Abstract
Objectives
Long non-coding RNA (lncRNA) associated with microvascular invasion in hepatocellular carcinoma (MVIH) has been recently reported to act as a predictor for the poor recurrence-free survival of hepatocellular carcinoma (HCC) after hepatectomy. However, the biological role of MVIH in the tumorigenesis of HCC is still unclear.
Methods
In the study reported here, MVIH expression levels were detected by real-time polymerase chain reaction (PCR) in tumor tissue of HCC patients and in HCC cells, including SMMC7721 and HepG2 cells. Cell viability and apoptosis were determined by MTT and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) methods, respectively. The model of transplantation tumor of HepG2 cells in nude mice was used to evaluate the effects of MVIH and miR-199a on HCC in vivo.
Results
MVIH expression was significantly increased and miR-199a expression was significantly decreased in tumor tissue and HCC cells. si-MVIH inhibited HCC cell viability and promoted cell apoptosis, but this effect was reversed by miR-199a inhibitor. Luciferase reporter assay and RNA immunoprecipitation experiment showed that miR-199a had a direct binding ability to MVIH RNA. In nude mice with transplantation, the tumor volume was reduced by si-MVIH, and miR-199a inhibitor canceled this decrease.
Conclusion
MVIH promoted cell growth and inhibited cell apoptosis of HCC via inhibiting miR-199a expression.
Keywords: long non-coding RNA, HCC, miR-199a, cell apoptosis
Introduction
Hepatocellular carcinoma (HCC) is one of the most common solid tumors worldwide. The occurrence of HCC in Asia and Africa accounts for the majority of all cases,1 and the annual increase of HCC incidence was 3.7% and 2.9% in men and women, respectively, in the USA in the last decade.2 More than half of the HCC burden has been attributed to HBV infection,3 and the epidemic of chronic HCV is closely linked to the increase of HCC incidence in developed countries, including Japan, Italy, and the USA.4 Although significant progress has been made in the treatment of HCC, it is the third leading cause of cancer-related deaths worldwide.5 Therefore, it is necessary to determine the regulatory network underlying the development and progression of HCC, which remains to be elucidated, for exploring new therapeutic strategies.
Emerging evidence has revealed that non-coding RNAs (ncRNAs) are frequently regulated in various pathological processes, especially in tumorigenesis.5 The two major classes of ncRNAs are long ncRNAs (lncRNAs) and small ncRNAs.6 microRNAs (miRNAs), a class of small ncRNAs, normally direct RNA-induced silencing complex (RISC) to the 3′UTR of target genes and regulate their expression. Mounting evidence has demonstrated that miRNAs take part in all stages and aspects of cancer cell development, differentiation, apoptosis, and adhesion,7 and hence their possible use as diagnostic, prognostic, and predictive biomarkers for cancers has been extensively studied.8 The new discovery of lncRNAs discloses a new understanding of gene expression in cancers. The recent studies have showed that lncRNAs, including HOTIR, MALTA-1, MEG3, and HULC, are dysregulated in HCC and involved in cell invasion, apoptosis, tumorigenesis, metastasis, prognosis, and diagnosis.9–12
Microvascular invasion in hepatocellular carcinoma (MVIH), located in the intron of the RPS24 gene and encoding a protein that belongs to the S24E family of ribosomal proteins, is an lncRNA associated with microvascular invasion of HCC.13,14 The present knowledge of MVIH is limited. A recent study demonstrated that MVIH is generally overexpressed in HCC and serves as a predictor for the recurrence-free survival of HCC after hepatectomy.14 An increase in MVIH expression has also been observed in non-small cell lung cancer.15 However, the molecular mechanism of MVIH in HCC remains unknown.
Previous studies indicate that lncRNA may regulate miRNA expression, subsequently modulating the target genes of miRNAs at the post-transcriptional level.15,16 miR-199a has been reported to function as tumor suppressor. Bioinformatics analysis reveals MVIH contains one conserved target site of miR-199a. In the study reported here, we investigated the role of MVIH in expression of FZD7, which is a downstream gene of miR-199a, providing a new insight to elaborate the molecular mechanism of cancer development.
Materials and methods
Specimen collection
A total of 15 patients diagnosed with HCC who had undergone routine hepatic resection in the Affiliated Hospital of Xuzhou Medical College from October 2013 to October 2014 were included in this study. Written informed consent was obtained from all the participants, and the study was approved by the Ethics Committee of the Affiliated Hospital of Xuzhou Medical College. None of the patients had received preoperative radiotherapy or chemotherapy prior to surgical resection. The diagnoses of HCC were evaluated with conventional clinical, histological diagnosis, differentiation, or cytological criteria. The tumor tissues and corresponding adjacent non-tumor tissues were collected in liquid nitrogen and stored at −80°C immediately after resection.
Real-time polymerase chain reaction
The total RNA was isolated from tumor tissues or cells using TRIzol® Reagent (Thermo Fisher Scientific, Waltham, MA, USA). The produced cDNA was evaluated and quantified by spectrophotometry. Total RNA (1 μg) was used to reverse transcribe to cDNA using an ImProm-II™ Reverse Transcription System (Promega Corporation, Fitchburg, WI, USA). The expression levels of MVIH and miR-199a were determined on an Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific) with a Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific). GAPDH or U6 was used to act as a reference gene. The relative expression levels of MVIH and miR-199a were calculated with the 2−ΔΔCT method. The specific sequences of primers were as follows:
MVIH
Forward, 5′-AATTTTGCACATCTGAACAGCC-3′ and Reverse, 5′-TTCAAAATCCCACTACGCCCA-3′
GAPDH
Forward, 5′-GTCAACGGATTTGGTCTGTATT-3′ and Reverse, 5′-AGTCTTCTGGGTGGCAGTGAT-3′
miR-199a
Forward, 5′-GCCACAGTAGTCTGCACAT-3′ and Reverse, 5′-CAGTGCGTGTCGTGGAGT-3′
U6
Forward, 5′-GGGCAGGAAGAGGGCCTAT-3′ and Reverse, 5′-AAAAATATGGAACGCTTCACGAATTTG-3′
Cell culture
Human liver cancer cell lines (HepG2 and SMMC7721) and a normal liver cell line (HL-7702) were obtained from the Institute of Biochemistry and Cell Biology, BIBS, CAS (Shanghai, People’s Republic of China). No ethics statement was required from the institutional review board for the use of these cell lines. Cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% fetal bovine serum (Gibco®; Thermo Fisher Scientific), 100 U/mL penicillin, and 100 mg/mL streptomycin in humidified air at 37°C with 5% CO2.
Vector construction
The constructed small interfering RNA-MVIH (si-MVIH) vector and empty vector (negative control [NC]) were synthesized by Inovogen Tech Co, Ltd (Beijing, People’s Republic of China). The luciferase reporter vectors (pMIR-MVIH-WT, pMIR-MVIH-Mut) were constructed by Takara Biotechnology (Dalian) Co, Ltd (Dalian, People’s Republic of China). The MVIH cDNA fragment containing the predicted potential miR-199a binding sites or mutant sites was amplified by real-time polymerase chain reaction (PCR) method, and cloned into p-MIR-REPORT™ Luciferase vector (Promega Corporation) according to the manufacturer’s instructions. The pcDNA-MVIH and pcDNA-NC plasmids were prepared by Life Technologies (Beijing, People’s Republic of China). The MVIH sequence was cloned into the pcDNA3.1 vector by PCR method.
Cell transfection
When the HepG2 and SMMC7721 cells were cultured to 60%–70% confluence, the si-MVIH (or si-NC) was transfected into the cells using Lipofectamine® 2000 (Thermo Fisher Scientific) to co-incubate for 24 hours, and then the cells were harvested to detect the cell viability and apoptosis. The plasmids pcDNA-MVIH (or empty pcDNA) were transfected into cells for 48 hours, and the cells were collected to perform real-time PCR and Western blot experiments. Luciferase reporter plasmid, wild-type luciferase reporter plasmid (p-MIR-MVIH-WT), or mutant luciferase reporter plasmid (pMIR-MVIH-Mut), was co-transfected with miR-199a mimic into SMMC7721 or HepG2 cells for 48 hours, and the luciferase activity was determined by GloMax® 96 Microplate Luminometer with Dual-Luciferase® Reporter Assay System (Promega Corporation) according to the manufacturer’s protocol.
MTT assay
The viability of SMMC7721 and HL-7702 cells was determined by MTT assay kit (Beyotime Biotechnology, People’s Republic of China). The cells were incubated in 96-well culture plates at a concentration of 1×104 cells/well. si-MVIH was transfected into HepG2 or SMMC7721 cells for 48 hours, and then the miR-199a inhibitor or NC was transfected into cells for another 48 hours. After the transfection, 40 μL MTT stock solution was supplemented into medium for 4 hours, and then the supernatant was discarded and 200 μL dimethyl sulfoxide (DMSO) was administrated. The absorbance was recorded at 570 nm with a Multiskan® Spectrum (Thermo Fisher Scientific).
Terminal deoxynucleotidyl transferase dUTP nick end labeling method
The apoptosis of SMMC7721 and HL-7702 cells was detected using a Colorimetric TUNEL Apoptosis Assay Kit (Beyotime Biotechnology) according to the instructions. In brief, the cells adherent on the slides were pretreated with CDMC, and then fixed with 4% fresh paraformaldehyde for 30 minutes at room temperature. After washing once with phosphate-buffered saline (PBS), the fixed cells were immersed in 0.1% Triton™ X-100 for 2 minutes at 4°C. H2O2 (0.3%) in methanol was used to rinse the cells for 20 minutes and PBS was used to rinse them three times. The slides were incubated with a mixed solution contained TdT and Bio-16-dUTP in TdT buffer at 37°C for 60 minutes. The cells were rinsed with PBS and incubated with streptavidin-horseradish peroxidase conjugate to stop the reaction. The slides were stained with diaminobenzidine for 10 minutes. The terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL)-positive cells were calculated.
RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) was performed with a Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (EMD Millipore, Billerica, MA, USA). In brief, the cells were lysed with RIP lysis buffer to obtain the cell lysis. The protein was quantified using a Bradford Protein Assay Kit (Beyotime Biotechnology). The whole cell extract was then co-incubated with RIP buffer containing A + G magnetic beads conjugated with human anti-Ago2 antibody (EMD Millipore) or NC normal mouse immunoglobulin G (IgG) (EMD Millipore). Ago2 is a key component of the RISC complex. An EZ-ECL kit was used to visualize Ago2 and IgG protein expression. The immunoprecipitated RNA was isolated, and real-time PCR was conducted to measure MVIH and miR-199a in the precipitates.
Western blot analysis
The total protein from HepG2 and SMMC7721 cells were lysed with RIPA lysis buffer (Beyotime Biotechnology), which contained protease inhibitors. The protein quantity was measured by bicinchoninic acid protein assay kit (Beyotime Biotechnology). The protein extracts were separated using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to nitrocellulose membranes and blocked in 5% nonfat milk prior to incubation with the primary antibodies anti-FZD7 overnight at 4°C. After being washed, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody for 2 hours. The protein expression was visualized using a BeyoECL Plus ECL Kit (Beyotime Biotechnology). Beta-actin served as the control protein.
Xenotransplant nude mice
The animal study was approved by the Committee on the Ethics of Animal Experiments of the Affiliated Hospital of Xuzhou Medical College. A total of 32 BALB/c nude mice aged 6 weeks old were used to establish xenotransplant mice with HepG2 HCC cells. The HepG2 cells were pretreated with stable transfection of si-MVIH or si-control, and then injected subcutaneously into the flanks of mice. Two weeks after the injection, the miR-199a inhibitor or NC was injected into the tumor sites. The tumor volume was recorded every week, and it was calculated by the formula as follows: tumor volume = πAB2/6 (in which A is the largest diameter, B is the perpendicular diameter).
Statistical analysis
All statistical analyses were performed with SPSS (v 18.0) software. The measurement data were presented as mean ± standard deviation (SD). The statistical difference was analyzed by independent-samples t-test between two groups. A P-value <0.05 was considered to indicate significant difference.
Results
MVIH expression is elevated in HCC
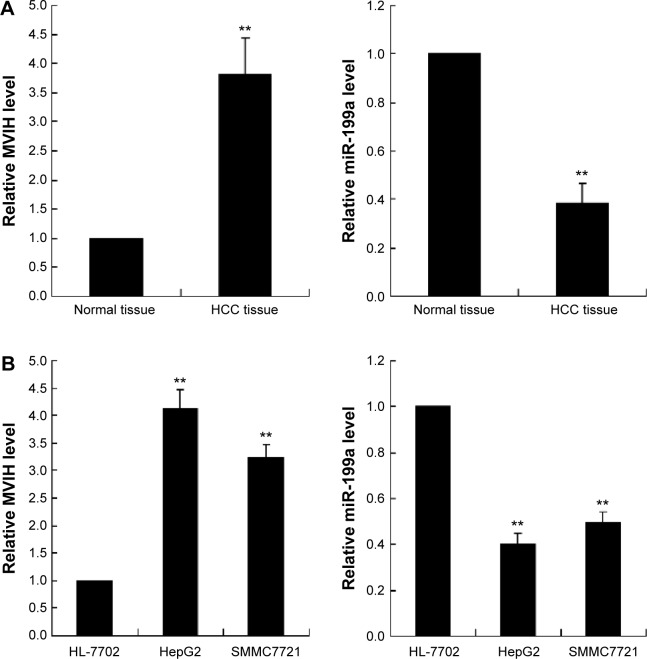
MVIH and miR-199a expression was detected by real-time PCR. As shown in Figure 1A, the MVIH level in HCC tissue was 3.75-fold than that of normal tissue, and the miR-199a level was 0.39-fold than that of normal tissue. The MVIH and miR-199a expression levels were also detected in HCC cells. As indicated in Figure 1B, SMMC7721 and HepG2 HCC cells had a higher expression of MVIH and lower expression of miR-199a compared with normal hepatocyte HL-7702 cells.
Figure 1.
The microvascular invasion in hepatocellular carcinoma (MVIH) and miR-199a expression in hepatocellular carcinoma (HCC) tissue and cells. The relative MVIH and miR-199a expression levels were quantified by real-time polymerase chain reaction (PCR) in HCC tissue (n=15, A) and cells (n=3, B).
Note: **P<0.01, versus normal tissue or normal hepatocyte HL-7702 cells.
Downregulation of MVIH inhibits cell viability and promotes cell apoptosis of HCC cells
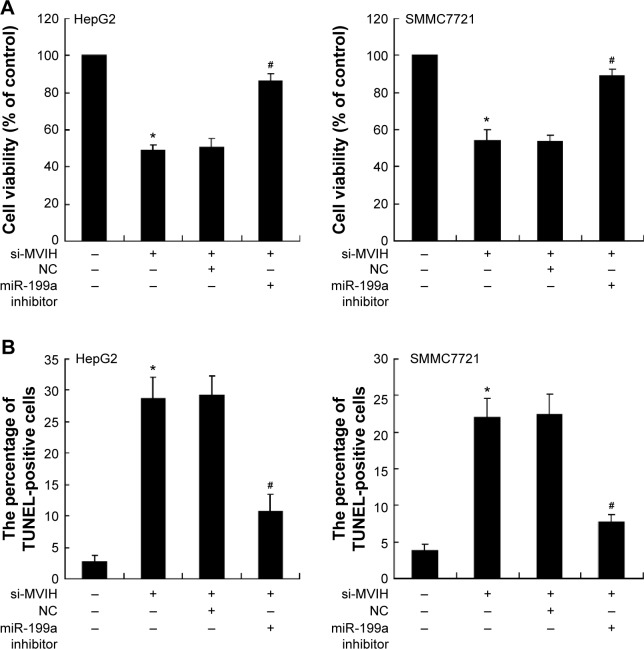
To investigate the role of MVIH in HCC, cells were transfected with si-MVIH to downregulate MVIH expression. The results showed that si-MVIH significantly inhibited the cell viability of HepG2 and SMMC7721 cells (Figure 2A and B), while markedly promoted the cell apoptosis (Figure 2C and D). HCC cells supplemented with miR-199a inhibitor, the decrease of cell viability and increase of cell apoptosis induced by si-MVIH were reversed.
Figure 2.
Downregulation of microvascular invasion in hepatocellular carcinoma (MVIH) inhibits cell viability and promotes cell apoptosis of hepatocellular carcinoma cells. siRNA-MVIH was transfected into hepatocellular carcinoma cells (HepG2 and SMMC7721) for 48 hours and then the miR-199a inhibitor or negative control (NC) was transfected into hepatocellular carcinoma cells for another 48 hours. The cell viability was detected by MTT assay (A); the cell apoptosis was measured by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) method (B).
Notes: *P<0.05, versus with cells transfected with si-MVIH; #P<0.05, versus with cells transfected with si-MVIH and NC.
miR-199a targets MVIH in HCC cells
Bioinformatics analysis revealed MVIH contains one conserved target site of miR-199a. To confirm this prediction, the wild-type sequence of MVIH (MVIH-WT) or its mutant sequence (MVIH-Mut) was sub-cloned into the pMIR luciferase reporter and then co-transfected with miR-199a mimic into SMMC7721 or HepG2 cells. The relative luciferase activity of pMIR-UCA1-WT was significantly reduced after the transfection of miR-199a mimic. However, no effect was observed in pMIR-UCA1-Mut (Figure 3A). The RIP experiments was used to confirm whether both MVIH and miR-199a might be in the RISC complex. The results showed that MVIH and miR-199a were enriched to more than 100 times in Ago2 pellets relative to control IgG immunoprecipitates (Figure 3B), which demonstrated that MVIH is present in Ago-2-containing RISC through association with miR-199a. In miR-199a mimic co-transfected with pcDNA-MVIH (or pcDNA-NC) in SMMC7721 or HepG2 cells, the miR-199a expression level was significantly inhibited by pcDNA-MVIH in a dose-dependent manner (Figure 3C). However, FZD7, the target gene of miR-199a, was upregulated by pcDNA-MVIH (Figure 3D).
Figure 3.
miR-199a targets 3′UTR of microvascular invasion in hepatocellular carcinoma (MVIH) in hepatocellular carcinoma cells. Luciferase reporter plasmid, wild-type luciferase reporter plasmid (p-MIR-MVIH-WT) or mutant luciferase reporter plasmid (pMIR-MVIH-Mut), was co-transfected with miR-199a mimic into SMMC7721 or HepG2 cells; the relative luciferase activity was detected by luciferase reporter assay (A). HepG2 cells lysate was treated with antibody Ago2 or immunoglobulin (Ig) G, and then the RNA immunoprecipitation was performed to detect the relative MVIH and miR-199a enrichment (B). Co-transfection of miR-199a mimic with pcDNA-MVIH or pcDNA-NC into SMMC7721 or HepG2 cells; the concentrations of pcDNA-MVIH were 1 μg, 2 μg, and 4 μg; the relative miR-199a level was quantified by real-time polymerase chain reaction (C). The protein expression of FZD7 was detected by Western blot (D).
Note: **P<0.01, versus the group treated with negative control (NC; A) or anti-IgG (B) or transfection of miR-199a with empty pcDNA (C).
Abbreviations: IP, immunoprecipitation; RIP, RNA immunoprecipitation; WB, western blot.
MVIH and miR-199a participate in the tumor growth in vivo
HepG2 cells were injected into nude mice to establish tumor xenograft model. HepG2 cells were pretreated with stable transfection with si-MVIH or si-control. As shown in Figure 4, the tumor growth of mice with si-MVIH transfection was slower than that in mice transfected with si-control. In addition, the tumor volume in the si-MVIH group was 26.4% of that in si-control group. To inject miR-199a inhibitor into tumor sites during the period of tumor growth, it was found that the decrease in tumor volume induced by si-MVIH was reversed by miR-199a inhibitor.
Figure 4.
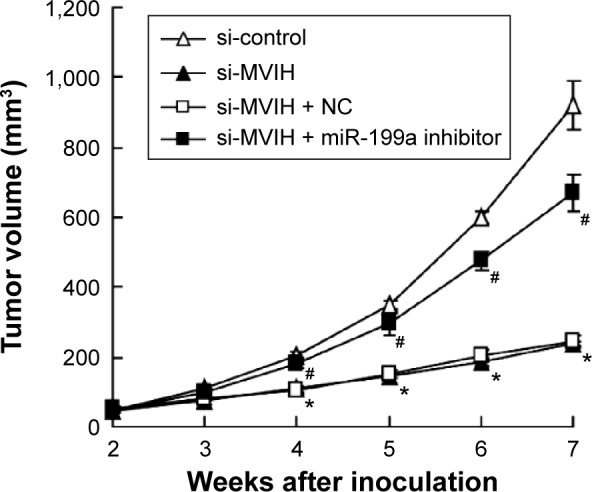
Microvascular invasion in hepatocellular carcinoma (MVIH) and miR-199a participate in the tumor growth in vivo. HepG2 cells were stably transfected with small interfering RNA-microvascular invasion in hepatocellular carcinoma (si-MVIH) or siRNA-control. Two weeks after the injection with HepG2 cells (HepG2 cells were pretreated with different treatments), the mouse model of transplantation tumor was established. miR-199a inhibitor or negative control (NC) was injected into the tumor sites (n=8).
Notes: *P<0.05, versus the mice with si-control treatment; #P<0.05, versus the mice with si-MVIH + NC.
Abbreviation: si-MVIH+NC, siRNA-MVIH + negative control.
Discussion
The new discovery of lncRNA transcripts has dramatically increased our understanding of cell biology, especially in cancers. More than 200 nucleotides long, lncRNAs are closely related to the regulation of protein-coding genes at chromatin modification, transcriptional, and post-transcriptional levels in various physiological processes.17 Recently, extensive studies have been conducted on lncRNAs and their molecular functions,18–20 and the emerging evidences show numerous lncRNAs are altered in HCC.11,21,22 Yuan et al has demonstrated that the MVIH overexpression is associated with frequent microvascular invasion, higher tumor node metastasis stage, and decreased recurrence-free survival.14 In the study reported here, we found MVIH was overexpressed in tumor tissue and cells of HCC. In addition, the downregulation of MVIH could reduce the tumor volume in vivo and decrease the cancer cell viability in vitro. These findings are consistent with previous study.
Although the biological roles of lncRNAs in human cancers have been identified, the molecular mechanisms underlying cancers are complicated and still little known. A growing number of studies have demonstrated that lncRNAs may act as miRNA sponges or antagomirs to be involved in the regulation of miRNAs and their target genes.23,24 Kallen et al illustrated that the H19 lncRNA harbors both canonical and noncanonical binding sites for let-7 miRNA, and acts as a natural sponge for the regulation of let-7 availability.23 It is still unknown whether MVIH takes part in cancer biology via regulation of miRNA expression. Previously, abundant reports showed that miR-199a functions as a tumor suppressor, promoting cancer cell apoptosis and preventing cancer migration in cancers.25–27 Bioinformatics analysis also revealed MVIH contains one conserved target site of miR-199a. We detected the expression levels of miR-199a in tumor tissue and cells of HCC, and the results showed that miR-199a levels were significantly decreased. We proposed MVIH might regulate HCC cells viability by inhibiting the expression of miR-199a, which functions as an miR-199a sponge. In in vitro experiments in which HCC cells were supplemented with miR-199a inhibitor, the downregulation of MVIH-induced decrease in cell viability and increase in cell apoptosis were reversed. This result partly indicated that MVIH interacted with miR-199a to play the role in regulating cancer cell viability and apoptosis. To further prove our hypothesis, the luciferase reporter assay was performed. As expected, a direct binding ability of the miR-199a response elements on the MVIH RNA was confirmed. Ago2 is a key component of the RISC, and it can captures miRNAs which are working as guide molecules for interaction with target mRNAs. The RIP experiment produced even more evidence for the association between MVIH and miR-199a in Ago2-containing RISC. In the transplantation tumor mice, it was found that the decrease of tumor volume induced by si-MVIH was also reversed by the miR-199a inhibitor, which suggested there was a reciprocity relationship between MVIH and miR-199a. Taken together, these findings provide evidence of MVIH acting as a miRNA sponge, linking the regulation of gene expression in HCC pathogenesis.
To further investigate the effects of MVIH on the regulation of miR-199a and its target mRNA expression, FZD7 was studied. FZD7 is a member of the “frizzled” gene family encode 7-transmembrane domain proteins that are receptors for Wnt signaling proteins.28 The alteration of FZD7 expression has been reported to play a critical role in numerous human cancers, including gastric cancer, breast cancer, colorectal cancer, and so on.29–31 Recently, FZD7 has been identified to be a functional target of miR-199a and participate in cell proliferation and cell cycle regulation of HCC.32 In the study reported here, miR-199a expression was inhibited by pcDNA-MVIH, and FZD7 protein expression was upregulated after the co-transfection of miR-199a mimic with pcDNA-MVIH into HCC cells. Therefore, the alteration of FZD7 expression mediated by MVIH and miR-199a was believed to be involved in the tumorigenesis process of HCC.
Conclusion
The study found that MVIH was overexpressed in vitro and in vivo of HCC, and MVIH acted as an miR-199a sponge to promote HCC cell growth and inhibit cell apoptosis. Further studies are needed to elucidate the precise mechanism underlying the regulation of miRNAs and their target genes by lncRNAs.
Acknowledgments
This work was supported by scientific research project from bureau of Science and Technology in Xuzhou, Jiangsu province (XM09B092) and the Talents of High Level Scientific Research Foundation of Jiangsu province (2013-wsw-058), People’s Republic China.
Footnotes
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118(12):3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 4.Dhanasekaran R, Limaye A, Cabrera R. Hepatocellular carcinoma: consensus recommendations of the National Cancer Institute Clinical Trials Planning Meeting. Hepat Med. 2012;4:19–37. [Google Scholar]
- 5.Thomas MB, Jaffe D, Choti MM, et al. Hepatocellular carcinoma: consensus recommendations of the National Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol. 2010;28(25):3994–4005. doi: 10.1200/JCO.2010.28.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12(2):136–149. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomaru Y, Hayashizaki Y. Cancer research with non-coding RNA. Cancer Sci. 2006;97(12):1285–1290. doi: 10.1111/j.1349-7006.2006.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4(3):143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Zhou L, Wu LM, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18(5):1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 10.Lai MC, Yang Z, Zhou L, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29(3):1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 11.Braconi C, Kogure T, Valeri N, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30(47):4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Meng XM, Huang C, et al. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344(1):20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 14.Yuan SX, Yang F, Yang Y, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology. 2012;56(6):2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- 15.Nie FQ, Zhu Q, Xu TP, et al. Long non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumour Biol. 2014;35(8):7587–7594. doi: 10.1007/s13277-014-2009-7. [DOI] [PubMed] [Google Scholar]
- 16.Wang K, Long B, Zhou LY. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 17.Gibb EA, Vucic EA, Enfield KS, et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6(10):e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nadal-Ribelles M, Solé C, Xu Z, Steinmetz LM, de Nadal E, Posas F. Control of Cdc28 CDK1 by a stress-induced lncRNA. Mol Cell. 2014;53(4):549–561. doi: 10.1016/j.molcel.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Chen Z, Wang X, Huang Z, He Z, Chen Y. Long non-coding RNA: a new player in cancer. J Hematol Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan JH, Yang F, Wang F, et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Sun J, Bie B, Zhang S, Yang J, Li Z. Long non-coding RNAs: critical players in hepatocellular carcinoma. Int J Mol Sci. 2014;15(11):20434–20448. doi: 10.3390/ijms151120434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kallen AN, Zhou XB, Xu J, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52(1):101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsang FH, Au SL, Wei L, et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int. 2015;35(5):1597–1606. doi: 10.1111/liv.12746. [DOI] [PubMed] [Google Scholar]
- 25.Kinose Y, Sawada K, Nakamura K, et al. The hypoxia-related microRNA miR-199a-3p displays tumor suppressor functions in ovarian carcinoma. Oncotarget. 2015;6(13):11342–11356. doi: 10.18632/oncotarget.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian Y, Zhang YZ, Chen W. MicroRNA-199a-3p and microRNA-34a regulate apoptosis in human osteosarcoma cells. Biosci Rep. 2014;34(4) doi: 10.1042/BSR20140084. pii: e00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang SH, Zhou JD, He QY, Yin ZQ, Cao K, Luo CQ. MiR-199a inhibits the ability of proliferation and migration by regulating CD44-Ezrin signaling in cutaneous squamous cell carcinoma cells. Int J Clin Exp Pathol. 2014;7(10):7131–7141. [PMC free article] [PubMed] [Google Scholar]
- 28.von Maltzahn J, Bentzinger CF, Rudnicki MA. Wnt7a-Fzd7 signalling directly activates the Akt/mTOR anabolic growth pathway in skeletal muscle. Nat Cell Biol. 2012;14(2):186–191. doi: 10.1038/ncb2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirikoshi H, Sekihara H, Katoh M. Up-regulation of Frizzled-7 (FZD7) in human gastric cancer. Int J Oncol. 2001;19(1):111–115. [PubMed] [Google Scholar]
- 30.Yang L, Wu X, Wang Y, et al. FZD7 has a critical role in cell proliferation in triple negative breast cancer. Oncogene. 2011;30(43):4437–4446. doi: 10.1038/onc.2011.145. [DOI] [PubMed] [Google Scholar]
- 31.Vincan E, Flanagan DJ, Pouliot N, Brabletz T, Spaderna S. Variable FZD7 expression in colorectal cancers indicates regulation by the tumour microenvironment. Dev Dyn. 2010;239(1):311–317. doi: 10.1002/dvdy.22045. [DOI] [PubMed] [Google Scholar]
- 32.Song J, Gao L, Yang G, et al. MiR-199a regulates cell proliferation and survival by targeting FZD7. PloS One. 2014;9(10):e110074. doi: 10.1371/journal.pone.0110074. [DOI] [PMC free article] [PubMed] [Google Scholar]