Abstract
Background
In contrast to mammals, the zebrafish has the remarkable capacity to regenerate its pancreatic beta cells very efficiently. Understanding the mechanisms of regeneration in the zebrafish and the differences with mammals will be fundamental to discovering molecules able to stimulate the regeneration process in mammals. To identify the pancreatic cells able to give rise to new beta cells in the zebrafish, we generated new transgenic lines allowing the tracing of multipotent pancreatic progenitors and endocrine precursors.
Results
Using novel bacterial artificial chromosome transgenic nkx6.1 and ascl1b reporter lines, we established that nkx6.1-positive cells give rise to all the pancreatic cell types and ascl1b-positive cells give rise to all the endocrine cell types in the zebrafish embryo. These two genes are initially co-expressed in the pancreatic primordium and their domains segregate, not as a result of mutual repression, but through the opposite effects of Notch signaling, maintaining nkx6.1 expression while repressing ascl1b in progenitors. In the adult zebrafish, nkx6.1 expression persists exclusively in the ductal tree at the tip of which its expression coincides with Notch active signaling in centroacinar/terminal end duct cells. Tracing these cells reveals that they are able to differentiate into other ductal cells and into insulin-expressing cells in normal (non-diabetic) animals. This capacity of ductal cells to generate endocrine cells is supported by the detection of ascl1b in the nkx6.1:GFP ductal cell transcriptome. This transcriptome also reveals, besides actors of the Notch and Wnt pathways, several novel markers such as id2a. Finally, we show that beta cell ablation in the adult zebrafish triggers proliferation of ductal cells and their differentiation into insulin-expressing cells.
Conclusions
We have shown that, in the zebrafish embryo, nkx6.1+ cells are bona fide multipotent pancreatic progenitors, while ascl1b+ cells represent committed endocrine precursors. In contrast to the mouse, pancreatic progenitor markers nkx6.1 and pdx1 continue to be expressed in adult ductal cells, a subset of which we show are still able to proliferate and undergo ductal and endocrine differentiation, providing robust evidence of the existence of pancreatic progenitor/stem cells in the adult zebrafish. Our findings support the hypothesis that nkx6.1+ pancreatic progenitors contribute to beta cell regeneration. Further characterization of these cells will open up new perspectives for anti-diabetic therapies.
Electronic supplementary material
The online version of this article (doi:10.1186/s12915-015-0179-4) contains supplementary material, which is available to authorized users.
Keywords: nkx6.1, ascl1, pancreas, duct, centroacinar cells, beta cells, stem cells, lineage tracing, multipotent progenitors, regeneration, diabetes, zebrafish, Notch, Wnt
Background
The pancreas is composed of an exocrine compartment with acinar and ductal cells, which secrete and transport digestive enzymes into the gut, and an endocrine compartment, which regulates glucose homeostasis by secreting pancreatic hormones into the bloodstream. Loss of pancreatic insulin-producing cells (beta cells) is a hallmark of diabetes. An attractive therapeutic approach to cure this disease is to stimulate beta cell regeneration in vivo from another pancreatic cell type or from progenitors. However, the regenerative capacity of beta cells is very limited in mammals and the cellular and molecular mechanisms involved need to be well understood before we will be able to stimulate this process. The zebrafish (Danio rerio), owing to its phenomenal capacity to restore beta cells after targeted cell ablation [1–3], has become an attractive model organism for the study of the regeneration process. To this end, tools need to be developed, especially to define the source of the new beta cells. So far, no zebrafish transgenic lines have been available to allow a lineage tracing of either multipotent pancreatic progenitors, giving rise to both exocrine and endocrine tissues, or pancreatic endocrine precursor cells.
The pancreas develops from two domains, called the dorsal bud and the ventral bud, which emerge from the foregut endoderm [4, 5]. In zebrafish, the dorsal bud generates the first wave of endocrine cells, which cluster at 24 hours post fertilization (hpf) to form the principal islet [6]. The ventral bud emerges anteriorly to the dorsal bud at 32 hpf and gives rise to acinar, ductal, and to a second wave of endocrine cells [4, 7, 8]. These late endocrine cells originate either from the extra-pancreatic ducts (EPDs) and contribute to the expansion of the principal islet [9–11] or from the intra-pancreatic ducts (IPDs) and form small secondary islets all along them. These IPDs contain pancreatic Notch-responsive cells (PNCs), which represent a population of progenitors of endocrine cells and ductal cells but not of acinar cells [7, 12].
Notch signaling pathway controls the differentiation of pancreatic cells both in zebrafish and mice (reviewed by [13]). One of its functions is to maintain a pool of progenitors in an undifferentiated state through the repression of genes of the achaete scute-like (ASCL) family or of the atonal-related protein (ARP) family. In the murine pancreas, Notch signaling prevents endocrine cell differentiation through the repression of neurog3 [14]. In zebrafish, neurog3 is not expressed in the pancreas and therefore the control of endocrine cell fate is fulfilled by other ASCL/ARP factors, namely Ascl1b and Neurod1, which are both repressed by Notch signaling [15]. Exactly like the inactivation of murine Neurog3, their simultaneous inactivation completely prevents endocrine cell differentiation leading to the loss of all hormone-secreting cells [15]. ascl1b is the earliest pancreatic marker identified during zebrafish development, its expression starting at the end of gastrulation in the prospective pancreatic region (10 hpf). ascl1b is transiently expressed during the formation of the dorsal bud (10–17 hpf) and, like murine Neurog3, is not detected in hormone-expressing cells. Later, in the ventral bud, ascl1b expression is turned on when the endocrine cell differentiation program is induced through the blocking of Notch signaling [7, 12, 16]. This Notch inactivation triggers a massive expression of ascl1b in IPDs [15]. These data suggest that ascl1b expression is restricted to the committed endocrine precursors. However, the observation that the onset of ascl1b expression in the prospective pancreatic region precedes all other known pancreatic progenitor markers raises the possibility of the multipotency of the first ascl1b+ cells.
Another key factor for pancreatic development is the homeobox transcription factor Nkx6.1. In the mouse, it is expressed in the multipotent progenitors during early pancreatic development [17], and, in the zebrafish, nkx6.1 is expressed early in the pancreatic primordium of the dorsal bud (from 11.5 hpf onwards) [18]. At later developmental stages in the mouse embryo, Nkx6.1 becomes restricted to the endocrine/duct bipotential trunk domain [19]. Similarly, nkx6.1 is first broadly expressed in the zebrafish pancreatic ventral bud primordium [20], then segregates from the ptf1a+ acinar cells to persist in the primitive ducts [20–22] that will give rise to the mature ducts and to secondary islets [7]. In the mouse, Nkx6.1 is expressed in the differentiated beta cells [23] while in the zebrafish, nkx6.1 is never expressed in beta cells nor in the other pancreatic hormone-expressing cells [18]. These data suggest that in zebrafish nkx6.1 also marks multipotent pancreatic progenitors. However, previous findings suggested that the early ventral bud primordium was composed of a heterogeneous population of pancreatic cells comprising Notch-responsive cells, giving rise to ductal and endocrine cells, separated from the ptf1a+ cells, which generate the acinar cells [7]. This study raises the question of the identity of the multipotent pancreatic progenitors in the zebrafish ventral pancreatic bud and its derivatives.
Here, we show that nkx6.1 labels multipotent pancreatic progenitors giving rise to all of the different pancreatic cell types (endocrine, ductal, and acinar) while ascl1b marks endocrine precursors leading to the different endocrine cell types. For this purpose, we have generated two novel bacterial artificial chromosome (BAC) transgenic nkx6.1 and ascl1b reporter lines, Tg(nkx6.1:eGFP) and Tg(ascl1b:eGFP-2A-creERT2), that both faithfully recapitulate the expression of the nkx6.1 and ascl1b endogenous genes. Using these novel transgenic tools, we were able to analyze in detail the interdependency between these two factors and their relationship with the Notch signaling pathway. We also demonstrate that nkx6.1 expression persists in the adult ductal tree, notably in the centroacinar/terminal end duct cells (CACs), for which we show that they are able to differentiate into insulin-expressing cells in vivo. By isolating nkx6.1:eGFP+ cells from the dissected pancreases of adult fish, we determined the transcriptome of adult pancreatic ductal cells, which revealed the expression of several regulatory genes potentially involved in endocrine regeneration. Finally, we provide evidence that regenerating beta cells also originate from ductal cells.
Results
The bacterial artificial chromosome reporter Tg(nkx6.1:eGFP) recapitulates in vivo the expression of the endogenous nkx6.1 gene
To label the nkx6.1-expressing cells, we generated a transgenic line driving the expression of the enhanced green fluorescent protein (eGFP) under the control of nkx6.1 regulatory regions. We engineered a BAC spanning from 55 kb upstream to 95 kb downstream of the nkx6.1 gene and inserted the eGFP coding regions into exon 1, replacing the beginning of the nkx6.1 open reading frame (Additional file 1: Fig. S1A). This BAC reporter construct was introduced into the zebrafish genome using the Tol2 transposon system [24, 25] and the stable transgenic line Tg(nkx6.1:eGFP) obtained showed expression of green fluorescent protein (GFP) in the nervous system and in the pancreas, which mirrors the endogenous Nkx6.1 protein expression (Additional file 1: Fig. S1B). Detailed comparison of the localization of these two proteins in the pancreas during development confirmed that GFP is indeed co-expressed with Nkx6.1 (Fig. 1). Indeed, together with the endogenous Nkx6.1 protein [18], GFP is expressed at the base of the endocrine islet at 24 and 30 hpf (Fig. 1b, c), in the ventral bud at 38 and 48 hpf (Fig. 1d, e), and in IPDs and EPDs at 4 days post fertilization (dpf) (Fig. 1f, f'). In contrast, at earlier stages, GFP was detected in a subset of Nkx6.1+ cells, probably due to the delay of GFP expression compared to Nkx6.1. Indeed, at 17 hpf, about 75 % of the Nkx6.1+ cells showed detectable GFP expression (Fig. 1a, a') and at 14 hpf, this proportion dropped even further to about 25–30 % (data not shown). Conversely, a few hours after the onset of nkx6.1 gene expression, some GFP+/Nkx6.1– cells were also detected (green arrows, Fig. 1b'–e'). This GFP labeling is not the result of an ectopic expression of the gfp transcript, as double fluorescent whole-mount in situ hybridization (WISH) showed that the gfp transcripts are present in the same pancreatic domain as nkx6.1 transcripts (data not shown) and importantly, like nkx6.1, gfp transcripts were not found in hormone-expressing cells (Additional file 1: Fig. S1C–C'', D–D''). Hence, prolonged GFP detection is rather due to the well-known high stability of GFP (±24 h half-life [26]), which persists in cells where Nkx6.1 protein is no longer found. This is nicely illustrated at 30 hpf, where strong GFP expression is detected at the base of the forming islet where Nkx6.1+ pancreatic progenitors are located, while weak GFP labeling is found dorsally within the islet, where differentiated endocrine cells, devoid of Nkx6.1, are clustered (Fig. 1c). As expected, this prolonged GFP detection will gradually fade away, finally to disappear completely in the differentiated endocrine cells (Fig. 1f).
Fig. 1.
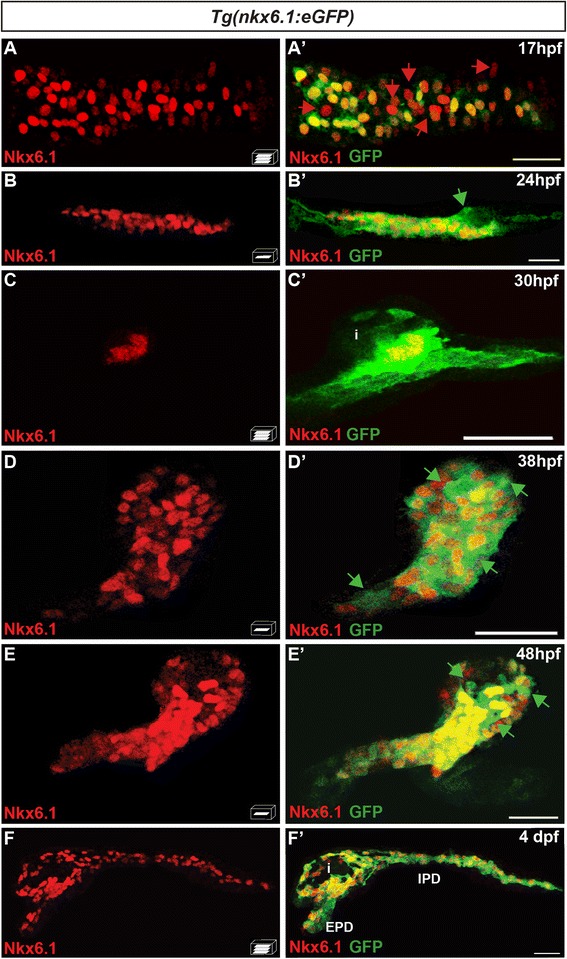
The bacterial artificial chromosome reporter line Tg(nkx6.1:eGFP) mirrors the expression of the endogenous nkx6.1 gene. Immunodetection of endogenous Nkx6.1 (red) and GFP (green) in Tg(nkx6.1:eGFP) embryos of the indicated stages. Green arrows point to Nkx6.1–/GFP+ cells and red arrows to Nkx6.1+/GFP- cells. All views are either lateral (b, b', c, and c') or ventral (a, a', d, d', f, and f') with the anterior part to the left. They represent either z-plane confocal images (b, d, e) or confocal projection images (a, c, f). Scale bars = 30 μm. EPD extra-pancreatic duct, IPD intra-pancreatic duct, i islet
The high stability of GFP allowed us to perform short-term lineage tracing to follow the immediate progeny arising from nkx6.1+ cells.
nkx6.1-expressing cells are multipotent progenitors giving rise to all pancreatic cell lineages
Using short-term lineage tracing, we first assessed if nkx6.1+ cells can give rise to the first wave of endocrine cells of the dorsal bud using the Isl1 marker, which labels all mature endocrine cells. In contrast to the endogenous nkx6.1 and the nkx6.1:GFP transcripts, which are not co-expressed with isl1 ([18] and Additional file 1: Fig. S1D–D"), GFP was detected in 40 ± 3.8 % of Isl1+ cells in Tg(nkx6.1:eGFP) embryos (n = 10) at 30 hpf indicating that nkx6.1 cells can give rise to endocrine cells (Fig. 2a–a"). Also, we found GFP in all different endocrine cell types, i.e. in 35 ± 18.8 % of insulin+ (Ins+) cells (n = 21) (Fig. 2b–b''), 42 ± 12.5 % somatostatin+ (Sst+) cells (n = 4) (Fig. 2c–c"), and 77 ± 3.7 % of glucagon+ (Gcg+) cells (n = 5) (Fig. 2d–d"). The percentage of GFP+ cells in the different endocrine subtypes appeared to depend on the onset of expression for each hormone, which is from 15 hpf onward for ins, 17 hpf for sst2 and 21 hpf for gcga. Therefore, when the first hormone-expressing cells differentiate from the pool of nkx6.1+ progenitors, only a minority of them have accumulated enough GFP to be detected, as explained above (see Fig. 1a and data not shown).
Fig. 2.
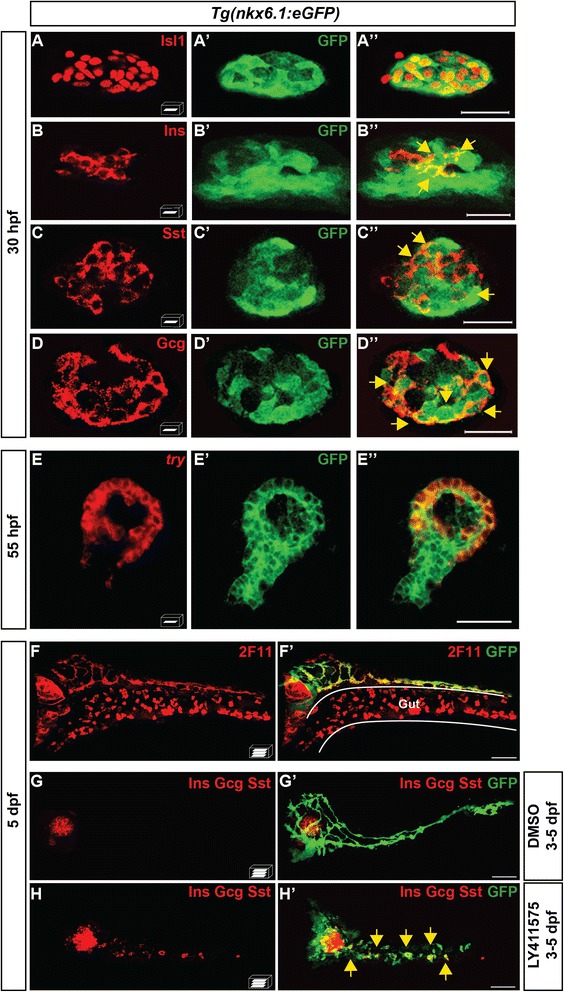
nkx6.1-expressing cells give rise to all pancreatic cell types. a–d" Immunodetection in 30-hpf Tg(nkx6.1:eGFP) embryos of GFP with Isl1 (a–a"), Ins (b–b"), Sst (c–c"), or Gcg (d–d"). Yellow arrows point to cells co-expressing GFP and the respective hormones. e–e" Fluorescent whole-mount in situ hybridization (WISH) of 55-hpf Tg(nkx6.1:eGFP) embryos using a try probe followed by GFP immunodetection. f, f' Immunodetection in 5-dpf Tg(nkx6.1:eGFP) embryos of GFP and of the hepato-pancreatic duct marker 2F11. g–h' Immunodetection of GFP and of Ins, Gcg, and Sst hormones in 5-dpf Tg(nkx6.1:eGFP) embryos treated from 3 to 5 dpf with dimethyl sulfoxide (DMSO) (g) or with the Notch signaling inhibitor, LY411575 (h). Yellow arrows point to secondary endocrine GFP+/hormones+ cells found in IPDs. All views are ventral with the anterior part to the left and represent either z-plane confocal images (a-e) or confocal projection images (f-h). Scale bars = 20 μm (a-e) or 40 μm (f-h)
Next, we analyzed whether the nkx6.1+ cells also contribute to the cells originating from the ventral bud (ductal, acinar, and secondary islets). With Nkx6.1 being expressed in all pancreatic ductal cells (as shown in Fig. 1e), we detected accordingly an expression of GFP in all pancreatic ducts labeled by 2F11 antibody (Fig. 2f, f'). In contrast, while endogenous nkx6.1 never co-localizes with trypsin (data not shown), a marker of mature acinar cells, GFP was detected in a large majority of acinar cells at 55 hpf (70 ± 25 % of trypsin+ cells (n = 8)), the stage when acinar cells have just begun to differentiate (Fig. 2e, e'). Here again, the prolonged GFP detection in the acinar cells gradually disappears and, from 3 dpf, the acinar cells are no longer labeled with GFP (shown at 5 dpf in Fig. 2f'). And finally, to determine whether nkx6.1+ cells can give rise to the secondary islets, emerging from IPDs, we treated Tg(nkx6.1:eGFP) larvae with the Notch-signaling inhibitor LY411575 from 3 to 5 dpf to increase the number of late endocrine cells and thereby facilitate their detection [7, 12, 16]. In LY411575-treated larvae, we observed an increase of the endocrine cells in the principal islet and the appearance of numerous endocrine cells in the pancreatic tail (yellow arrows, Fig. 2h, h'), as previously reported. All these endocrine cells are co-labeled with GFP (n = 4) indicating that the nkx6.1+ ductal cells can also give rise to the secondary islets.
These data indicate that nkx6.1-expressing cells are multi-lineage pancreatic progenitors, which can differentiate into endocrine, acinar, and ductal cells.
ascl1b-expressing cells give rise exclusively to the endocrine lineage
To determine whether ascl1b is expressed in the multipotent pancreatic progenitors or in the endocrine precursors, we determined the cell fate of the ascl1b-expressing cells. To that end, a BAC reporter Tg(ascl1b:eGFP-2A-creERT2) was engineered where the bicistronic transcript eGFP-2A-creERT2 is under the control of the promoter and regulatory sequences of ascl1b. Thus, we replaced the beginning of the ascl1b open reading frame with an eGFP-2A-creERT2 cassette (Additional file 2: Fig. S2A). The expression profile of GFP in the stable transgenic line Tg(ascl1b:eGFP-2A-creERT2) faithfully recapitulates the expression of the endogenous ascl1b transcript (Additional file 2: Fig. S2B–D).
Cell fate experiments were performed with Cre/loxP-based lineage tracing approaches by crossing the Tg(ascl1b:eGFP-2A-creERT2) with Cre-responder transgenic lines, either Tg(ubi:loxP-AmCyan-loxP-ZsYellow), termed Tg(ubi:CSY) [27], or Tg(ubi:loxP-eGFP-LoxP-mCherry), termed Tg(ubi:Switch) [28] (Fig. 3a). The double-transgenic embryos were treated five times with 4-hydroxytamoxifen (4OHT) from 11 to 15 hpf, the period when ascl1b expression reaches its maximal level [15], and the embryos were analyzed at 48 or 72 hpf. With these five 4OHT treatments, many ascl1b-expressing cells have undergone CRE recombination while no recombination was detected in the treated single-transgenic embryos used as control (data not shown). The CRE-mediated recombination (rec) marker (standing for either ZsYellow or mCherry) analyzed in double-transgenic embryos at 48 hpf was detected in 38 ± 4.3 % of Isl1+ cells (n = 5), indicating that ascl1b+ cells give rise to the endocrine cells of the dorsal bud (Fig. 3b–b"). In a similar way, the rec marker was detected in 58 ± 7.1 % of the Ins+ cells (n = 9) (Fig. 3c–c") and in 59 ± 3.7 % of the Gcg+ cells (n = 9) (Fig. 3d–d"). In contrast, the rec marker, clearly visible in the endocrine islet, was not detected at 72 hpf in the ductal cells, labeled by Nkx6.1, nor in the acinar cells, which surround them (Fig. 3e, e'), indicating that ascl1b+ cells cannot give rise to exocrine cells. Finally, we determined whether the ascl1b+ cells give rise to the secondary islets emerging from IPDs, either artificially induced by inhibiting the Notch pathway or naturally occurring in 20-dpf larvae. As the combined treatment of LY411575 with 4OHT was lethal, we performed short-term lineage tracing based on GFP expression (instead of Cre/loxP-based lineage tracing analyses). As shown in Fig. 3f, g, LY411575 treatment from 3 to 5 days of the Tg(ascl1b:eGFP-2A-creERT2) larvae led to the appearance of GFP cells all along the IPDs, most of these cells being also positive for glucagon or insulin hormones (yellow arrows, Fig. 3g, g'), indicating that ascl1b+ cells give rise to induced secondary islets. To trace the naturally occurring endocrine cells, we treated Tg(ascl1b:eGFP-2A-creERT2); Tg(ubi:Switch) larvae with 4OHT at 13, 14, and 17 dpf and analyzed the larvae at 20 dpf. The rec marker was detected within the principal islet (Fig. 3h, h') as well as in secondary islets (Fig. 3i, i') confirming that ascl1b+ cells can give rise to secondary islets.
Fig. 3.
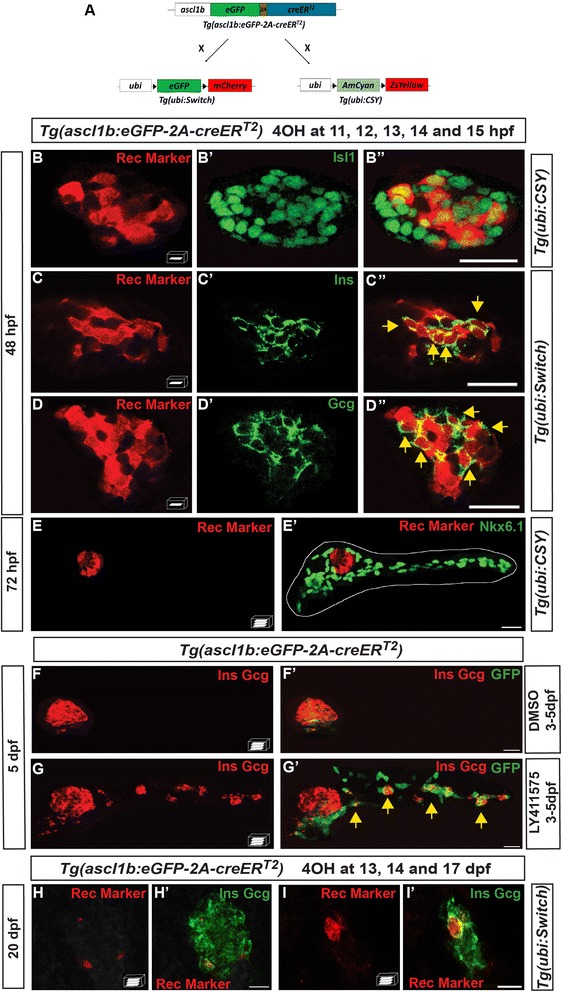
ascl1b-expressing cells give rise exclusively to endocrine cells of both dorsal and ventral bud. a–e', h–i' Genetic lineage tracing using the Cre-loxP system. a Schematic representation of the genetic lineage tracing experiments. The transgenic line Tg(ascl1b:eGFP-creER T2 ) was crossed with the Tg(ubi:loxP-eGFP-loxP-mCherry) line, abbreviated Tg(ubi:Switch), or with the Tg(ubi:loxP-AmCyan-loxP-ZsYellow) line, abbreviated Tg(ubi:CSY), treated with 4-hydroxytamoxifen (4OHT) at 11, 12, 13, 14, and 15 hpf (b–e') or at 13, 14, and 17 dpf (h–i') and fixed for analysis at the indicated times. Black triangles in a represent loxP sites. b–e' Immunodetection of CRE-mediated rec markers (ZsYellow or mCherry, red) and Isl1 (b–b"), Ins (c–c"), Gcg (d–d"), or Nkx6.1 (e, e') in 4OHT-treated embryos (green). The dotted white line delimits the pancreas (e'). Yellow arrows point to cells co-expressing rec marker (ZsYellow or mCherry) and the respective hormones (Ins or Gcg). f–g' Short-term lineage tracing: immunodetection of GFP and the Ins and Gcg hormones in 5-dpf Tg(ascl1b:eGFP-creER T2 ) embryos treated from 3 to 5 dpf with DMSO (f, f') or with the Notch signaling inhibitor, LY411575 (g, g'). Yellow arrows in g' point to GFP+/Ins+/Gcg+ secondary endocrine cells found in the IPDs. h–i' Immunodetection at 20 dpf of the CRE-mediated rec marker mCherry together with Ins and Gcg in 4OHT-treated larvae. All views are ventral with the anterior part to the left and represent z-plane confocal images (b–d") or confocal projection images (e–i'). Scale bars = 20 μm
In conclusion, our data demonstrate that ascl1b+ cells exclusively give rise to the endocrine cells originating from both the dorsal and ventral buds.
nkx6.1 and ascl1b are first co-expressed in the endocrine precursors of the dorsal bud but rapidly their expression domain segregates
As presented above, nkx6.1 is expressed in the multipotent pancreatic progenitors and ascl1b in the endocrine precursors; we therefore analyzed the relationship between these two populations by comparing the Nkx6.1 and GFP proteins in Tg(ascl1b:eGFP-2A-creERT2) embryos. At 14 hpf, the ascl1b:eGFP cells delineate two lines adjacent to the midline (Fig. 4a–a"). These cells correspond to the most medial endodermal cells (indicated by M in Fig. 4a'), reported to give rise mostly to pancreatic endocrine cells [29, 30]. At this stage, all of these ascl1b:eGFP cells also express Nkx6.1. In contrast, the Nkx6.1 expression domain is larger and, in addition to its expression in the hypochord (indicated by H in Fig. 4a), it is also expressed in the lateral cells, reported to give rise to exocrine and intestinal cells [29]. Rapidly, these two domains segregate since, as early as 1 hour later, the majority of ascl1b:eGFP+ cells no longer express Nkx6.1 (Fig. 4b–b"). Thus, separation of the two domains is largely completed when hormone-expressing cells start differentiating.
Fig. 4.
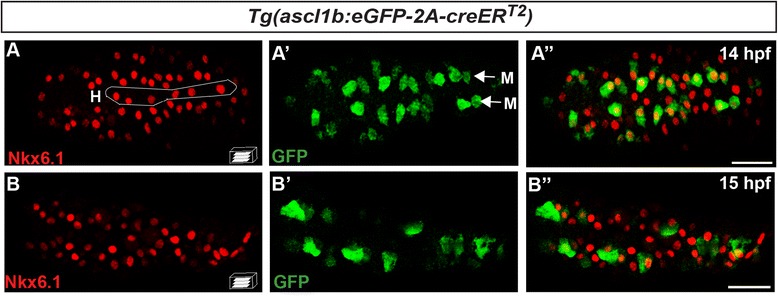
nkx6.1 and ascl1b are first co-expressed in the endocrine precursors of the dorsal bud but rapidly their expression domain moves apart. Immunodetection of endogenous Nkx6.1 and GFP in Tg(ascl1b:eGFP-creER T2 ) embryos at 14 hpf (a–a") and 15 hpf (b–b"). All views are ventral with the anterior part to the left and represent confocal projection images. Scale bars = 40 μm. H hypochord, M medial cells
To determine whether such segregation results from a mutual repression, we examined whether the loss of ascl1b leads to an increase of nkx6.1 expression and vice versa. We generated ascl1b and nkx6.1 loss-of-function mutants using the CRISPR/cas9 genome editing technology [31] (see “Methods” and Additional file 3: Fig. S3A, A' and Additional file 4: Fig. S4A, A'). As shown in Additional file 3: Fig. S3, the loss of ascl1b does not affect the expression of nkx6.1 (Additional file 3: Fig. S3D, E) while it significantly reduces the number of sst2+ cells (Additional file 3: Fig. S3B, C), as reported for the ascl1b morphants [15]. In the same way, ascl1b expression does not increase in nkx6.1 loss-of-function mutant embryos (Additional file 4: Fig. S4F, G), for which the effective loss of nkx6.1 expression was confirmed by immunodetection of Nkx6.1 (Additional file 4: Fig. S4B, C) and by the drastic reduction in the number of Gcg+ cells (Additional file 4: Fig. S4D, E), as reported for the nkx6.1 morphants [18].
ascl1b and nkx6.1 are regulated in an opposite way by the Notch signaling pathway
We then tested whether the segregation of the ascl1b and nkx6.1 expression domains results from a different response to the Delta/Notch signaling pathway. To analyze the impact of Notch signaling on nkx6.1 and ascl1b expression, we performed Notch loss- and gain-of- function analyses. We first analyzed the expression of nkx6.1 and ascl1b in mind bomb mutants (mibta52b) in which Notch signaling is disrupted [32]. As previously reported [15], expression of ascl1b is strongly increased in mib embryos at 15 hpf (Fig. 5a, b), in both the pancreas and the nervous system. The opposite effect was observed for nkx6.1 whose expression is reduced at the same stage (Fig. 5c, d) and completely lost at 18 hpf (Fig. 5e, f). This suggests that the Notch signaling pathway is essential for maintaining nkx6.1 expression but not for its initiation. This was confirmed by the finding that, at 13 hpf, nkx6.1 expression was unchanged in mib mutants while ascl1b was already upregulated (Additional file 5: Fig. S5A–D).
Fig. 5.
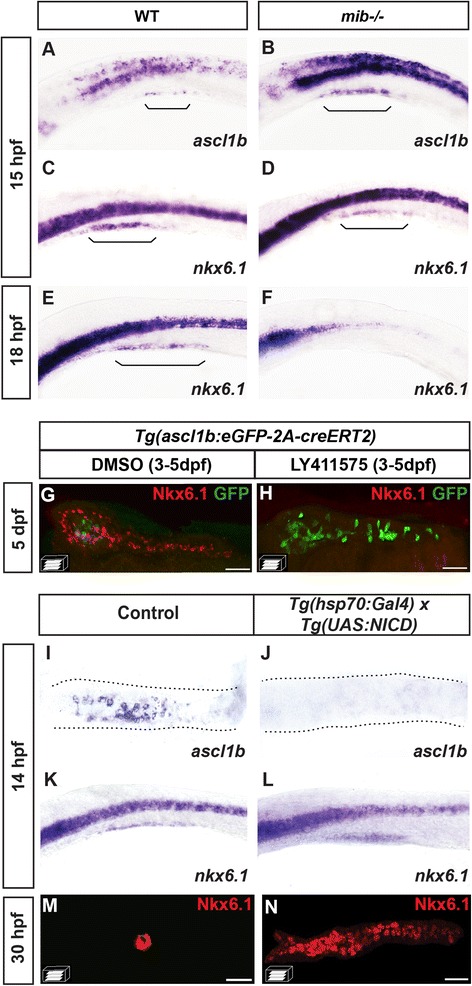
ascl1b and nkx6.1 are regulated in an opposite way by the Notch signaling pathway. a–f WISH on wild-type (WT) embryos (a, c, e) or mind bomb (mib -/-) mutants (b, d, f) with ascl1b (a, b) or nkx6.1 (c–f) probes. Brackets delimit the pancreatic domain. g, h Immunodetection of GFP and endogenous Nkx6.1 in 5-dpf Tg(ascl1b:eGFP-creER T2 ) embryos treated from 3 to 5 dpf with DMSO (g) or with the Notch signaling inhibitor, LY411575 (h). i–n WISH with ascl1b (i, j) or nkx6.1 (k, l) probes or immunodetection of endogenous Nkx6.1 (m, n) of double-transgenic Tg(hsp70:Gal4) x Tg(UAS:NICD) embryos (j, l, n) or of control simple-transgenic embryos (i, k, m), both heat-shocked at 11 hpf for 20 min. Lateral views (a–f, k, l) or ventral views (g–j, m, n) of embryos of the indicated stages with the anterior to the left. Scale bars = 40 μm
The same conclusion was drawn for the ventral bud where treatment for 3–5 dpf with the LY411575 Notch inhibitor led to a complete loss of Nkx6.1 expression and a drastic increase of ascl1b:eGFP at 5 dpf (Fig. 5g, h). Like the dorsal bud, the initiation of nkx6.1 expression in the ventral bud is not dependent on Notch signaling as its expression at 34 hpf was not perturbed in the mib mutant (Additional file 5: Fig. S5E, F).
For gain-of-function approaches, we crossed the Tg(hsp70:Gal4) with Tg(UAS:NICD) [33] and heat-shocked the embryos at 11 hpf to overexpress the Notch intracellular domain (NICD). At 3 hours after the heat-shock, we observed a complete loss of ascl1b expression (Fig. 5i, j) concomitant with an increase of nkx6.1 in the NICD overexpressed embryos (Fig. 5k, l). This increase was even more important at 30 hpf, when a drastic expansion in the number of nkx6.1+ cells was observed in the embryos overexpressing NICD (Fig. 5n) compared to the control (Fig. 5m).
In conclusion, these data show that Notch signaling represses ascl1b expression while it is essential for maintaining nkx6.1 expression. By contrast, the initiation of nkx6.1 expression is independent of Notch activity, both in the dorsal and the ventral buds.
Most, but not all, Nkx6.1+ cells are Notch-responsive cells
nkx6.1 being dependent on Notch signaling for maintaining its own expression, this prompted us to compare the location of the Notch-responsive cells and the Nkx6.1+ cells using the Tg(Tp1:VenusPest) [16] or Tg(Tp1:eGFP) [12] lines in which fluorescent markers are under the control of Notch-responsive elements (Tp1). In the prospective dorsal bud, we could detect Venus labeling in a subset of Nkx6.1 cells at 14 hpf (Fig. 6a, a') while 3 hours later (17 hpf), Venus was found in the vast majority of Nkx6.1+ cells (Fig. 6b, b'). Similarly, at the beginning of the formation of the ventral bud (38 hpf), only a subpopulation of Nkx6.1+ cells present some Notch activity (Fig. 6c, c') while at 65 hpf, the vast majority of Nkx6.1+ cells show Notch activity with the exception of the EPD anlagen (Fig. 6d, d'), known to be Notch inactive [34]. At 4 days, virtually all Nkx6.1+ cells are Notch-responsive in the IPDs (but the EPDs are still devoid of Notch activity) (Fig. 6e, e').
Fig. 6.
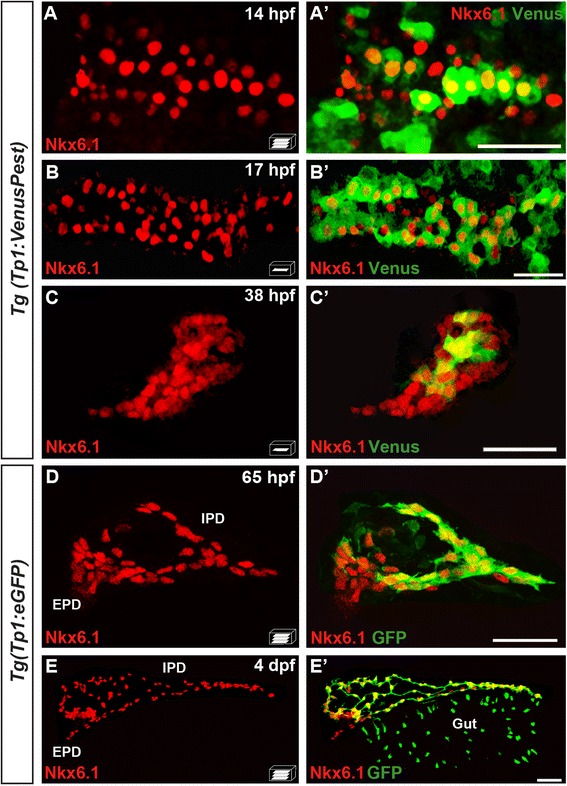
The pancreatic expression domain of Nkx6.1 includes the Notch-responsive cells. Immunodetection of endogenous Nkx6.1 (red) and Venus (revealed with anti-GFP, green) in Tg(TP1:VenusPest) (a–c') or GFP in Tg(Tp1:eGFP) embryos (d–e') at the indicated stages. All views are ventral with the anterior part to the left and represent either z-plane confocal images (b–c') or confocal projection images (a, a', d, d', e, e'). Scale bars = 40 μm. EPD extra-pancreatic duct, IPD intra-pancreatic duct
In conclusion, these observations indicate that, in both the dorsal and ventral buds, nkx6.1+ cells progressively acquire Notch signaling activity, essential for maintaining nkx6.1 expression.
nkx6.1 expression persists in ductal cells in the pancreas of adult zebrafish
We then wanted to characterize nkx6.1 expression in the pancreas of adult zebrafish. Immunodetection on paraffin sections through the pancreas of Tg(nkx6.1:eGFP) fish that were 6 to 9 months old revealed that nkx6.1:eGFP expression persists in adult zebrafish (Fig. 7a). Comparison of endogenous Nkx6.1 protein and GFP shows that, in the adult too, Tg(nkx6.1:eGFP) recapitulates the pattern of Nkx6.1 expression (data not shown). nkx6.1:eGFP expression is confined to the ducts and to isolated cells scattered throughout the exocrine tissue and was not detected in beta cells (Fig. 7a, a') or acinar cells. These nkx6.1:eGFP+ cells dispersed within the exocrine pancreas exhibit long cellular extensions characteristic of CACs (inset in Fig. 7a) [12]. In the adult zebrafish pancreas, as in mammals, the CACs can also be identified by Notch signaling activity [12]. Thus, to confirm the expression of nkx6.1 in CACs, we used the Notch reporter line Tg(Tp1:VenusPest) [16], in which the destabilized Venus fluorescent protein (VenusPest) highlights cells harboring ongoing Notch activity, and which labels the CACs, as expected (Fig. 7b and inset). All Venus+ cells in the pancreas were found exclusively in the ductal system, and more particularly in the CACs and not within the ductular structures (Additional file 6: Fig. S6). Comparison of Venus with Nkx6.1 confirmed that Nkx6.1 is indeed expressed in all Venus+ CACs (Fig. 7c, c'). In contrast, while ducts also display nkx6.1 expression as revealed with either nkx6.1:eGFP (Fig. 7a, a') or with the endogenous Nkx6.1 protein (Fig. 7d), they are devoid of Notch ongoing activity (white arrows pointing at Venus– ducts in Fig. 7b, d; see also Fig. 8a and Additional file 6: Fig. S6).
Fig. 7.
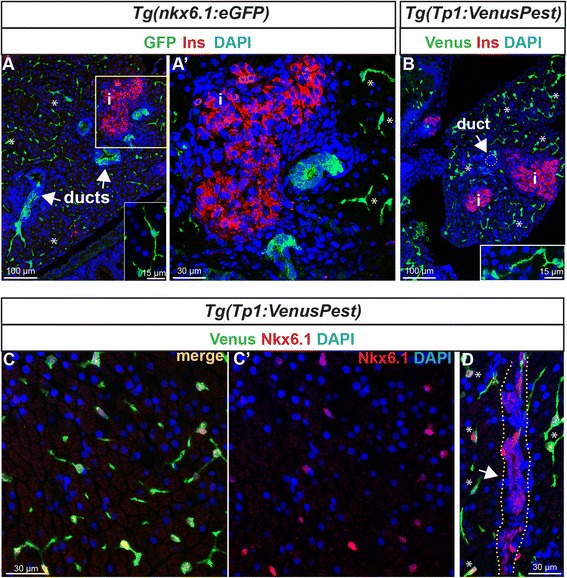
Expression of nkx6.1 persists in duct cells in the adult pancreas. a GFP and insulin (Ins, red) immunodetection on section through the pancreas of adult Tg(nkx6.1:eGFP) zebrafish. White arrows point to pancreatic ducts and asterisks show cells (presumably centroacinar/ terminal end duct cells (CACs) dispersed throughout the exocrine tissue). a' Close-up of the islet highlighted with Ins. b Venus (green) and Ins (red) immunodetection in Tg(Tp1:VenusPest) showing the presence of Venus in CACs (asterisks) as previously reported [12], but not in duct cells within ductular structures (white arrow). Insets in (a) and (b) show isolated CACs. c–d Immunodetection of Venus (green) and of endogenous Nkx6.1 (red) in Tg(Tp1:VenusPest) revealing co-labeling of both markers in CACs (c, c') while Nkx6.1 alone, but not Venus, labels the ducts. The white arrow in (d) points to a duct and asterisks indicate CACs. Dotted yellow lines delimit the duct. c' Same as (c) showing Nkx6.1 (red) and 4',6-diamidino-2-phenylindole (DAPI) only. i, islet
Fig. 8.
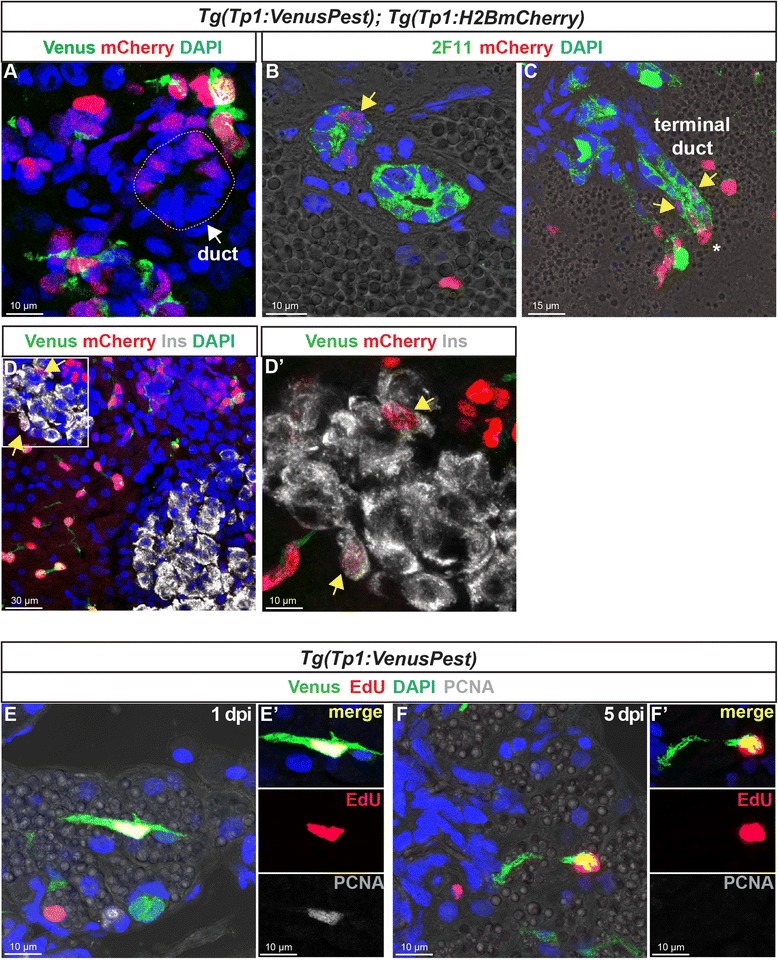
In the adult pancreas, Notch-responsive CACs give rise to other ductal and endocrine cells and have the capacity to replicate. Immunodetection on sections through the pancreas of adult Tg(Tp1:VenusPest); Tg(Tp1:H2BmCherry). a Comparison of Venus (green) and H2BmCherry (red) labeling showing a small duct containing H2BmCherry+ cells that have lost Venus. b Comparison of H2BmCherry (red) with the ductal marker 2F11 (green) showing some 2F11+ cells within a small duct co-expressing the stable H2BmCherry marker (yellow arrow). c Weak H2BmCherry labeling near the extremity of a ductular structure (terminal or intercalated duct) (yellow arrows); a CAC (intense H2BmCherry) at the tip of the terminal duct is indicated by an asterisk. d and d' (close-up) Some H2BmCherry+ cells, devoid of Venus (Notch off) co-express the beta cell marker Ins (white) (yellow arrows). Sections acquired in the head of the pancreas, at the level of the main endocrine islet. e–f' Detection of 5-ethynyl-2′-deoxyuridine (EdU) (red), Venus (green), and proliferating cell nuclear antigen (PCNA) (white) in Tg(Tp1:VenusPest) adult fish injected with EdU. The pancreas was analyzed 20 hours (1 day post injection) (e, e') and 5 days (f, f') after EdU injection
Adult centroacinar cells display progenitor capacity in physiological conditions
We then asked whether CACs could generate other pancreatic cell types in adult zebrafish under physiological conditions by using the double Tg(Tp1:VenusPest); Tg(Tp1:H2BmCherry) in which the stable H2BmCherry protein labels cells harboring ongoing Notch activity (Venus+ mCherry+) and cells having previously experienced Notch activity (Venus– mCherry+) [16]. This tool has been previously exploited to characterize and follow the fate of the Notch-responsive progenitors in the IPDs of larvae [16, 35]. We thus characterized the Venus– H2BmCherry+ cells in adult fish to identify the pancreatic cell types derived from CACs. In about 30 % of all H2BmCherry+ cells, Notch activity was switched off (Venus–). Many of these cells, with weak H2BmCherry labeling, were identified within small ducts (Fig. 8a–c), which can be identified by the ductal marker 2F11 (Fig. 8b, c and Additional file 7: Fig. S7A-C for the separated colors and additional example) [11, 22, 36] or by Nkx6.1 (Additional file 7: Fig. S7D), and at the tip of which reside CACs (intense H2BmCherry, asterisks in Fig. 8c and Additional file 7: Fig. S7). Furthermore, low levels of H2BmCherry were also identified in some insulin-expressing cells (Fig. 8d, d'). About 4.9 ± 2.6 % (n = 4) of the H2BmCherry+/Notch off (Venus–) display Ins labeling. These findings show that mCherry+ terminal end duct cells and insulin-expressing cells originate from Notch positive CACs. Overall, this reveals that a subset of the Nkx6.1+ ductal cells, the Notch-responsive CACs, can generate ductal and endocrine cells.
To determine the capacity of CACs to replicate, their proliferative status was analyzed using 5-ethynyl-2′-deoxyuridine (EdU) labeling and proliferating cell nuclear antigen (PCNA) immunodetection in Tg(Tp1:VenusPest) adult fish. One day after EdU injection, a small fraction of CACs (5.8 ± 2.6 %, out of 600 counted CACs) had incorporated EdU. All these EdU+ Venus+ cells also express the proliferation marker PCNA (Fig. 8e, e'). In contrast, 5 days post injection, Venus+ cells that still display EdU labeling were no longer PCNA+ (Fig. 8f, f'), but still harbor characteristics of CACs, suggesting that they became post-mitotic CACs. The capacity of CACs to replicate together with their ability to undergo ductal and endocrine differentiation indicate that they can behave as adult pancreatic progenitor/stem cells in vivo.
nkx6.1-expressing ductal cells contribute to beta cell regeneration in adult zebrafish
The capacity of CACs to differentiate into beta cell in the normal adult zebrafish raises the question whether similar plasticity exists for regeneration. To induce beta cell regeneration, we used the Tg(ins:NTR-mCherry) line [1] and treatment with metronidazole (MTZ). The metronidazole is converted into a cytotoxic compound by the nitroreductase enzyme (NTR), which thereby triggers selective beta cell death by apoptosis [3]. In adult zebrafish, the ablation of beta cells causes dramatic hyperglycemia within 3 days rapidly followed by spontaneous normalization within 2 weeks and beta cell regeneration [2]. To analyze the ductal cells in the setting of beta cell regeneration, Tg(nkx6.1:eGFP); Tg(ins:NTR-mCherry) adult fish were treated with MTZ (day 1) and sacrificed at different time points during regeneration to analyze GFP and Ins on tissue sections at the level of the principal islet. The blood glucose level was measured just before sacrifice. At 3 days post treatment (dpt), ablation was total and effective as reported [2, 37]. MTZ-treated fish displayed severe hyperglycemia as expected (>500 mg/dl, n = 10 fish) while the glycemia of non-treated fish was at normal values of 61 ± 19 mg/dl (n = 11). Immunolabeling at 3 dpt indicated total beta cell ablation as manifested by the absence of insulin staining in the pancreas of MTZ-treated Tg(nkx6.1:eGFP); Tg(ins:NTR-mCherry) fish (Fig. 9a, b). At 9 dpt, the first new beta cells have started to reappear (Fig. 9c). Glycemia was still above normal values but nonetheless had decreased (145 ± 31 mg/dl, n = 2). The principal islet in the treated fish still showed very weak insulin expression but a few new beta cells (about 5 % of the islet cells, compared to 40–70 % in control fish) can be detected in the principal islet as well as throughout the exocrine tissue as isolated cells or as small clusters of Ins-expressing cells next to CACs and ductal cells marked by GFP (Fig. 9c, yellow arrows). The presence of insulin-expressing cells at 9 dpt is indicative of beta cell regeneration. Interestingly, some of the regenerating insulin+ cells displayed weak GFP staining (7.9 ± 3.2 % of the Ins+ cells, 452 counted cells, n = 2 fish) (Fig. 9f). These cells were found next to strongly GFP-labeled ductal nkx6.1:eGFP cells. GFP+ Ins+ cells were also detected at 21 dpt (11.1 ± 4.3 % of the Ins+ cells, 940 counted cells, n = 2 fish) (Fig. 9g). At this stage, a large number of beta cells have recovered (about 30–50 % of islet cells, Fig. 9d) [2, 37] and glycemia was normalized (57 ± 9 mg/dl). In contrast, insulin+ cells never harbor GFP labeling in non-treated fish (Fig. 9e). Thus, by using the same approach of short-term tracing of nkx6.1:eGFP+ cells as in embryos, this finding strongly suggests that ductal nkx6.1:eGFP+ cells contribute to regenerated beta cells.
Fig. 9.
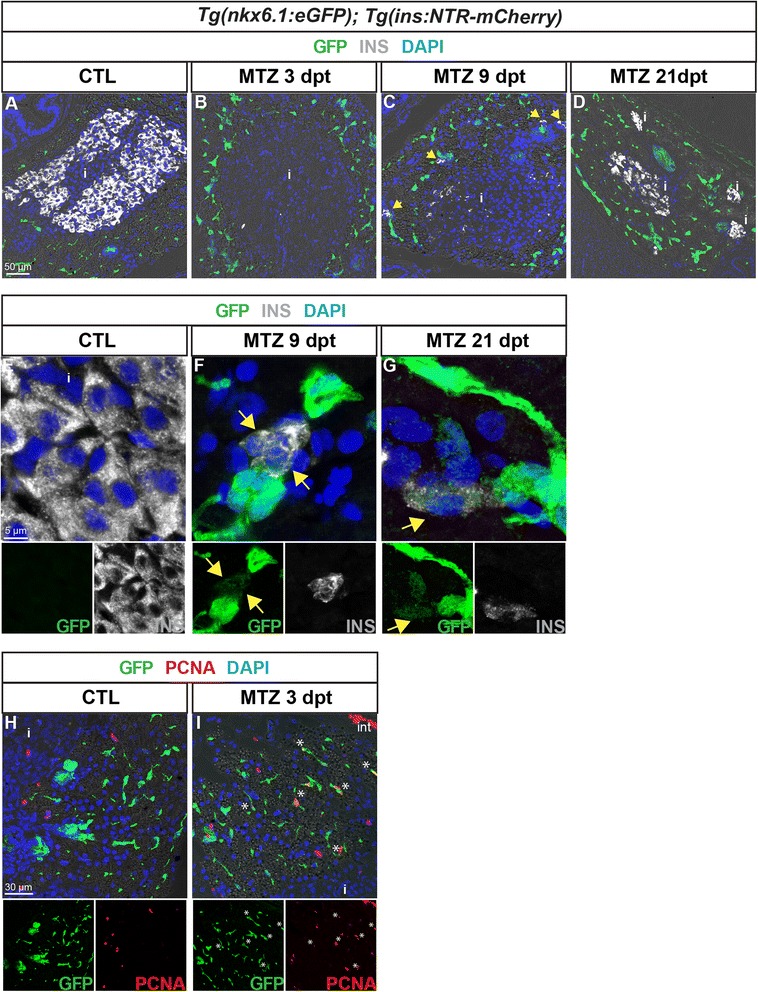
nkx6.1:eGFP+ cells proliferate and differentiate into new insulin-expressing cells after beta cell specific ablation. GFP (green) and Ins (white) labeling in the pancreas of Tg(nkx6.1:eGFP); Tg(ins:NTR-mCherry) adult fish. a, b Non-treated fish (CTL, a) show intense Ins staining in beta cells, while Ins+ cells are not detected 3 dpt with MTZ, indicating efficient ablation (b). Note that debris of one beta cell (Ins+) is observed in the islet. c At 9 dpt, Ins-expressing cells start to be detected in the principal islet and in extra-insular locations close to ductal GFP+ cells (yellow arrows). d At 21 dpt, islets show intense Ins staining consistent with beta cell recovery. e–g (and separate channels) While GFP is never detected in beta cells of control fish (e), some regenerating Ins+ cells display weak GFP labeling at both stages of regeneration analyzed, i.e. at 9 dpt (f) and 21 dpt (g). h, i (and separate channels) Beta cell ablation triggers proliferation of CACs as shown at 3 dpt (h) compared to CTL (i) (asterisks). i islet
Next we assessed proliferation in Tg(nkx6.1:eGFP); Tg(ins:NTR-mCherry) in response to beta cell ablation. Three days post treatment, nkx6.1:eGFP ductal cells showed increased proliferation as illustrated with PCNA (Fig. 9h, i). This was observed not only within ductal structures as previously described ([2] and not shown) but also for CACs. These observations suggest that ductal cells with pancreatic progenitor properties activate proliferation prior to differentiation into beta cells during regeneration.
Transcriptomic analysis of adult nkx6.1+ pancreatic ductal cells
To get a comprehensive characterization of pancreatic nkx6.1+ ductal cells in adult zebrafish, we determined their transcriptome landscape. Ductal GFP+ cells were isolated from dissected pancreases of adult Tg(nkx6.1:eGFP) fish (with ~95 % purity) and used in RNA-seq experiments. The expression of 15,888 genes could be detected in the ductal transcriptome (complete data available at [38]), in which many genes already known to be expressed in pancreatic ducts in either zebrafish or mammals were found at high expression level, such as sox9b, hnf1ba, onecut1/hnf6, cftr, cdh17, ca2, and ctgfa in addition to nkx6.1 (Fig. 10). We also detected expression of fgfr4 and sdc4, recently proposed as novel ductal markers in the murine embryonic pancreas [39]. In contrast to these ductal genes, the acinar markers ptf1a and rbpjl, the pan-endocrine markers pax6b and isl1, and the lineage specific genes mnx1 and arx were either not detected or detected at extremely low levels in the nkx6.1:eGFP+ ductal cells transcriptome (Fig. 10), underscoring the accuracy of our fluorescence-activated cell sorting (FACS) cell preparations. In contrast to mouse or human adults in which the embryonic pancreatic progenitor marker Pdx1 is not expressed in the pancreatic ducts in normal condition, nkx6.1:eGFP+ duct cells of healthy zebrafish display a robust expression of pdx1. Comparison of the duct transcriptome with those of pancreatic acinar and endocrine cells (manuscript in preparation) highlighted 3,684 genes with preferential expression in duct cells. Among them, 293 duct-specific genes were identified with strong enrichment (≥16-fold) and low expression in the other pancreatic cells (Additional file 8: Table S1), in which we find sox9b, onecut1/hnf6, cdh17, ctgfa, and nkx6.1, corroborating their status as duct-specific markers. Various components of the Notch signaling pathway could also be identified, namely notch2, and different Hairy and enhancer of split-related genes (her6, her9, and her15.1), confirming that a subpopulation of nkx6.1+ cells (the CACs) experiences Notch activity. In addition to genes involved in the Notch signaling cascade, our analyses also identified novel duct-specific markers such as id2a, encoding for a HLH transcription factor, which we also detected co-expressed with sox9b by WISH in 3-dpf larvae (Additional file 9: Fig. S8), and several components of the Wnt pathways such as the Wnt and SFRP ligands sfrp5, sfrp3/frzb, and wnt7bb (Additional file 8: Table S1). At a lower expression level, we could also detect the Wnt receptor fzd7a, which was the only Wnt receptor significantly expressed in the adult pancreas (565 ± 106 normalized counts with 68-fold enrichment in the ducts). These observations suggest that Wnt signaling may play an important role in adult ducts.
Fig. 10.
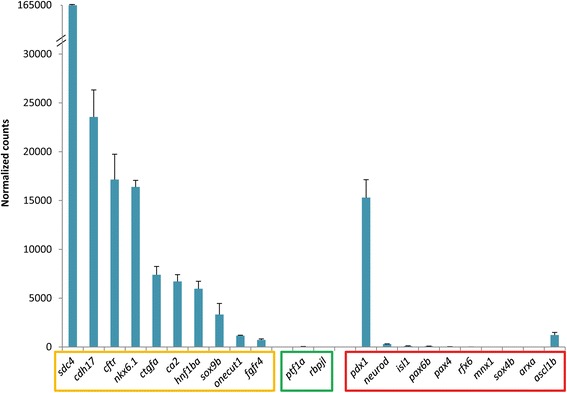
Expression of known pancreatic markers in the transcriptome of adult nkx6.1:eGFP cells. Expression values are expressed as normalized counts. High expression of known ductal genes (orange box) is detected while acinar (green box) and endocrine (red box) markers display low abundance, with the exception of pdx1 and ascl1b (see text)
Strikingly, the only endocrine transcription factor that displayed substantial expression (>1000 counts) and enrichment in the ductal transcriptome was the pro-endocrine gene ascl1b (Fig. 10). As ascl1b specifically marks the endocrine precursors during pancreas development, these data support our observation that, within the nkx6.1:eGFP+ cell domain, some cells activate a pro-endocrine differentiation program in normal adult zebrafish.
Discussion
In this study, we determined during development the fate of two pancreatic cell populations marked by nkx6.1 and ascl1b and found that nkx6.1+ cells are bona fide multipotent pancreatic progenitors while ascl1b+ cells represent committed endocrine precursors. We found also that nkx6.1 is maintained in adult zebrafish in ducts and CAC/terminal end duct cells, which we show still have the potential to give rise to endocrine cells in normal (non-diabetic) animals. The progenitor potency of adult nkx6.1+ cells is also reflected in their transcriptome through the expression of several pancreatic progenitor markers and by their capacity to generate new beta cells after beta cell ablation.
Although ascl1b marks the endocrine precursor cells, as opposed to nkx6.1, which is the first multipotent pancreatic progenitor marker known to date, it is surprising to note that this transcription factor begins to be expressed in the prospective pancreatic region at 10 hpf [15], i.e. more than 1 hour before the appearance of nkx6.1 (11.5 hpf) [18]. Pdx1, known in the mouse to be also expressed in the multipotent pancreatic progenitors, appears even later (14 hpf [6]). This brings the interesting concept that the first cells in the pancreatic anlagen acquire an endocrine identity before acquiring a pancreatic identity, suggesting that the mechanisms controlling pancreatic and endocrine identity are not necessary linked and can act in parallel. This situation seems to be restricted to the dorsal bud as, later, nkx6.1 is first expressed during the formation of the ventral bud and ascl1b is then detected in endocrine committed cells. This peculiar situation could be related to the different lineage potential of the dorsal versus ventral bud cells. Indeed the dorsal bud gives rise exclusively to endocrine cells while the ventral bud is able to give rise to all pancreatic cell types [4, 40].
After a transient overlapping expression in the dorsal pancreatic anlagen, nkx6.1- and ascl1b-expressing domains segregate progressively. Cross-repressive interactions between lineage-determining transcription factors have been proposed as a molecular mechanism for establishing lineage allocation in several tissues [41–43]. We show that such a mechanism does not occur here, as we could not observe any cross-repression between nkx6.1 and ascl1b in the dorsal bud when analyzing both mutants. The segregation can instead be explained by the opposite effects of the Notch signaling pathway on nkx6.1 and ascl1b expression. By loss- and gain-of-function experiments, we definitely prove that ascl1b is repressed, while nkx6.1 is maintained, by Notch signaling in both the dorsal and ventral buds. The importance of Notch signaling in regulating nkx6.1 expression has been also shown in mice where disruption of Notch signaling results in the loss of the pro-trunk determinant Nkx6.1 and the acquisition of pro-acinar identity [44]. Direct binding of RBPJ-K to the Nkx6.1 promoter supports a direct role of Notch signaling in the expression of Nkx6.1. In contrast, the initiation of nkx6.1 expression is independent of Notch signaling in both zebrafish pancreatic buds. This is in accordance with our findings that Nkx6.1+ cells progressively become Notch-responsive PNCs being found only in a subpopulation of the Nkx6.1+ cells at the beginning of the formation of both the dorsal and ventral buds. This could explain why the PNCs, found in a subdomain of the ventral bud at an early stage, can differentiate only into ductal and endocrine cells, while nkx6.1-expressing cells give rise to all pancreatic cell types. As nkx6.1 and ptf1a are initially co-expressed in the ventral bud primordium ([20] and our unpublished data), we can hypothesize that the nkx6.1+/ptf1a+/Notch on cells give rise to the ductal and late endocrine cells while the nkx6.1+/ptf1a+/Notch off cells will give rise to the acinar cells. This model appears to contradict the data of Wang et al. [7] showing that the ventral bud primordium consists of two non-overlapping cell populations: a ptf1-expressing domain and a Notch-responsive progenitor core. It is possible that this discrepancy is due to the tools used to label the Notch-responsive cells: in our study, we used the Tp1bglob:VenusPest transgenic line allowing the detection of cells with current Notch activity, while Wang and collaborators used the Tp1bglob:hgmb1-mCherry line, which could show a delay in mCherry detection.
In adult zebrafish, nkx6.1 expression persists in the pancreas where it is specifically restricted to the ducts. This situation is different in the mouse as Nkx6.1 persists in beta cells but not in adult ducts. In zebrafish, it is nkx6.2, an nkx6.1 homolog with functional equivalence [18, 45], which is expressed specifically in beta cells [18], suggesting that the Nkx6 function in beta cells is fulfilled by Nkx6.2 in zebrafish. Persistence of nkx6.1 expression in the zebrafish ducts is also associated with persistence of another pancreatic progenitor marker, pdx1, whose expression is also restricted to beta cells in mammals. Combined with other hallmarks of embryonic pancreatic progenitors such as sox9b, hnf1ba, and Notch signaling components, the expression of genes nkx6.1 and pdx1 suggests that at least some ductal cells behave as pancreatic progenitors in adult zebrafish. Moreover, detection of the endocrine precursor marker ascl1b in the adult nkx6.1:eGFP+ ductal cell transcriptome is consistent with some ductal cells initiating an endocrine differentiation program and transiently expressing ascl1b, even in physiological conditions. The progenitor potential of ductal cells is fully revealed in the setting of beta cell regeneration where nkx6.1:eGFP+ cells show increased proliferation as well as the ability to differentiate into new beta cells. The adult pancreatic progenitors contributing to beta cell regeneration could be the CACs, as supported by our observation that they are able to replicate and to generate other ductal cells as well as endocrine beta cells. On the other hand, we cannot exclude the possibility that other ductal cells also contribute to regeneration based on Tg(nkx6.1:eGFP), which labels more broadly the ducts. Nevertheless, although the tool we used here to monitor lineage tracing presents some limitations (based on the persistence of GFP in assessing the short-term lineage tracing of nkx6.1-expressing cells), during the revision of our manuscript, a study by Delaspre et al. [37] established through CRE-based genetic labeling that Notch-responsive cells give rise to regenerated beta cells in adult zebrafish. Our data are in full accordance with their findings, and support the conclusion that ductal cells, possibly CACs, possess regenerative capacity. To determine whether CACs only or other ductal cells contribute to beta cell regeneration will require other genetic lineage tracings with markers expressed only in ductular structures and not in CACs.
In contrast to mammals, the adult zebrafish has the remarkable capacity to regenerate its beta cells rapidly and spontaneously following their selective destruction [2]. Our findings showing that ductal cells, such as CACs, behave as pancreatic progenitors/stem cells in normal (non-diabetic) adult animals and during regeneration strongly suggest that they could also constitute a source of regenerated beta cells in diabetic mammalian models. In the mouse, adult murine CACs display endocrine and exocrine progenitor potential in vitro with self-renewing ability [46], but evidences of their potential in vivo as progenitors of endocrine cells are missing. Indeed, CRE-based cell tracing of Hes1+ terminal duct cells/CACs in adult mice failed to show any islet progenitor capacity while these cells seemed to contribute to the ductal tree [47]. One explanation for this difference would be that while zebrafish CACs present pancreatic progenitor activity, mammalian CACs have retained a very limited capacity in vivo, which could not be evidenced in the mouse model of beta cell regeneration used. Another explanation would be that CACs form a heterogeneous cell population. This latter hypothesis could be verified by the analysis of the expression of different ductal markers identified in our transcriptome, which could help determine the existence of different ductal cell subpopulations.
A perspective of our work would be to examine thoroughly in zebrafish ductal cells in physiological conditions and during beta regeneration to identify mechanisms that could then be harnessed to promote beta cell regeneration in mammals. A first clue is provided by the fact that Nkx6.1 and Pdx1 are normally not expressed in mammalian duct cells, in contrast to zebrafish. It is therefore tempting to hypothesize that inducing the expression of these two factors in the pancreatic ducts in the adult mouse or human could enhance their progenitor potential. Consistent with this hypothesis, driving ectopic Pdx1 expression is already harnessed for transdifferentiating different cells into beta cells in vivo (liver cells, pancreatic acinar cells, …) [48–51] and for transdifferentiating pancreatic duct cells into beta cells in vitro [52]. Interestingly, the ability of Pdx1 to reprogram liver cells into beta cells is substantially increased by the combined action of Nkx6.1 [53], which potentiates induction of the early pancreatic master genes Neurog3 and Isl1.
In the zebrafish larvae, two sources have been described for the regeneration of beta cells: progenitor cells in the developing ducts [35] and alpha cells [54]. From our data in adults, we cannot rule out the contribution by other cell types than ductal cells. Indeed, in addition to increased proliferation of ductal cells, some islet cells show also increased proliferation after beta cell destruction ([2] and our data not shown) suggesting the involvement of other pancreatic, and possibly endocrine, cell types in regeneration. These possibilities remain to be validated in the adult zebrafish.
Our transcriptomic characterization of nkx6.1+ cells in normal non-regenerating adult zebrafish identified many of the ductal markers known throughout species, showing that many of these genes and their expression are conserved between zebrafish and mammals, as for example, different markers of Notch signaling [55–57], and sdc4 and fgfr4. These latter two genes have been previously proposed as ductal markers in the mouse embryo [39]. Additional markers were also identified here, notably id2a. Id2 has been shown to be expressed in ductal epithelial cells of IFNγNOD mice, a model of a regenerating pancreas harboring hyperplasic ducts, and to be involved in their expansion [58]. We demonstrate here its expression in ducts in larvae and in healthy adult pancreas, which may correlate with a role in development and in ductal constitutive homeostasis. Our findings also reveal that, besides the Notch pathway, ductal cells specifically express various components of Wnt signaling pathways. The expression of the fzd7a receptor, of two secreted Wnt antagonists, sfrp5 and frzb/sfrp3, and of the agonist wnt7bb suggests a complex control of the activity of the Wnt pathway(s) in adult pancreatic ducts. Whether and how these different factors orchestrate pancreatic duct development, homeostasis, and function remains to be determined. Numerous cross-talks between the Notch and the Wnt/beta-catenin pathways occur during development, tissue homeostasis, and disease, notably by regulating the balance of stem cells and differentiated cells (reviewed in [59]). During pancreas development, beta-catenin controls the patterning of multipotent versus bipotent embryonic pancreatic progenitors in the mouse, in part, by inhibiting Notch signaling [60]. Our findings raise the question whether similar interactions shape the fate decision of progenitors/stem cells in the adult pancreas.
Conclusions
We have developed transgenic tools enabling the characterization of nkx6.1+ and ascl1b+ progenitor cell populations and showed that, in the zebrafish embryo, nkx6.1+ cells are multipotent pancreatic progenitors, while ascl1b+ cells represent committed endocrine precursors. In adult zebrafish, nkx6.1 expression persists exclusively in the ductal tree, notably in CACs. Transcriptomic profiling of adult nkx6.1+ ductal cells reveals hallmarks of embryonic pancreatic progenitors and identifies novel ductal markers. Our data also strongly suggest that adult zebrafish ductal cells, possibly CACs, possess regenerative capacity. Further characterization of ductal cells in this animal model should bring new insight into regeneration in mammals and open up new perspectives for anti-diabetic therapies.
Methods
Zebrafish maintenance, mutant and transgenic lines, and LY411575 treatment
Zebrafish (Danio rerio) were raised and cared for according to standard protocols [61]. All animal work has been conducted according to national guidelines and all animal experiments described herein were approved by the ethical committee of the University of Liège (protocol numbers 371, 1285, and 1662). Wild-type embryos from the AB strain were used and staged according to Kimmel [62]. Homozygous mind bomb mutants were obtained by mating heterozygous fish for the (mibta52b) allele [63]. The following transgenic lines were used: Tg(hsp70l:Gal4)1.5kca4 abbreviated Tg(hsp:Gal4) and Tg(UAS:myc-Notch1a-intra)kca3 abbreviated Tg(UAS:NICD) [33], Tg(Tp1bglob:eGFP)um14 abbreviated Tg(Tp1:eGFP) [12], Tg(TP1bglob:VenusPest)S940 abbreviated Tg(TP1:VenusPest) [16], Tg(Tp1bglob:H2BmCherry)S939 abbreviated Tg(Tp1:H2BmCherry) [16], Tg(ubi:loxP-EGFP-loxP-mCherry) abbreviated ubi:Switch [28], Tg(ubi:loxP-AmCyan-loxP-ZsYellow) abbreviated ubi:CSY [27], Tg(ins:NTR-mCherry) [1], and Tg(nkx6.1:eGFP); Tg(ins:NTR-mCherry).
The LY411575 treatment was performed by incubating the embryos during the indicated period with a 10-μm LY411575 solution (Medchemexpress), replaced every day.
Generation of BAC transgenic lines
The polymerase chain reaction (PCR) primers used to generate the constructs are listed in Additional file 10: Table S2. The BAC:nkx6.1 (Imagenes, DKEY-173 K2) DNA was introduced by electroporation into SW102 E. coli (derived from DY380) [64]. These bacteria contain the lambda prophage recombineering system and a galactose operon where the galactokinase gene (galK) has been deleted. The eGFP gene was inserted into exon 1 of nkx6.1, replacing the beginning of the nkx6.1 open reading frame (amino acids (aa) 1 to 149) using a two-step positive and negative galk selection [25, 64, 65]. During the first step, the cassette containing the galk gene was amplified by PCR with the pair of primers O180F and O253R, containing at the 5′ end 50 bases identical to the nkx6.1 sequence to allow homologous recombination and electroporated into the bacteria SW102 containing BAC:nkx6.1. Only recombinant bacteria are able to grow on minimal medium containing galactose as carbon source. During the second step, the galK gene was replaced by the eGFP gene. The eGFP cassette was amplified by PCR with the primers O186F and O256R containing at the 5′ end the same 50 bases identical to the nkx6.1 sequence to allow homologous recombination and the eGFP sequences to anneal to the eGFP cassette. After electroporation, the bacteria were plated on minimal medium containing two-deoxy-D-galactose (DOG), a galactose analogue that after phosphorylation by GalK, becomes toxic. Only bacteria that have lost the galk gene survived on DOG-containing medium. To facilitate the insertion of the BAC in the genome of zebrafish, the iTol2 cassette was also inserted into the backbone of BAC:nkx6.1-eGFP [24, 25, 65]. The iTol2 cassette was amplified by PCR with the pair of primers O215F and O216R. The final construct (BAC:nkx6.1-eGFP) was purified with Nucleobond® BAC100 (Macherey-Nagel) and injected into the cytoplasm of one-cell-stage zebrafish embryos together with the mRNA for the transposase. The embryos and larvae were screened for GFP expression and the fluorescent injected fish were raised to adulthood and the offspring were screened for fluorescence. The transgenic line obtained was abbreviated to Tg(nkx6.1:eGFP) in the article.
To generate the (ascl1b:eGFP-2A-creERT2) transgenic line, we used BAC:ascl1b (Imagenes, DKEY-265 N18) spanning from 61 kb upstream and 89 kb downstream of the ascl1b gene. The GFP-2A-creERT2 cassette was inserted into exon 1 of ascl1b, replacing the beginning of the ascl1b open reading frame (aa 1 to 163) using the same two-step positive and negative galk selection as described above. For the first step, the cassette containing the galk gene was obtained by PCR using the primers O275F and O276R and for the second step, the GFP-2A-creERT2 cassette was amplified using the primers O277F and O278R.
4OHT treatment for creERT2 induction
4-Hydroxytamoxifen (4OHT, Sigma H7904) was dissolved in DMSO as a stock solution of 10 mM and kept in single-use aliquots in the dark at –70 °C. A working concentration of 10 μM 4OHT was demonstrated to lead optimally to Cre-mediated recombination without causing deleterious development defects. Embryos were treated five times from 11 to 15 hpf in E3 containing 10 μM 4OHT and kept in the dark at 28 °C. After the treatments, the embryos were washed in fresh E3 and fixed at 48 hpf or 72 hpf.
CRISPR/cas9 genome mutagenesis
The nkx6.1 and ascl1b mutant lines were generated by CRISPR/Cas9 technology essentially as described previously [31, 66]. The targeted sites were selected using the ZiFiT software package [67] in the first exon of nkx6.1 (CCAAACCCCTGACAGAGCTTC) before the homeodomain coding region and in the first exon of ascl1b (GGAGACGCTGCGCTCCGCCGTGG) corresponding to the helix-loop-helix coding domain. The selected oligonucleotides were inserted into the plasmid DR274 (Addgene) and the gRNA (guide RNA) was synthesized by in vitro transcription using T7 RNA polymerase. Fertilized zebrafish eggs were injected with about 1 nl of a solution containing 50 ng of gRNA and 300 ng of nls-zCas9-nls mRNA obtained by transcription of the plasmid pT3TS-nCas9n (Addgene). The efficiency of mutagenesis was verified by genotyping using Heteroduplex Migration Assays [68] after amplification of targeted genomic sequences. Injected embryos were raised until adulthood and crossed with wild-type fish to generate heterozygote mutant F1 fish. Fish harboring frame-shift mutations were kept and used to raise F2 mutant lines, i.e. the ascl1bulg-M2C and nkx6.1ulg-M5 lines carrying, respectively, 11 and seven nucleotide deletions.
Whole mount in situ hybridization, whole mount immunohistochemistry, and immunohistochemistry on paraffin sections
Double fluorescent and visible WISH were performed as previously described [69, 70] with the following probes: ascl1b [15, 71], nkx6.1 [18, 72], somatostatin 2 (sst2) [73], and try [6].
Immunohistochemistry (IHC) on whole-mount embryos was performed as described [15]. For IHC on paraffin sections with adult tissues, adult fish between 6 and 9 months old were fixed in 4 % PFA (paraformaldehyde) overnight at 4° after euthanasia and opening of the abdominal skin. The digestive tract was then dissected and embedded in paraffin following standard procedures. Then, 5-μm sections were collected through the head of the pancreas at the level of the main pancreatic islet. Immunodetection was performed after standard antigen retrieval.
The antibodies used were: polyclonal rabbit anti-mCherry/dsRed (Living Colors DsRed Polyclonal Antibody, Clontech) 1:500, polyclonal rabbit anti-ZsYellow (Living Colors anti-RCFP polyclonal pan from Clontech) 1:300, chicken anti-GFP (Aves lab) 1:1000, mouse monoclonal anti-Nkx6.1 (clone F55A10) 1:20, mouse monoclonal anti-Isl1 (Hybridoma bank) 1:50, guinea pig anti-insulin (Dako) 1:500, mouse anti-glucagon (Sigma) 1:300, polyclonal rabbit anti-somatostatin (MP Biomedicals) 1:300, mouse anti-PCNA (Sigma) 1:1000, mouse 2F11 mAb (Abcam) 1:1000, and Alexa Fluor secondary antibodies (Invitrogen). Venus was detected with anti-GFP. Finally, 4',6-diamidino-2-phenylindole (DAPI) was used as nuclear staining.
Images were acquired with a Leica SP2 or SP5 confocal microscope and processed with Imaris 7.2.3 and Photoshop CS5. To count the Tp1:H2BmCherry+ cells expressing insulin, the total number of pancreatic mCherry+/Venus– and mCherry+/Ins+ cells was calculated for six sections every 15 μm for four fish.
EdU injection and detection in adult zebrafish
A 12.5-mM EdU solution in DPBS (Dulbecco's Phosphate Buffer Saline) containing 0.25 % DMSO was injected intraperitoneally in 6–9-month-old fish at 100 μg/g body weight after anesthesia in tricaine methane sulfonate. Fish were then sacrificed and fixed in 4 % PFA. EdU (Click-iT® Labeling Technologies, Life Technologies) incorporation was detected on paraffin sections with Alexa Fluor555 before proceeding to IHC detection.
To count EdU+ cells, the total number of pancreatic Venus+ CAC and Venus+/EdU+ cells was calculated in three to six sections every 15 μm for four fish.
Induction of beta cell ablation in adult zebrafish
Adult Tg(nkx6.1:eGFP); Tg(ins:NTR-mCherry) zebrafish between 6 and 9 months old were treated in fish water containing 10 mM MTZ (three to four fish per 500 ml, Sigma 3761) for 20 hours at 28 °C. Then the water was replaced twice before re-integration into the system.
Fish were anesthetized with tricaine and their glycemia was measured using the Accu-Chek Aviva glucometer system (Roche Diagnostics) with blood collected at the level of the tail. To minimize variations, the fish were fasted for 24 hours before measurement. After decapitation, the whole fish were fixed in 4 % PFA overnight at 4 °C. The digestive tract was then dissected prior to paraffin embedding.
FACS purification of ductal cells
The pancreas of three to five Tg(nkx6.1:eGFP) adult fish (6–9 months old) were dissected and collected in HBSS with calcium. Dissociation was performed in HBSS (Hank's Balanced Salt Solution) with Ca2+/Mg2+ supplemented with 1 mg/ml collagenase IV (Life Technologies 17104-019) and collagenase P (Roche 1121386501) and 1.5 mg/ml dispase II (Life Technologies 17105-041) for 20 min at 30 °C. After several washes in HBSS without Ca2+/Mg2+, a single-cell suspension was obtained by Tryple Select 1× incubation for 10 min. Dissociation was stopped by HBSS without Ca2+/Mg2+ containing 1 % BSA (bovine serum albumin) and 2 mM EDTA (ethylenediaminetetraacetic acid). GFP+ cells were isolated on FACS Aria II under purity mode and the purity of the sorted cells was confirmed for a small fraction with an epifluorescence microscope (~95 %). Cells were immediately lyzed in 3.5 μl of reaction buffer (SMARTer Ultra Low RNA kit for Illumina sequencing, Clontech) and stored at –80 °C. Three independent replicates were generated from 6–9-month-old fish.
cDNA synthesis and library preparation
cDNA synthesis was performed using the SMARTer Ultra Low RNA kit for Illumina sequencing (Clontech) according to the manufacturer’s recommendations. Then 3000–5000 sorted GFP+ cells were directly lyzed in 3.5 μl of reaction buffer and immediately frozen at –80 °C. cDNAs were synthesized, purified with Ampure XP beads and then amplified with 13 PCR cycles with Advantage 2 Polymerase Mix (50×, Clontech). The PCR products were purified on SPRI AMPure XP beads (Beckman Coulter), and the size distribution was checked on a high-sensitivity DNA chip (Agilent Bioanalyzer). cDNA libraries were prepared with TruSeq Nano DNA kit or Nextera XT DNA (Illumina). For TruSeq Nano libraries, 20–30 ng cDNA was sheared by sonication (parameters adjusted to obtain fragments from 350 to 450 bp). For Nextera libraries, 1 ng was fragmented by tagmentation. Then cDNA libraries were prepared according to the manufacturer’s recommendations. Samples were sequenced on an Illumina HiSeq 2000 at an average of 72.3 million 100-bp paired-end reads. RNA-seq data have been deposited in the European Nucleotide Archive from EMBL-EBI [38].
Data analysis of the duct transcriptome
Before mapping, the first 30 bases of each read were trimmed to remove the adapters incorporated by the cDNA synthesis process. Trimmed reads were mapped to the zebrafish genome (Zv9, Ensembl genes version 75, ensembl.org) using the Tophat v.2.0.9 software [74]. For the three replicates, the total number of reads for Duct 1 was 40,336,250 (79.9 % mapped), for Duct 2 was 86,308,531 (82.5 % mapped), and for Duct 3 was 91,980,011 (67.6 % mapped).
HT-Seq count was used to estimate the expression level by counting how many reads align to each gene of the annotation (gene set, Ensembl.org) [75]. The expression of 15,888 genes was detected with at least one read in all three replicates.
To describe the set of genes enriched or preferentially expressed in ductal tissue, the ductal transcriptome was compared with acinar and endocrine transcriptomes prepared following the same methodology (composed of alpha, beta, and delta cells, not shown here, manuscript in preparation). The R package EBSeq was used to call differential expressed genes [76]. Ductal genes were identified based on their posterior probability (adjusted by false discovery rate) of being differentially expressed from the other two cell types. It was found that 3,684 genes were preferentially expressed in the duct transcriptome. Stricter thresholds were applied to identify genes with highly specific ductal expression (see “Results”).
Acknowledgments
We are grateful to Christian Mosiman for advice on tamoxifen treatment and Marielle Lebrun and Alain Vanderplasschen for advice on galk recombineering. We thank Koichi Kawakami for the itol2 cassette [25]. We are very grateful to Didier Stainier and Nicolay Ninov for the transgenic lines Tg(Tp1:VenusPest) and Tg(Tp1:H2BmCherry) [16], Francesco Argenton for Tg(Tp1glob:eGFP) [12], Carole Wilson (UCL fish facility, UK) for Tg(ubi:Switch) [28], Geoffrey Burns for Tg(ubi:loxP-AmCyan-loxP-ZsYellow), and Jean-Claude TWIZERE for Tg(hsp70:Gal4) and Tg(UAS:myc-notch1a-intra) [33]. We thank the following technical platforms: GIGA-Zebrafish (M Winandy and H Pendeville), GIGA-Cell Imaging and Flow Cytometry platform (S Ormenese and S Raafat), GIGA-Genotranscriptomic (B Hennuy, W Coppieters, and L Karim), and GIGA-Immunohistochemistry (C Humblet and E Dortu).
APG and DB were supported by FRIA (Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture) and the Léon Fredericq fund, ETS by WBI (Wallonie-Bruxelles International) and the Léon Fredericq fund, and VVB by the Belgian State’s Interuniversity Attraction Poles Program (SSTC, PAI). BP, IM, and MLV are Chercheur qualifié FNRS (Fonds National pour la Recherche Scientifique). This work was funded by the FNRS-FRS, the Belgian State’s Interuniversity Attraction Poles Program (SSTC, PAI), and the Fonds Speciaux from the ULg (University of Liège).
Abbreviations
- 4OHT
4-hydroxytamoxifen
- aa
amino acid
- ARP
atonal related protein
- ASCL
achaete-scute like
- BAC
bacterial artificial chromosome
- bHLH
basic helix-loop-helix
- bp
base pair
- CAC
centroacinar/terminal end duct cell
- creERT2
4OHT inducible Cre-recombinase
- DMSO
dimethyl sulfoxide
- dpf
days post fertilization
- DOG
two-deoxy-D-galactose
- dpt
days post treatment
- EdU
5-ethynyl-2′-deoxyuridine
- eGFP
enhanced green fluorescent protein
- EPD
extra-pancreatic duct
- FACS
fluorescence-activated cell sorting
- Gcg
glucagon
- GFP
green fluorescent protein
- hpf
hours post fertilization
- IHC
immunohistochemistry
- Ins
insulin
- IPD
intra-pancreatic duct
- kb
kilobyte
- MTZ
metronidazole
- NICD
Notch intracellular domain
- NTR
nitroreductase enzyme
- PCNA
proliferating cell nuclear antigen
- PCR
polymerase chain reaction
- PNC
pancreatic Notch-responsive cell
- rec
recombinant
- Sst
somatostatin
- VenusPest
Venus fluorescent protein
- WISH
whole-mount in situ hybridization
- WT
wild type
Additional files
The bacterial artificial chromosome (BAC) reporter line Tg(nkx6.1:eGFP) mirrors the expression of the endogenous nkx6.1 gene. A Schematic representation of the −55 to +95 kb nkx6.1:eGFP BAC transgene. This BAC includes sequence blocks that are highly conserved amongst vertebrates (black boxes) located from 11 to 77 kb downstream of the nkx6.1 gene. By BAC recombineering using galK selection, the eGFP cassette (green box) was introduced into exon 1, replacing the beginning of the nkx6.1 open reading frame (aa 1 to 149). B Epifluorescence microscopy images of the immunodetection of endogenous Nkx6.1 (red) and GFP (green) in Tg(nkx6.1:eGFP) embryos. Lateral views of 48 hpf embryos with the anterior part to the left. C Confocal projection images of whole mount fluorescent in situ hybridization on a Tg(nkx6.1:eGFP) embryo showing that the gfp transcripts are not expressed in the insulin+ cells at 20 hpf. D Confocal projection images of whole mount fluorescent in situ hybridization on a Tg(nkx6.1:eGFP) embryo showing that the gfp transcripts are not expressed in the isl1+ cells at 30 hpf. Scale bar = 15 μm. P pancreas. (PNG 2609 kb)
The bacterial artificial chromosome (BAC) reporter line Tg (ascl1b:eGFP-creER T2 ) mirrors the expression of the endogenous ascl1b gene. A Schematic representation of the −61 to +89 kb ascl1b:eGFP-2A-creER T2 BAC transgene. By BAC recombineering using galK selection, the eGFP-2A-creER T2 cassette is introduced into exon 1, replacing the beginning of the ascl1b open reading frame (aa 1 to 163). B Epifluorescence microscopy images of the immunodetection of GFP in 17-hpf Tg(ascl1b:eGFP-creER T2 ) embryos. C, D Visible WISH showing expression of endogenous ascl1b in a wild-type (WT) embryo (C) and of GFP in Tg(ascl1b:eGFP-creER T2 ) embryos (D) at 15 hpf. Lateral views with the anterior part to the left. P pancreas. (PNG 1068 kb)
nkx6.1 expression is not repressed by Ascl1b. A Schematic representation of wild-type (WT) and mutant Ascl1bulg-M2C proteins. The basic domain (b) (+70/+78) is represented by a blue box and the helix loop helix (HLH) domain (+79/+123) by a green box. The coding region of the mutant Ascl1bulg-M2C protein contains an 11-bp deletion after the aa 107 codon, leading to a frameshift and the production of an aberrant region of 48 aa instead of the second helix, known to be essential for the function of the bHLH proteins. A' Table showing part of the nucleotide and protein sequence of the ascl1b gene and of the mutated form in the Ascl1bulg-M2C mutant. B, C WISH showing the drastic reduction of somatostatin expression in the ascl1b-/- mutant compared to the WT embryo at 30 hpf. This phenotype is identical to the one of embryos injected with a translation-blocking morpholino, targeting the translation start site of ascl1b mRNA [15, 77, 78], suggesting that the mutant Ascl1bulg-M2C is effectively a null mutant. D, E Confocal projection images of Nkx6.1 immunodetection showing equivalent number of nkx6.1+ cells in WT and ascl1b -/- mutants at 16 hpf. All views are ventral with the anterior part to the left. Scale bars = 40 μm. (PNG 1321 kb)
ascl1b expression is not repressed by Nkx6.1. A Schematic representation of wild-type (WT) and mutant Nkx6.1ulg-M5 proteins with the yellow box representing the homeodomain and the red box representing the NK domain. The coding region of the mutant Nkx6.1ulg-M5 protein contains a 7-bp deletion after the aa 141 codon, leading to the apparition of a STOP codon just after the deletion. The black line represents the region of the protein recognized by the Nkx6.1 antibody. A' Table showing part of the nucleotide and protein sequence of the nkx6.1 gene and of the mutated form in the Nkx6.1ulg-M5 mutant. B, C Nkx6.1 immunodetection showing the loss of the full-length Nkx6.1 protein in the nkx6.1 -/- mutant. D, E Glucagon immunodetection showing a drastic reduction of the number of glucagon-expressing cells in the nkx6.1 -/- mutant compared to the WT embryo at 30 hpf. This phenotype is the same as the one of embryos injected with the MO1 translation-blocking morpholino, targeting the translation start site of nkx6.1 mRNA [18], shown to prevent nkx6.1 expression efficiently in the neural tube [72]. This strongly suggests that the mutant Nkx6.1ulg-M5 is a null mutant. F, G Confocal projection images of fluorescent WISH showing equivalent number of ascl1b+ cells in WT and nkx6.1 -/- mutants at 15 hpf. All views are ventral with the anterior part to the left. Scale bars = 40 μM. (PNG 1527 kb)
The initiation of nkx6.1 expression is independent of Notch signaling. A–D Fluorescent WISH showing a drastic increase in the pancreatic dorsal bud of ascl1b expression in the mind bomb (mib -/- ) mutants while nkx6.1 expression is not affected at 13 hpf. E, F Fluorescent WISH showing that the pancreatic expression of nkx6.1 in the ventral bud is not perturbed at 34 hpf in mib -/- mutants. The ventral (A, B, E, F) or lateral (C, D) views represent confocal projection images with the anterior to the left. Scale bars = 40 μM. NS nervous system. (PNG 968 kb)
All Notch-responsive cells in the adult pancreas are CACs. Notch active cells in Tg(Tp1:VenusPest) adult fish were labeled with anti-GFP to visualize VenusPest (green) and the ductal marker 2F11 (red). VenusPest labeling is detected throughout the exocrine tissue (see asterisks highlighting examples) and where it highlights the CACs together with 2F11. In contrast, ductal structures with intense 2F11 staining do not exhibit any Venus+ (Notch on) cells. Note also the presence of 2F11 in endocrine islet cells as previously reported [79]. (PNG 2854 kb)
Notch-responsive terminal end duct cells/CACs give rise to other ductal cells. Immunodetection of H2BmCherry and ductal markers in the pancreas of adult Tg(Tp1:VenusPest); Tg(Tp1:H2BmCherry) zebrafish. A–C H2BmCherry+ cells (red) in ductular structures labeled with the ductal (ducts and CACs) marker 2F11 (green). A, B separate channels of Fig. 8b–c. B, C Weak H2BmCherry labeling is present near the extremity of a terminal (or intercalated) duct (yellow arrows). The asterisks identify a CAC at the tip of the duct (strong H2BmCherry labeling). D Comparison of H2BmCherry (red) with endogenous Nkx6.1 (green) showing ductal Nkx6.1+ cells co-expressing H2BmCherry (yellow arrows). (PNG 4967 kb)
Duct-specific genes listed by decreased expression level. We found 293 genes corresponding to the following “specificity” criteria: gene expression level normalized counts >1000 in duct and <1000 in non-ductal (averaged endocrine and acinar) transcriptome and ≥16 fold enrichment in ducts versus non-duct (averaged endocrine and acinar), expressed in log2 ratio (≥4). Ratio is mean of normalized counts in duct transcriptomes/mean of normalized counts in endocrine and in acinar transcriptomes. Known ductal markers and genes involved in the Notch pathway are highlighted in red. Novel markers and genes involved in the Wnt pathway are highlighted in green. (DOCX 26 kb)
Ductal expression of id2a during zebrafish development. Whole mount fluorescent in situ hybridization showing id2a (green) and the ductal marker sox9b (red) in a 3-dpf larva. The expression of both genes overlaps as highlighted by yellow labeling. In addition to expression in the intrapancreatic ducts (IPD), id2a is also expressed in the intrahepatic (IHD). HPD hepatopancreatic ducts. (PNG 537 kb)
List of the primers used in this study. The 50 nucleotides added to allow homologous recombination are indicated in capital letters. (DOCX 12 kb)
Footnotes
Aurélie P. Ghaye David Bergemann Marianne L. Voz and Isabelle Manfroid contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
APG and DB participated in the design of the study, generated the data and participated in their interpretation, and in the drafting of the manuscript. ETS carried out the RNA-seq analyses. LCF initiated the study on ascl1b (design and experiments). VVB participated in the generation of experimental data. BP participated in the design of the study and in data interpretation, and helped draft the manuscript. MLV and IM conceived the study, participated in the interpretation of the data, and wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Aurélie P. Ghaye, Email: aurelieghaye@outlook.be
David Bergemann, Email: david.bergemann@student.ulg.ac.be.
Estefania Tarifeño-Saldivia, Email: etarifeno@student.ulg.ac.be.
Lydie C. Flasse, Email: lydie.flasse@sund.ku.dk
Virginie Von Berg, Email: vonbergvirg@hotmail.com.
Bernard Peers, Email: bpeers@ulg.ac.be.
Marianne L. Voz, Email: mvoz@ulg.ac.be
Isabelle Manfroid, Email: isabelle.manfroid@ulg.ac.be.
References
- 1.Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124:218–29. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moss JB, Koustubhan P, Greenman M, Parsons MJ, Walter I, Moss LG. Regeneration of the pancreas in adult zebrafish. Diabetes. 2009;58:1844–51. doi: 10.2337/db08-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–35. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 4.Field HA, Dong PD, Beis D, Stainier DY. Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev Biol. 2003;261:197–208. doi: 10.1016/S0012-1606(03)00308-7. [DOI] [PubMed] [Google Scholar]
- 5.Kumar M, Melton D. Pancreas specification: a budding question. Curr Opin Genet Dev. 2003;13:401–7. doi: 10.1016/S0959-437X(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 6.Biemar F, Argenton F, Schmidtke R, Epperlein S, Peers B, Driever W. Pancreas development in zebrafish: early dispersed appearance of endocrine hormone expressing cells and their convergence to form the definitive islet. Dev Biol. 2001;230:189–203. doi: 10.1006/dbio.2000.0103. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Rovira M, Yusuff S, Parsons MJ. Genetic inducible fate mapping in larval zebrafish reveals origins of adult insulin-producing beta-cells. Development. 2011;138:609–17. doi: 10.1242/dev.059097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hesselson D, Anderson RM, Beinat M, Stainier DY. Distinct populations of quiescent and proliferative pancreatic beta-cells identified by HOTcre mediated labeling. Proc Natl Acad Sci U S A. 2009;106:14896–901. doi: 10.1073/pnas.0906348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimmel RA, Onder L, Wilfinger A, Ellertsdottir E, Meyer D. Requirement for Pdx1 in specification of latent endocrine progenitors in zebrafish. BMC Biol. 2011;9:75. doi: 10.1186/1741-7007-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung WS, Andersson O, Row R, Kimelman D, Stainier DY. Suppression of Alk8-mediated Bmp signaling cell-autonomously induces pancreatic beta-cells in zebrafish. Proc Natl Acad Sci U S A. 2010;107:1142–7. doi: 10.1073/pnas.0910205107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong PD, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, et al. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat Genet. 2007;39:397–402. doi: 10.1038/ng1961. [DOI] [PubMed] [Google Scholar]
- 12.Parsons MJ, Pisharath H, Yusuff S, Moore JC, Siekmann AF, Lawson N, et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech Dev. 2009;126:898–912. doi: 10.1016/j.mod.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Afelik S, Jensen J. Notch signaling in the pancreas: patterning and cell fate specification. Wiley Interdiscip Rev Dev Biol. 2013;2:531–44. doi: 10.1002/wdev.99. [DOI] [PubMed] [Google Scholar]
- 14.Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, et al. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–81. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 15.Flasse LC, Pirson JL, Stern DG, Von Berg V, Manfroid I, Peers B, et al. Ascl1b and Neurod1, instead of Neurog3, control pancreatic endocrine cell fate in zebrafish. BMC Biol. 2013;11:78. doi: 10.1186/1741-7007-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ninov N, Borius M, Stainier DY. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139:1557–67. doi: 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007;13:103–14. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Binot AC, Manfroid I, Flasse L, Winandy M, Motte P, Martial JA, et al. Nkx6.1 and nkx6.2 regulate alpha- and beta-cell formation in zebrafish by acting on pancreatic endocrine progenitor cells. Dev Biol. 2010;340:397–407. doi: 10.1016/j.ydbio.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 19.Schaffer AE, Freude KK, Nelson SB, Sander M. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev Cell. 2010;18:1022–9. doi: 10.1016/j.devcel.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hesselson D, Anderson RM, Stainier DY. Suppression of Ptf1a activity induces acinar-to-endocrine conversion. Curr Biol. 2011;21:712–17. doi: 10.1016/j.cub.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong PD, Provost E, Leach SD, Stainier DY. Graded levels of Ptf1a differentially regulate endocrine and exocrine fates in the developing pancreas. Genes Dev. 2008;22:1445–50. doi: 10.1101/gad.1663208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manfroid I, Ghaye A, Naye F, Detry N, Palm S, Pan L, et al. Zebrafish sox9b is crucial for hepatopancreatic duct development and pancreatic endocrine cell regeneration. Dev Biol. 2012;366:268–78. doi: 10.1016/j.ydbio.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, et al. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development. 2000;127:5533–40. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 24.Suster ML, Sumiyama K, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics. 2009;10:477. doi: 10.1186/1471-2164-10-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suster ML, Abe G, Schouw A, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish. Nat Protoc. 2011;6:1998–2021. doi: 10.1038/nprot.2011.416. [DOI] [PubMed] [Google Scholar]
- 26.Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–40. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Cashman TJ, Nevis KR, Obregon P, Carney SA, Liu Y, et al. Latent TGF-beta binding protein 3 identifies a second heart field in zebrafish. Nature. 2011;474:645–8. doi: 10.1038/nature10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–77. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung WS, Shin CH, Stainier DY. Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Dev Cell. 2008;15:738–48. doi: 10.1016/j.devcel.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung WS, Stainier DY. Intra-endodermal interactions are required for pancreatic beta cell induction. Dev Cell. 2008;14:582–93. doi: 10.1016/j.devcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–9. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/S1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- 33.Scheer N, Groth A, Hans S, Campos-Ortega JA. An instructive function for Notch in promoting gliogenesis in the zebrafish retina. Development. 2001;128:1099–107. doi: 10.1242/dev.128.7.1099. [DOI] [PubMed] [Google Scholar]
- 34.Lorent K, Moore JC, Siekmann AF, Lawson N, Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev Dyn. 2010;239:855–64. doi: 10.1002/dvdy.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ninov N, Hesselson D, Gut P, Zhou A, Fidelin K, Stainier DY. Metabolic regulation of cellular plasticity in the pancreas. Curr Biol. 2013;23:1242–50. doi: 10.1016/j.cub.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuda H, Parsons MJ, Leach SD. Aldh1-expressing endocrine progenitor cells regulate secondary islet formation in larval zebrafish pancreas. PLoS One. 2013;8 doi: 10.1371/journal.pone.0074350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delaspre F, Beer RL, Rovira M, Huang W, Wang G, Gee S, et al. Centroacinar cells are progenitors that contribute to endocrine pancreas regeneration. Diabetes. 2015. Epub ahead of print. [DOI] [PMC free article] [PubMed]
- 38.European Nucleotide Archive. RNAseq from mature ductal cells from nkx6.1:GFP zebrafish lines. http://www.ebi.ac.uk/ena/data/view/PRJEB10137
- 39.Shih HP, Kopp JL, Sandhu M, Dubois CL, Seymour PA, Grapin-Botton A, et al. A Notch-dependent molecular circuitry initiates pancreatic endocrine and ductal cell differentiation. Development. 2012;139:2488–99. doi: 10.1242/dev.078634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zecchin E, Mavropoulos A, Devos N, Filippi A, Tiso N, Meyer D, et al. Evolutionary conserved role of ptf1a in the specification of exocrine pancreatic fates. Dev Biol. 2004;268:174–84. doi: 10.1016/j.ydbio.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 41.Briscoe J, Pierani A, Jessell TM, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–45. doi: 10.1016/S0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 42.Olguin HC, Yang Z, Tapscott SJ, Olwin BB. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J Cell Biol. 2007;177:769–79. doi: 10.1083/jcb.200608122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–66. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 44.Afelik S, Qu X, Hasrouni E, Bukys MA, Deering T, Nieuwoudt S, et al. Notch-mediated patterning and cell fate allocation of pancreatic progenitor cells. Development. 2012;139:1744–53. doi: 10.1242/dev.075804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson SB, Schaffer AE, Sander M. The transcription factors Nkx6.1 and Nkx6.2 possess equivalent activities in promoting beta-cell fate specification in Pdx1+ pancreatic progenitor cells. Development. 2007;134:2491–500. doi: 10.1242/dev.002691. [DOI] [PubMed] [Google Scholar]
- 46.Rovira M, Scott SG, Liss AS, Jensen J, Thayer SP, Leach SD. Isolation and characterization of centroacinar/terminal ductal progenitor cells in adult mouse pancreas. Proc Natl Acad Sci U S A. 2010;107:75–80. doi: 10.1073/pnas.0912589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kopinke D, Brailsford M, Shea JE, Leavitt R, Scaife CL, Murtaugh LC. Lineage tracing reveals the dynamic contribution of Hes1+ cells to the developing and adult pancreas. Development. 2011;138:431–41. doi: 10.1242/dev.053843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–32. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferber S, Halkin A, Cohen H, Ber I, Einav Y, Goldberg I, et al. Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat Med. 2000;6:568–72. doi: 10.1038/75050. [DOI] [PubMed] [Google Scholar]
- 50.Shternhall-Ron K, Quintana FJ, Perl S, Meivar-Levy I, Barshack I, Cohen IR, et al. Ectopic PDX-1 expression in liver ameliorates type 1 diabetes. J Autoimmun. 2007;28:134–42. doi: 10.1016/j.jaut.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 51.Ber I, Shternhall K, Perl S, Ohanuna Z, Goldberg I, Barshack I, et al. Functional, persistent, and extended liver to pancreas transdifferentiation. J Biol Chem. 2003;278:31950–7. doi: 10.1074/jbc.M303127200. [DOI] [PubMed] [Google Scholar]
- 52.Miyashita K, Miyatsuka T, Matsuoka TA, Sasaki S, Takebe S, Yasuda T, et al. Sequential introduction and dosage balance of defined transcription factors affect reprogramming efficiency from pancreatic duct cells into insulin-producing cells. Biochem Biophys Res Commun. 2014;444:514–9. doi: 10.1016/j.bbrc.2014.01.083. [DOI] [PubMed] [Google Scholar]
- 53.Gefen-Halevi S, Rachmut IH, Molakandov K, Berneman D, Mor E, Meivar-Levy I, et al. NKX6.1 promotes PDX-1-induced liver to pancreatic beta-cells reprogramming. Cell Reprogram. 2010;12:655–64. doi: 10.1089/cell.2010.0030. [DOI] [PubMed] [Google Scholar]
- 54.Ye L, Robertson MA, Hesselson D, Stainier DY, Anderson RM. Glucagon is essential for alpha cell transdifferentiation and beta cell neogenesis. Development. 2015;142:1407–17. doi: 10.1242/dev.117911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–76. doi: 10.1016/S1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 56.Strobel O. In vivo lineage tracing defines the role of acinar-to-ductal transdifferentiation in inflammatory ductal metaplasia. Gastroenterology. 2007;133:1999–2009. doi: 10.1053/j.gastro.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stanger BZ, Stiles B, Lauwers GY, Bardeesy N, Mendoza M, Wang Y, et al. Pten constrains centroacinar cell expansion and malignant transformation in the pancreas. Cancer Cell. 2005;8:185–95. doi: 10.1016/j.ccr.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 58.Hua H, Zhang YQ, Dabernat S, Kritzik M, Dietz D, Sterling L, et al. BMP4 regulates pancreatic progenitor cell expansion through Id2. J Biol Chem. 2006;281:13574–80. doi: 10.1074/jbc.M600526200. [DOI] [PubMed] [Google Scholar]
- 59.Collu GM, Hidalgo-Sastre A, Brennan K. Wnt-Notch signalling crosstalk in development and disease. Cell Mol Life Sci. 2014;71:3553–67. doi: 10.1007/s00018-014-1644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baumgartner BK, Cash G, Hansen H, Ostler S, Murtaugh LC. Distinct requirements for beta-catenin in pancreatic epithelial growth and patterning. Dev Biol. 2014;391:89–98. doi: 10.1016/j.ydbio.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio) 3. Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
- 62.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 63.Haddon C, Jiang YJ, Smithers L, Lewis J. Delta-Notch signalling and the patterning of sensory cell differentiation in the zebrafish ear: evidence from the mind bomb mutant. Development. 1998;125:4637–44. doi: 10.1242/dev.125.23.4637. [DOI] [PubMed] [Google Scholar]
- 64.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bussmann J, Schulte-Merker S. Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development. 2011;138:4327–32. doi: 10.1242/dev.068080. [DOI] [PubMed] [Google Scholar]
- 66.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–9. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.ZiFiT Targeter Version 4.2. http://zifit.partners.org/ZiFiT/
- 68.Ota S, Hisano Y, Muraki M, Hoshijima K, Dahlem TJ, Grunwald DJ, et al. Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays. Genes Cells. 2013;18:450–8. doi: 10.1111/gtc.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mavropoulos A, Devos N, Biemar F, Zecchin E, Argenton F, Edlund H, et al. sox4b is a key player of pancreatic alpha cell differentiation in zebrafish. Dev Biol. 2005;285:211–23. doi: 10.1016/j.ydbio.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 70.Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-T. [DOI] [PubMed] [Google Scholar]
- 71.Allende ML, Weinberg ES. The expression pattern of two zebrafish achaete-scute homolog (ash) genes is altered in the embryonic brain of the cyclops mutant. Dev Biol. 1994;166:509–30. doi: 10.1006/dbio.1994.1334. [DOI] [PubMed] [Google Scholar]
- 72.Cheesman SE, Layden MJ, Von Ohlen T, Doe CQ, Eisen JS. Zebrafish and fly Nkx6 proteins have similar CNS expression patterns and regulate motoneuron formation. Development. 2004;131:5221–32. doi: 10.1242/dev.01397. [DOI] [PubMed] [Google Scholar]
- 73.Argenton F, Zecchin E, Bortolussi M. Early appearance of pancreatic hormone-expressing cells in the zebrafish embryo. Mech Dev. 1999;87:217–21. doi: 10.1016/S0925-4773(99)00151-3. [DOI] [PubMed] [Google Scholar]
- 74.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–43. doi: 10.1093/bioinformatics/btt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nikolaou N, Watanabe-Asaka T, Gerety S, Distel M, Koster RW, Wilkinson DG. Lunatic fringe promotes the lateral inhibition of neurogenesis. Development. 2009;136:2523–33. doi: 10.1242/dev.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amoyel M, Cheng YC, Jiang YJ, Wilkinson DG. Wnt1 regulates neurogenesis and mediates lateral inhibition of boundary cell specification in the zebrafish hindbrain. Development. 2005;132:775–85. doi: 10.1242/dev.01616. [DOI] [PubMed] [Google Scholar]
- 79.Zhang D, Golubkov VS, Han W, Correa RG, Zhou Y, Lee S, et al. Identification of Annexin A4 as a hepatopancreas factor involved in liver cell survival. Dev Biol. 2014;395:96–110. doi: 10.1016/j.ydbio.2014.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]


