Abstract
The molecular genetic relationship between esophageal adenocarcinoma (EAC) and its precursor lesion, Barrett’s esophagus, is poorly understood. Using whole-genome sequencing on 23 paired Barrett’s esophagus and EAC samples, together with one in-depth Barrett’s esophagus case-study sampled over time and space, we have provided new insights on the following aspects: i) Barrett’s esophagus is polyclonal and highly mutated even in the absence of dysplasia; ii) when cancer develops, copy number increases and heterogeneity persists such that the spectrum of mutations often shows surprisingly little overlap between EAC and adjacent Barrett’s esophagus; and iii) despite differences in specific coding mutations the mutational context suggests a common causative insult underlying these two conditions. From a clinical perspective, the histopathological assessment of dysplasia appears to be a poor reflection of the molecular disarray within the Barrett’s epithelium and a molecular Cytosponge™ technique overcomes sampling bias and has capacity to reflect the entire clonal architecture.
Introduction
Most epithelial cancers present de novo but have progressed from a clinically silent pre-invasive state or so-called intra-epithelial neoplasia. There is growing interest in understanding the life-history of cancers at a molecular level1 so that more focused cancer prevention strategies can be implemented which circumvent the current problems of over-diagnosis inherent in mass screening programs2. Barrett’s esophagus is the precursor lesion to the aggressive cancer, esophageal adenocarcinoma (EAC). In a minority of patients, estimated at 0.33% per year3, Barrett’s esophagus can progress from non-dysplastic Barrett’s esophagus, through intermediate stages of low-grade dysplasia (LGD) and high-grade dysplasia (HGD) to adenocarcinoma. This condition is a classic example of a disease in which clinical strategies characterized by endoscopic random biopsy sampling and pathological diagnosis have arguably failed to improve outcomes for patients4,5.
Historically studies have focused on mutation, methylation and/or loss of heterozygosity of specific target genes, most commonly p16 and TP53, to try to understand the natural history of Barrett’s esophagus and to predict patients at high-risk of cancer6-10. However, there are conflicting interpretations of these data concerning the clonal evolution of this disease. One model formulated by Maley and colleagues7,11 proposed that a mutation (most commonly inactivation of p16), that confers a selective advantage to a cell will sweep across the Barrett’s segment resulting in this mutation being present in the majority of the cells in that Barrett’s segment, a so-called selective sweep. As additional advantageous mutations arise (commonly TP53 loss), these cell clones can then also expand across the Barrett’s segment. This results in a cancer with a serial accumulation of mutations, including drivers and hitchhikers, a proportion of which would be present in all Barrett’s epithelium. Leedham et al.6 have subsequently described a more heterogeneous model whereby multiple independent clones arise, some of which die out, and some of which are maintained. Hence, genetic aberrations present within these clones would not necessarily sweep across the whole Barrett’s segment, depending on the competitive advantage of the individual clones. This scenario could lead to a far more heterogeneous Barrett’s esophagus and cancer tissue. Although the two different models are not mutually exclusive12, the limited resolution of the molecular genetic alterations that stems from analysis of a small number of candidate genes, has made it difficult to resolve the issue.
Recent DNA sequencing studies have demonstrated a high mutational burden within EAC13-16, and described distinct mutational signatures observed within this cancer type13,16,17. TP53 is by far the most commonly mutated gene within EAC, followed by a plethora of genes that are mutated in a smaller proportion of cases (<25%), such as ARID1A, SMARCA4, SMAD4 and SYNE115,16. In a study using exome sequencing data from two matched Barrett’s and EAC samples, Agrawal and colleagues14 found that approximately 80% of the cancer mutations were already present in DNA from the adjacent Barrett’s epithelium. However, the grade of dysplasia and the spatial relationship of these samples were not reported. We recently demonstrated that putative esophageal driver genes, such as ARID1A and SMARCA4, were also recurrently mutated in patients with a very stable phenotype who had never shown any evidence of dysplasia within their Barrett’s segment over multiple years of follow-up (median 58 months)13. This would argue against such genes having a causal role in cancer progression. In contrast, TP53 and SMAD4 mutations were highly specific to patients with HGD and EAC, respectively. However, though informative, these studies were not designed to deduce the clonal architecture over time and space for Barrett’s esophagus carcinogenesis.
The aims of this study were therefore to examine the clonal ordering and heterogeneity of Barrett’s esophagus (including various degrees of dysplasia) that had progressed to EAC. We were able to examine this to a higher level of detail than had been previously possible through virtue of some highly characterized sample-sets and the recent technological developments which permit genome-wide sequencing from minute pieces of paraffin-embedded archival material, as well as fresh-frozen tissues. Thus we interrogated the mutational landscape, on a genome-wide scale, of 23 paired Barrett’s esophagus and EAC samples as well as 73 samples taken over a three-year period from one patient’s Barrett’s esophagussegment (displaying all the different stages of progression, from non-dysplastic Barrett’s to intramucosal adenocarcinoma). From these data we were able to determine the mutational load, the mutational context as well as the clonal composition and heterogeneity in pathologically defined steps of Barrett’s esophagus carcinogenesis.
Results
Barrett’s esophagus is highly mutated and polyclonal
Paired Barrett’s and cancer samples taken at the same time point from 23 EAC patients (see Supplementary Table 1 and 2 for demographic and tumor information) with macroscopically visible Barrett’s esophagus were whole-genome sequenced. All Barrett’s and EAC samples were sequenced to a minimum of 54-fold coverage and the germline normal comparison (blood or normal esophageal squamous) to a minimum of 31-fold coverage (Supplementary Table 3). The median number of single nucleotide variants (SNVs) present within the EAC samples was 18,786 (interquartile range (IQR) 15,007-32,034) and 12,714 (IQR 6,604-21,559) for the Barrett’s esophagus samples. The average somatic mutation rate for the Barrett’s samples was 6.76/Mb which is higher than for multiple myeloma (2.9 SNVs/Mb)18, luminal breast cancer (1.1 SNVs/Mb)19, hepatocellular carcinoma (3.69 SNVs/Mb)20 and colorectal adenocarcinoma (5.9 SNVs/Mb)21. Although Barrett’s esophagus adjacent to EAC was found to be highly mutated and contain thousands of SNVs, even for samples with no histological dysplasia, there were significantly more SNVs called in the tumor samples (average somatic mutation rate 10.02/Mb) compared to the Barrett’s esophagus samples, (Wilcoxon Signed Rank test, p<0.001)(Figure 1a). Furthermore, this was apparently not driven by a difference in the purity between the Barrett’s and the tumor samples as there was no significant correlation between the number of SNVs called and the purity of the samples (Spearman’s Rho, r=0.108, p=0.475) (Supplementary Table 3). Of note, the Barrett’s esophagus samples with dysplasia (either LGD, HGD or indefinite for dysplasia called by two independent expert pathologists), did not have significantly more mutations than the Barrett’s esophagus samples with no dysplasia (Mann Whitney test, p=0.271).
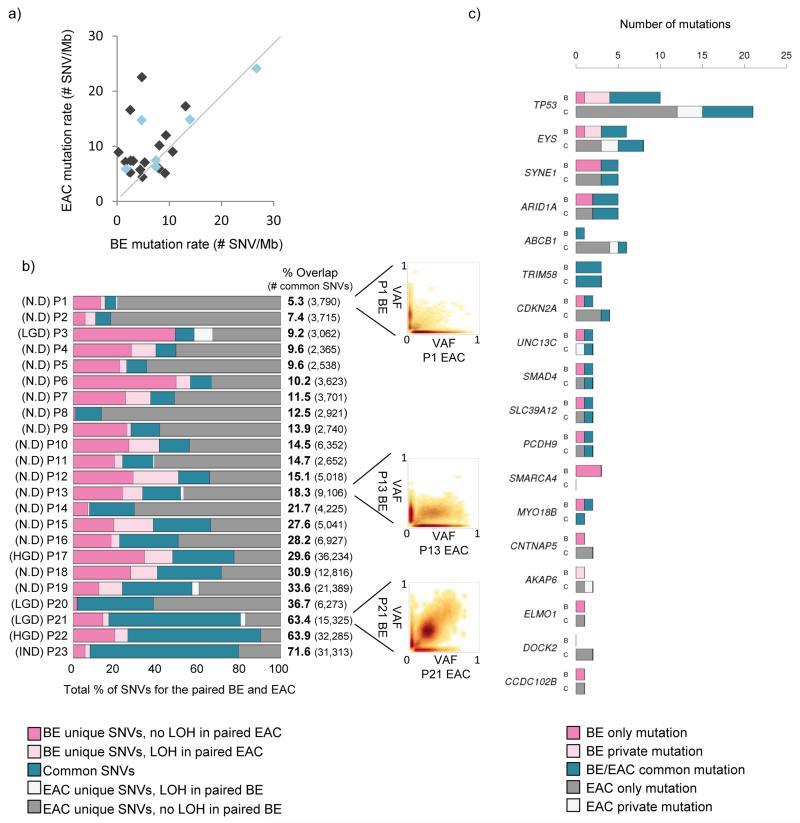
Figure 1. Paired Barrett’s and EAC samples have a varied overlap.
a) Scatter plot comparing the mutation rate between paired Barrett’s and EAC samples. Light blue dots indicate Barrett’s samples with some degree of dysplasia present. b) Diagram showing percentage overlap between SNVs in paired Barrett’s and EAC samples, including highlighting SNVs that lie in areas of LOH in the reciprocal sample. Barrett’s unique SNVs that lie in an area of LOH in the paired EAC sample are shown in light pink and EAC unique SNVs that lie in a region of LOH in the paired Barrett’s sample are shown in white. Samples are ranked according to their degree of overlap, from poor to good. Scatter plots illustrating an example of a poor overlap (patient P1), a fair overlap (patient P13) and a good overlap (patient P21) are shown. c) Bar graph showing genes that were found to be recurrently mutated in previous EAC sequencing studies13,16 and are mutated in at least two patients in our Barrett’s-EAC cohort (SNVs or indels). For each gene of interest the data for the Barrett’s samples are in the bar labelled “B” and for cancer samples in the bar labelled “C”. Mutations that are common to both paired Barrett’s and EAC samples (shown in teal), as well as mutations that are Barrett’s unique (dark pink) or EAC unique (dark grey) are shown. Also shown are mutations in the same gene that have a different base pair change between paired Barrett’s and EAC samples (called “private mutations”).
Surprisingly, when we looked at the percentage of SNVs that were common to both the paired Barrett’s esophagus and EAC samples, after performing additional filtering and only considering SNVs with good coverage in both paired samples (see online methods for more information), we found a lower degree of overlap than we would have first expected (<20% overlap between the SNVs in the paired Barrett’s esophagus and the EAC samples in 13/23 (57%) of the samples), and this low degree of overlap was not primarily due to loss of heterozygosity (LOH) in the paired sample (Figure 1b) or differences in purity or sequencing depth between the paired samples. The Barrett’s esophagus samples that had a better overlap with their paired EAC sample were more likely to be dysplastic (Mann-Whitney test, p=0.019). We did not find any other association between samples with a poor overlap and any clinical features, such as stage or differentiation status of the tumor, age or gender of the patient as well as the length of the Barrett’s segment or the tumor (p>0.5 for all comparisons). As expected TP53 was the most recurrently-mutated gene occurring in 19/23 (82.6%) EAC samples. Barrett’s esophagus samples harbored TP53 mutations less commonly (9/23 (39.1%) of which 5 were dysplastic). Of note, TP53 mutations present within Barrett’s esophagus adjacent to EAC were not always present within the paired EAC sample (4/9 cases), however, out of these four cases, three of them had a different TP53 mutation present in the paired EAC sample (Barrett’s esophagus private mutations in Figure 1c). Other previously reported putative EAC driver genes, such as EYS, ARID1A and ABCB1, were mutated less commonly (≤30%) and if mutated within a patient’s tissue (either Barrett’s esophagus or EAC), were seldom shared (21/73, (28.8%)) between the paired Barrett’s and EAC samples (Figure 1c).
In order to be sure that the poor overlap was not due to sampling bias we performed whole-genome sequencing (WGS) on additional samples taken from 5/23 cases. These data showed a very similar degree of overlap to the paired Barrett’s and EAC data (Figure 1), regardless of whether you consider all samples together or any two samples from the Barrett’s esophagus compared with the EAC from the same patient (Supplementary Figure 1). Hence, taken together these data show that there is significant heterogeneity in the spectrum of mutations with surprisingly little overlap in the molecular genetics between EAC and adjacent Barrett’s esophagus.
Copy number increases as EAC develops
Although Barrett’s esophagus adjacent to EAC was found to be highly mutated, we observed a stark contrast between the copy number aberrations within the Barrett’s samples compared to the EAC samples (Figure 2a and b and Supplementary Figure 2). With the exception of two Barrett’s esophagus samples (from patients P3 and P21) both of which showed features in keeping with LGD, the Barrett’s esophagus samples, even those with HGD (patients P17 and P22), contained very few copy number changes, with the vast majority of their genomes being diploid (median percentage genome copy number 2 = 99.7% (range 62.8-100.0%)). This was significantly different from the EAC samples (Wilcoxon Signed Rank test, p<0.001), which showed a range of copy numbers including some highly amplified regions (15-20 copies), with a median percentage of the genome not diploid = 37.6% (range 2.1-87.8%).
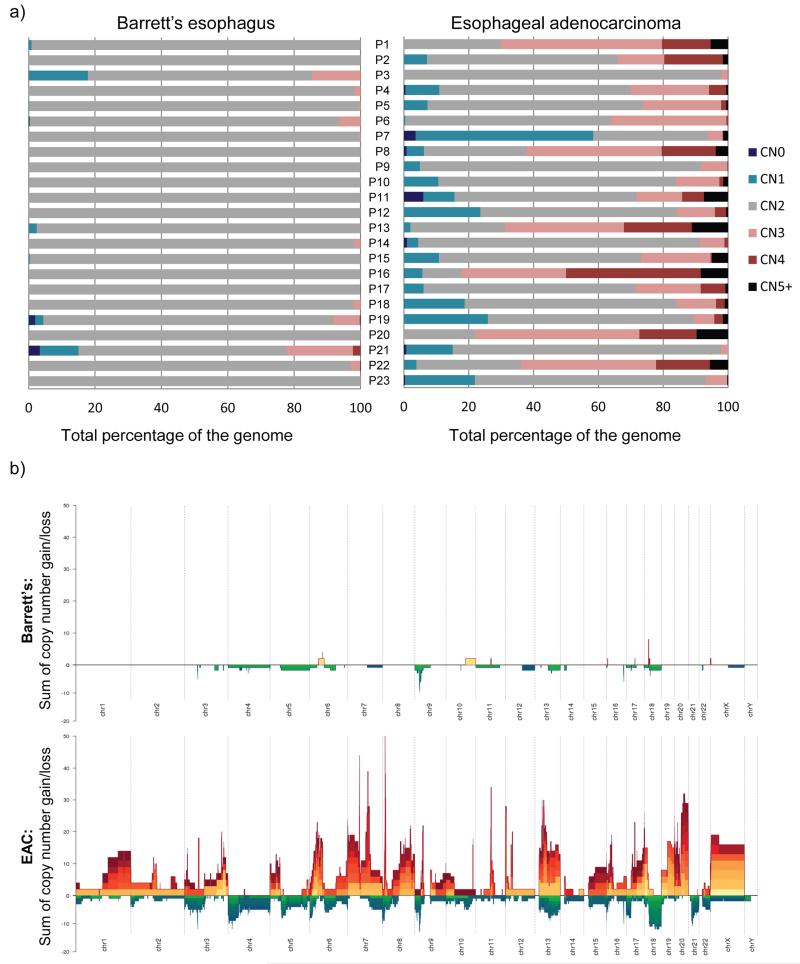
Figure 2. EAC samples display multiple copy number changes compared to paired Barrett’s esophagus samples.
a) Graphs showing the percentage of the genome at different copy number states for each patient (P) in turn (P1-23) for their paired Barrett’s esophagus (left hand graph) and EAC samples (right hand graph). CN0 = copy number 0, CN1 = copy number 1, CN2 = copy number 2, CN3 = copy number 3, CN4 = copy number 4, CN5+ = at least copy number 5. b) Stacked mountain plots summarizing copy number variation within all of the 23 Barrett’s esophagus and EAC samples. Gains of at least two copies on top of the normal copy number in that region are illustrated by yellow-orange-red mountains and deletions are represented by green-blue valleys. The height or depth of the mountain or valley indicates the summed copy number status across all patients for that region. The colors represent different samples so that the higher the number of different colors in a region, the higher the number of samples that display that copy number change.
The only common copy number change within the Barrett’s samples was 9p LOH which was present in 11/23 (48%) samples. The majority of EAC samples (16/23 (70%)) also had 9p LOH. However, of the 11 Barrett’s samples with 9p LOH, only 7 also had the same alteration in their cancer sample. Within the EAC samples, when considering more focal copy number changes (i.e. less than half of the particular chromosome arm) we found 17 commonly amplified (at least four copies) and 18 deleted regions that were present in at least 3/23 (13%) samples. The commonly amplified regions included previously reported amplified regions in EAC22, namely GATA4, KLF5, MYB, PRKCI, CCND1, FGF3, FGF4, FGF19 and VEGFA, some of which are potential therapeutic targets. The deleted regions included some known fragile sites (FHIT and WWOX) as well as some previously reported22 and potentially interesting targets, namely A2BP1, CDKN2A, PDE4D, PTPRD and PARK2.
Mutational context is unchanged between Barrett’s and EAC
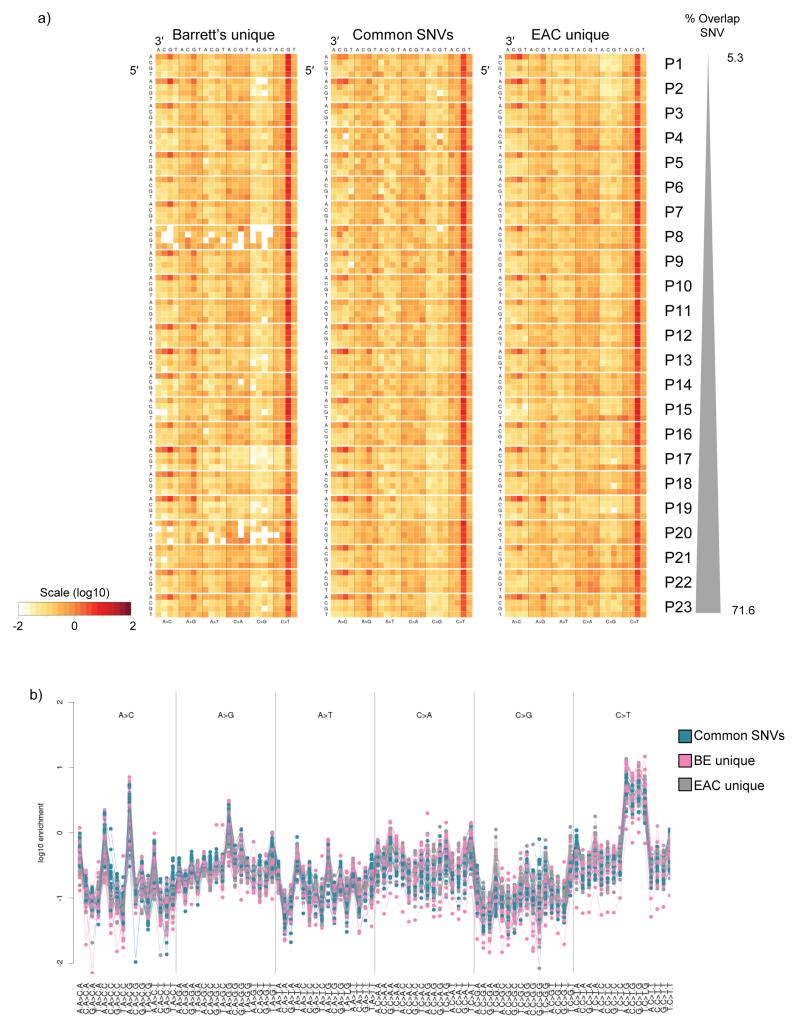
From the SNV overlap data we could infer the early SNVs (i.e. those present in both the paired Barrett’s and EAC samples) as well as the later events (i.e. SNVs unique to either the Barrett’s or the tumor samples). From these categorizations, we wanted to know whether the mutagenic stimulus was common between the early and the late events. The previously identified EAC mutational signature (A:T>C:G specifically at AAG trinucleotides)13,16 was similarly enriched in both the early and the late SNVs (both Barrett’s and EAC unique), (Figure 3a and b). There was a good correlation when comparing the mutational context between all the Barrett’s esophagus unique, the EAC unique and the common SNVs (average correlation = 0.83, 0.86 and 0.86, respectively), however, there appeared to be some difference between the early and the late SNVs, albeit small (6.9% variance in the second component), when using principal component analysis (Supplementary Figure 3a). This difference seems to be driven mainly by four different mutational contexts; A>C at CAC, C>G at CCG, A>T at CAC and C>G at TCA (Supplementary Figure 3b).
Figure 3. The mutational context is similar in both early and late SNVs.
a) Heat map showing the log-transformed values representing the fraction of each mutation type at each trinucleotide mutation context corrected for the frequency of each trinucleotide in the reference genome, as described in Nik-Zainal et al27. The mutational contexts were calculated separately for the three subsets of SNVs per patient, i.e. 1. Barrett’s unique and not in EAC LOH, 2. common to Barrett’s esophagus and EAC, and 3. EAC unique and not in Barrett’s esophagus LOH. b) Mutational context plotted as a dot plot showing the enrichment of the trinucleotide mutational context information for every possible option for all 23 paired Barrett’s esophagus and EAC samples.
Spatial and temporal characterization of a Barrett’s segment
As we observed such heterogeneity in the genetic landscape between the paired Barrett’s and EAC samples, we sought to characterize, in precise detail, the Barrett’s esophagus segment from a single patient (AHM1051) in order to better understand the clonal evolution and how this relates to cancer development. To do this we studied multiple samples from a 58-year-old male patient who displayed all the stages of the Barrett’s esophagus progression series; gastric metaplasia, intestinal metaplasia, LGD, HGD and intramucosal adenocarcinoma. Following a tertiary referral this patient underwent five endoscopies between May 2009 to March 2012, and samples were available across the full length of the patient’s 10 cm Barrett’s segment for this time period (outlined in Figure 4). This patient was also selected as he had previously swallowed a Cytosponge, which is a non-endoscopic cell sampling device that collects cells from the gastro-esophageal junction, the entire length of the esophagus, as well as the oropharynx, providing an opportunity to assess the clonal architecture from a single sample, which may have clinical application.
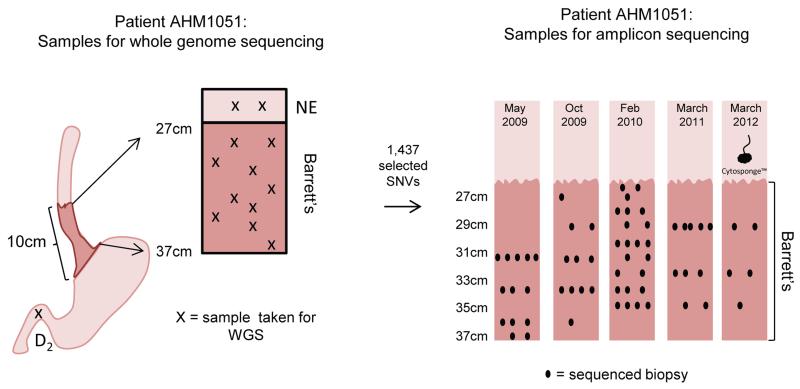
Figure 4. Summary of the samples sequenced for patient AHM1051.
Ten Barrett’s esophagus samples (depicted by crosses on patient AHM1051’s Barrett’s segment) representing all stages along the progression sequence from Barrett’s esophagus with no dysplasia to intramucosal adenocarcinoma, as well as two normal esophageal squamous (NE) and one duodenum (D2) sample were sequenced (whole-genome sequencing, WGS). From the WGS data, 1,437 SNVs were assessed on additional FFPE samples taken from multiple different endoscopies across the full length of the 10cm Barrett’s esophagus segment between May 2009 and March 2012. The biopsy samples selected for amplicon sequencing are shown using black dots (positions of the biopsies along the x-axis are for illustration only and hold no extra information). One Cytosponge sample taken in March 2012 was also included in the amplicon sequencing.
Somatic SNVs (both coding (variant allele fraction (VAF)>0.08) and non-coding present at the highest VAFs within the different Barrett’s esophagus WGS libraries) were selected for targeted re-sequencing, with the expectation that these SNVs would define the clones present within this patient’s Barrett’s segment and could be assessed across a larger number of samples. Using a panel of 1,443 targets, sequencing was performed on 73 individual samples (Figure 4). We used a custom-made data browser to investigate this complex dataset (available with Supplementary Material). All but two of the 1,443 SNVs gave useable data with sufficient sequencing depth for the 73 samples with a median coverage of 7,760× (IQR 6,015-10,100) for the samples and a median coverage of 6,504× (IQR 3,285-10,940) for the 1,441 targets. 99.7% (1,437/1,441) of the selected SNVs were verified as real and somatic (VAF > 0.01).
Clonal heterogeneity within non-dysplastic Barrett’s
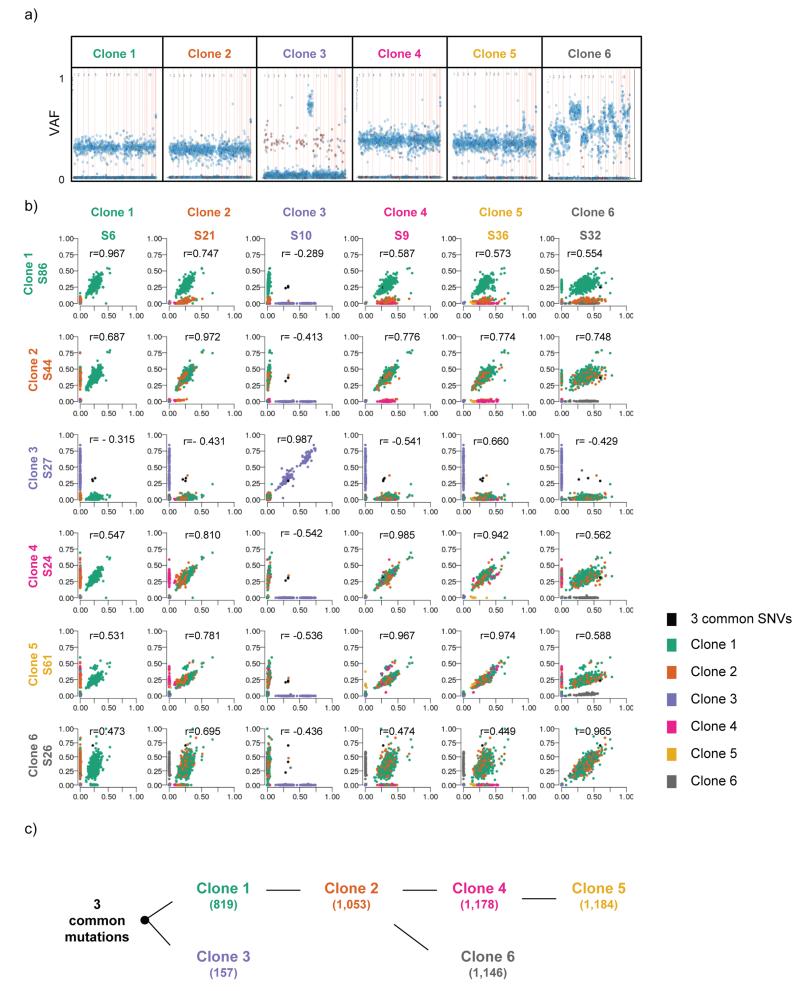
Within the patient’s sequenced Barrett’s samples, 22 biopsies showed no evidence of dysplasia. These samples were particularly interesting as we were interested to see the clonal ordering within low-risk Barrett’s epithelium that looked cytologically normal. Of these biopsies, one had gastric metaplasia and all the rest contained intestinal metaplasia. An initial analysis was performed using the Pearson correlation of the VAF values from the targeted amplicon sequencing. This method allowed us identify clones based on their overall mutation patterns, taking advantage of the large proportion of VAF that are affected by the extensive copy number changes in the EAC samples. Because it groups based on VAF patterns, this method is more resistant to variable clonality and purity. The analysis of these samples revealed six different groups and pairwise comparison of the samples (Figures 5a and b and Figure 6a) allowed us to derive the clonal hierarchy in the Barrett’s segment (Figure 5c).
Figure 5. Six distinct clones are present within patient AHM1051’s non-dysplastic Barrett’s esophagus.
a) Variant allele fraction (VAF) plot representing an example sample of the six distinct clones. The 1,437 assessed SNVs are represented by dots within the chart. The SNVs are ordered on the x-axis according to their genomic location (from chr1 to sex chromosomes). The y-axis represents the VAF for each mutation for that sample. The blue dots represent intergenic or intronic SNVs and the red dots represent coding SNVs. b) Scatter plots showing the correlation between two different representative samples within the same clone as well as compared to two different samples from every other clone. The sample names (e.g. S6 (sample 6)) are given under the different clone headings. Each dot within the scatter plot represents one of the 1,437 assessed SNVs and the dots are coloured according to the different clones the SNVs belong to. The r value represents the Pearson correlation. c) Diagram representing the clonal ordering within patient AHM1051’s Barrett’s esophagus segment. The numbers in brackets represent the number of SNVs (out of a total of 1,437 that were assessed) that are present in each of the clones.
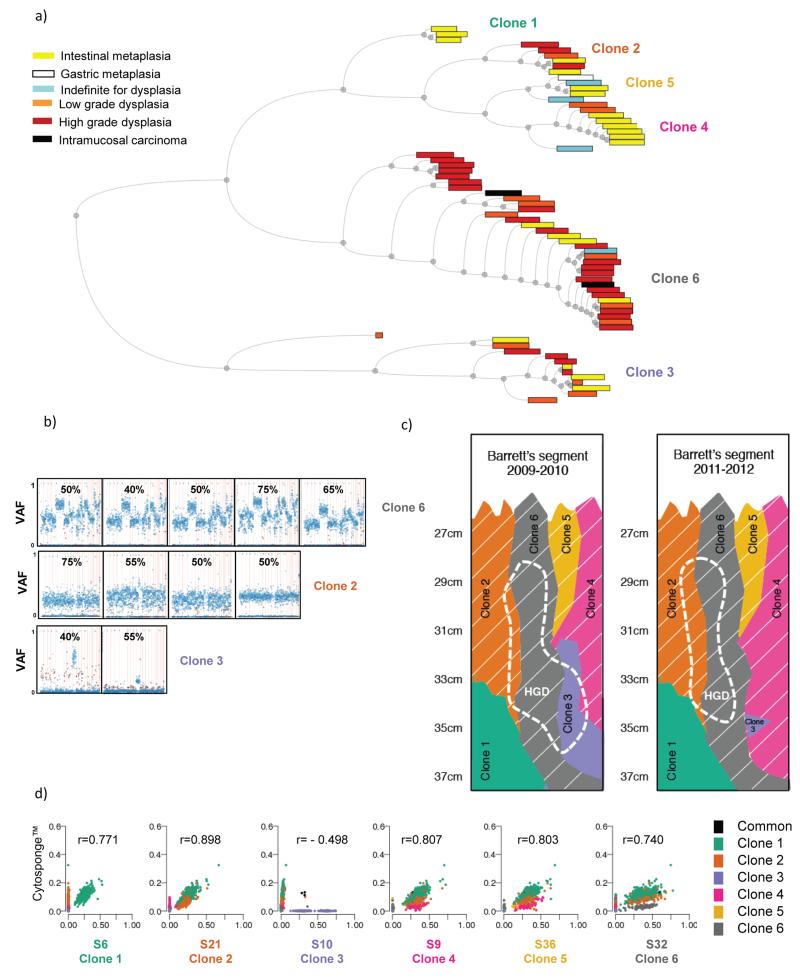
Figure 6. Multiple different clones can give rise to dysplasia.
a) Hierarchical tree showing all samples processed for amplicon re-sequencing. Within the tree, samples are paired according to their Pearson correlation score. The distance between pairs is represented by the horizontal distance between the pair and their parent node. There is no information embedded in the vertical distance of this tree. The colors of the boxes represent the histopathological grade for each of the Barrett’s esophagus samples. The width of the boxes is proportional to the number of SNVs with VAF>0.01 in each sample. The clones represented by the six different branches are noted. b) VAF plots as described in Figure 5a showing examples of samples that contain ≥40% high grade dysplasia (as determined by the average between two expert upper gastrointestinal pathologists) within the sample and grouped according to which clones they represent. The number at the top of each graph indicates the percentage of high grade dysplasia in that specific sample, as assessed by two expert gastrointestinal pathologists. c) Illustration depicting the clonal arrangement in patient AHM1051’s Barrett’s esophagus segment before (2009-2010) and after (2011-2012) endoscopic treatment. The region corresponding to high grade dysplasia, before and after treatment, is shown using a dashed line. The clones containing the widely-spread TP53 mutation are indicated by thin, slanted, white lines. d) Scatter plots indicating the correlation between SNVs identified in the DNA from cells collected using the Cytosponge compared with the six different clones. The r value represents the Pearson correlation.
Three SNVs, all occurring in non-coding regions and representing the most-recent common ancestor (chr4:33658353 T>C, chr13:88285478_A>C and chr18:11483749_A>C), were found in all six clones providing evidence for an initial clonal sweep of the patient’s Barrett’s segment. After this initial clonal sweep two very different clones arose, Clone 1 and Clone 3 (Figure 5b and c) displaying many more genome-wide SNVs (Clone 1: 819/1,437 assessed SNVs and Clone 3: 157/1,437 assessed SNVs). Clone 3 had 9p LOH, a common event observed in non-dysplastic Barrett’s esophagus and one that has been previously shown to impart selective advantage to Barrett’s esophagus cells7, however this clone did not appear to seed further clones within this patient’s Barrett’s segment (Figure 5c). Unlike Clone 3, Clone 1 was able to seed a daughter clone, which we have called Clone 2. Clone 2 contained 234 additional SNVs, compared to Clone 1 (colored in orange in Figure 5b), and subsequently this gave rise to two different clones, namely Clone 4 and Clone 6. Clone 4, containing 1,178/1,437 assessed SNVs, subsequently gave rise to an additional clone, Clone 5 (1,184/1,437 SNVs assessed as well as a large-scale deletion/amplification on chromosome 15). Clone 6 (1,146/1,437 assessed SNVs), in addition to the 93 extra SNVs compared to Clone 2, displayed a large number of copy number changes, specifically large-scale deletions in chromosomes 5, 11, 13 and 18, as evidenced by the jagged appearance of the VAF plots in Figure 5a and supported by the WGS data (Supplementary Figure 4). This is especially interesting as all of the three samples that are classified as Clone 6 were assigned a histopathological diagnosis of non-dysplastic Barrett’s esophagus although they contain a large number of somatic mutations and copy number changes. More information on how the clonal hierarchy was derived can be found in online methods. It should be noted that whether or not the sample sequenced was fresh-frozen or paraffin-embedded did not lead to any systematic bias that affected the clonal assignments.
Dysplasia can develop from multiple different clones
In addition to multiple regions of Barrett’s esophagus with no dysplasia, patient AHM1051 has multiple areas with cellular abnormalities which were graded according to the degree of dysplasia by consensus review by two expert gastrointestinal pathologists. These samples allowed us to assess the clonal architecture within the Barrett’s esophagus progression sequence. Clone 6 (Figure 5a and Supplementary Figure 3) displayed the highest number of copy number changes and would be the obvious culprit for seeding HGD. However, interestingly, Clones 2, 3 and 6 were all found to give rise to HGD (Figure 6a, b and c). Similarly clones 2, 3, 4 and 6 all appeared to give rise to LGD. Clones 1 and 5 were the exceptions for LGD; however, as there are few samples available from these two clones, this may be a result of sampling bias.
Stability of the Barrett’s esophagus segment over time
Taken together, these data demonstrate a genetically stable clonal pattern within this patient’s 10-cm Barrett’s segment. All six identified clones were present in 2009, the year that this patient was diagnosed as having Barrett’s esophagus with dysplasia. Almost three years later, the same six clones can be identified (Figure 6c). The only alteration to the Barrett’s esophagus segment appears to be as a result of this patient’s clinically ineffective endoscopic treatment which resulted in the shrinking of Clone 3 (only partially visible in one biopsy post-treatment). This decrease in Clone 3 is further evident using a non-endoscopic, cell sampling device, the Cytosponge. Amplicon sequencing of cells collected using the Cytosponge taken after endoscopic treatment (March 2012), showed no evidence of Clone 3 (Figure 6d) suggesting that this clone had substantially shrunk in response to endoscopic therapy. However, SNVs defining clones 1, 2, 4, 5 and 6 were all present within the Cytosponge sample and clone 6 was the most abundant in keeping with the large size of this clone ascertained form the biopsy data (Figure 6d). This suggests that the Cytosponge could simultaneously sample all five clones present within the patient’s Barrett’s segment and provide some indication of relative clone size.
Given the pathogenic importance of TP53, in a more targeted approach we also mapped mutations at this locus within this patient’s Barrett’s segment. TP53 sequencing on the Cytosponge sample13, identified a dominant TP53 mutation (chr17:7577538 C>T) which was also identified in the WGS. This particular TP53 mutation established itself in Clone 2 (VAF between 0.28 – 0.56) and was therefore also present in all the daughter clones (i.e. Clones 4, 5 and 6) (shown by slanted white lines in Figure 6c), however, with a much higher VAF in Clone 6 (between 41-73%), probably due to LOH. The presence of a TP53 mutation in non-dysplastic Barrett’s esophagus from a patient with early cancer is similar to that seen in the WGS data which showed that 4/17 (23.5%) non-dysplastic Barrett’s samples taken adjacent to EAC contained TP53 mutations. These data from patients who have progressed to cancer should not be confused with studies that look at the TP53 mutation prevalence in non-dysplastic Barrett’s esophagus from patients who never progress to dysplasia and/or EAC13,23,24 .
Discussion
In summary, the application of powerful sequencing technology to cases of Barrett’s associated carcinogenesis has led to new insights about the history of the disease. Firstly, there are numerous somatic point mutations as well as small insertion and deletions which occur in all pathological stages of the disease progression. The specific SNVs generally overlap poorly between paired Barrett’s and EAC samples, however, the mutational context of these SNVs is mostly common between the ends of the disease spectrum (non-dysplastic Barrett’s esophagus and EAC) suggesting exposure to common mutagens throughout the progression sequence. Although the mutational context is consistent along the progression sequence, there is a marked increase in the copy number changes in EAC, which is rarely seen in Barrett’s epithelium. From a clinical perspective, the histopathological assessment of dysplasia appears to be a poor reflection of the molecular disarray within the Barrett’s epithelium as the same aberrant genetic profile was seen in both dysplastic and non-dysplastic Barrett’s tissue.
The variation in the overlap between the paired Barrett’s and EAC samples in this study, especially the high proportion of patients’ samples that showed such heterogeneity (13/23 Barrett’s-EAC pairs with <20% overlap), was surprising. However, these data do demonstrate how much the Barrett’s segment has evolved since seeding the tumor giving an indication as to the long natural history of the condition. One way to capture this genetic heterogeneity in patients with a patent lumen is using the non-biased Cytosponge which we have shown can sample all five remaining clones in patient AHM1051’s Barrett’s esophagus segment after endoscopic therapy. This is a proof-of principle experiment and further work is required to demonstrate the clinical utility of this sampling approach for detecting mutations representative of the entire Barrett’s esophagus segment.
The in-depth study of the Barrett’s esophagus segment of patient AHM1051 led to a detailed map of the clonal ordering and heterogeneity within a 10-cm Barrett’s segment. All six clones identified in this Barrett’s esophagus segment contain multiple SNVs and have varying abilities to seed further daughter clones. Furthermore, not all of the six clones span the whole length of the Barrett’s esophagus segment, but some appear to remain more localized. The clonal pattern present in this patient has aspects of both models that have previously been proposed for the clonal evolution of Barrett’s esophagus6,7. There is evidence of a common ancestor prior to branched evolution within the patient’s Barrett’s segment, indicated by the three non-coding, common mutations present in all Barrett’s esophagus samples. This is also supported by the common SNVs identified between the 23 paired Barrett’s esophagus and EAC samples. However there is also evidence for the emergence of distinct clones, such as Clone 1 and 3, neither of which were able to sweep across the whole Barrett’s segment. Furthermore, deep sequencing of individual Barrett’s esophagus biopsies has allowed the identification of multiple clones within the same biopsy, demonstrating that this approach is sufficient to identify the clonal heterogeneity within Barrett’s esophagus without requiring further microdissection.
This study is only one step in further understanding clonal evolution and heterogeneity in Barrett’s esophagus. The holy grail is still to be able to predict which patients will develop dysplasia and ultimately cancer. Approaches, such as the one used here, have the ability to further improve our understanding of the clonal structure within Barrett’s esophagus before and after the development of dysplasia, with the hope of being able to predict which patients will progress to cancer. Li et al25 recently reported that Barrett’s esophagus patients who do not progress to cancer have stable genomes, whereas patients who progressed to cancer had unstable and diverse genomes which evolved at least four years before developing a cancer. However, our data suggest a more complex situation since HGD can arise from multiple different clones, some of which appear to have very few copy number aberrations. The clinical implication of these findings is that when endoscopic therapies, such as mucosal resection or ablation, are undertaken there is a need to treat the whole Barrett’s esophagus segment to ensure that no residual Barrett’s epithelium remains. This is important as meta-analysis of 18 studies demonstrated that Barrett’s esophagus with intestinal metaplasia was still present in 22% of individuals (95% confidence interval: 14-30%) who were treated with radiofrequency ablation26.
In conclusion, this study has shed more light on the similarities and differences in the genetic aberrations in the progression from Barrett’s esophagus to EAC. In the future it will be important to integrate transcriptomic and epigenetic data with genome-wide DNA sequence in patients who span the disease spectrum. This approach, which incorporates state of the art technology on fresh and archival specimens, paves the way for further studies in other tissue types aiming to chart the progress from intra-epithelial to invasive cancer.
Online methods
Patients, clinical material and consent
This study was approved by the Institutional Ethics Committees (REC Ns 07/H0305/52, 10/H0305/1 and 10/H0308/71) and all patients gave individual informed consent. Patients with EAC were recruited prospectively at six different centers (Addenbrookes Hospital, Cambridge, Royal Surrey County Hospital, Guildford, St Thomas Hospital, London, Gloucester Royal Hospital, Gloucester, Edinburgh Royal Infirmary, Edinburgh and Salford Royal Infirmary, Manchester) and samples (both cancer and Barrett’s esophagus) were obtained at the same time either from surgical resection, endoscopic ultrasound or endoscopic mucosal resection. Blood (for 22/23 EAC patients) or frozen normal squamous esophageal samples at least 5cm distant from the tumor (for 1/23 patients), were used as a germline reference. For the Barrett’s esophagus patient (AHM1051) who was studied in detail, a frozen duodenum sample was used as the germline control. Tissue samples were either snap-frozen in liquid nitrogen immediately after collection and stored at −80°C or formalin-fixed and paraffin embedded (FFPE) as per the usual clinical protocol. Prior to DNA extraction, one section was cut from each esophageal tissue sample and H&E staining was performed to assess the exact histopathology for every individual sample. EAC samples were deemed suitable for DNA extraction only after consensus review by two expert gastrointestinal pathologists confirmed the tumor cellularity to be ≥70%. Where blood was not available for the germline reference the same review process was applied to the normal esophageal samples to ensure that only squamous epithelium was present. Barrett’s esophagus samples, both frozen and FFPE, were reviewed by two expert gastrointestinal pathologist (M O’D and SM) to identify any dysplasia or cancer cells. For the whole-genome sequencing, Barrett’s esophagus samples with any evidence of cancer cells or cancer samples with any evidence of Barrett’s esophagus, were excluded.
DNA extraction from clinical material
For the frozen normal, Barrett’s esophagus and EAC samples, DNA was extracted from the tissue using either the DNeasy kit (Qiagen) or the AllPrep DNA/RNA Mini Kit (Qiagen), according to the manufacturer’s instructions. DNA was extracted from blood samples using the Nucleon™ Genomic Extraction kit (Gen-Probe,) according to the manufacturer’s instructions. For the Barrett’s esophagus FFPE biopsy samples, genomic DNA was extracted from the rest of the diagnostic biopsy and each diagnostic biopsy was extracted separately. For the Cytosponge sample, genomic DNA was extracted from 8 × 10 μm sections of the processed Cytosponge FFPE clot. The FFPE samples were extracted using Deparaffinization Buffer (Qiagen) and the QIAamp FFPE DNA Tissue Kit (Qiagen). The protocol was followed as described by the manufacturer with the exception that samples were incubated at 56°C for 24 hours instead of the described 1 hour, and 10 μl of extra Proteinase K was added to the samples roughly half way through the 24 hour incubation. For all sample types the DNA was quantified using the Qubit™ dsDNA BR or HS Assay Kits (Life Technologies).
Whole-genome sequencing analysis
For the patients with EAC; a normal, a Barrett’s esophagus and a cancer sample were sequenced for each of the 23 patients. For 5/23 patients, additional Barrett’s esophagus and EAC samples were also sequenced. For the Barrett’s esophagus patient, AHM1051, 13 samples were sequenced of which 10 were from fresh-frozen biopsies and 3 were from FFPE diagnostic biopsies. The 13 samples included two normal esophageal squamous, one duodenum, three Barrett’s esophagus with intestinal metaplasia, two Barrett’s esophagus with gastric metaplasia, two Barrett’s esophagus with LGD, two Barrett’s esophagus with HGD, and one with intramucosal adenocarcinoma. The 13 samples were collected between May 2009 and March 2012.
For all fresh-frozen samples, libraries were constructed with ~300bp insert length. For all FFPE derived samples, library preparation followed a modified TruSeq PCR Free protocol designed to retain more fragments of low molecular weight an abundance of which are often present in FFPE derived DNA as a result of degradation of the sample. Whole genome sequencing (WGS) was performed on the Illumina HiSeq2000 instrument. We generated 100 bp paired-end sequence reads using v3 clustering and sequencing chemistry. Alignment to human GRCh37.1 and quality control was performed using the Illumina CASAVA v1.8 pipeline for the AHM1051patient WGS data and using ISAAC28 for all samples of the patients with EAC. Identification of somatic single nucleotide variants (SNVs) and somatic small indels (<50bp) was performed by Strelka29 in the 12 Barrett’s esophagus samples from patient AHM1051 using the duodenum sample as the matching normal sample and in both cancer and Barrett’s esophagus samples using the matched normal for the patients with EAC.
The overlap of SNV calls between a matching Barrett’s esophagus and EAC sample was calculated as: at the union of SNV positions, the base counts for variant and reference allele were retrieved. Positions that did not achieve at least 18× coverage in both samples, that have more than 1500× coverage or that have more than 10 reads filtered out because of poor alignment score in either sample were discarded from the following counts. Positions were considered in the intersection (overlap) if there were high quality reads in one sample for a mutation to be called with high confidence. In this case we would look at the other paired sample to determine if there was any evidence to support the somatic variant in that sample. Since it was highly unlikely that the exact same base pair change would occur in another sample from that patient by chance, a single read was considered to be sufficient supporting the variant allele; SNVs were considered unique in Barrett’s esophagus or EAC if the corresponding EAC or Barrett’s esophagus sample contained only reference bases covering the position. The unique SNVs were further separated into two categories depending on whether or not they fell in regions of LOH in the paired sample, as one cannot tell if those SNVs are truly unique to the sample or have been lost in the other sample. Half of the coding SNVs (47/89) as well as all of the indels (9/9) shown in Figure 1c were manually reviewed to ensure that the observations were robust and not a result of any analysis artefact. All of the inspected 47 SNVs and 9 indels were found to be real and somatic. 101 SNVs (of which 49 were classed as Barrett’s esophagus unique, 41 EAC unique and 11 common between the Barrett’s esophagus and the EAC sample) from Patient 3, representing a patient with poor overlap between the paired Barrett’s esophagus and EAC samples, were manually reviewed. 99/101 (98%) were confirmed to be real and somatic with the other two being classed as inconclusive.
The mutational context of SNVs was calculated as in Nik-Zainal et al.27 for each of the three subsets of SNVs per patient: (i) unique to Barrett’s esophagus not in EAC LOH, (ii) unique to EAC not in Barrett’s esophagus LOH, and (iii) common to Barrett’s esophagus and EAC. Pairwise correlations for each of the comparisons were performed and then averaged to give an average correlation value per comparison.
Copy number and loss of heterozygosity analysis
A two-step workflow was used for the detection of copy number and LOH changes (data not shown). First, GC-normalized coverage and B-allele ratios were derived from tumour and normal samples and jointly segmented using an unbalanced Haar wavelet transform. Next, given the location of change points in the genome, a least-squares model was applied to derive purity and ploidy that were used to assign a copy number to each segment. For this last step, the observed coverage, germline and somatic B-allele ratios were compared with the expected ones for each copy number, purity and ploidy combination to identify the model with the smallest least-squares value. Since this method is unpublished we also ran CNAnorm for case P12 EAC and demonstrated the same results30.
Amplicon sequencing material and analysis for patient AHM1051
In order to assess any clonal heterogeneity within patient AHM1051’s Barrett’s esophagus segment, SNVs with the highest variant allele fraction (VAF) in each Barrett’s esophagus WGS library (all with a VAF >0.25), as well as all coding SNVs with a VAF > 0.08, were selected for amplicon sequencing. This resulted in the selection of 1,801 SNVs which were deemed to have the potential to define clones within this patient’s Barrett’s esophagus segment. After running Illumina’s Design Studio software for the TruSeq Custom Amplicon (TSCA) Protocol, 1,443 loci were deemed fit for the TSCA Protocol (Illumina).
Targeted sequencing, using the TSCA protocol followed by high throughput sequencing on the HiSeq2000, was performed on 73 individual samples (70 Barrett’s esophagus samples and 3 normal samples) including 10 of the original 13 samples used for WGS. Of the 73 samples, 64 were FFPE and 9 were fresh-frozen. For three of the original Barrett’s esophagus WGS samples there was not enough material remaining. The 70 Barrett’s esophagus samples represented the whole length of the Barrett’s esophagus segment and were taken from five different endoscopy visits, three which were before any endoscopic treatment (05/2009, 10/2009, 02/2010) and two which were post treatment (03/2011, 03/2012); endoscopic mucosal resection performed on the 22/04/2010 and radiofrequency ablation performed on the 8/12/2010. Three normal control samples (the same as used for WGS) were also included in the amplicon sequencing experiment. The TSCA protocol was performed as described by the manufacturer followed by data analysis using ISIS 2.4 Custom Amplicon Workflow for processing the alignments. For downstream analysis the read depth and VAF of each target base was extracted for all samples. Individual positions with read depth ≤20 were considered as zero depth (ie. no VAF value at those positions).
Visualization of the targeted amplicon-sequencing data
A custom bioinformatics tool was designed and constructed using the JavaScript library d3.js, to allow interrogation and exploration of the TSCA data. The tool, available in supplementary data, loads 3 data files: the sample metadata (date of sampling, location down the esophagus, histology report, DNA concentration and TSCA average depth), the sequencing metadata (genomic location, base change and annotation, including gene and consequence when relevant) and the VAF values for all targets in all samples (null values were converted to 0). The tool allows three main interactive explorations of the data: (i) visualization of TSCA data for individually selected samples in a Manhattan-like plot where the Y-axis displays the VAF values of the targets within each given sample ordered on the X-axis according to genomic coordinate; (ii) a pairwise scatterplot of VAF values between two selected samples and (iii) a hierarchical tree of the samples. The tree display is dynamic and allows the user to remove samples and/or targets and to choose between five distance metrics between samples.
The five distance metrics (binary, Euclidean, Manhattan, Max and Pearson) are described in the help page of the tool. All analysis results were given using the Pearson metric
where D(A,B) is the Pearson distance between samples A and B and is the variant read fraction of mutation i in sample A.
A distance matrix is generated by calculating all pairwise distances between samples. This distance matrix is then used to generate the tree using hierarchical clustering using the complete methodology for linkage of the nodes.
Determining the clonal hierarchy within the Barrett’s segment of patient AHM1051
Clone 3 is distinctly different from the five other identified clones and only shares 3 SNVs in common with the other five clones. Clone 1 is the oldest clone out of the other five clones and contains 819/1,437 assessed SNVs. Clone 2 then arose from Clone 1 as Clone 1 and 2 share the same 819 SNVs and in addition Clone 2 contains 234 SNVs (Clone 2 has a total of 1,053/1,437 SNVs assessed). Clone 4 must have arisen from Clone 2 as it contains the same 1,053 SNVs as Clones 2 as well as 125 more SNVs which are not present in either Clone 1 or 2 (Clone 4 has a total of 1,178/1,437 SNVs assessed). Clone 5 then arose from Clone 4 as it contains the same 1,178 SNVs as Clone 4 as well as 6 additional SNVs and a large scale deletion in chromosome 15 (evidenced by the increased VAF in the VAF plot in Figure 5a). Clone 6 must have arisen from Clone 2 as Clones 4 and 5 do not share all their SNVs with Clone 6. Clone 1 and 2 do share the majority of its SNVs with Clone 6 (with the exception of some SNVs which are probably lost through copy number changes present in Clone 6). Clone 6 contains an additional 93 SNVs compared to Clone 2, as well as multiple copy number changes in chromosomes 5, 11, 13 and 18.
Statistical analysis
The Mann-Whitney test was used to compare continuous variables between groups and a Fisher’s Exact Test was used to compare counts between categorical variables. All reported p values were two sided.
Supplementary Material
Acknowledgements
We thank the Human Research Tissue Bank which is supported by the NIHR Cambridge Biomedical Research Centre. This study was partly funded by a project grant from Cancer Research UK. Rebecca Fitzgerald has programmatic funding from the Medical Research Council and infrastructure support from the Biomedical Research Centre and the Experimental Medicine Centre. We would like to thank all the patients who took part in the study. We thank Mark Dunning for his bioinformatics assistance. We thank the Edinburgh Experimental Cancer Medicine Centre.
The OCCAMS consortium members
Stephen J Hayes7,8, Yeng Ang7, Anne-Marie Lydon7, Soney Dharmaprasad7, Sandra Greer9, Shaun Preston10, Sarah Oakes10, Vicki Save11, Simon Paterson-Brown11, Olga Tucker12,13, Derek Alderson12, Philippe Taniere12, Jamie Kelly14, James Byrne14, Donna Sharland14, Nina Holling14, Lisa Boulter14, Fergus Noble14, Bernard Stacey14, Charles Crichton13, Hugh Barr15, Neil Shepherd15, L. Max Almond15, Oliver Old15, James Gossage16,17,18, Andrew Davies16,17,18, Robert Mason16,17,18, Fuju Chang16,17, Janine Zylstra16,17, Grant Sanders19, Tim Wheatley19, Richard Berrisford19, Tim Bracey19, Catherine Harden19, David Bunting19, Tom Roques20, Jenny Nobes20, Suat Loo20, Mike Lewis20, Ed Cheong20, Oliver Priest20, Simon L Parsons21, Irshad Soomro21, Philip Kaye21, John Saunders21, Vincent Pang21, Neil T Welch21, James A Catton21, John P Duffy21, Krish Ragunath21, Laurence Lovat22, Rehan Haidry22, Haroon Miah22, Sarah Kerr22, Victor Eneh22, Rommel Butawan22, Laszlo Igali23, Hugo Ford24, David Gilligan24, Peter Safranek24, Andy Hindmarsh24, Vijayendran Sudjendran24, Andy Metz24, Nick Carroll24, Michael Scott25, Alison Cluroe3, Ahmad Miremadi3, Betania Mahler-Araujo3, Olga Knight1, Barbara Nutzinger1, Chris Peters16, Zarah Abdullahi1, Irene Debriram-Beecham1, Jason Crawte1, Shona MacRae1, Ayesha Noorani1, Rachael Fels Elliott1, Xiaodun Li1, Lawrence Bower26, Achilleas Achilleos26, Paul Edwards26, Simon Tavare26, Matthew Eldridge26, Jan Bornschein1, Sebastian Zeki1, Hamza Chettouh1, Maria Secrier26, Nadeera de Silva1, Eleanor Gregson1, Tsun-Po Yang1, J. Robert O’Neill27, Mariagnese Barbera1, Pierre Lao-Sirieix1, Nicola Grehan1, Chin-Ann J. Ong1, Laura Smith1, Shaun Preston28, Sarah Oakes28 and Suzy Lishman29.
7. Salford Royal NHS Foundation Trust, Salford, UK
8. Faculty of Medical and Human Sciences, University of Manchester, UK
9. Wigan and Leigh NHS Foundation Trust, Wigan, Manchester, UK
10. Royal Surrey County Hospital NHS Foundation Trust, Guildford, UK
11. Edinburgh Royal Infirmary, Edinburgh, UK
12. University Hospitals Birmingham NHS Foundation Trust, Birmingham, UK
13. Department of Computer Science, University of Oxford, UK
14. Southampton General Hospital, Southampton, UK
15. Gloucester Royal Hospital, Gloucester, UK
16. St Thomas’s Hospital, London, UK
17. King’s College London, London, UK
18. Karolinska Institutet, Stockholm, Swedan
19. Plymouth Hospitals N H S Trust, Plymouth, UK
20. Norfolk and Norwich University Hospital NHS Foundation Trust, Norwich, UK
21. Nottingham University Hospitals NHS Trust, Nottingham, UK
22. University College London, London, UK
23. Norfolk and Waveney Cellular Pathology Network, Norwich, UK
24. Cambridge University Hospitals NHS Foundation Trust, Cambridge, UK
25. Department of Pathology, Wythenshawe Hospital, Manchester, UK
26. CRUK Cambridge Institute, University of Cambridge, Cambridge, UK
27. Edinburgh University, Edinburgh, UK
28. Royal Surrey County Hospital, Guildford
29. Peterborough Hospitals NHS Trust, Peterborough
Footnotes
Accession codes
WGS data can be found at EGAD00001001394. Supplementary table 4 provides the information to match the sample identifiers to the patients presented in this manuscript.
COMPETING FINANCIAL INTERESTS
RCF developed the Cytosponge technology which has been licensed by MRC-Technology to Covidien. RCF has no direct pecuniary interest. JB, AW, RKC, HN, SI, MH, ZK, MR, SH, DB are employees of Illumina.
References
- 1.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esserman LJ, et al. Addressing overdiagnosis and overtreatment in cancer: a prescription for change. Lancet Oncol. 2014;15:e234–42. doi: 10.1016/S1470-2045(13)70598-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desai TK, et al. The incidence of oesophageal adenocarcinoma in non-dysplastic Barrett’s oesophagus: a meta-analysis. Gut. 2012;61:970–6. doi: 10.1136/gutjnl-2011-300730. [DOI] [PubMed] [Google Scholar]
- 4.Corley DA, et al. Impact of endoscopic surveillance on mortality from Barrett’s esophagus-associated esophageal adenocarcinomas. Gastroenterology. 2013;145:312–9 e1. doi: 10.1053/j.gastro.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaheen NJ, Hur C. Garlic, silver bullets, and surveillance upper endoscopy for Barrett’s esophagus. Gastroenterology. 2013;145:273–6. doi: 10.1053/j.gastro.2013.06.028. [DOI] [PubMed] [Google Scholar]
- 6.Leedham SJ, et al. Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human Barrett’s oesophagus. Gut. 2008;57:1041–8. doi: 10.1136/gut.2007.143339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maley CC, et al. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barrett’s esophagus. Cancer Res. 2004;64:3414–27. doi: 10.1158/0008-5472.CAN-03-3249. [DOI] [PubMed] [Google Scholar]
- 8.Schulmann K, et al. Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett’s-associated neoplastic progression and predicts progression risk. Oncogene. 2005;24:4138–48. doi: 10.1038/sj.onc.1208598. [DOI] [PubMed] [Google Scholar]
- 9.Reid BJ, et al. Predictors of progression in Barrett’s esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am J Gastroenterol. 2001;96:2839–48. doi: 10.1111/j.1572-0241.2001.04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kastelein F, et al. Aberrant p53 protein expression is associated with an increased risk of neoplastic progression in patients with Barrett’s oesophagus. Gut. 2013;62:1676–83. doi: 10.1136/gutjnl-2012-303594. [DOI] [PubMed] [Google Scholar]
- 11.Maley CC. Multistage carcinogenesis in Barrett’s esophagus. Cancer Lett. 2007;245:22–32. doi: 10.1016/j.canlet.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Fitzgerald RC. Dissecting out the genetic origins of Barrett’s oesophagus. Gut. 2008;57:1033–4. doi: 10.1136/gut.2008.151530. [DOI] [PubMed] [Google Scholar]
- 13.Weaver JM, et al. Ordering of mutations in preinvasive disease stages of esophageal carcinogenesis. Nat Genet. 2014 doi: 10.1038/ng.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agrawal N, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chong IY, et al. The genomic landscape of oesophagogastric junctional adenocarcinoma. J Pathol. 2013;231:301–10. doi: 10.1002/path.4247. [DOI] [PubMed] [Google Scholar]
- 16.Dulak AM, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013 doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis MJ, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–60. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kan Z, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422–33. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bass AJ, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43:964–8. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dulak AM, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012;72:4383–93. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider PM, et al. Mutations of p53 in Barrett’s esophagus and Barrett’s cancer: a prospective study of ninety-eight cases. J Thorac Cardiovasc Surg. 1996;111:323–31. doi: 10.1016/s0022-5223(96)70441-5. discussion 331-3. [DOI] [PubMed] [Google Scholar]
- 24.Dolan K, Walker SJ, Gosney J, Field JK, Sutton R. TP53 mutations in malignant and premalignant Barrett’s esophagus. Dis Esophagus. 2003;16:83–9. doi: 10.1046/j.1442-2050.2003.00302.x. [DOI] [PubMed] [Google Scholar]
- 25.Li X, et al. Temporal and spatial evolution of somatic chromosomal alterations: a case-cohort study of Barrett’s esophagus. Cancer Prev Res (Phila) 2014;7:114–27. doi: 10.1158/1940-6207.CAPR-13-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orman ES, Li N, Shaheen NJ. Efficacy and durability of radiofrequency ablation for Barrett’s Esophagus: systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2013;11:1245–55. doi: 10.1016/j.cgh.2013.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–93. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 28.Raczy C, et al. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics. 2013;29:2041–3. doi: 10.1093/bioinformatics/btt314. [DOI] [PubMed] [Google Scholar]
- 29.Saunders CT, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–7. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 30.Gusnanto A, Wood HM, Pawitan Y, Rabbitts P, Berri S. Correcting for cancer genome size and tumour cell content enables better estimation of copy number alterations from next-generation sequence data. Bioinformatics. 2012;28:40–7. doi: 10.1093/bioinformatics/btr593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.