Abstract
There is an increasing interest in using three-dimensional (3D) spheroids for modeling cancer and tissue biology to accelerate translation research. Development of higher throughput assays to quantify phenotypic changes in spheroids is an active area of investigation. The goal of this study was to develop higher throughput high-content imaging and analysis methods to characterize phenotypic changes in human cancer spheroids in response to compound treatment. We optimized spheroid cell culture protocols using low adhesion U-bottom 96- and 384-well plates for three common cancer cell lines and improved the workflow with a one-step staining procedure that reduces assay time and minimizes variability. We streamlined imaging acquisition by using a maximum projection algorithm that combines cellular information from multiple slices through a 3D object into a single image, enabling efficient comparison of different spheroid phenotypes. A custom image analysis method was implemented to provide multiparametric characterization of single-cell and spheroid phenotypes. We report a number of readouts, including quantification of marker-specific cell numbers, measurement of cell viability and apoptosis, and characterization of spheroid size and shape. Assay performance was assessed using established anticancer cytostatic and cytotoxic drugs. We demonstrated concentration–response effects for different readouts and measured IC50 values, comparing 3D spheroid results to two-dimensional cell cultures. Finally, a library of 119 approved anticancer drugs was screened across a wide range of concentrations using HCT116 colon cancer spheroids. The proposed methods can increase performance and throughput of high-content assays for compound screening and evaluation of anticancer drugs with 3D cell models.
Introduction
In vitro aggregates of stem cells and tumor cells called spheroids have been used for decades as models to recapitulate the in vivo tissue environment.1–3 Spheroids can be established from a single cell type or created from mixtures of multiple cell types such as tumor, stromal, and immune cells. Spheroids are believed to mimic tumor behavior more effectively than conventional two-dimensional (2D) cell cultures because spheroids, much like tumors, contain both surface-exposed and deeply buried cells, proliferating and nonproliferating cells, and well-oxygenated and hypoxic cells.4 In three-dimensional (3D) growth conditions, spheroids reproduce important parameters of tumor microenvironments, including oxygen and nutrient gradients.5 Spheroids have been used for breast cancer,6,7 colon,8 prostate, and other cancer disease modeling to identify novel anticancer therapeutics.1,5 3D models have also been employed for developmental biology research and toxicity screening.9
There has been significant progress in development of 3D cell models and techniques during the last several years.10,11 Hanging drop techniques use specialized plates12 and liquid handlers for media change and compound addition processes. Cells are then transferred to separate plates for analysis. Engineered microenvironments aid the formation of 3D structures in extracellular matrices (e.g., Matrigel) for screening of anticancer agents.13,14 Photolithographic techniques can form multiple spheroids on micropatterned surfaces9,15,16 or more complex organ-on-a-chip structures.17 Recently, spheroid formation in low-attachment round-bottom plates has become popular as the method offers a more simple workflow and is suitable for high-content imaging.18
Different biological readouts from spheroid assays have been described. Common methods include disruption of spheroids and analysis of cell lysates or suspensions for adenosine triphosphate or other viability markers with microplate readers.1 High-content imaging methods have been demonstrated as promising techniques for characterizing the effects of chemical compounds and siRNA libraries on spheroids.5,19,20 Imaging has advantages over other analytical methods, such as plate reader-based luminescence or fluorescence, in that no disruption of the spheroids is required and multiple biological readouts are possible.1,21 Transmitted light (TL) imaging can be used to ascertain spheroid size and, to some extent, density. Fluorescence imaging can be used with viability dyes, DNA-binding dyes, apoptosis dyes, or other fluorescence markers.22 Alternatively, cells can be transfected with fluorescent proteins before spheroid formation to enable detection of changes in spheroid size or intensity by fluorescence.23 This method can be extended to more complex multicellular models that express a plurality of fluorescent markers.
Typically, low-magnification imaging is used to capture a full spheroid in one image and provide enough depth of field to sample farther into the center of the object. This approach limits assay readouts to changes in spheroid diameters or average fluorescence intensities or intensity ratios.19 The use of higher magnification provides single-cell resolution and the collection of more information about cellular content and spheroid structure. Higher magnification imaging also allows derivation of multiparametric outputs for characterizing complex phenotypes of spheroids treated with compounds.19 However, increased complexity introduces new assay development challenges, especially in a screening environment. The system must capture a relatively large object in one image and resolve individual cells in a thick solid object. Stains must penetrate inside spheroids. The analysis must enable comparisons between different spheroid phenotypes, derive IC50 values for compounds, show good assay reproducibility, and allow sufficient throughput.
The goal of this study was to develop and characterize high-magnification high-content imaging and image analysis methods suitable for high-throughput compound screening. We have optimized live cell protocols for spheroid assays using 10x and 20x objectives, 96-well or 384-well plates, and three commonly used cell lines: HCT116 (colon cancer), DU145 (prostate cancer cell line), and HepG2 (hepatocellular carcinoma). Sample handling steps for cell culture, treatment, and staining have been reduced to minimize spheroid disturbances and increase assay reproducibility. We have optimized and compared imaging methods and described measurements for multiparametric characterization of different spheroid phenotypes and determination of IC50 values. Furthermore, we characterized the assay using six benchmark cytotoxic compounds and compared IC50 values for 3D and 2D cultures. Finally, we have tested a small library of approved anticancer drugs (NIH-Approved Oncology Drugs Plated Set) in a wide range of concentrations and ranked compounds by potency. This method can enhance development of disease-relevant cell-based assays for efficient assessment of chemicals and drug candidates by high-throughput quantitative screening.
Methods
Cell Models
Three cell lines were used: HCT116, DU145, and HepG2 (ATCC, Manassas, VA). Cryopreserved cells were thawed and maintained according to provided protocols. HCT116 cells were grown in McCoy's 5A (Corning, Manassas, VA) supplemented with 10% fetal bovine serum (Gemini Bio-Products, Sacramento, CA). DU145 and HepG2 cells were grown in Minimum Essential Medium (Corning) supplemented with 10% fetal bovine serum. For the concentration response studies and phenotypic assay, cells were plated into 96-well or 384-well, ultralow-attachment, Corning U-shaped, black clear-bottom plates (Corning 4520 and 3830, respectively) at densities of 1,500 cells/well in the appropriate media at 37°C and 5% CO2. Media were changed every 3 days.
Chemicals and Treatments
For compound screening, test agents were prepared as 10–100 mM stock solutions in tissue culture-grade dimethyl sulfoxide (DMSO; Sigma-Aldrich, St. Louis, MO). The final concentration of DMSO in media was 0.1%. Compounds (Sigma-Aldrich) were tested in quadruplicates or octuplets (384-well format) or triplicates (96-well format) in an eight-point dilution series. For the library screen, compounds were tested in duplicates. For assays, spheroids were plated 24 h before initiation of the experiment. Cells were then exposed to the indicated concentrations of compounds for 7 days. For the caspase activation assay, spheroids were cultured for 2 days, then treated with compounds for 48 h.
Multiparametric Live Cell Toxicity Assay
Following incubation with test compounds, spheroids were stained with a mixture of three dyes: 2 μM calcein AM, 3 μM of EthD-1, and 33 μM Hoechst 33342 (Life Technologies, Carlsbad, CA). Dyes were prepared in sterile phosphate-buffered saline (PBS; Corning). In separate experiments, CellEvent Caspase 3/7 reagent (Life Technologies) was used to evaluate compound ability to trigger apoptosis signaling. 7.5 μM CellEvent was added with 10 μM Hoechst nuclear dye diluted in sterile Hank's Balanced Salt Solution (Corning). Dye solutions were prepared immediately before use and added directly to media without aspiration. Since dye penetration is slower into spheroids than 2D cultures, spheroids were incubated with dye for 3 h before imaging. Dye solution was not washed out, and care was taken during pipetting to avoid spheroid loss, disintegration, or displacement.
Image Acquisition
Images were acquired using either a widefield (ImageXpress® Micro XLS) or confocal (ImageXpress® Micro Confocal) automatic imaging system (Molecular Devices, Sunnyvale, CA), with a 20× Plan Apo (NA = 0.45) or 10× Plan Fluor (NA = 0.3) objective. The confocal system was equipped with a 60-μm pinhole. Typical image acquisition settings are given in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/adt). A stack of 7–11 images separated by 10–35 μm was acquired, starting at the well bottom and covering approximately the lower half of each spheroid. Typically, a Z-stack of images covered 100–200 μm for HCT116 or HepG2 spheroids and 100–170 μm for DU145 spheroids. Although all individual images can be saved, we stored and analyzed 2D projection images only to reduce data storage space and image analysis time. Image analysis was performed using the maximum projection (MaxPro) image of confocal image stacks and best focus projection of widefield image stacks. TL images were used for cell culture protocol optimization.
Image Analysis
Images were analyzed using MetaXpress® 6 software (Molecular Devices). Count Nuclei and Cell Scoring application modules were used for nuclear count and live/dead assessment, respectively. A customized analysis for additional multiparametric outputs was done using a protocol created in the MetaXpress Custom Module Editor (CME). The custom module analysis first identified the spheroid using Hoechst staining. Then, viable cells were identified by the presence of calcein AM or by the absence of EthD-1 signal and dead cells by the presence of EthD-1 signal. Output measurements included spheroid width, spheroid area, average intensity for calcein AM or EthD-1, counts of all nuclei, and evaluation of average nuclear size and average intensity. Calcein AM-positive cells were counted, and their area and intensity values were recorded. These data were also recorded for Live (EthD-1 negative) and Dead (EthD-1 positive) cells. TL images were also characterized by CME. Images were smoothed with a Gaussian filter and inverted. Spheroids were identified using the Find Blobs function. Spheroid object sizes were characterized and compared by width, height, and area. EC50 values were determined using the 4-parameter curve fit from SoftMax® Pro 6 software (Molecular Devices).
Results
Development and Optimization of the Live Cell High-Content Assay with 3D Spheroid Cultures
The goal of this study was to develop and evaluate fast, accurate, and reproducible high-content imaging methods to investigate effects of anticancer compounds on the morphology and viability of 3D spheroid cultures using live and fixed cells. We used low-attachment, U-shaped, black clear-bottom plates to simplify cell culture, compound addition, and imaging. These plates eliminate spheroid transfer steps and center the spheroids in the wells, facilitating capture of an entire spheroid in one 10× or 20× image. Cells aggregate at the bottom of the U-shaped wells and form spheroids within 24 h (Supplementary Fig. S1). The thin plastic bottom of each well aids focusing and image acquisition with standard automated imaging systems.
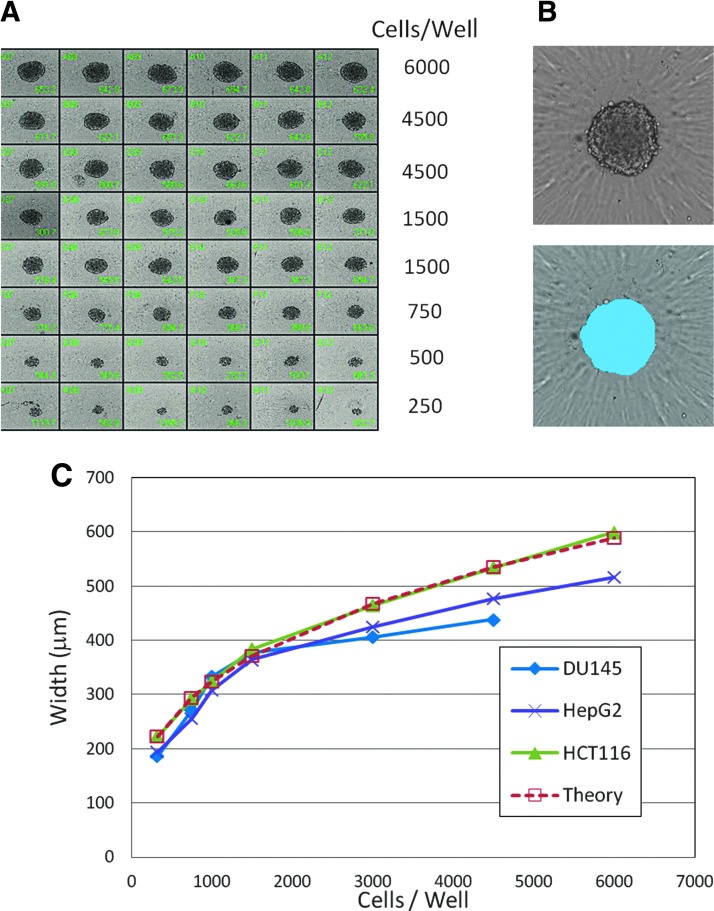
We studied reproducibility of spheroid formation and dependence of the spheroid size on the number of plated cells using three common cell lines: HCT116, DU145, and HepG2. Cells were plated at different densities (250–6,000 cells/well), incubated for 48 h, and then imaged using TL imaging. For HCT116 cells, we found by visual assessment that a plating density of 1,000–1,500 cells/well resulted in consistent spheroid size and shape, with sizes suitable for image acquisition and analysis (Fig. 1A, B). The dependence of spheroid diameter on the number of plated cells for the three cell lines is shown in Figure 1C. The response agreed well with a simple, spherical cell volume model. A plating density of 1,500 cells/well was used for subsequent assays. At this density, the average spheroid maximum diameter after 2 days was consistent as measured by TL imaging with a value of 557 ± 50 μm (n = 96) yielding a coefficient of variation of 9.0%.
Fig. 1.
(A) Spheroids from HCT116 cells plated at different densities. Images were acquired 48 h postplating. Smaller spheroids (500 cells/well or less) were observed to have less consistent spheroidal shape. (B) Example of image analysis and spheroid mask (blue) using the TL image. (C) Dependence of spheroid width as determined by image analysis on cell plating density (n = 12) for three cell types: HCT116 cells (green); HepG2 cells (purple); DU145 cells (blue). The response was modeled using a spherical volume model with a single-cell diameter variable. A fit of the HCT116 data to the theoretical spherical volume model using a cell diameter of 32 μm is shown in red. Error bars not shown; SDs were less than 5% of values.
Compound Treatment
We studied the dependence of compound treatment duration on spheroid size to determine an optimal time window for compound incubation. Spheroids were treated 24 h postplating with mitomycin C (1 μM) and staurosporine (1 μM) and incubated for 3, 5, or 7 days. TL images were taken and analyzed using MetaXpress CME. Small differences in spheroid width between treated and untreated samples were observed on days 3 and 5, while more than 2× decrease in spheroid width was measured on day 7 (staurosporine = 2.9-fold and mitomycin C = 2.1-fold, data not shown). Therefore, assays were performed using 7-day compound treatment.
Staining
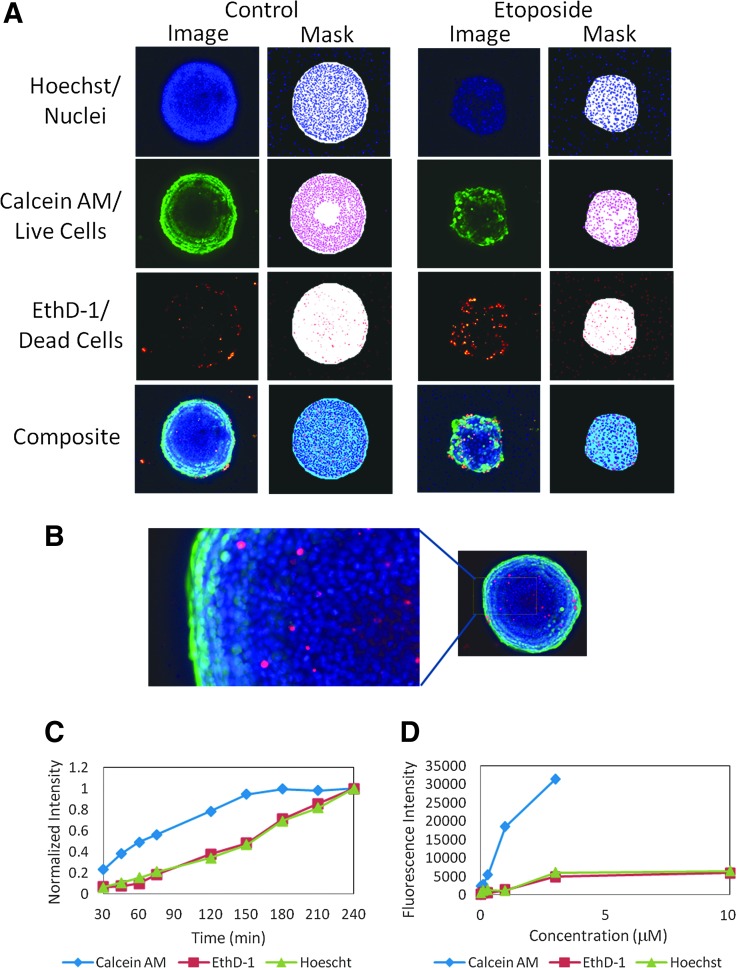
Assay staining protocols must minimize disturbances that would cause spheroid degradation or displacement from the well center. We used a previously developed staining protocol with one-step dye mixture addition that eliminates the need for fixing cells or repeated washes.24 Images of representative spheroids stained with a mix of three dyes are shown in Figure 2A and B. Calcein AM was used to measure metabolically active cells, viability, and a variety of morphological parameters. Hoechst was utilized to measure total cell count and nuclear shape. EthD-1 selectively penetrates cells with damaged outer membranes and was used to measure dead or necrotic cells. We optimized dye incubation times and concentrations; results from those studies are shown in Figure 2C and D.
Fig. 2.
(A) Untreated and treated spheroids were stained with a combination of three dyes: Hoechst 15 μM, EthD-1 3 μM, and calcein AM 1 μM. Images of Hoechst, calcein AM, and EthD-1 were taken using DAPI, FITC, and Texas Red channels, respectively. A composite image of all three channels is shown at the bottom. Images were generated using maximum projection from a Z-stack of seven images 30 μm apart. The resulting object masks from image analyses are shown to the right of each image. (B) Zoomed region of an untreated spheroid showing mixed population of cells. (C). Average fluorescence intensities from spheroids (n = 3) versus incubation time for all three stains. The data are normalized to the 240-min time point. (D) Average fluorescence intensity for spheroids versus dye concentration for all three stains. Error bars not shown; SDs were less than 5% of values.
For this study, we selected dyes known to diffuse efficiently into spheroids.25 A time course of Hoechst, EthD-1, and calcein AM staining from 30 min to 4 h was performed. Fluorescence intensity for calcein AM stabilized after 3 h of incubation, while other dye intensities continually increased. A staining time of 3 h was selected for further assays. Spheroid imaging can be accomplished for up to 24 h if plates are stored at 4°C (data not shown).
Imaging 3D Spheroids
Issues that affect spheroid assay quality include poor light penetration into or out of the spheroid center, light scattering by cells, and high background from out-of-plane fluorescence.26 These issues can have significant effects on larger objects (>600 μm). Confocal microscopy can provide better background rejection and sharper images. In these studies, images were taken using an automated confocal or widefield fluorescence microscope with a large field of view camera. The system can effectively capture spheroids in a single image using 10× or 20× objectives. Images acquired with a 20× objective allowed better visualization of details, but did not provide a significant difference in cell count (data not shown).
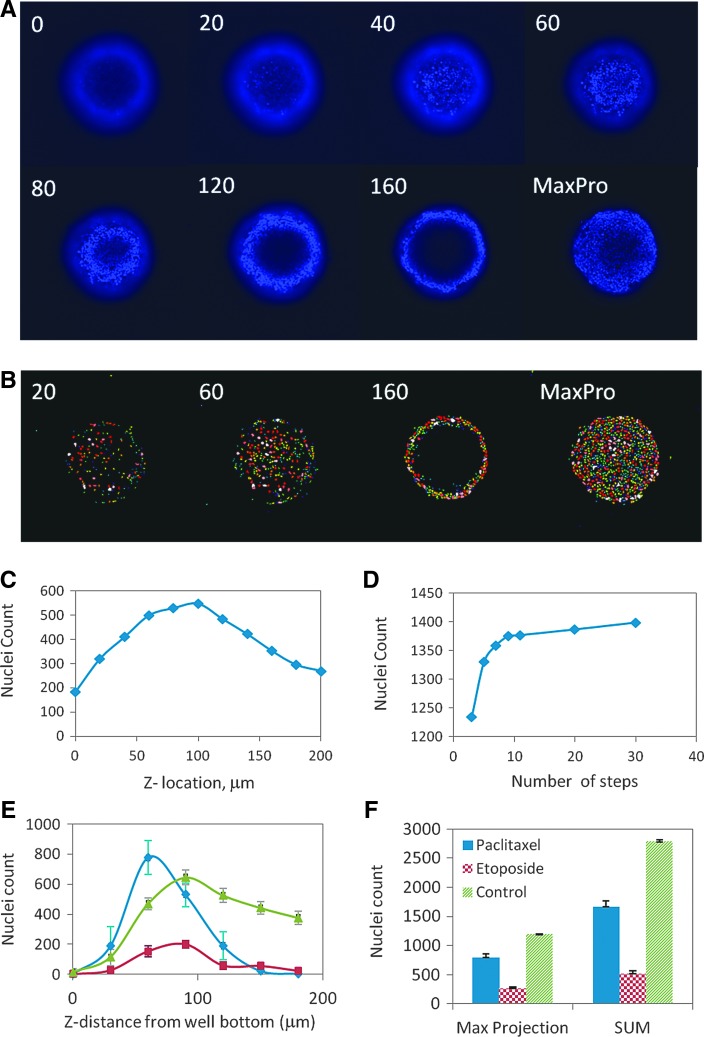
At the magnifications used in these studies, taking only one image with a fixed offset does not allow adequate comparison of spheroids of different sizes or shapes, making acquisition of images at multiple focal planes necessary. Imaging protocols were studied using Hoechst-stained spheroids. A series of images was acquired at different planes along the focal axis (Z-stack) and combined into a MaxPro image to enable efficient segmentation and counting of distinct nuclei. The MaxPro algorithm composes a single image by combining maximum intensity pixels from an image stack along lines orthogonal to the image planes. A representative Z-stack of images is shown in Figure 3A. In each image, only a fraction of nuclei are resolved, while other nuclei are either out of focus, too dim, or otherwise obscured (Fig. 3B). As a result, the nuclear count represents only a fraction of the actual nuclei present in each slice. The total cell count from each image in a Z-stack is shown in Figure 3C.
Fig. 3.
(A) Confocal images of a spheroid stained with Hoechst taken at indicated distances from the well bottom. As the image plane moves up from the well bottom toward the spheroid center, regions of distinct nuclei appear first in the center (at the surface of spheroid in contact with the plate bottom), then form a ring pattern that increases in diameter until it reaches some maximum associated with the outer surface of the spheroid. The MaxPro image is generated from 11 individual images taken 20 μm apart. (B) Nuclei segmentation shown for corresponding images in A. Individual nuclei are given a random false color. (C) Nuclei counts for a single spheroid measured from the 11 different Z-images taken at indicated distances from the plate bottom. (D) Dependence of nuclei count in the MaxPro image from a single spheroid on the number of individual images in a Z-stack taken over a 200 μm distance. The Z-stack setup was varied from three images taken 100 μm apart to 30 images taken 5 μm apart. (E) Nuclei counts measured from the separate z-images for three representative phenotypes of spheroids: Control (untreated, green), etoposide (200 μM, red), and paclitaxel (400 nM, blue). (F) Comparison between the nuclei counts obtained from the MaxPro images and sum of the nuclei count from all individual images for the three representative phenotypes of spheroids. Error bars in E and F represent ±1 SD (n = 6).
We have studied dependence of the number of counted nuclei using MaxPro with the number of images in a Z-stack for a 200-μm-thick region (Fig. 3D). An increase in the number of z images results in a larger number of nuclei counted in the MaxPro image. However, there is an inflection point around seven to nine slices. This represents a practical compromise between better sampling of cells within a given spheroid and increased acquisition time.
We further studied the use of this analysis with a variety of spheroid phenotypes. The response of nuclei count versus z-distance is significantly different for the three phenotypes (Fig. 3E). However, there is a good correlation between MaxPro cell count and the sum of individual slices (Fig. 3F). Analysis using MaxPro gives comparable data to image summation for the spheroid types encountered in these assays and decreases analysis time and data storage requirements, thus providing efficient comparison of spheroid sizes and phenotypes.
Phenotypic Analysis of the Effects of Anticancer Drugs
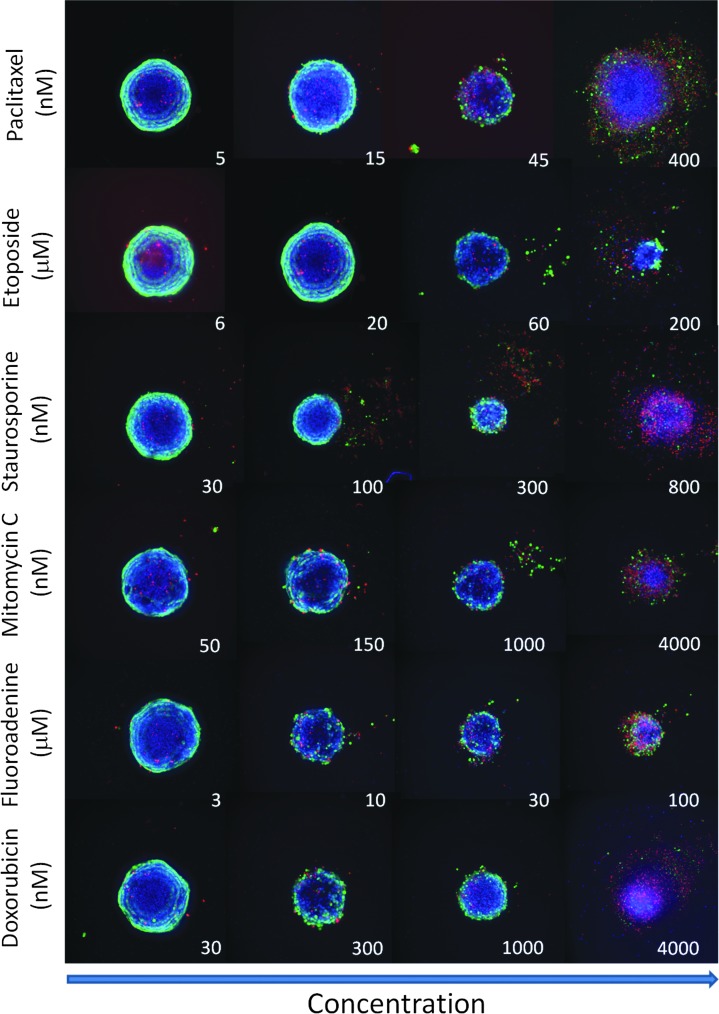
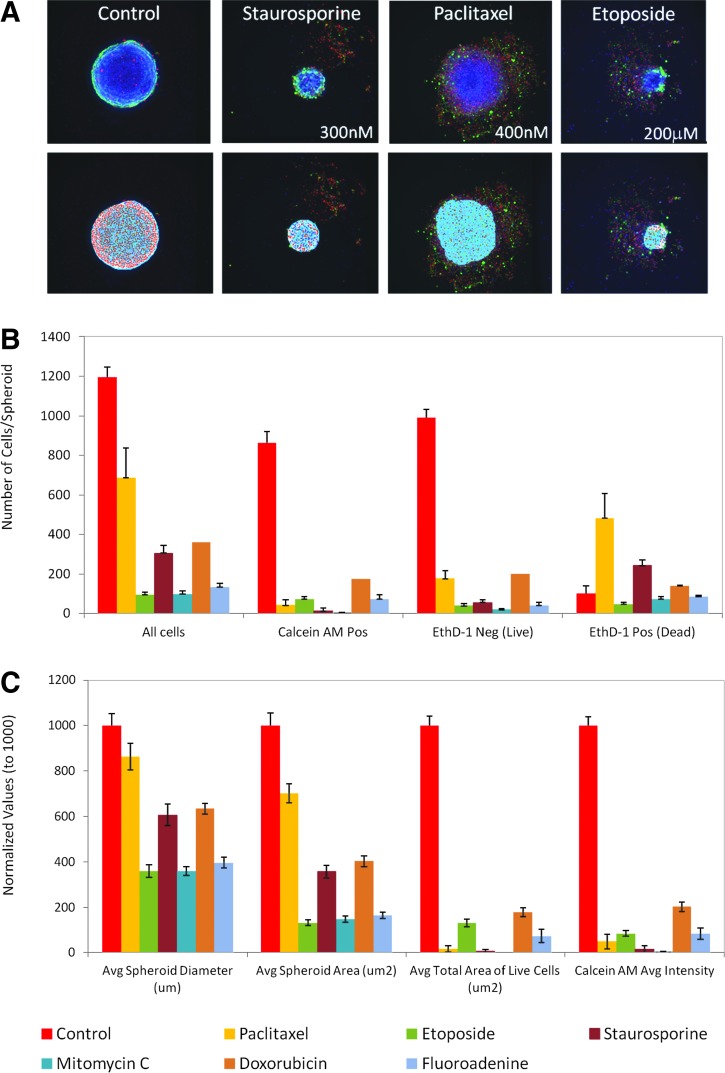
Spheroid assay performance was characterized using several compounds representing different classes of anticancer drugs: paclitaxel (interferes with microtubule formation during cell division),27 etoposide (topoisomerase inhibitor),28,29 staurosporine (kinase inhibitor),27,30 mitomycin C (DNA cross-linker),31,32 doxorubicin (anthracycline, DNA cross-linker),28 and fluoroadenine (purine analog, interferes with DNA synthesis). We have studied phenotypic changes of spheroids after treatment with the compounds across a wide range of concentrations (Fig. 4). Complex phenotypes are observed with treatment. Some spheroids lose their spherical shape, appear disintegrated, loose, flat, or irregular, have cells detached from the main body, or exhibit condensed nuclei due to cell death. In such cases, spheroid width, area, or total count of nuclei may not demonstrate expected concentration-dependent responses. Additional stains and other readouts such as counting numbers of live/dead or apoptotic cells in a well could improve compound effect quantification.
Fig. 4.
Representative images of spheroids treated with different compound concentrations. Composite images of Hoechst (blue), calcein AM (green), and EthD-1 (red). Note the dose-dependent decrease of spheroid size and also increase in the number of dead cells (in red) across all treatments. Spheroids treated with high concentrations of some compounds (e.g., paclitaxel and staurosporine) appear to disintegrate.
Quantitative analysis of the images included derivation of parameters to assess morphological features of spheroids and cell content and complexity (Fig. 5). Compound treatment typically decreases spheroid size or width. Analysis of whole spheroids with viability dyes has been described previously.25 In this study, we show that higher resolution spheroid imaging enables counting and scoring of individual cells. We counted the total number of cells in the image, the number of calcein AM-positive cells, the number of EthD-1-negative cells (Live), and the number of EthD-1- positive cells (Dead), as well as the average areas and intensities of cells expressing different markers.
Fig. 5.
(A) Representative spheroid phenotypes (top) and their corresponding masks (bottom) showing image analysis and segmentation. (B) Image analysis readouts derived as a result of Nuclei Count and Cell Scoring analysis for the following compounds: control (0.1% DMSO), paclitaxel 150 nM, etoposide 200 μM, staurosporine 300 nM, mitomycin C 1 μM, doxorubicin 1 μM, and fluoroadenine 100 μM. (C) Image analysis readouts were derived from geometric or average intensity readouts. The values in C are normalized to DMSO controls (set to 1,000). Error bars in B and C represent ± 1 SD (n = 8).
We have used MetaXpress CME for multiparametric image analysis to quantify different biological outputs. The analysis algorithm defined spheroids in the Hoechst image, scored cells as calcein AM-positive using colocalization of calcein AM and Hoechst staining, as Live by the presence of Hoechst and absence of EthD-1, and scored cells as Dead using colocalization of EthD-1 and Hoechst (Supplementary Fig. S2). Figure 5 illustrates the effects of different compounds on the various readouts, including number of total or marker-positive cells, spheroid area, or width. A majority of the parameters present sufficient assay performance for quantification of compound effects (Supplementary Table S2).
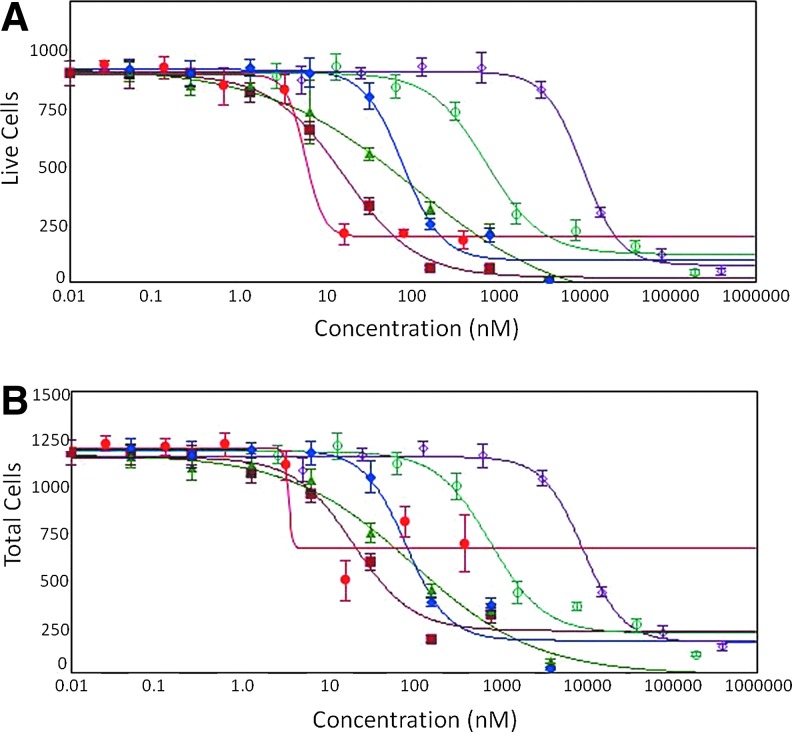
A concentration-dependent decrease of spheroid sizes can be observed as a result of compound treatment as well as decrease of cell number and a loss of cell viability. We determined IC50 values for the six compounds for different readouts (Table 1). Concentration responses were observed with all parameters. The number of metabolically active calcein AM-positive cells, or EtHD-1 excluded Live cells, was most sensitive to cytotoxicity as indicated by the fold change between control and treated samples and had the best 4-parameter curve fits (Fig. 6). Other readouts may provide important phenotypic information. For example, shape factor that characterizes circularity of spheroids can be used to measure spheroids with irregular shapes. It is important to note that the analysis used here is based on cell count within a spheroid body and excludes detached cells. The number of Dead cells has typically shown poor 4-parameter fit, except for some compounds (paclitaxel and staurosporine). However, this information could potentially be used to determine whether compounds cause cytostatic or cytotoxic effects.
Table 1.
IC50 Values for Select Compounds Using Different Output Parameters
| IC50 Values (nM)a | Total cells | Live cells (EthD-1 Neg) | Calcein AM-Pos cells | Dead cells (EthD-1 Pos) | Total spheroid area (μm2) | Calcein AM-Pos cell area (μm2) | Spheroid width (μm) | Calcein AM total intensity |
|---|---|---|---|---|---|---|---|---|
| Paclitaxel | 2.6 ± 2.0 | 4.0 ± 0.3 | 2.6 ± 1.3 | 8.2 ± 1.0 | 2.69 ± 0.62 | no fit | no fit | 2.1 ± 2.4 |
| Staurosporine | 10.5 ± 1.1 | 8.9 ± 0.4 | 9.8 ± 2.4 | 3.2 ± 1.7 | 10.4 ± 1.8 | 0.4 ± 1.3 | 12.1 ± 3.2 | 12.2 ± 1.3 |
| Doxorubicin | 26.7 ± 5.0 | 25.2 ± 3.7 | 27.1 ± 6.5 | No fit | 37 ± 11 | 22.4 ± 5.4 | 43 ± 24 | 21.8 ± 7.1 |
| Mitomycin C | 34 ± 10 | 34.9 ± 4.4 | 35.9 ± 7.2 | No fit | 70 ± 15 | 20.8 ± 25 | 109 ± 27 | 14 ± 10 |
| Etoposide | 312 ± 14 | 280 ± 10 | 292 ± 18 | No fit | 392 ± 43 | 288 ± 81 | 567 ± 106 | 556 ± 28 |
| Fluoroadenine (μM) | 6.1 ± 1.0 | 10.2 ± 0.50 | 6.97 ± 0.93 | No fit | 12.9 ± 2.6 | 7.5 ± 1.7 | 17.1 ± 8.6 | 5.1 ± 0.37 |
aIC50 values in nM, except where indicated as μM.
Error limits are one Standard Error of parameter estimate, n = 8.
Pos, positive; Neg, negative.
Fig. 6.
Concentration-dependent effects and 4-parameter curve fits of selected compounds in 3D spheroid culture using (A) Live cells (EthD-1 negative) per spheroid or (B) total cells per spheroid as determined by Cell Scoring analysis of MaxPro image. Red circles—paclitaxel; dark red squares—staurosporine; blue diamonds—doxorubicin; green triangles—mitomycin C; teal open circles—etoposide; purple open diamonds—fluoroadenine. Error bars represent ±1 SD (n = 8).
We have also tested compound effects on spheroids made from other cell lines. HepG2 spheroids had similar size and morphology in comparison with HCT116 cells. DU145 formed spheroids of smaller size (300–400 μm in diameter). However, similar cell staining, imaging, and analysis protocols were applicable for determining effects of anticancer drugs and IC50 values. The IC50 values for etoposide, mitomycin C, and staurosporine for 3D spheroid cultures across three cell types are given in Table 2.
Table 2.
IC50 Values for Select Compounds for Various Cell Lines Using Live Cell Readout
| IC50 Values (nM) | HCT116 | DU145 | HepG2 |
|---|---|---|---|
| Staurosporine | 16.2 ± 2.1 | 7.1 ± 4.5 | 29 ± 34 |
| Mitomycin C | 107 ± 63 | 819 ± 374 | 332 ± 183 |
| Etoposide | 746 ± 206 | 1464 ± 850 | 2421 ± 850 |
Error limits are one standard error of parameter estimate, n = 3.
We compared compound response measurements from 3D cultures versus those obtained from 2D cultures. HCT116 cells were plated simultaneously into 384-well regular or U-shaped plates and treated with the same concentrations of six compounds. Comparison of IC50 values is presented in the Table 3.33 For 2D culture, IC50 values were very similar for 7- and 3-day treatments. Comparison of IC50 values between 2D and 3D cultures shows that IC50 values for 3D spheroid cultures were 2–2.5× smaller than the corresponding values for 2D culture. Rank order for the potency of six selected compounds was the same in either 2D or 3D cultures.
Table 3.
Comparison of IC50 Values Between 3D and 2D Cell Cultures
| 3D, 7 days | 2D, 3 days | 2D, 7 days | 2D Literature | |
|---|---|---|---|---|
| IC50 Values (nM)a | Live cells | Live cells | Live cells | Viability |
| Paclitaxel | 4.0 ± 0.3 | 8.27 ± 0.51 | 8.1 ± 0.2 | 3.933 |
| Staurosporine | 8.9 ± 0.4 | 20 ± 2 | 23.5 ± 1.8 | 7023 |
| Doxorubicin | 25.2 ± 3.7 | 50.7 ± 1.3 | 42 ± 12 | 34033 |
| Mitomycin C | 34.9 ± 4.4 | 98 ± 15 | 119 ± 14 | |
| Etoposide | 280 ± 9.7 | 704 ± 251 | 507 ± 234 | 41033 |
| Fluoroadenine (μM) | 10.2 ± 0.5 | 18.0 ± 4.6 | 14.9 ± 1.5 |
aIC50 values in nM, except where indicated as μM.
Error limits are one standard error of parameter estimate, n = 6.
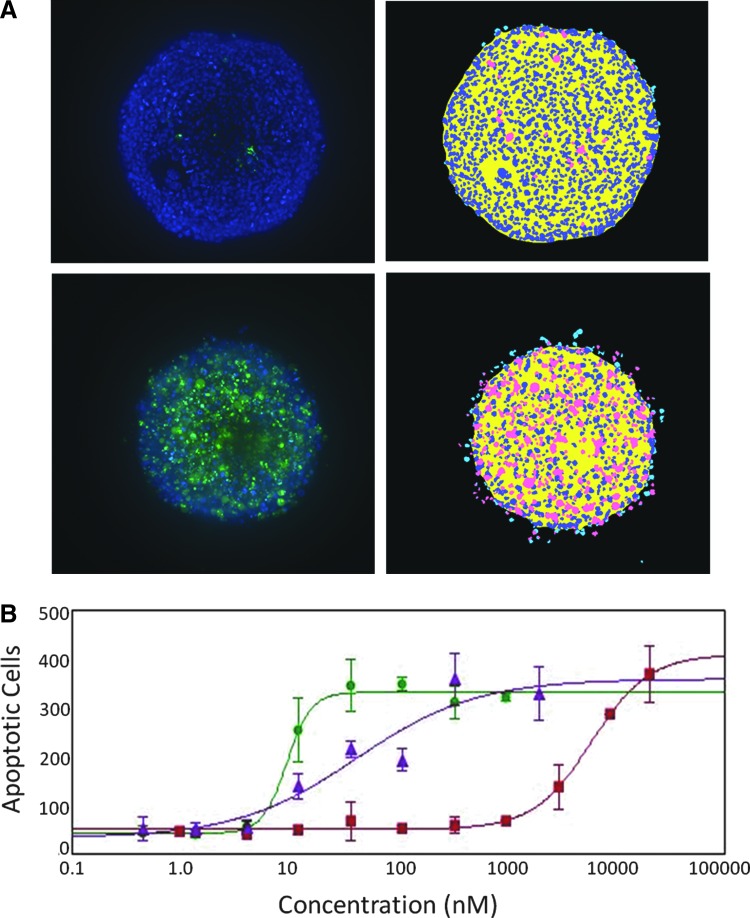
To investigate potential cytotoxic mechanisms, we evaluated the apoptosis phenotype. Activation of apoptosis using CellEvent Caspase 3/7 dye was measured 48 h after compound treatment. Typical activated caspase 3/7 staining patterns for control cells and those treated with the apoptotic agent, staurosporine, are shown in Figure 7A. Treatment with compounds causing apoptosis resulted in a concentration-dependent increase of caspase 3/7 staining intensity and the number of caspase 3/7-positive (apoptotic) cells (Fig. 7B). Treatment of spheroids with apoptosis compounds, staurosporine and paclitaxel, resulted in much lower IC50 values when compared with treatment with the cytostatic agent, mitomycin C.
Fig. 7.
Apoptosis assay. Spheroids were treated with indicated compounds for 42 h, then stained with Hoechst (blue) and caspase 3/7 (green) reagents. (A) Representative images and analysis of control and sample treated with 1 μM staurosporine. Nuclei are identified by a blue mask; Apoptotic cells are identified by a pink mask. (B) Dose-dependent increase of number of apoptotic cells in spheroids treated with 3 compounds: paclitaxel (green circles, IC50 = 9.5 nM), staurosporine (purple triangles, IC50 = 41.5 nM), and mitomycin C (red squares, IC50 = 6.01 μM). The content of apoptotic cells was ∼50% in the treated samples. Error bars represent ± 1 SD (n = 3).
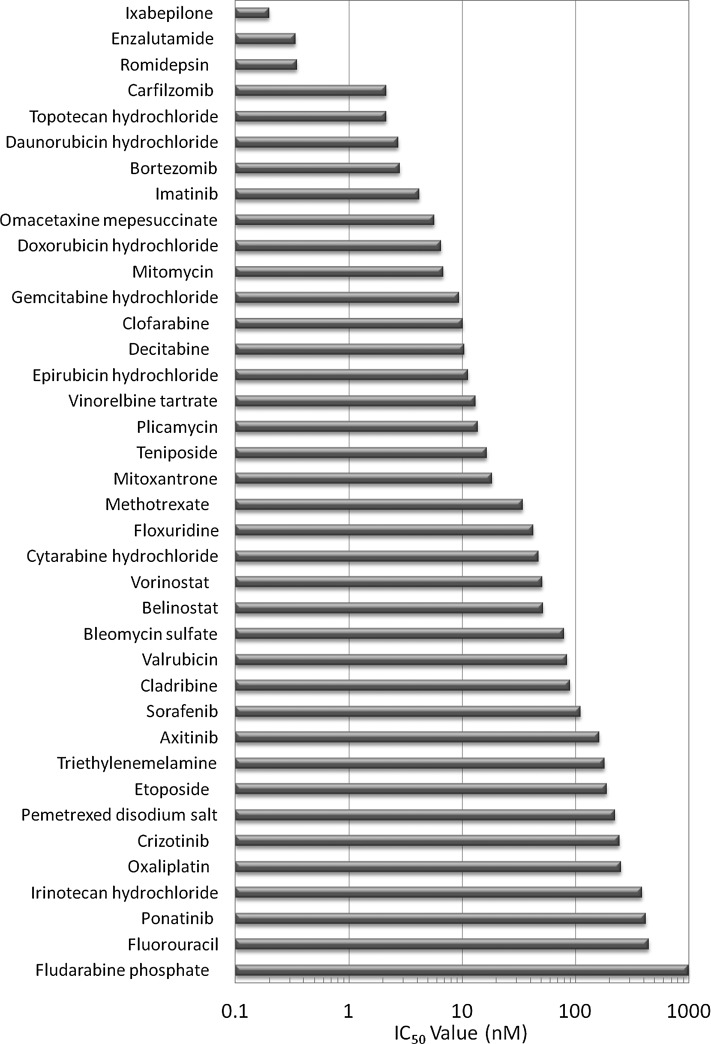
To demonstrate utility of the assay, we tested a library of 119 approved anticancer drugs and 20 control compounds across six concentrations from 0.1 nM to 10 μM. Compound effects were evaluated using the parameter calcein AM-positive cells per spheroid, and results are shown in Supplementary Table S3. Of the test compounds, 74 caused a significant decrease (>3 SD from DMSO control) in the number of calcein AM-positive cells, while 45 compounds showed no effect over the range of concentrations. The 20 negative controls also showed no effect at the tested concentrations. Of the positive compounds, seven showed an effect at all concentrations (<1 nM), 38 showed good concentration response and IC50 values were determined by a 4-parameter fit, and 29 compounds had a significant effect only at 10 μM (>1,000 nM). The latter compounds were ranked by fold decrease in the number of calcein AM-positive cells. The IC50 values for the 38 compounds that had good fits are shown in Figure 8. The assay sensitivity from this limited set of compounds was calculated to be 62% with a specificity of 100%.
Fig. 8.
IC50 values of 38 compounds from the small library screen of 119 approved anticancer drugs obtained from 4-parameter fits to calcein AM-positive cell readout over the range of tested concentrations.
Discussion
Human 3D tissue models can provide systems for analyzing the pathogenesis of diseases, including cancer and other disorders, and have been used for therapeutic screening.1,34,35 Although in vivo validation remains the gold standard, 3D models can accelerate the process by quickly identifying the most promising targets and establishing mechanisms of action. While the number of 3D cell culturing publications is increasing rapidly, the current limited characterization of 3D assays diminishes the confidence necessary to drive adoption of new methods. Three-dimensional methods have not yet replaced 2D culturing on a large scale, including in the drug development process. There are still important limitations to 3D systems, including scalability, reproducibility, sensitivity, and compatibility with screening systems.
The aim of the present study was to develop and characterize imaging methods that would be compatible with automated instruments and show feasibility of using 3D spheroid cultures for screening new drug candidates. We have addressed common obstacles related to spheroid imaging and analysis, and optimized cell culture, compound treatment, and staining protocols for 3D spheroid cultures.
In contrast to previous studies that employed lower resolution imaging of spheroids and analysis of size, intensity, or ratio of intensities of different fluorescent markers, we have focused on optimizing higher resolution imaging and cell population analysis that allows multiparametric quantification of different phenotypes. In this work, we have employed U-shaped plates for culturing spheroids to streamline 3D culture and facilitate high-resolution imaging. We have detailed, characterized, and compared different imaging protocols and demonstrated the MaxPro algorithm for comparative analysis of spheroid size and morphology. These methods of imaging and analysis can be extended to other 3D assay systems. Importantly, we demonstrate that live cell protocols provide sufficient assay quality while increasing reproducibility and throughput.
We have shown here that image analysis at single-cell resolution delivers a number of informative phenotypic readouts that enable screening for deleterious effects of test compounds on cell morphology and viability from a simple workflow assay protocol. Using a set of representative cytotoxic compounds, we demonstrated how different readouts can be used in combination to characterize complex cytostatic and cytotoxic effects on spheroid growth, compactness, viability, apoptosis, and other parameters to assess different effects of anticancer drugs.
We provided comprehensive assay characterization using a representative set of anticancer compounds. We characterized IC50 values for different readouts and showed that counting Live cells or calcein AM-positive cells in spheroids provides the greatest sensitivity to compound effects and best curve fit and can be efficiently used for ranking compound potencies. Other parameters would provide additional information about specific phenotypes, for example, spheroid size, integrity, or content of dead cells. Further assessment of spatial morphology features will be a subject of future assay development.
There are factors that should be considered when interpreting these assay results. First, there is a possibility of limited penetration of compounds inside spheroids due to spheroid compactness. This would create a concentration gradient that could affect compound cytotoxicity. Second, cells are cultured in suspension before they form spheroids (∼24 h). While the majority of cancer cells are viable in the spheroids as evidenced by calcein AM staining, there is the potential that their metabolic activity could be compromised during the suspension phase, which could also affect their toxicity response.
Compound response in 3D cultures may be different from 2D due to several factors: insufficient penetration of drugs into the core of spheroid quiescent cells and hypoxic areas inside of spheroids, altered gene expression, and other tumor-like features.36,37 It has been shown previously that 3D cultures can be more or less sensitive than 2D cultures to compound effects and that these relative sensitivities correlate with compound mechanisms of action.1,12,18,38 For example, it has been reported that kinase inhibitors are more sensitive in 3D models,18 while cell cycle inhibitors are more sensitive in 2D models.12 The studies here found that IC50 values for six selected compounds were smaller in 3D spheroids than 2D culture, while the rank order of compound potency was the same. Although we have obtained different IC50 values in 3D cultures, the limited set of compounds and relatively modest differences are not sufficient to characterize differences between 3D versus 2D cultures. A broader comparison should be a subject of further investigation.
Finally, we have demonstrated the feasibility of the assay for screening by testing a compound library of approved anticancer drugs using 3D spheroid cultures of HCT116 cells. The obtained data demonstrate greater potency of compounds that are currently used for treatment of cancer, suggesting utility of the assay system for testing novel anticancer drugs and drug combinations.
We anticipate successful applications of the live cell imaging method described here to high-content assays utilizing other cell types and additional markers for readouts such as hypoxia, mitochondria, cytoskeleton, and kinase activation. The approach may also be extensible to more complex 3D systems, such as cultures containing multiple spheroids per well or multiple cell types.
In summary, the 3D spheroid cell model combined with high-content imaging-based assays presented here shows promise as a sensitive and reproducible screening tool for assessing changes in cell proliferation, viability, and morphology. While predictivity of the assay in comparison with clinical data still needs to be established, further development of the methods and models will increase the utility of this in vitro tool for screening.
Supplementary Material
Abbreviations Used
- 2D
two-dimensional
- 3D
three-dimensional
- CME
Custom Module Editor
- DAPI
4',6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- EthD-1
ethidium homodimer-1
- FITC
fluorescein isothiocyanate
- MaxPro
maximum projection
- NA
numerical aperture.
- PBS
phosphate-buffered saline
- SD
standard deviation
- TL
transmitted light
- TRITC
tetramethyl rhodamine isothiocyanate
Acknowledgment
The authors would like to thank Bill Godfrey for his helpful discussions.
Disclosure Statement
O. Sirenko, T. Mitlo, J. Hesley, and S. Luke are employed by Molecular Devices, LLC, which manufactures the imaging instruments and software used in these studies.
References
- 1.Kunz-Schughart LA, Freyer JP, Hofstaedter F, et al. : The use of 3-D cultures for high-throughput screening: the multicellular spheroid model. J Biomol Screen 2004;9:273–285 [DOI] [PubMed] [Google Scholar]
- 2.Mueller-Klieser W. Three-dimensional cell cultures: from molecular mechanisms to clinical applications. Am J Physiol 1997;273:C1109–C1123 [DOI] [PubMed] [Google Scholar]
- 3.Wartenberg M, Donmez F, Ling FC, et al. : Tumor-induced angiogenesis studied in confrontation cultures of multicellular tumor spheroids and embryoid bodies grown from pluripotent embryonic stem cells. FASEB J 2001;15:995–1005 [DOI] [PubMed] [Google Scholar]
- 4.Frieboes HB, Zheng X, Sun CH, et al. : An integrated computational/experimental model of tumor invasion. Cancer Res 2006;66:1597–1604 [DOI] [PubMed] [Google Scholar]
- 5.Wenzel C, Riefke B, Grundemann S, et al. : 3D high-content screening for the identification of compounds that target cells in dormant tumor spheroid regions. Exp Cell Res 2014;323:131–143 [DOI] [PubMed] [Google Scholar]
- 6.Calcagno AM, Salcido CD, Gillet JP, et al. : Prolonged drug selection of breast cancer cells and enrichment of cancer stem cell characteristics. J Natl Cancer Inst 2010;102:1637–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponti D, Costa A, Zaffaroni N, et al. : Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res 2005;65:5506–5511 [DOI] [PubMed] [Google Scholar]
- 8.Thoma CR, Stroebel S, Rosch N, et al. : A high-throughput-compatible 3D microtissue co-culture system for phenotypic RNAi screening applications. J Biomol Screen 2013;18:1330–1337 [DOI] [PubMed] [Google Scholar]
- 9.Ramsden D, Tweedie DJ, Chan TS, et al. : Bridging in vitro and in vivo metabolism and transport of faldaprevir in human using a novel cocultured human hepatocyte system, HepatoPac. Drug Metab Dispos 2014;42:394–406 [DOI] [PubMed] [Google Scholar]
- 10.Bhadriraju K, Chen CS. Engineering cellular microenvironments to improve cell-based drug testing. Drug Discov Today 2002;7:612–620 [DOI] [PubMed] [Google Scholar]
- 11.Lin RZ, Chang HY. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol J 2008;3:1172–1184 [DOI] [PubMed] [Google Scholar]
- 12.Tung YC, Hsiao AY, Allen SG, et al. : High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 2011;136:473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Wang J. Mixed hydrogel bead-based tumor spheroid formation and anticancer drug testing. Analyst 2014;139:2449–2458 [DOI] [PubMed] [Google Scholar]
- 14.Montanez-Sauri SI, Sung KE, Berthier E, et al. : Enabling screening in 3D microenvironments: probing matrix and stromal effects on the morphology and proliferation of T47D breast carcinoma cells. Integr Biol (Camb) 2013;5:631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda J, Khademhosseini A, Yeo Y, et al. : Micromolding of photocrosslinkable chitosan hydrogel for spheroid microarray and co-cultures. Biomaterials 2006;27:5259–5267 [DOI] [PubMed] [Google Scholar]
- 16.Wang W, Itaka K, Ohba S, et al.: 3D spheroid culture system on micropatterned substrates for improved differentiation efficiency of multipotent mesenchymal stem cells. Biomaterials 2009;30:2705–2715 [DOI] [PubMed] [Google Scholar]
- 17.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol 2014;32:760–772 [DOI] [PubMed] [Google Scholar]
- 18.Vinci M, Gowan S, Boxall F, et al. : Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol 2012;10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krausz E, de Hoogt R, Gustin E, et al. : Translation of a tumor microenvironment mimicking 3D tumor growth co-culture assay platform to high-content screening. J Biomol Screen 2013;18:54–66 [DOI] [PubMed] [Google Scholar]
- 20.Horman SR, To J, Orth AP. An HTS-compatible 3D colony formation assay to identify tumor-specific chemotherapeutics. J Biomol Screen 2013;18:1298–1308 [DOI] [PubMed] [Google Scholar]
- 21.Herrmann R, Fayad W, Schwarz S, et al. : Screening for compounds that induce apoptosis of cancer cells grown as multicellular spheroids. J Biomol Screen 2008;13:1–8 [DOI] [PubMed] [Google Scholar]
- 22.Waite CL, Roth CM. PAMAM-RGD conjugates enhance siRNA delivery through a multicellular spheroid model of malignant glioma. Bioconjug Chem 2009;20:1908–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drewitz M, Helbling M, Fried N, et al. : Towards automated production and drug sensitivity testing using scaffold-free spherical tumor microtissues. Biotechnol J 2011;6:1488–1496 [DOI] [PubMed] [Google Scholar]
- 24.Sirenko O, Hesley J, Rusyn I, et al. : High-content assays for hepatotoxicity using induced pluripotent stem cell-derived cells. Assay Drug Dev Technol 2014;12:43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Achilli TM, McCalla S, Meyer J, et al. : Multilayer spheroids to quantify drug uptake and diffusion in 3D. Mol Pharm 2014;11:2071–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hama H, Kurokawa H, Kawano H, et al. : Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci 2011;14:1481–1488 [DOI] [PubMed] [Google Scholar]
- 27.Takeda M, Mizokami A, Mamiya K, et al. : The establishment of two paclitaxel-resistant prostate cancer cell lines and the mechanisms of paclitaxel resistance with two cell lines. Prostate 2007;67:955–967 [DOI] [PubMed] [Google Scholar]
- 28.Hossein G, Zavareh VA, Fard PS. Combined Treatment of Androgen-Independent Prostate Cancer Cell Line DU145 with Chemotherapeutic Agents and Lithium Chloride: Effect on Growth Arrest and/or Apoptosis. Avicenna J Med Biotechnol 2012;4:75–87 [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Qubaisi M, Rozita R, Yeap SK, et al. : Selective cytotoxicity of goniothalamin against hepatoblastoma HepG2 cells. Molecules 2011;16:2944–2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fallon RJ. Staurosporine inhibits a tyrosine protein kinase in human hepatoma cell membranes. Biochem Biophys Res Commun 1990;170:1191–1196 [DOI] [PubMed] [Google Scholar]
- 31.Chan PK, Aldrich MB, Chakrabarty S. Assessment of tumor cell sensitivity to mitomycin C by “B23 translocation” assay. Cancer Lett 1988;40:143–149 [DOI] [PubMed] [Google Scholar]
- 32.Yang EB, Tang WY, Zhang K, et al. : Norcantharidin inhibits growth of human HepG2 cell-transplanted tumor in nude mice and prolongs host survival. Cancer Lett 1997;117:93–98 [DOI] [PubMed] [Google Scholar]
- 33.Smith V, Raynaud F, Workman P, et al. : Characterization of a human colorectal carcinoma cell line with acquired resistance to flavopiridol. Mol Pharmacol 2001;60:885–893 [DOI] [PubMed] [Google Scholar]
- 34.Khademhosseini A, Langer R, Borenstein J, et al. : Microscale technologies for tissue engineering and biology. Proc Natl Acad Sci U S A 2006;103:2480–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langer R, Tirrell DA. Designing materials for biology and medicine. Nature 2004;428:487–492 [DOI] [PubMed] [Google Scholar]
- 36.Desoize B, Jardillier J. Multicellular resistance: a paradigm for clinical resistance? Crit Rev Oncol Hematol 2000;36:193–207 [DOI] [PubMed] [Google Scholar]
- 37.Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell 2007;130:601–610 [DOI] [PubMed] [Google Scholar]
- 38.Hartung T. Toxicology for the twenty-first century. Nature 2009;460:208–212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.