided radiotherapy. Nat Rev Cancer. 2007;7:949–960. doi: 10.1038/nrc2288. [DOI] [PubMed] [Google Scholar]
- 52.Holthusen H. Erfahrungen über die Verträglichkeitsgrenze für Röntgenstrahlen und deren nutzanwendung zur verhütung von schäden [German] Strahlentherapie. 1936;57:254–269. [Google Scholar]
- 53.Valentini V, et al. Nomograms for predicting local recurrence, distant metastases, and overall survival for patients with locally advanced rectal cancer on the basis of European randomized clinical trials. J Clin Oncol. 2011;29:3163–3172. doi: 10.1200/JCO.2010.33.1595. [DOI] [PubMed] [Google Scholar]
- 54.Belderbos J, et al. Randomised trial of sequential versus concurrent chemo-radiotherapy in patients with inoperable non-small cell lung cancer (EORTC 08972–22973) Eur J Cancer. 2007;43:114–121. doi: 10.1016/j.ejca.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 55.Lambin P, et al. The ESTRO Breur Lecture 2009. From population to voxel-based radiotherapy: exploiting intra-tumour and intra-organ heterogeneity for advanced treatment of non-small cell lung cancer. Radiother Oncol. 2010;96:145–152. doi: 10.1016/j.radonc.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 56.Graham MV, et al. Clinical dose-volume histogram analysis for pneumonitis after 3D treatment for non-small cell lung cancer (NSCLC) Int J Radiat Oncol Biol Phys. 1999;45:323–329. doi: 10.1016/s0360-3016(99)00183-2. [DOI] [PubMed] [Google Scholar]
- 57.Emami B, et al. Tolerance of normal tissue to therapeutic irradiation. Int J Radiat Oncol Biol Phys. 1991;21:109–122. doi: 10.1016/0360-3016(91)90171-y. [DOI] [PubMed] [Google Scholar]
- 58.Ottolenghi A, Smyth V, Trott KR. The risks to healthy tissues from the use of existing and emerging techniques for radiation therapy. Radiat Prot Dosimetry. 2011;143:533–535. doi: 10.1093/rpd/ncq524. [DOI] [PubMed] [Google Scholar]
- 59.Beetz I, et al. NTCP models for patient-rated xerostomia and sticky saliva after treatment with intensity modulated radiotherapy for head and neck cancer: the role of dosimetric and clinical factors. Radiother Oncol. doi: 10.1016/j.radonc.2012.03.004. http://dx.doi.org/10.1016/j.radonc.2012.03.004. [DOI] [PubMed]
- 60.van der Schaaf A, et al. Multivariate modeling of complications with data driven variable selection: guarding against overfitting and effects of data set size. Radiother Oncol. doi: 10.1016/j.radonc.2011.12.006. http://dx.doi.org/10.1016/j.radonc.2011.12.006. [DOI] [PubMed]
- 61.Xu CJ, van der Schaaf A, van’t Veld AA, Langendijk JA, Schilstra C. Statistical validation of normal tissue complication probability models. Int J Radiat Oncol Biol Phys. 2012;84:e123–e129. doi: 10.1016/j.ijrobp.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 62.Nijsten SM, Mijnheer BJ, Dekker AL, Lambin P, Minken AW. Routine individualised patient dosimetry using electronic portal imaging devices. Radiother Oncol. 2007;83:65–75. doi: 10.1016/j.radonc.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 63.van Elmpt W, Petit S, De Ruysscher D, Lambin P, Dekker A. 3D dose delivery verification using repeated cone-beam imaging and EPID dosimetry for stereotactic body radiotherapy of non-small cell lung cancer. Radiother Oncol. 2010;94:188–194. doi: 10.1016/j.radonc.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 64.van Elmpt W, et al. 3D in vivo dosimetry using megavoltage cone-beam CT and EPID dosimetry. Int J Radiat Oncol Biol Phys. 2009;73:1580–1587. doi: 10.1016/j.ijrobp.2008.11.051. [DOI] [PubMed] [Google Scholar]
- 65.Rodrigues G, Lock M, D’Souza D, Yu E, Van Dyk J. Prediction of radiation pneumonitis by dose–volume histogram parameters in lung cancer—a systematic review. Radiother Oncol. 2004;71 :127–138. doi: 10.1016/j.radonc.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 66.Werner-Wasik M, Yorke E, Deasy J, Nam J, Marks LB. Radiation dose-volume effects in the esophagus. Int J Radiat Oncol Biol Phys. 2010;76 (Suppl 3):S86–S93. doi: 10.1016/j.ijrobp.2009.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saunders M, Rojas AM, Dische S. CHART revisited: a conservative approach for advanced head and neck cancer. Clin Oncol (R Coll Radiol) 2008;20:127–133. doi: 10.1016/j.clon.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 68.Turner N, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–2023. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Begg AC, et al. The value of pretreatment cell kinetic parameters as predictors for radiotherapy outcome in head and neck cancer: a multicenter analysis. Radiother Oncol. 1999;50:13–23. doi: 10.1016/s0167-8140(98)00147-9. [DOI] [PubMed] [Google Scholar]
- 70.Taguchi F, et al. Mass spectrometry to classify non-small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst. 2007;99:838–846. doi: 10.1093/jnci/djk195. [DOI] [PubMed] [Google Scholar]
- 71.Hessel F, et al. Impact of increased cell loss on the repopulation rate during fractionated irradiation in human FaDu squamous cell carcinoma growing in nude mice. Int J Radiat Biol. 2003;79:479–486. doi: 10.1080/0955300031000107871. [DOI] [PubMed] [Google Scholar]
- 72.Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer. 2008;8:545–554. doi: 10.1038/nrc2419. [DOI] [PubMed] [Google Scholar]
- 73.Ben-Josef E, et al. Impact of overall treatment time on survival and local control in patients with anal cancer: a pooled data analysis of Radiation Therapy Oncology Group trials 87–04 and 98–11. J Clin Oncol. 2010;28:5061–5066. doi: 10.1200/JCO.2010.29.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thames HD, et al. The role of overall treatment time in the outcome of radiotherapy of prostate cancer: an analysis of biochemical failure in 4839 men treated between 1987 and 1995. Radiother Oncol. 2010;96:6–12. doi: 10.1016/j.radonc.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 75.Fass L. Imaging and cancer: A review. Mol Oncol. 2008;2:115–152. doi: 10.1016/j.molonc.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Torigian DA, Huang SS, Houseni M, Alavi A. Functional imaging of cancer with emphasis on molecular techniques. CA Cancer J Clin. 2007;57:206–224. doi: 10.3322/canjclin.57.4.206. [DOI] [PubMed] [Google Scholar]
- 77.Eadie LH, Taylor P, Gibson AP. A systematic review of computer-assisted diagnosis in diagnostic cancer imaging. Eur J Radiol. 2012;81:e70–e76. doi: 10.1016/j.ejrad.2011.01.098. [DOI] [PubMed] [Google Scholar]
- 78.Gillies RJ, Anderson AR, Gatenby RA, Morse DL. The biology underlying molecular imaging in oncology: from genome to anatome and back again. Clin Radiol. 2010;65:517–521. doi: 10.1016/j.crad.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lambin P, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Velazquez ER, Aerts HJ, Oberije C, De Ruysscher D, Lambin P. Prediction of residual metabolic activity after treatment in NSCLC patients. Acta Oncol. 2010;49:1033–1039. doi: 10.3109/0284186X.2010.498441. [DOI] [PubMed] [Google Scholar]
- 81.Cangir AK, et al. Prognostic value of tumor size in non-small cell lung cancer larger than five centimeters in diameter. Lung Cancer. 2004;46:325–331. doi: 10.1016/j.lungcan.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 82.Lam JS, et al. Prognostic relevance of tumour size in T3a renal cell carcinoma: a multicentre experience. Eur Urol. 2007;52:155–162. doi: 10.1016/j.eururo.2007.01.106. [DOI] [PubMed] [Google Scholar]
- 83.Pitson G, et al. Tumor size and oxygenation are independent predictors of nodal diseases in patients with cervix cancer. Int J Radiat Oncol Biol Phys. 2001;51:699–703. doi: 10.1016/s0360-3016(01)01662-5. [DOI] [PubMed] [Google Scholar]
- 84.Thomas F, et al. Radical radiotherapy alone in non-operable breast cancer: The major impact of tumor size and histological grade on prognosis. Radiother Oncol. 1988;13:267–276. doi: 10.1016/0167-8140(88)90222-8. [DOI] [PubMed] [Google Scholar]
- 85.Steenbakkers RJ, et al. Observer variation in target volume delineation of lung cancer related to radiation oncologist-computer interaction: a ‘Big Brother’ evaluation. Radiother Oncol. 2005;77:182–190. doi: 10.1016/j.radonc.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 86.Greco C, Rosenzweig K, Cascini GL, Tamburrini O. Current status of PET/CT for tumour volume definition in radiotherapy treatment planning for non-small cell lung cancer (NSCLC) Lung Cancer. 2007;57:125–134. doi: 10.1016/j.lungcan.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 87.Caldwell CB, et al. Observer variation in contouring gross tumor volume in patients with poorly defined non-small-cell lung tumors on CT: the impact of 18FDG-hybrid PET fusion. Int J Radiat Oncol Biol Phys. 2001;51:923–931. doi: 10.1016/s0360-3016(01)01722-9. [DOI] [PubMed] [Google Scholar]
- 88.Bowden P, et al. Measurement of lung tumor volumes using three-dimensional computer planning software. Int J Radiat Oncol Biol Phys. 2002;53 :566–573. doi: 10.1016/s0360-3016(02)02783-9. [DOI] [PubMed] [Google Scholar]
- 89.Nishino M, et al. CT tumor volume measurement in advanced non-small-cell lung cancer: performance characteristics of an emerging clinical tool. Acad Radiol. 2011;18:54–62. doi: 10.1016/j.acra.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marcus CD, et al. Imaging techniques to evaluate the response to treatment in oncology: current standards and perspectives. Crit Rev Oncol Hematol. 2009;72:217–238. doi: 10.1016/j.critrevonc.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 91.Schwartz LH, Mazumdar M, Brown W, Smith A, Panicek DM. Variability in response assessment in solid tumors: effect of number of lesions chosen for measurement. Clin Cancer Res. 2003;9:4318–4323. [PubMed] [Google Scholar]
- 92.Erasmus JJ, et al. Interobserver and intraobserver variability in measurement of non-small-cell carcinoma lung lesions: implications for assessment of tumor response. J Clin Oncol. 2003;21:2574–2582. doi: 10.1200/JCO.2003.01.144. [DOI] [PubMed] [Google Scholar]
- 93.Therasse P. Measuring the clinical response. What does it mean? Eur J Cancer. 2002;38:1817–1823. doi: 10.1016/s0959-8049(02)00182-x. [DOI] [PubMed] [Google Scholar]
- 94.Nehmeh SA, Erdi YE. Respiratory motion in positron emission tomography/computed tomography: a review. Semin Nucl Med. 2008;38:167–176. doi: 10.1053/j.semnuclmed.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 95.Sonke JJ, Belderbos J. Adaptive radiotherapy for lung cancer. Semin Radiat Oncol. 2010;20:94–106. doi: 10.1016/j.semradonc.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 96.van Baardwijk A, et al. PET-CT-based auto-contouring in non-small-cell lung cancer correlates with pathology and reduces interobserver variability in the delineation of the primary tumor and involved nodal volumes. Int J Radiat Oncol Biol Phys. 2007;68:771–778. doi: 10.1016/j.ijrobp.2006.12.067. [DOI] [PubMed] [Google Scholar]
- 97.Wu K, et al. PET CT thresholds for radiotherapy target definition in non-small-cell lung cancer: how close are we to the pathologic findings? Int J Radiat Oncol Biol Phys. 2010;77:699–706. doi: 10.1016/j.ijrobp.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 98.Wanet M, et al. Gradient-based delineation of the primary GTV on FDG-PET in non-small cell lung cancer: a comparison with threshold-based approaches, CT and surgical specimens. Radiother Oncol. 2011;98:117–125. doi: 10.1016/j.radonc.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 99.Strassmann G, et al. Atlas-based semiautomatic target volume definition (CTV) for head-and-neck tumors. Int J Radiat Oncol Biol Phys. 2010;78:1270–1276. doi: 10.1016/j.ijrobp.2010.01.029. [DOI] [PubMed] [Google Scholar]
- 100.Nestle U, et al. Comparison of different methods for delineation of 18F-FDG PET-positive tissue for target volume definition in radiotherapy of patients with non-small cell lung cancer. J Nucl Med. 2005;46:1342–1348. [PubMed] [Google Scholar]
- 101.Daisne JF, et al. Tumor volume in pharyngolaryngeal squamous cell carcinoma: comparison at CT, MR imaging, and FDG PET and validation with surgical specimen. Radiology. 2004;233:93–100. doi: 10.1148/radiol.2331030660. [DOI] [PubMed] [Google Scholar]
- 102.van Loon J, et al. Therapeutic implications of molecular imaging with PET in the combined modality treatment of lung cancer. Cancer Treat Rev. 2011;37:331–343. doi: 10.1016/j.ctrv.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 103.Wood KA, Hoskin PJ, Saunders MI. Positron emission tomography in oncology: a review. Clin Oncol. 2007;19:237–255. doi: 10.1016/j.clon.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 104.O’Connor JP, et al. Quantitative imaging biomarkers in the clinical development of targeted therapeutics: current and future perspectives. Lancet Oncol. 2008;9:766–776. doi: 10.1016/S1470-2045(08)70196-7. [DOI] [PubMed] [Google Scholar]
- 105.van Baardwijk A, et al. Time trends in the maximal uptake of FDG on PET scan during thoracic radiotherapy. A prospective study in locally advanced non-small cell lung cancer (NSCLC) patients. Radiother Oncol. 2007;82:145–152. doi: 10.1016/j.radonc.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 106.Rodney JH. PET for therapeutic response monitoring in oncology. PET Clinics. 2008;3:89–99. doi: 10.1016/j.cpet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 107.Chung HH, et al. Prognostic value of metabolic tumor volume measured by FDG-PET/CT in patients with cervical cancer. Gynecol Oncol. 2011;120:270–274. doi: 10.1016/j.ygyno.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 108.Borst GR, et al. Standardised FDG uptake: a prognostic factor for inoperable non-small cell lung cancer. Eur J Cancer. 2005;41:1533–1541. doi: 10.1016/j.ejca.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 109.Mac Manus MP, et al. Metabolic (FDG–PET) response after radical radiotherapy/ chemoradiotherapy for non-small cell lung cancer correlates with patterns of failure. Lung Cancer. 2005;49:95–108. doi: 10.1016/j.lungcan.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 110.Hoekstra CJ, et al. Prognostic relevance of response evaluation using [18F]-2-fluoro-2-deoxy-d-glucose positron emission tomography in patients with locally advanced non-small-cell lung cancer. J Clin Oncol. 2005;23:8362–8370. doi: 10.1200/JCO.2005.01.1189. [DOI] [PubMed] [Google Scholar]
- 111.Soto DE, Kessler ML, Piert M, Eisbruch A. Correlation between pretreatment FDG–PET biological target volume and anatomical location of failure after radiation therapy for head and neck cancers. Radiother Oncol. 2008;89:13–18. doi: 10.1016/j.radonc.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lambrecht M, et al. The use of FDG–PET/CT and diffusion-weighted magnetic resonance imaging for response prediction before, during and after preoperative chemoradiotherapy for rectal cancer. Acta Oncol. 2010;49:956–963. doi: 10.3109/0284186X.2010.498439. [DOI] [PubMed] [Google Scholar]
- 113.Janssen MHM, et al. Evaluation of early metabolic responses in rectal cancer during combined radiochemotherapy or radiotherapy alone: Sequential FDG–PET–CT findings. Radiother Oncol. 2010;94:151–155. doi: 10.1016/j.radonc.2009.12.033. [DOI] [PubMed] [Google Scholar]
- 114.Ceulemans G, et al. Can 18-FDG-PET during radiotherapy replace post-therapy scanning for detection/demonstration of tumor response in head-and-neck cancer? Int J Radiat Oncol Biol Phys. 2011;81:938–942. doi: 10.1016/j.ijrobp.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 115.van Loon J, et al. Early CT and FDG-metabolic tumour volume changes show a significant correlation with survival in stage I-III small cell lung cancer: a hypothesis generating study. Radiother Oncol. 2011;99:172–175. doi: 10.1016/j.radonc.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bussink J, Kaanders JH, van der Graaf WT, Oyen WJ. PET–CT for radiotherapy treatment planning and response monitoring in solid tumors. Nat Rev Clin Oncol. 2011;8:233–242. doi: 10.1038/nrclinonc.2010.218. [DOI] [PubMed] [Google Scholar]
- 117.Boellaard R. Need for standardization of 18F-FDG PET/CT for treatment response assessments. J Nucl Med. 2011;52 (Suppl 2):93–100. doi: 10.2967/jnumed.110.085662. [DOI] [PubMed] [Google Scholar]
- 118.Boellaard R, et al. The Netherlands protocol for standardisation and quantification of FDG whole body PET studies in multi-centre trials. Eur J Nucl Med Mol Imaging. 2008;35:2320–2333. doi: 10.1007/s00259-008-0874-2. [DOI] [PubMed] [Google Scholar]
- 119.Boellaard R, et al. FDG PET and PET/CT: EANM procedure guidelines for tumour PET imaging: version 1.0. Eur J Nucl Med Mol Imaging. 2010;37:181–200. doi: 10.1007/s00259-009-1297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bayouth JE, et al. Image-based biomarkers in clinical practice. Semin Radiat Oncol. 2011;21:157–166. doi: 10.1016/j.semradonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Harry VN, Semple SI, Parkin DE, Gilbert FJ. Use of new imaging techniques to predict tumour response to therapy. Lancet Oncol. 2010;11:92–102. doi: 10.1016/S1470-2045(09)70190-1. [DOI] [PubMed] [Google Scholar]
- 122.Heijmen L, et al. Tumour response prediction by diffusion-weighted MR imaging: ready for clinical use? Crit Rev Oncol Hematol. 2012;83:194–207. doi: 10.1016/j.critrevonc.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 123.Lambrecht M, et al. The prognostic value of pretherapeutic diffusion-weighted MRI in oropharyngeal carcinoma treated with (chemo-) radiotherapy. Cancer Imaging. 2011;11:S112–S113. [Google Scholar]
- 124.Vandecaveye V, et al. Diffusion-weighted magnetic resonance imaging early after chemoradiotherapy to monitor treatment response in head-and-neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys. 2012;82:1098–1107. doi: 10.1016/j.ijrobp.2011.02.044. [DOI] [PubMed] [Google Scholar]
- 125.Kim SY, et al. Malignant hepatic tumors: short-term reproducibility of apparent diffusion coefficients with breath-hold and respiratory-triggered diffusion-weighted MR imaging. Radiology. 2010;255:815–823. doi: 10.1148/radiol.10091706. [DOI] [PubMed] [Google Scholar]
- 126.Sinkus R, Van Beers BE, Vilgrain V, Desouza N, Waterton JC. Apparent diffusion coefficient from magnetic resonance imaging as a biomarker in oncology drug development. Eur J Cancer. 2012;48:425–431. doi: 10.1016/j.ejca.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 127.Kierkels RG, et al. Comparison between perfusion computed tomography and dynamic contrast-enhanced magnetic resonance imaging in rectal cancer. Int J Radiat Oncol Biol Phys. 2010;77 :400–408. doi: 10.1016/j.ijrobp.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 128.Shukla-Dave A, et al. Dynamic contrast-enhanced magnetic resonance imaging as a predictor of outcome in head and neck squamous cell carcinoma patients with nodal metastases. Int J Radiat Oncol Biol Phys. 2012;82:1837–1844. doi: 10.1016/j.ijrobp.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yaromina A, et al. Co-localisation of hypoxia and perfusion markers with parameters of glucose metabolism in human squamous cell carcinoma (hSCC) xenografts. Int J Radiat Biol. 2009;85:972–980. doi: 10.3109/09553000903232868. [DOI] [PubMed] [Google Scholar]
- 130.Mörchel P, et al. Correlating quantitative MR measurements of standardized tumor lines with histological parameters and tumor control dose. Radiother Oncol. 2010;96:123–130. doi: 10.1016/j.radonc.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 131.Quennet V, et al. Tumor lactate content predicts for response to fractionated irradiation of human squamous cell carcinomas in nude mice. Radiother Oncol. 2006;81:130–135. doi: 10.1016/j.radonc.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 132.Kim YI, et al. Multiphase contrast-enhanced CT imaging in hepatocellular carcinoma correlation with immunohistochemical angiogenic activities. Acad Radiol. 2007;14:1084–1091. doi: 10.1016/j.acra.2007.06.015. [DOI] [PubMed] [Google Scholar]
- 133.Miles KA. Perfusion CT for the assessment of tumour vascularity: which protocol? Br J Radiol. 2003;76 :S36–S42. doi: 10.1259/bjr/18486642. [DOI] [PubMed] [Google Scholar]
- 134.Miles KA. Molecular imaging with dynamic contrast-enhanced computed tomography. Clin Radiol. 2010;65:549–556. doi: 10.1016/j.crad.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 135.Petralia G, et al. CT perfusion in oncology: how to do it. Cancer Imaging. 2010;10:8–19. doi: 10.1102/1470-7330.2010.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Asselin MC, O’Connor JP, Boellaard R, Thacker NA, Jackson A. Quantifying heterogeneity in human tumours using MRI and PET. Eur J Cancer. 2012;48:447–455. doi: 10.1016/j.ejca.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 137.Eary JF, O’Sullivan F, O’Sullivan J, Conrad EU. Spatial heterogeneity in sarcoma 18F-FDG uptake as a predictor of patient outcome. J Nucl Med. 2008;49:1973–1979. doi: 10.2967/jnumed.108.053397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tixier F, et al. Intratumor heterogeneity characterized by textural features on baseline 18F-FDG PET images predicts response to concomitant radiochemotherapy in esophageal cancer. J Nucl Med. 2011;52:369–378. doi: 10.2967/jnumed.110.082404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Diehn M, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci USA. 2008;105:5213–5218. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kuo MD, Gollub J, Sirlin CB, Ooi C, Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J Vasc Interv Radiol. 2007;18 :821–831. doi: 10.1016/j.jvir.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 141.Segal E, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 142.Rutman AM, Kuo MD. Radiogenomics: creating a link between molecular diagnostics and diagnostic imaging. Eur J Radiol. 2009;70:232–241. doi: 10.1016/j.ejrad.2009.01.050. [DOI] [PubMed] [Google Scholar]
- 143.Lindegaard JC, Overgaard J, Bentzen SM, Pedersen D. Is there a radiobiologic basis for improving the treatment of advanced stage cervical cancer? J Natl Cancer Inst Monogr. 1996:105–112. [PubMed] [Google Scholar]
- 144.Slonina D, Gasińska A. Intrinsic radiosensitivity of healthy donors and cancer patients as determined by the lymphocyte micronucleus assay. Int J Radiat Biol. 1997;72:693–701. doi: 10.1080/095530097142852. [DOI] [PubMed] [Google Scholar]
- 145.Fertil B, Malaise EP. Intrinsic radiosensitivity of human cell lines is correlated with radioresponsiveness of human tumors: analysis of 101 published survival curves. Int J Radiat Oncol Biol Phys. 1985;11:1699–1707. doi: 10.1016/0360-3016(85)90223-8. [DOI] [PubMed] [Google Scholar]
- 146.West CM, Davidson SE, Roberts SA, Hunter RD. The independence of intrinsic radiosensitivity as a prognostic factor for patient response to radiotherapy of carcinoma of the cervix. Br J Cancer. 1997;76:1184–1190. doi: 10.1038/bjc.1997.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Björk-Eriksson T, West C, Karlsson E, Mercke C. Tumor radiosensitivity (SF2) is a prognostic factor for local control in head and neck cancers. Int J Radiat Oncol Biol Phys. 2000;46:13–19. doi: 10.1016/s0360-3016(99)00373-9. [DOI] [PubMed] [Google Scholar]
- 148.Bartelink H, et al. Towards prediction and modulation of treatment response. Radiother Oncol. 1999;50:1–11. doi: 10.1016/s0167-8140(99)00009-2. [DOI] [PubMed] [Google Scholar]
- 149.Begg AC. Predicting recurrence after radiotherapy in head and neck cancer. Semin Radiat Oncol. 2012;22:108–118. doi: 10.1016/j.semradonc.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 150.Menegakis A, et al. Prediction of clonogenic cell survival curves based on the number of residual DNA double strand breaks measured by γ-H2AX staining. Int J Radiat Biol. 2009;85:1032–1041. doi: 10.3109/09553000903242149. [DOI] [PubMed] [Google Scholar]
- 151.Olive PL, Banáth JP. Phosphorylation of histone H2AX as a measure of radiosensitivity. Int J Radiat Oncol Biol Phys. 2004;58:331–335. doi: 10.1016/j.ijrobp.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 152.Höckel M, et al. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Cancer Res. 1996;56:4509–4515. [PubMed] [Google Scholar]
- 153.Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 154.Chouaib S, et al. Hypoxia promotes tumor growth in linking angiogenesis to immune escape. Front Immunol. 2012;3:21. doi: 10.3389/fimmu.2012.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kaanders JH, et al. Pimonidazole binding and tumor vascularity predict for treatment outcome in head and neck cancer. Cancer Res. 2002;62:7066–7074. [PubMed] [Google Scholar]
- 156.Nordsmark M, et al. The prognostic value of in human cervix pimonidazole and tumour pO2 carcinomas after radiation therapy: a prospective international multi-center study. Radiother Oncol. 2006;80:123–131. doi: 10.1016/j.radonc.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 157.Rouschop KMA, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Krause BJ, Beck R, Souvatzoglou M, Piert M. PET and PET/CT studies of tumor tissue oxygenation. Q J Nucl Med Mol Imaging. 2006;50 :28–43. [PubMed] [Google Scholar]
- 159.Dubois LJ, et al. Preclinical evaluation and validation of [18F]HX4, a promising hypoxia marker for PET imaging. Proc Natl Acad Sci USA. 2011;108:14620–14625. doi: 10.1073/pnas.1102526108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.van Loon J, et al. Selective nodal irradiation on basis of (18)FDG–PET scans in limited-disease small-cell lung cancer: a prospective study. Int J Radiat Oncol Biol Phys. 2010;77:329–336. doi: 10.1016/j.ijrobp.2009.04.075. [DOI] [PubMed] [Google Scholar]
- 161.West CM, Cooper RA, Loncaster JA, Wilks DP, Bromley M. Tumor vascularity: a histological measure of angiogenesis and hypoxia. Cancer Res. 2001;61:2907–2910. [PubMed] [Google Scholar]
- 162.Maciejewski B, Withers HR, Taylor JM, Hliniak A. Dose fractionation and regeneration in radiotherapy for cancer of the oral cavity and oropharynx: tumor dose-response and repopulation. Int J Radiat Oncol Biol Phys. 1989;16:831–843. doi: 10.1016/0360-3016(89)90503-8. [DOI] [PubMed] [Google Scholar]
- 163.Suzuki Y, et al. Prognostic impact of mitotic index of proliferating cell populations in cervical cancer patients treated with carbon ion beam. Cancer. 2009;115:1875–1882. doi: 10.1002/cncr.24189. [DOI] [PubMed] [Google Scholar]
- 164.Turesson I, Nyman J, Holmberg E, Odén A. Prognostic factors for acute and late skin reactions in radiotherapy patients. Int J Radiat Oncol Biol Phys. 1996;36:1065–1075. doi: 10.1016/s0360-3016(96)00426-9. [DOI] [PubMed] [Google Scholar]
- 165.Johansen J, Bentzen SM, Overgaard J, Overgaard M. Evidence for a positive correlation between in vitro radiosensitivity of normal human skin fibroblasts and the occurrence of subcutaneous fibrosis after radiotherapy. Int J Radiat Biol. 1994;66:407–412. doi: 10.1080/09553009414551361. [DOI] [PubMed] [Google Scholar]
- 166.West CM, et al. Lymphocyte radiosensitivity is a significant prognostic factor for morbidity in carcinoma of the cervix. Int J Radiat Oncol Biol Phys. 2001;51:10–15. doi: 10.1016/s0360-3016(01)01575-9. [DOI] [PubMed] [Google Scholar]
- 167.Peacock J, et al. Cellular radiosensitivity and complication risk after curative radiotherapy. Radiother Oncol. 2000;55:173–178. doi: 10.1016/s0167-8140(00)00173-0. [DOI] [PubMed] [Google Scholar]
- 168.Russell NS, et al. Low predictive value of intrinsic fibroblast radiosensitivity for fibrosis development following radiotherapy for breast cancer. Int J Radiat Biol. 1998;73:661–670. doi: 10.1080/095530098141915. [DOI] [PubMed] [Google Scholar]
- 169.Russell NS, Arlett CF, Bartelink H, Begg AC. Use of fluorescence in situ hybridization to determine the relationship between chromosome aberrations and cell survival in eight human fibroblast strains. Int J Radiat Biol. 1995;68:185–196. doi: 10.1080/09553009514551091. [DOI] [PubMed] [Google Scholar]
- 170.Kiltie AE, et al. A correlation between residual radiation-induced DNA double-strand breaks in cultured fibroblasts and late radiotherapy reactions in breast cancer patients. Radiother Oncol. 1999;51:55–65. doi: 10.1016/s0167-8140(99)00030-4. [DOI] [PubMed] [Google Scholar]
- 171.Dileto CL, Travis EL. Fibroblast radiosensitivity in vitro and lung fibrosis in vivo: comparison between a fibrosis-prone and fibrosis-resistant mouse strain. Radiat Res. 1996;146:61–67. [PubMed] [Google Scholar]
- 172.Azria D, et al. Concurrent or sequential adjuvant letrozole and radiotherapy after conservative surgery for early-stage breast cancer (CO-HO-RT): a phase 2 randomised trial. Lancet Oncol. 2010;11:258–265. doi: 10.1016/S1470-2045(10)70013-9. [DOI] [PubMed] [Google Scholar]
- 173.Bentzen SM. Preventing or reducing late side effects of radiation therapy: radiobiology meets molecular pathology. Nat Rev Cancer. 2006;6:702–713. doi: 10.1038/nrc1950. [DOI] [PubMed] [Google Scholar]
- 174.Rodemann HP, Bamberg M. Cellular basis of radiation-induced fibrosis. Radiother Oncol. 1995;35:83–90. doi: 10.1016/0167-8140(95)01540-w. [DOI] [PubMed] [Google Scholar]
- 175.Andreassen CN, Alsner J, Overgaard M, Sørensen FB, Overgaard J. Risk of radiation-induced subcutaneous fibrosis in relation to single nucleotide polymorphisms in TGFB1, SOD2, XRCC1, XRCC3, APEX and ATM—a study based on DNA from formalin fixed paraffin embedded tissue samples. Int J Radiat Biol. 2006;82:577–586. doi: 10.1080/09553000600876637. [DOI] [PubMed] [Google Scholar]
- 176.Chang-Claude J, et al. Association between polymorphisms in the DNA repair genes, XRCC1, APE1, and XPD and acute side effects of radiotherapy in breast cancer patients. Clin Cancer Res. 2005;11:4802–4809. doi: 10.1158/1078-0432.CCR-04-2657. [DOI] [PubMed] [Google Scholar]
- 177.Barnett GC, et al. Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: a prospective analysis study. Lancet Oncol. 2012;13:65–77. doi: 10.1016/S1470-2045(11)70302-3. [DOI] [PubMed] [Google Scholar]
- 178.Cammann H, Jung K, Meyer HA, Stephan C. Avoiding pitfalls in applying prediction models, as illustrated by the example of prostate cancer diagnosis. Clin Chem. 2011;57:1490–1498. doi: 10.1373/clinchem.2011.166959. [DOI] [PubMed] [Google Scholar]
- 179.Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26 :1364–1370. doi: 10.1200/JCO.2007.12.9791. [DOI] [PubMed] [Google Scholar]
- 180.Dehing-Oberije C, et al. Development and validation of a prognostic model using blood biomarker information for prediction of survival of non-small-cell lung cancer patients treated with combined chemotherapy and radiation or radiotherapy alone (NCT00181519, NCT00573040, and NCT00572325) Int J Radiat Oncol Biol Phys. 2011;81:360–368. doi: 10.1016/j.ijrobp.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 181.Egelmeer AG, et al. Development and validation of a nomogram for prediction of survival and local control in laryngeal carcinoma patients treated with radiotherapy alone: a cohort study based on 994 patients. Radiother Oncol. 2011;100:108–115. doi: 10.1016/j.radonc.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 182.van Stiphout RG, et al. Development and external validation of a predictive model for pathological complete response of rectal cancer patients including sequential PET–CT imaging. Radiother Oncol. 2011;98:126–133. doi: 10.1016/j.radonc.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 183.Marko NF, Xu Z, Gao T, Kattan MW, Weil RJ. Predicting survival in women with breast cancer and brain metastasis: a nomogram outperforms current survival prediction models. Cancer. 2011;118:3749–3757. doi: 10.1002/cncr.26716. [DOI] [PubMed] [Google Scholar]
- 184.Rudloff U, et al. Nomogram for predicting the risk of local recurrence after breast-conserving surgery for ductal carcinoma in situ. J Clin Oncol. 2010;28:3762–3769. doi: 10.1200/JCO.2009.26.8847. [DOI] [PubMed] [Google Scholar]
- 185.Adjuvant! Inc. Adjuvant! Online [online] 2011 http://www.adjuvantonline.com.
- 186.Hajage D, et al. External validation of Adjuvant! Online breast cancer prognosis tool. Prioritising recommendations for improvement. PLoS ONE. 2011;6:e27446. doi: 10.1371/journal.pone.0027446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kuo YL, Chen DR, Chang TW. Accuracy validation of Adjuvant! Online in Taiwanese breast cancer patients—a 10-year analysis. BMC Med Inform Decis Mak. 2012;12:108. doi: 10.1186/1472-6947-12-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.MAASTRO Clinic. Cancer Prediction Models [online] 2012 http://www.predictcancer.org.
- 189.Ginsburg GS, Staples J, Abernethy AP. Academic medical centers: ripe for rapid-learning personalized health care. Sci Transl Med. 2011;3:101cm127. doi: 10.1126/scitranslmed.3002386. [DOI] [PubMed] [Google Scholar]
- 190.Deasy JO, et al. Improving normal tissue complication probability models: the need to adopt a “data-pooling” culture. Int J Radiat Oncol Biol Phys. 2010;76 (Suppl 3):S151–S154. doi: 10.1016/j.ijrobp.2009.06.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Roelofs E, et al. Design of and technical challenges involved in a framework for multicentric radiotherapy treatment planning studies. Radiother Oncol. 2010;97:567–571. doi: 10.1016/j.radonc.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 192.Euregional Computer Assisted Theragnostics (EuroCAT) project [online] 2012 http://www.eurocat.info.
- 193.De Ruysscher D, et al. First report on the patient database for the identification of the genetic pathways involved in patients over-reacting to radiotherapy: GENEPI–II. Radiother Oncol. 2010;97:36–39. doi: 10.1016/j.radonc.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 194.West C, et al. Establishment of a radiogenomics consortium. Int J Radiat Oncol Biol Phys. 2010;76:1295–1296. doi: 10.1016/j.ijrobp.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 195.Kessel KA, et al. Connection of European particle therapy centers and generation of a common particle database system within the European ULICE-framework. Radiat Oncol. 2012;7:115. doi: 10.1186/1748-717X-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 197.Wulfkuhle JD, Liotta LA, Petricoin EF. Proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3:267–275. doi: 10.1038/nrc1043. [DOI] [PubMed] [Google Scholar]
- 198.Pinkel D, Albertson DG. Array comparative genomic hybridization and its applications in cancer. Nat Genet. 2005;37 (Suppl):S11–S17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 199.Quackenbush J. Microarray analysis and tumor classification. N Engl J Med. 2006;354:2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- 200.Lu Y, et al. Kinome siRNA-phosphoproteomic screen identifies networks regulating AKT signaling. Oncogene. 2011;30:4567–4577. doi: 10.1038/onc.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 204.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Best T, et al. Variants at 6q21 implicate PRDM1 in the etiology of therapy-induced second malignancies after Hodgkin’s lymphoma. Nat Med. 2011;17:941–943. doi: 10.1038/nm.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kerns SL, et al. Genome-wide association study to identify single nucleotide polymorphisms (SNPs) associated with the development of erectile dysfunction in African-American men after radiotherapy for prostate cancer. Int J Radiat Oncol Biol Phys. 2010;78:1292–1300. doi: 10.1016/j.ijrobp.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.de Jong MC, et al. CD44 expression predicts local recurrence after radiotherapy in larynx cancer. Clin Cancer Res. 2010;16:5329–5338. doi: 10.1158/1078-0432.CCR-10-0799. [DOI] [PubMed] [Google Scholar]
- 208.Kitahara O, Katagiri T, Tsunoda T, Harima Y, Nakamura Y. Classification of sensitivity or resistance of cervical cancers to ionizing radiation according to expression profiles of 62 genes selected by cDNA microarray analysis. Neoplasia. 2002;4:295–303. doi: 10.1038/sj.neo.7900251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Torres-Roca JF, et al. Prediction of radiation sensitivity using a gene expression classifier. Cancer Res. 2005;65:7169–7176. doi: 10.1158/0008-5472.CAN-05-0656. [DOI] [PubMed] [Google Scholar]
- 210.Wong YF, et al. Gene expression pattern associated with radiotherapy sensitivity in cervical cancer. Cancer J. 2006;12:189–193. doi: 10.1097/00130404-200605000-00006. [DOI] [PubMed] [Google Scholar]
- 211.Yang S, et al. Identification of prognostic biomarkers for response to radiotherapy by DNA microarray in nasopharyngeal carcinoma patients. Int J Oncol. 2012;40:1590–1600. doi: 10.3892/ijo.2012.1341. [DOI] [PubMed] [Google Scholar]
- 212.Starmans MH, et al. The prognostic value of temporal in vitro and in vivo derived hypoxia gene-expression signatures in breast cancer. Radiother Oncol. 2012;102:436–443. doi: 10.1016/j.radonc.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 213.Winter SC, et al. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007;67:3441–3449. doi: 10.1158/0008-5472.CAN-06-3322. [DOI] [PubMed] [Google Scholar]
- 214.Starmans MH, et al. Robust prognostic value of a knowledge-based proliferation signature across large patient microarray studies spanning different cancer types. Br J Cancer. 2008;99:1884–1890. doi: 10.1038/sj.bjc.6604746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Koboldt DC, Ding L, Mardis ER, Wilson RK. Challenges of sequencing human genomes. Brief Bioinform. 2010;11:484–498. doi: 10.1093/bib/bbq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99:147–157. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 217.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Teo YY. Common statistical issues in genome-wide association studies: a review on power, data quality control, genotype calling and population structure. Curr Opin Lipidol. 2008;19:133–143. doi: 10.1097/MOL.0b013e3282f5dd77. [DOI] [PubMed] [Google Scholar]
- 219.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 220.International Cancer Genome Consortium. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.van Elmpt W, Ollers M, Dingemans AM, Lambin P, De Ruysscher D. Response assessment using 18F-FDG PET early in the course of radiotherapy correlates with survival in advanced-stage non-small cell lung cancer. J Nucl Med. 2012;53:1514–1520. doi: 10.2967/jnumed.111.102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mayr NA, et al. Characterizing tumor heterogeneity with functional imaging and quantifying high-risk tumor volume for early prediction of treatment outcome: cervical cancer as a model. Int J Radiat Oncol Biol Phys. 2012;83:972–979. doi: 10.1016/j.ijrobp.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.European Commission. Digital Agenda for Europe [online] 2012 http://ec.europa.eu/information_society/digital-agenda/index_en.htm.
- 224.European Commission Information Society. ICT for better healthcare in Europe [online] 2012 http://ec.europa.eu/information_society/activities/health/index_en.htm.






