Abstract
Scutellaria baicalensis has a wide range of biological activities and has been considered as an important traditional drug in Asia and North America for centuries. A partial-length cDNA clone encoding phytoene synthase (SbPSY) and full-length cDNA clonesencoding phytoene desaturase (SbPDS), ξ-carotene desaturase (SbZDS), β-ring carotene hydroxylase (SbCHXB), and zeaxanthin epoxidase (SbZEP)were identifiedin S. baicalensis. Sequence analyses revealed that these proteins share high identity and conserved domains with their orthologous genes. SbPSY, SbPDS, SbZDS, SbCHXB, and SbZEP were constitutively expressed in the roots, stems, leaves, and flowers of S.baicalensis. SbPSY, SbPDS, and SbZDS were highly expressed in the stems, leaves, and flowers and showed low expression in the roots, where only trace amounts of carotenoids were detected. SbCHXB and SbZEP transcripts were expressed at relatively high levels in the roots, stems, and flowers and were expressed at low levels in the leaves, where carotenoids were mostly distributed. The predominant carotenoids in S.baicalensiswere lutein and β-carotene, with abundant amounts found in the leaves (517.19 and 228.37 μg g-1 dry weight, respectively). Our study on the biosynthesis of carotenoids in S. baicalensis will provide basic data for elucidating the contribution of carotenoids to the considerable medicinal properties of S. baicalensis.
Keywords: Carotenoids, gene characterization, lutein, Scutellaria baicalensis, ß-carotene
Abbreviations
DEPC, diethylpyrocarbonate; HPLC, high-performance liquid chromatography; GGDP, geranylgeranyl diphosphate; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ξ-carotene desaturase; LCYB, lycopene ß-cyclase; LCYE, lycopene ε-cyclase; CHXB, ß-ring carotene hydroxylase; CHXE, ε-ring carotene hydroxylase; ZEP, zeaxanthin epoxidase; CCD, carotenoid cleavage dioxygenase; NCED, 9-cis epoxycarotenoid dioxygenase; ABA, abscisic acid
Introduction
Carotenoids are widely distributed in nature and represent the largest pigment group, with over 600 members identified to date (Cunningham and Gantt, 1998[5]). In plants, carotenoids play several important functions, such as the stabilization of lipid membranes (Havaux, 1998[10]), light collection for photosynthesis, and protection of the photosystem from photo-oxidation (Frank and Cogdell, 1996[7]; Ledford and Niyogi, 2005[19]). Carotenoids provide the yellow, orange, and red colors in flowers and fruits to attract pollinators and agents of seed dispersal (Howitt and Pogson, 2006[12]). Furthermore, carotenoids are precursors of apocarotenoids, which act as hormones, signaling compounds, chromophores, and scent or aroma constituents (Giuliano et al., 2003[9]). In animals and humans, carotenoids are essential nutrients and health-promoting compounds that are not synthesized de novo and must be acquired from the diet (Parker, 1996[23]). ß-Carotene is the primary dietary source of vitamin A, the deficiency of which leads to xerophthalmia, blindness, and premature death (Mayne, 1996[21]). Carotenoids also have strong antioxidant properties that may prevent degenerative diseases and reduce the risk of certain forms of cancer (Giovannucci, 1999[8]; Mayne, 1996[21]).
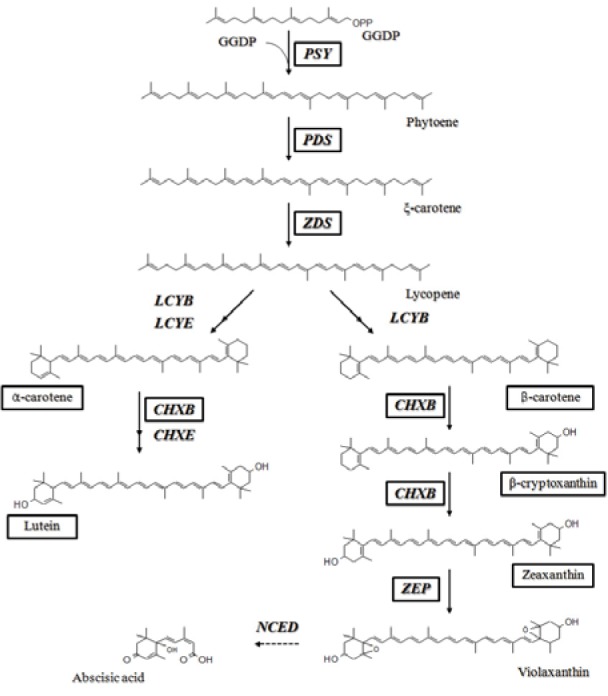
The carotenoid biosynthetic pathway has been extensively studied in higher plants (Cunningham and Gantt, 1998[5]). While carotenoids are synthesized in plastids, the corresponding genes are located in the nucleus, and their protein products are imported into the plastids. The most important step in the carotenoid biosynthetic pathway is the condensation of 2 geranylgeranyl diphosphate (GGDP) molecules to form phytoene; this process is catalyzed by phytoene synthase (PSY) (Figure 1(Fig. 1)). Phytoene undergoes a series of 4 desaturations to form lycopene via ξ-carotene, which is catalyzed by 2 enzymes, phytoene desaturase (PDS) and ξ-carotene desaturase (ZDS). The cyclization of lycopene is a branching point in the pathway: one leads to a-carotene and the other leads to ß-carotene. The formation of α-carotene requires the actions of lycopene ß-cyclase (LCYB) together with lycopene ε-cyclase (LCYE), and LCYB converts lycopene to ß-carotene in 2 reactions. Thereafter, α-carotene and ß-carotene are hydroxylated to produce lutein and zeaxanthin, respectively; this reaction is catalyzed by ß-ring carotene hydroxylase (CHXB) and ε-ring carotene hydroxylase (CHXE). Further epoxidation of zeaxanthin by zeaxanthin epoxidase produces violaxanthin, which is used to synthesize plant hormone abscisic acid (ABA) through oxidative cleavage catalyzed by 9-cis epoxycarotenoid dioxygenase (NCED) (Schwartz et al., 1997[27]). Elucidation of these biosynthetic molecules has increased our understanding of the mechanisms through which carotenoids function.
Figure 1. Carotenoid biosynthetic pathway in plants. The enzymes and carotenoids in black boxes were analyzed in this study. GGDP, geranylgeranyl diphosphate; PSY, phytoene synthase; PDS, phytoene desaturase; ZDS, ξ-carotene desaturase; LCYB, lycopene ß-cyclase; LCYE, lycopene ε-cyclase; CHXB, ß-ring carotene hydroxylase; CHXE, ε-ring carotene hydroxylase; ZEP, zeaxanthin epoxidase; NCED, 9-cis epoxycarotenoid dioxygenase.
Scutellaria baicalensis has been used as an important traditional drug for the treatment of dysentery, pyrexia, jaundice, and carbuncles in Asia and North America for centuries (Joshee et al., 2002[15]; Tang and Eisenbrand, 1992[28]). Pharmacological reports have indicated that S. baicalensis has a multitude of medicinal properties, including anti-inflammatory, antidiabetic, antiviral, antihypertension, antioxidant, and anticancer effects (Chan et al., 2000[4]; Huang et al., 2006[14]; Waisundara et al., 2008[33]). The biological activities of S. baicalensis are likely to be related to the variety of flavones, phenylethanoids, amino acids, sterols and essential oils found in this plant (Joshee et al., 2002[15]; Tang and Eisenbrand, 1992[28]). However, except for flavonoids, which are believed to be the major component of S. baicalensis, very few studies have described the health benefits of other compounds of S. baicalensis.
The goal of this study was to investigate the relationship between the expression levels of carotenoid biosynthetic genes and carotenoid accumulation in S. baicalensis. Here, partial-length cDNA encoding PSY and full-length cDNAs encoding PDS, ZDS, CHXB, and ZEP were isolated. Furthermore, transcript levels of carotenoid biosynthetic genes and carotenoid accumulation were analyzed in different organs of S. baicalensis using real-time PCR (qRT-PCR) and high performance liquid chromatography (HPLC), respectively.
Materials and Methods
Plant materials
S. baicalensis Georgi plants were grown under greenhouse conditions for 1 month and then transferred to the experimental farm of Chungnam National University (Daejeon, Korea). After flowering, the plants were dissected into the roots, stems, leaves, and flowers. All samples were immediately frozen in liquid nitrogen and then stored at 80 °C and/or freeze-dried for RNA isolation and HPLC analysis.
RNA isolation and cDNA synthesis
The samples were ground in a mortar with liquid nitrogen and total RNA was extracted from frozen powder using a Plant Total RNA Mini Kit (Geneaid, Taiwan) according to the manufacturer’s instructions. Gel electrophoresis and spectrophotometer were performed to test the quality and concentration of total extracted RNA, respectively. For first-strand cDNA synthesis, 1 µg of high-quality total RNA was used for reverse transcription using the ReverTra Ace-R kit (Toyobo, Japan). A 20-fold dilution of the 20 µL resulting cDNA was used as template qRT- PCR.
Isolation of cDNAs encoding enzymes involved in carotenoid biosynthetic pathway
In another study (unpublished data), we obtained 39,581 different genes from S. baicalensis using next generation DNA sequencing platforms (Roche/454 GS_FLX+ and Illumina/Solexa HiSeq2000). A partial-length cDNA encoding PSY and full-length cDNAs encoding PDS, ZDS, CHXB, and ZEP were indentified from this database. These proteins were then analyzed for homologies with known sequences and designed as SbPSY, SbPDS, SbZDS, SbCHXB, and SbZEP (GenBank accession numbers: KC417312, KC417313, KC417314, KC417315, and KC417316, respectively).
Sequence analysis
The deduced amino acid sequences of carotenoid biosynthetic genes from S. baicalensis were analyzed for homology using the BLAST program at the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/ BLAST). Sequence alignments were carried out using BioEdit Sequence Alignment Editor, version 5.0.9 (Department of Microbiology, North Carolina State University, Raleigh, NC, USA). The predicted molecular mass of protein was calculated by online website (http://www.sciencegateway.org/ tools/proteinmw.htm).
Quantitative real-time PCR
QRT-PCR primers amplified five target carotenoid biosynthetic genes and Actin housekeeping gene (HQ847728) of S. baicalensis were designed using the Primer3 website (http://frodo.wi.mit.edu/primer3/) (Table 1(Tab. 1)) and then tested for the specificity by PCR. For quantification of standard, the PCR products amplified from cDNA were purified, and the concentration of the products was measured to calculate the number of cDNA copies. The expression of five carotenoid biosynthetic genes was calculated by the method of relative quantification to Actin as the reference. Real-time PCR reactions were carried out in a 20-µl reaction mix containing 5 µl of template cDNA, 10 µl of 2× SYBR Green Realtime PCR Master Mix (Toyobo, Japan), 0.5 µl of each primer (10 µM), and DEPC water. Thermal cycling conditions were as follows: 95 °C for 5 min; 40 cycles of 95 °C for 15 s, 56 °C for 15 s, 72 °C for 20 s. PCR products were analyzed using Bio-Rad CFX Manager 2.0 software. Three replications for each sample were used for real-time analysis.
Table 1. Primers used for real-time PCR.
Carotenoid extraction and HPLC analysis
Extraction and measurement of carotenoids by high-performance liquid chromatography (HPLC) were performed as described by our group (Kim et al., 2012[16]). Briefly, carotenoids were released from the S. baicalensis samples (0.02 g) by adding 3 mL of ethanol containing 0.1 % ascorbic acid (w/v), vortex mixing for 20 s and placing in a water bath at 85 °C for 5 min. The carotenoid extract was saponified with potassium hydroxide (120 µL, 80 % w/v) at the 85 °C water bath for 10 min. After saponification, samples were placed immediately on ice, and cold deionised water (1.5 mL) was added. ß-Apo-8'-carotenal (0.2 mL, 25 g/mL) was added as an internal standard. Carotenoids were extracted twice with hexane (1.5 mL) by centrifugation at 1,200 × g to separate the layers. Aliquots of the extracts were dried under a stream of nitrogen and re-dissolved in 50:50 (v/v) dichloromethane/methanol before analysis by HPLC. The carotenoids were separated on a C30 YMC column (250 × 4.6 mm, 3 µm; Waters Corporation, Milford, MA, USA) by Agilent 1100 HPLC (Massy, France) equipped with a photodiode array (PDA) detector. Chromatograms were generated at 450 nm. Solvent A consisted of methanol/water (92:8 v/v) with 10 mM ammonium acetate. Solvent B consisted of 100 % methyl tert-butyl ether. Gradient elution was performed at 1 mL/min under the following conditions: 0 min, 90 % A/10 % B; 20 min, 83 % A/17 % B; 29 min, 75 % A/25 % B; 35 min, 30 % A/70 % B; 40 min, 30 % A/70 % B; 42 min, 25 % A/75 % B; 45 min, 90 % A/10 % B; and 55 min, 90 % A/10 % B. Carotenoid standards were purchased from CaroteNature (Lupsingen, Switzerland). For quantification purpose, calibration curves were drawn by plotting at four different concentrations of carotenoid standards according to the peak area ratios with ß-apo-8'-carotenal.
Results and Discussion
Sequence analyses of carotenoid biosynthetic genes from S. baicalensis
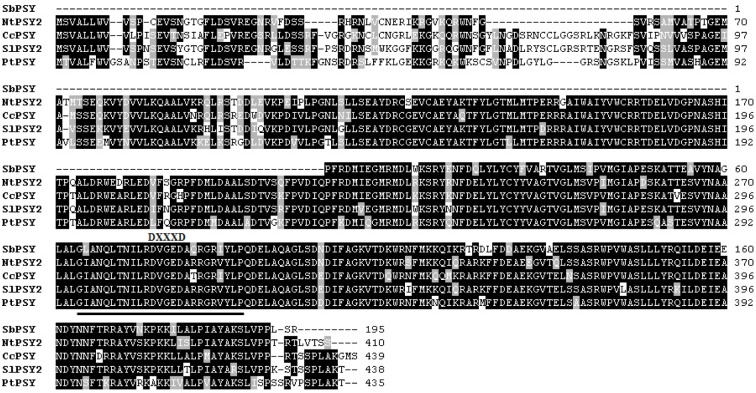
SbPSY was composed of 585 bp encoding a partial open reading frame (ORF) of 195 amino acids. A BLAST search at the amino acid level showed that SbPSY exhibited high homology to other PSYs (Figure 2(Fig. 2)). Specifically, SbPSY shared 86 % identity and 95 % similarity with Nicotiana tabacum PSY, 86 % identity and 93 % similarity with Coffea canephora PSY, 86 % identity and 93 % similarity with Solanum lycopersicum PSY, and 83 % identity and 93 % similarity with Populus trichocarpa PSY. The trans-isoprenyl diphosphate synthase, for the head-to-head condensation reaction (trans-IPPS-HH) domain and aspartate-rich region (DXXXD, where X encodes any amino acid) conserved in other PSY genes were found in SbPSY (Ohnuma et al., 1996[22]; Rasid et al., 2008[24]).
Figure 2. Multiple alignments of the amino acid sequences of SbPSY with other PSYs. Identical residues are indicated by a black background, and similar residues are shaded with a gray background. The solid underline is the trans-IPPS-HH domain. DXXXD, where X encodes for any amino acid, represents the aspartate-rich region. NtPSY2, Nicotiana tabacum (JX101474); CcPSY, Coffea canephora (DQ157164); SlPSY2, Solanum lycopersicum (NM_001247742); PtPSY, Populus trichocarpa (XM_002327528).
SbPDS was 2348 bp long and had an ORF of 1710, encoding a protein of 569 amino acids with a predicted molecular mass of 63.38 kDa (Figure 3(Fig. 3)). SbPDS was 85 %, 84 %, 83 %, and 83 % identical to PDS from Diospyros kaki, Vitis vinifera, N. benthamiana, and Prunus armeniaca, respectively. As shown in Figure 3(Fig. 3), SbPDS contained a conserved dinucleotide-binding motif (GXGX2GX3AX2LX3GX6EX5GG) and a carotenoid-binding domain also found in other orthologous genes (Yan et al., 2011[34]; Zhu et al., 2005[35]).
Figure 3. Multiple alignments of the amino acid sequences of SbPDS with other PDSs. Identical residues are indicated by a black background, and similar residues are shaded with a gray background. The solid underline indicates the dinucleotide-binding domain, and the dotted underline indicates the carotenoid-binding domain. DkPDS, Diospyros kaki (GU112527); VvPDS, Vitis vinifera (XM_002264231); NbPDS, Nicotiana benthamiana (DQ469932); PaPDS, Prunus armeniaca (AY822065).
SbZDS was composed of 2159 bp, with a 1725-bp ORF encoding a protein of 574 amino acids (predicted molecular mass of 63.55 kDa; Figure 4(Fig. 4)). The closest homolog of SbZDS was ZDS from Tagetes erecta (85 % identity and 91 % similarity) followed by ZDS from Helianthus annuus (84 % identity and 91 % similarity), ZDS from Chrysanthemum x morifolium (83 % identity and 90 % similarity), and ZDS from Daucus carota (84 % identity and 91 % similarity). Similar to SbPDS, SbZDS also contained a conserved dinucleotide-binding motif at the N-terminus and a carotenoid-binding domain at the C-terminus (Figure 4(Fig. 4)).
Figure 4. Multiple alignments of the amino acid sequences of SbZDS with other ZDSs. Identical residues are indicated by a black background, and similar residues are shaded with a gray background. The solid underline indicates the dinucleotide-binding domain, and the dotted underline indicates the carotenoid-binding domain. TeZDS, Tagetes erecta (AF251013); HaZDS, Helianthus annuus (AJ438587); CmZDS, Chrysanthemum x morifolium (AB205052); DcZDS1, Daucus carota (DQ222430).
SbCHXB consisted of 1410 bp, with a 939-bp ORF encoding a protein of 312 amino acids (predicted molecular mass of 34.48 kDa; Figure 5(Fig. 5)). SbCHXB exhibited 75 % identity and 84 % similarity with Ipomoea nil CHXB, 76 % identity and 83 % similarity with Vitis vinifera CHXB, 78 % identity and 85 % similarity with C. arabica CHXB, and 74 % identity and 82 % similarity with Lycopersicon esculentum CHXB. Four conservatively spaced histidine motifs proposed to be involved in iron binding during hydroxylation reactions are presented in Figure 5(Fig. 5) (Bouvier et al., 1998[3]).
Figure 5. Multiple alignments of the amino acid sequences of SbCHXB with other CHXBs. Identical residues are indicated by a black background, and similar residues are shaded with a gray background. HXXXH and HXXHH, where X encodes for any amino acid, represent 4 conservatively spaced histidine motifs. InCHXB, Ipomoea nil (AB499058); VvCHXB, Vitis vinifera (XM_002273545); CaCHXB, Coffea arabica (DQ157169); LeCHXB, Lycopersicon esculentum (DQ864755).
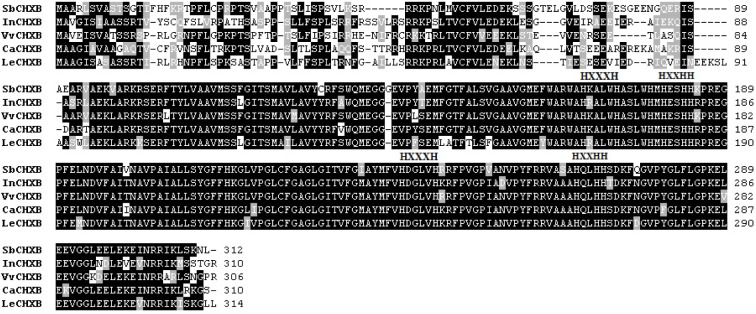
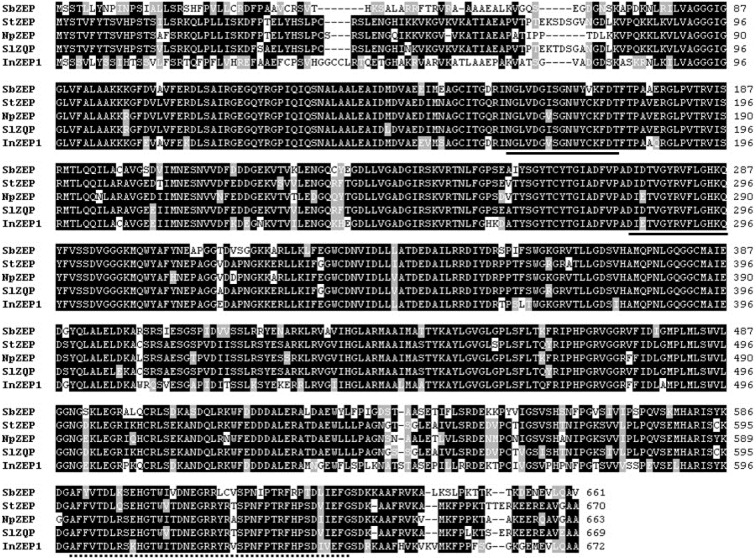
SbZEP was composed 2399 bp, with an ORF of 1986 bp encoding a protein of 661 amino acids with a predicted molecular mass of 72.09 kDa (Figure 6(Fig. 6)). SbZEP shared 76 % identity and 84 % similarity with Solanum tuberosum ZEP, 76 % identity and 83 % similarity with N. plumbaginifolia ZEP, 76 % identity and 84 % similarity with S. lycopersicum ZEP, and 75 % identity and 84 % similarity with Ipomoea nil ZEP. SbZEP displayed 2 short motifs typical of the lipocalin family of proteins and a phosphopeptide-binding domain (FHA domain), which are likely to be the common features of ZEP genes (Durocher and Jackson, 2002[6]; Hieber et al., 2000[11]).
Figure 6. Multiple alignments of the amino acid sequences of SbZEP with other ZEPs. Identical residues are indicated by a black background, and similar residues are shaded with a gray background. The solid underline indicates the short motifs typical of the lipocalin family of proteins, and the dotted underline indicates the FHA domain. StZEP, Solanum tuberosum (DQ206629); NpZEP, Nicotiana plumbaginifolia (X95732); SlZEP, Solanum lycopersicum (EU004202); InZEP1, Ipomoea nil (HQ827173).
Expression levels of carotenoid biosynthetic genes in different organs of S. baicalensis
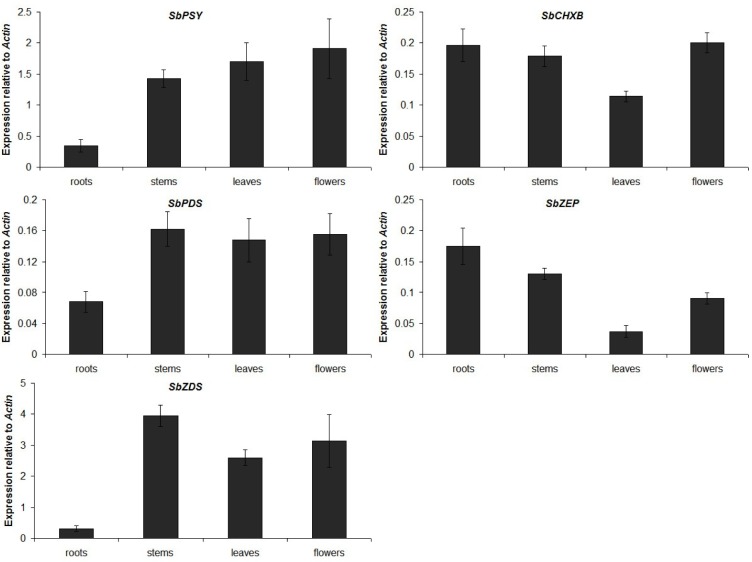
The expression of carotenoid biosynthetic genes was investigated in the roots, stems, leaves, and flowers of S. baicalensis by real-time PCR (Figure 7(Fig. 7)). One of the initial proteins involved in the carotenoid biosynthetic pathway, SbPSY, was expressed at the highest levels in the flowers and was found at lower levels in the leaves and stems, with the lowest expression observed in the roots. Similar to SbPSY, the expression levels of SbPDS and SbZDS were low in the roots and relatively high in the stems, leaves, and flowers. SbCHXB and SbZEP showed different transcriptional patterns with the first 3 enzymes in the carotenoid biosynthetic pathway. The mRNA transcript levels of SbCHXB and SbZEP were lowest in the leaves and higher in the roots, stems, and flowers.
Figure 7. Expression levels of carotenoid biosynthetic genes in different organs of S. baicalensis. The height of each bar and the error bars show the mean and standard error, respectively, from 3 independent measurements.
Analysis of carotenoid content in different organs of S. baicalensis
The same plant materials as those used for qRT-PCR were used to analyze the composition and content of carotenoids in S. baicalensis using HPLC (Table 2(Tab. 2)). In general, carotenoids were distributed mostly in the leaves, and the majority of carotenoids detected in S. baicalensis were lutein and ß-carotene. Specifically, highly abundant lutein content (517.19 µg g-1) was found in the leaves, whereas only a small amount of its precursor, α-carotene (2.23 µg g-1), was found in the same organ. The leaves also contained a significant amount of ß-carotene (228.37 µg g-1), which may explain the presence of considerable amounts of its cis isomers, 9-cis ß-carotene (21.36 µg g-1) and 13-cis ß-carotene (29.72 µg g-1). In contrast, accumulation of ß-carotene may cause poor accumulation of its derivatives, ß-cryptoxanthin (1.93 µg g-1) and zeaxanthin (1.94 µg g-1). In the stems, lutein (43.16 µg g-1) and ß-carotene (14.08 µg g-1) were also the major carotenoids. Only miniscule amounts of 9-cis ß-carotene (2.08 µg g-1), 13-cis ß-carotene (1.12 µg g-1), ß-cryptoxanthin (0.33 µg g-1), zeaxanthin (0.85 µg g-1), and no α-carotene were measured in the stems. The flowers accumulated lower amounts of lutein (13.18 µg g-1) and ß-carotene (6.99 µg g-1) than the stems. Other carotenoids were either expressed at very low levels or were absent in the flowers. In the roots, only trace amounts of lutein (0.88 µg g-1) and ß-carotene (0.35 µg g-1) could be found.
Table 2. Carotenoid composition and content in different organs of S. baicalensis (µg g-1 dry weight). The results have been expressed as mean (± standard errors of the mean) values (n = 3). N.D. = not detected.
In the present study, a partial-length cDNA encoding PSY and full-length cDNA encoding PDS, ZDS, CHXB, and ZEP were characterized, and carotenoid contents were analyzed in S. baicalensis. Carotenoid biosynthetic genes were constitutively expressed in all examined organs of S. baicalensis with different patterns between upstream genes and downstream genes. The transcription levels of SbPSY, SbPDS, and SbZDS were high in the stems, leaves, and flowers and low in the roots, while the transcription levels of SbCHXB and SbZDS were low in the leaves and relatively high in other organs. PSY, which catalyzes the first committed and rate-limiting step in carotenoid biosynthesis, is the major key regulator of carotenoid accumulation in plants (Rodríguez-Villalón et al., 2009[25]; Toledo-Ortiz et al., 2010[29]), suggesting that the low expression levels of SbPSY, SbPDS, and SbZDS caused only trace amounts of carotenoids to be found in the roots of S. baicalensis. However, SbPSY, SbPDS, and SbZDS were highly expressed in the stems and flowers, where only small amounts of carotenoids were detected. Further along in the pathway, carotenoids can be cleaved at various chain positions by carotenoid cleavage dioxygenases (CCDs) to form a broad range of apocarotenoids (Auldridge et al., 2006[2]; Huang et al., 2009[13]; Kloer and Schulz, 2006[17]). An inverse correlation between CCD expression and carotenoid content was observed in Arabidopsis (Auldridge et al., 2006[1]) and bitter melon (Tuan and Park, 2013[32]). Therefore, we hypothesize that the small amounts of carotenoids in the flowers and stems may be due to the high expression of CCDs. Further studies on CCDs in S. baicalensis are needed to clarify this hypothesis.
In the ß,ß-carotenoid branch, ß-carotene is hydroxylated by CHXB to produce zea-xanthin, which is then converted into viola-xanthin by ZEP. Low expression levels of SbCHXB and SbZEP probably result in the large accumulation of ß-carotene in the leaves of S. baicalensis. Unlike the findings for the ß,ß-carotenoid branch, in the ε,ß-carotenoid branch, lutein content was most abundant in the leaves, where its biosynthetic enzyme, SbCHXB, was expressed in low amounts. Indeed, SbCHXB has been proposed to have an active role in the flux of the ß,ß-carotenoid branch rather than the ε,ß-carotenoid branch of carotenoid biosynthesis in S. baicalensis.
Carotenoids are rarely biosynthesized in the underground organs (roots), but are very abundant in the photosynthetic organs (leaves) of S. baicalensis. This mechanism is similar to that in other plants, such as garlic (Tuan et al., 2011[30]) and bitter melon (Tuan et al., 2011[31]), and indicates the essential role of light in the accumulation of carotenoids in plants. Furthermore, we discovered that besides the roots (the most popular part of S. baicalensis), the leaves, which contain significant amounts of lutein (517.19 µg g-1) and ß-carotene (228.37 µg g-1), are also a potential medicinal part of this plant. For humans, lutein is believed to function as an important antioxidant and anti-inflammatory (Ma and Lin, 2010[20]; Sabour-Pickett et al., 2012[26]), and ß-carotene has been shown to have the capacity to reduce the risk of stroke, heart disease, and cancer (Kritchevsky, 1999[18]; Mayne, 1996[21]).
In conclusion, the molecular characterization of carotenoid biosynthetic genes along with carotenoid content may be useful to clarify the biosynthetic mechanisms of carotenoids in S. baicalensis in particular and in plants in general. In addition, these data will provide a foundation for the elucidation of the contribution of carotenoids to the great medicinal properties of S. baicalensis.
Notes
Pham Anh Tuan and Yeon Bok Kim have contributed equally to this work.
Naif Abdullah Al-Dhabi and Sang Un Park have contributed equally as corresponding authors to this work.
Acknowledgements
This project was supported by King Saud University, Deanship of Scientific Research, Addiriyah Chair for Environmental Studies.
References
- 1.Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, et al. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006;45:982–993. doi: 10.1111/j.1365-313X.2006.02666.x. [DOI] [PubMed] [Google Scholar]
- 2.Auldridge ME, McCarty DR, Klee HJ. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr Opin Plant Biol. 2006;9:315–321. doi: 10.1016/j.pbi.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 3.Bouvier F, Keller Y, d'Harlingue A, Camara B. Xanthophyll biosynthesis: molecular and functional characterization of carotenoid hydroxylases from pepper fruits (Capsicum annuum L.) Biochim Biophys Acta. 1998;1391:320–328. doi: 10.1016/s0005-2760(98)00029-0. [DOI] [PubMed] [Google Scholar]
- 4.Chan FL, Choi HL, Chen ZY, Chan PS, Huang Y. Induction of apoptosis in prostate cancer cell lines by a flavonoid, baicalin. Cancer Lett. 2000;160:219–228. doi: 10.1016/s0304-3835(00)00591-7. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham FX, Gantt E. Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- 6.Durocher D, Jackson SP. The FHA domain. FEBS Lett. 2002;513:58–66. doi: 10.1016/s0014-5793(01)03294-x. [DOI] [PubMed] [Google Scholar]
- 7.Frank HA, Cogdell RJ. Carotenoids in photosynthesis. Photochem Photobiol. 1996;63:257–264. doi: 10.1111/j.1751-1097.1996.tb03022.x. [DOI] [PubMed] [Google Scholar]
- 8.Giovannucci E. Tomatoes, tomato-based products, lycopene, and cancer: review of the epidemiologic literature. J Natl Cancer Inst. 1999;91:317–331. doi: 10.1093/jnci/91.4.317. [DOI] [PubMed] [Google Scholar]
- 9.Giuliano G, Al-Babili S, von Lintig J. Carotenoid oxygenases: cleave it or leave it. Trends Plant Sci. 2003;8:145–9. doi: 10.1016/S1360-1385(03)00053-0. [DOI] [PubMed] [Google Scholar]
- 10.Havaux M. Carotenoids as membrane stabilizers in chloroplasts. Trends Plant Sci. 1998;3:147–151. [Google Scholar]
- 11.Hieber AD, Bugos RC, Yamamoto HY. Plant lipocalins: Violaxanthin de-epoxidase and zeaxanthin epoxidase. Biochim Biophys Acta. 2000;1482:84–91. doi: 10.1016/s0167-4838(00)00141-2. [DOI] [PubMed] [Google Scholar]
- 12.Howitt CA, Pogson BJ. Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell Environ. 2006;29:435–445. doi: 10.1111/j.1365-3040.2005.01492.x. [DOI] [PubMed] [Google Scholar]
- 13.Huang FC, Molnar P, Schwab W. Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J Exp Bot. 2009;60:3011–22. doi: 10.1093/jxb/erp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang WH, Lee AR, Yang CH. Antioxidative and anti-inflammatory activities of polyhydroxyflavonoids of Scutellaria baicalensis Georgi. Biosci Biotechnol Biochem. 2006;70:2371–2380. doi: 10.1271/bbb.50698. [DOI] [PubMed] [Google Scholar]
- 15.Joshee N, Patrick TS, Mentreddy RS, Yadav AK. Skullcap: Potential medicinal crop. In: Janick J, Whipkey A, editors. Trends in new crops and new uses. Alexandria, VA: ASHS Press; 2002. pp. 580–586. [Google Scholar]
- 16.Kim JK, Ha SH, Park SY, Lee SM, Kim HJ, Lim SH, et al. Determination of lipophilic compounds in genetically modified rice using gas chromatography-time-of-flight-mass spectrometry. J Food Compos Anal. 2012;25:31–38. [Google Scholar]
- 17.Kloer DP, Schulz GE. Structural and biological aspects of carotenoid cleavage. Cell Mol Life Sci. 2006;63:2291–2303. doi: 10.1007/s00018-006-6176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kritchevsky SB. Beta-Carotene, carotenoids and the prevention of coronary heart disease. J Nutr. 1999;129:5–8. doi: 10.1093/jn/129.1.5. [DOI] [PubMed] [Google Scholar]
- 19.Ledford HK, Niyogi KK. Singlet oxygen and photooxidative stress management in plants and algae. Plant Cell Environ. 2005;28:1037–1045. [Google Scholar]
- 20.Ma L, Lin XM. Effects of lutein and zeaxanthin on aspects of eye health. J Sci Food Agric. 2010;90:2–12. doi: 10.1002/jsfa.3785. [DOI] [PubMed] [Google Scholar]
- 21.Mayne ST. Beta-carotene, carotenoids, and disease prevention in humans. FASEB J. 1996;10:690–701. [PubMed] [Google Scholar]
- 22.Ohnuma SI, Nakazawa T, Hemmi H, Hallberg AM, Koyama T, Ogura K, et al. Conversion from farnesyl disphosphate synthase to geranylgeranyl disphospate synthase by random chemical mutagenesis. J Biol Chem. 1996;271:10087–10095. doi: 10.1074/jbc.271.17.10087. [DOI] [PubMed] [Google Scholar]
- 23.Parker RS. Absorption, metabolism, and transport of carotenoids. FASEB J. 1996;10:542–551. [PubMed] [Google Scholar]
- 24.Rasid OA, Wan Nur Syuhada WS, Nor Hanin A, Masura SS, Zulqarnain M, Ho CL, et al. RT-PCR amplification and cloning of partial DNA sequence coding for oil palm (Elaeis oleifera) phytoene synthase gene. As Pac J Mol Biol Biotechnol. 2008;16:17–24. [Google Scholar]
- 25.Rodríguez-Villalón A, Gas E, Rodríguez-Concepción M. Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 2009;60:424–435. doi: 10.1111/j.1365-313X.2009.03966.x. [DOI] [PubMed] [Google Scholar]
- 26.Sabour-Pickett S, Nolan JM, Loughman J, Beatty S. A review of the evidence germane to the putative protective role of the macular carotenoids for age-related macular degeneration. Mol Nutr Food Res. 2012;56:270–286. doi: 10.1002/mnfr.201100219. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276:1872–4. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- 28.Tang W, Eisenbrand G. Chinese drugs of plant origin. New York: Springer Verlag; 1992. pp. 919–929. [Google Scholar]
- 29.Toledo-Ortiz G, Huq E, Rodríguez-Concepción M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc Natl Acad Sci USA. 2010;107:11626–11631. doi: 10.1073/pnas.0914428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tuan PA, Kim JK, Kim HH, Lee SY, Park NI, Park SU. Carotenoid accumulation and characterization of cDNAs encoding phytoene synthase and phytoene desaturase in garlic (Allium sativum) J Agric Food Chem. 2011;59:5412–5417. doi: 10.1021/jf2009827. [DOI] [PubMed] [Google Scholar]
- 31.Tuan PA, Kim JK, Park NI, Lee SY, Park SU. Carotenoid content and expression of phytoene synthase and phytoene desaturase genes in bitter melon (Momordica charantia) Food Chem. 2011;126:1686–1692. doi: 10.1016/j.foodchem.2010.12.058. [DOI] [PubMed] [Google Scholar]
- 32.Tuan PA, Park SU. Molecular cloning and characterization of cDNAs encoding carotenoid cleavage dioxygenase in bitter melon (Momordica charantia) J Plant Physiol. 2013;170:115–120. doi: 10.1016/j.jplph.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Waisundara VY, Hsu A, Huang D, Tan BKH. Scutellaria baicalensis: enhances the anti-diabetic activity of metformin in streptozotocin-induced diabetic wistar rats. Am J Chinese Med. 2008;36:517–540. doi: 10.1142/S0192415X08005953. [DOI] [PubMed] [Google Scholar]
- 34.Yan P, Gao XZ, Shen WT, Zhou P. Cloning and expression analysis of phytoene desaturase and Zeta-carotene desaturase genes in Carica papaya. Mol Biol Rep. 2011;38:785–791. doi: 10.1007/s11033-010-0167-2. [DOI] [PubMed] [Google Scholar]
- 35.Zhu YH, Jiang JG, Yan Y, Chen XW. Isolation and characterization of phytoene desaturase cDNA involved in the beta-carotene biosynthetic pathway in Dunaliella salina. J Agric Food Chem. 2005;53:5593–5597. doi: 10.1021/jf0506838. [DOI] [PubMed] [Google Scholar]