Abstract
Aims: Stress responsive signaling coordinated by nuclear factor erythroid 2-related factor 2 (Nrf2) provides an adaptive response for protection of cells against toxic insults, oxidative stress and metabolic dysfunction. Nrf2 regulates a battery of protective genes by binding to regulatory antioxidant response elements (AREs). The aim of this study was to examine how Nrf2 signals cell stress status and regulates transcription to maintain homeostasis. Results: In live cell microscopy we observed that Nrf2 undergoes autonomous translocational frequency-modulated oscillations between cytoplasm and nucleus. Oscillations occurred in quiescence and when cells were stimulated at physiological levels of activators, they decrease in period and amplitude and then evoke a cytoprotective transcriptional response. We propose a mechanism whereby oscillations are produced by negative feedback involving successive de-phosphorylation and phosphorylation steps. Nrf2 was inactivated in the nucleus and reactivated on return to the cytoplasm. Increased frequency of Nrf2 on return to the cytoplasm with increased reactivation or refresh-rate under stress conditions activated the transcriptional response mediating cytoprotective effects. The serine/threonine-protein phosphatase PGAM5, member of the Nrf2 interactome, was a key regulatory component. Innovation: We found that Nrf2 is activated in cells without change in total cellular Nrf2 protein concentration. Regulation of ARE-linked protective gene transcription occurs rather through translocational oscillations of Nrf2. We discovered cytoplasmic refresh rate of Nrf2 is important in maintaining and regulating the transcriptional response and links stress challenge to increased cytoplasmic surveillance. We found silencing and inhibition of PGAM5 provides potent activation of Nrf2. Conclusion: Frequency modulated translocational oscillations of Nrf2 mediate the ARE-linked cytoprotective transcriptional response. Antioxid. Redox Signal. 23, 613–629.
Introduction
Nuclear factor erythroid 2-related factor 2 (Nrf2) regulates the cellular expression of a battery of protective genes countering oxidative stress, environment toxic insults, lipid peroxidation, macromolecular damage, metabolic dysfunction and cell senescence (24, 38, 53, 67). It senses challenge to homeostasis in the cell cytoplasm and activates a protective transcriptional response. Such stress-responsive signaling is vital in resisting oxidative damage, cell dysfunction, cytotoxicity and mutagenesis, thereby contributing to resistance to drug toxicity, wound healing and decreasing risk of diabetes, vascular and neurodegenerative disease and ageing-related disease (5, 7, 15, 43, 44, 68). Improved manipulation of the Nrf2 system would likely lead to more effective deployment of micronutrient Nrf2 activators in functional foods and improved management and treatment of disease where Nrf2-mediated stress response is involved.
Innovation.
We suggest that an engineering principle might apply to nuclear factor erythroid 2-related factor 2 (Nrf2) to make it fit for purpose. The Nrf2 system is a cytoplasmic stress sensor as well as a mediator of increased protective gene transcription. Dynamical resolution of engineered sensors (e.g., in wireless sensor networks) relies on a high refresh rate because the sensor must reassess the relevant aspects of the environment at an appropriately high rate—a circumstance relevant to Nrf2. A process that repeatedly reassesses the environment is more reliable than one that relies on detecting changes in an equilibrium level of a signal that represents the environment. Such sensors can be linked to decision-making apparatus and can alter the refresh rate to match dynamical information. The functional pay-off and advantage achieved with this regulatory mechanism is the positive coupling of increased frequency of direct sensing of surveillance of cytoplasmic stress by Nrf2 to increased transcriptional response when cells are challenged by toxicants and metabolic disturbance. In clinical translation of this work it can be seen that employing a strategy of screening Nrf2 activators on the basis of maximizing residence time of Nrf2 in the nucleus may rather have selected for suboptimal if not poor inducers of the antioxidant response element-linked transcriptional response. A more effective approach may be to “tune” Nrf2 translocational oscillation frequency to maximize functionally active Nrf2 in the cell nucleus.
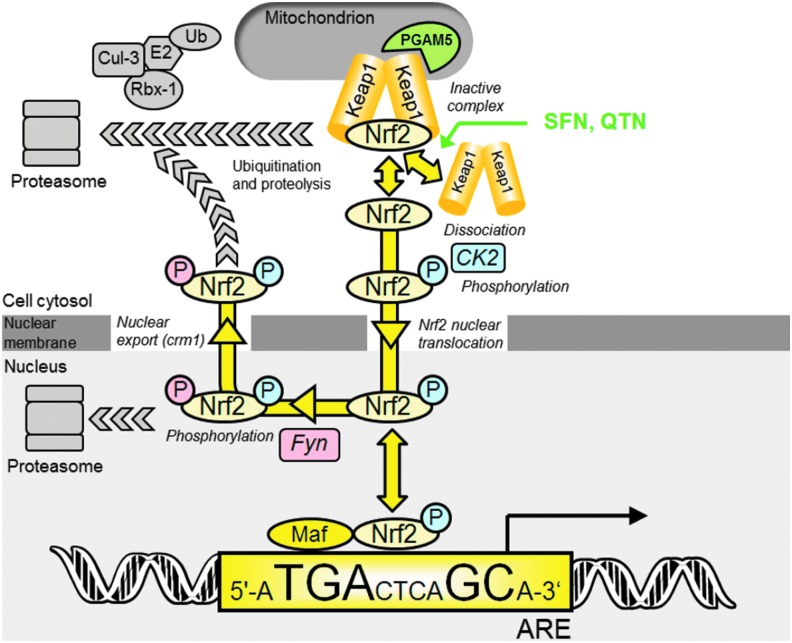
According to current understanding, under basal conditions, Nrf2 transactivational activity is repressed by binding to Kelch-like erythroid cell-derived protein with CNC homology-associated protein 1 (Keap1). This acts as an inhibitor holding Nrf2 in the cytoplasm and facilitating its degradation. Keap1 is a substrate adaptor protein for Cullin-3 (Cul3)-dependent E2 ubiquitin ligase complex, directing Nrf2 for degradation by the 26S proteasome (9). Keap1 and Nrf2 are held together with the protein threonine phosphatase, PGAM5, which tethers Keap1 and Nrf2 to the outer mitochondrial membrane (36).
The mechanisms of stress sensing and surveillance by the Nrf2 system are uncertain but under conditions of oxidative and electrophilic stress Nrf2 is released from Keap1. Nrf2 is phosphorylated by casein kinase-2 (CK2) (1). It translocates to the nucleus via importins α5 and β1 (58). In the nucleus, Nrf2 activates target genes by binding to regulatory antioxidant response elements (AREs) (45). Thereafter, Nrf2 is phosphorylated by Fyn kinase and expelled from the nucleus via the nuclear membrane export channel exportin-1/crm1, and degraded (20) (Fig. 1). Endogenous activators are lipid peroxidation and arachidonic acid oxidation products, 4-hydroxynonenal and J3-isoprostanes (13, 17), and exogenous activators are typically bioactive compounds from fruits and vegetables such as R-sulforaphane (SFN) and quercetin (QTN) (57, 59). The Nrf2/Keap1/ARE system regulates a large number of genes in mammals: basal expression of ca. 640 genes, inducible expression of 650 genes and basal and inducible expression of ca. 240 genes has been detected (38).
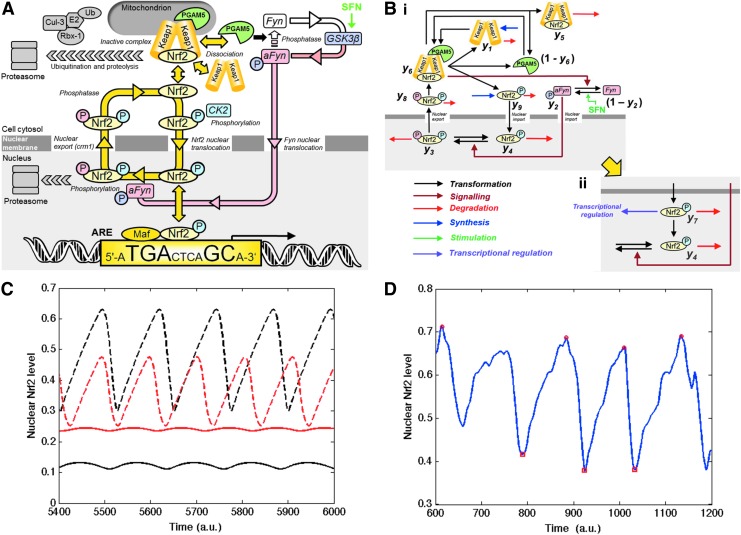
FIG. 1.
Current understanding of transcriptional control of ARE-linked gene expression by the Nrf2/Keap1 system. Key: P, phosphorylation (color-coded for CK2 or Fyn catalyzed modification). In quiescence, Nrf2 in the cytoplasm is complexed to Keap1 in association with Cul3-Rbx-1 attracting co-association of E2 ubiquitin ligase, Nrf2 ubiquitination and proteolysis. Upon activation (yellow arrows), Nrf2 dissociates from Keap1 and is phosphorylated by CK2; phosphorylated Nrf2 enters the nucleus and binds with accessory Maf protein to AREs. Thereafter, Nrf2 is phosphorylated by Fyn, exported from the nucleus and degraded by the proteasomal system. SFN and QTN are thought to activate Nrf2 by disrupting the Nrf2-Keap1 complex and stabilizing Nrf2 from proteolysis (57, 59)-shown in green. Currently unexplained features in the regulation of Nrf2 are: cytoplasmic binding of Nrf2, Keap1 and PGAM5 in a ternary complex tethered to mitochondria, proteasomal proteolysis of Nrf2 in the nucleus and potent inhibition of transcriptional activity by nuclear trapping of Nrf2 by short form estrogen-related receptor beta (73). The ARE shown is the mammalian consensus sequence (62). ARE, antioxidant response element; CK2, casein kinase-2; Cul3, Cullin-3; Keap1, Kelch-like erythroid cell-derived protein with CNC homology-associated protein 1; Nrf2, nuclear factor erythroid 2-related factor 2; QTN, quercetin; Rbx-1, ring-box 1; SFN, sulforaphane. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Research on the control of Nrf2 has focused on binding and chemical modification of its inhibitor, Keap1, by activators, stabilizing Nrf2 to proteolysis thus increasing cellular Nrf2 protein content (27). Our initial studies, however, showed that activation of cells to produce half-maximal ARE-linked transcriptional response elicited no significant change in Nrf2 protein concentration—although markedly higher and often toxic concentrations of activators did so (23, 57). Under physiological and health beneficial conditions, therefore, Nrf2 activation status may be encoded in the time dependence of subcellular Nrf2 location rather than its resistance to proteolysis. Herein we show, for the first time, that Nrf2 undergoes translocational oscillations from the cytoplasm to the nucleus in the basal state. When stimulated at physiological levels, the translocational oscillations of Nrf2 increase in frequency, decrease in amplitude and activate ARE-regulated genes. We present a mechanism to explain this where Nrf2 functionality is linked to reactivation on return to the cytoplasm or “refresh rate” which links protective transactivational response to increased frequency of direct surveillance of the cytoplasm.
Results
Nrf2 undergoes autonomous translocational oscillations and cell stimulation increases oscillation frequency, decreases oscillation amplitude and regulates ARE-linked gene transcription
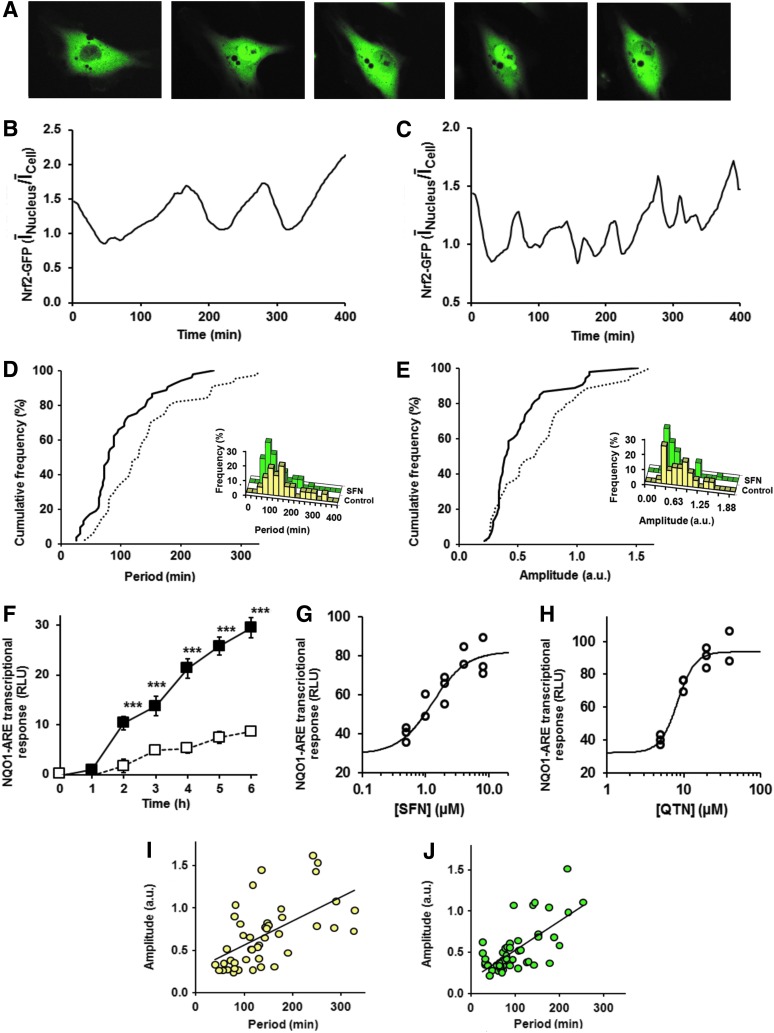
To examine Nrf2 stress response signaling between cytoplasmic and nuclear compartments we studied real-time changes in subcellular localization of Nrf2 in human HMEC-1 (human microvascular endothelial cell line-1) cells in vitro by live cell time-lapse fluorescence microscopy. HMEC-1 cells were transfected to express green fluorescent protein (GFP)-Nrf2 fusion protein. Dynamics of nucleus/cell GFP-Nrf2 fluorescence intensity ratio obtained from time-lapse image sequences revealed that Nrf2 undergoes oscillatory cytoplasm-nucleus translocation. Observation periods were for 400 min (Fig. 2A and Supplementary Video S1; Supplementary Data are available online at www.liebertpub.com/ars). Total cell fluorescence was unchanged over this period—in keeping with unchanged total cellular Nrf2 protein content (see Fig. 4B, L, and N). The fluorescence was quantified by computing the mean fluorescence per pixel in the nucleus and whole cell, ĪNucleus and ĪCell respectively. Average pixel intensities are used because they are less dependent upon cell size. The ratio ĪNucleus/ĪCell showed the periodicity of the translocational oscillations (Fig. 2B). The accumulation of Nrf2 into the nucleus was slow and, after a time delay, its expulsion from the nucleus was relatively rapid. Oscillations for individual cells were asynchronous with those of other cells. The period of oscillation, median (lower–upper quartile), was: 129 (81–175) min (n=44) with amplitude 0.65 (0.35–0.87) arbitrary units (Fig. 2D).
FIG. 2.
Cytoplasm-nucleus translocational oscillations of Nrf2 in human vascular endothelial HMEC-1 cells in vitro and response to functional stimulation. (A) Fluorescence microscopy image frames of HMEC-1 cells expression GFP-Nrf2 showing oscillation of cytoplasm-nucleus translocation. Images frames taken at time 46, 168, 218, 280 and 316 min poststimulation of HMEC-1 cells with 2 μM SFN. (B, C) Typical time course changes in GFP-Nrf2 nuclear/cellular localization ratio for control cells and cells stimulated with 2 μM SFN, respectively. The ratio plotted is ĪNucleus/ĪCell where ĪNucleus and ĪCell are average pixel intensities of GFP-Nrf2 fluorescence of the nucleus and whole cell in the field of view, respectively. NB ĪNucleus/ĪCell relates to the ratio of total nucleus/cell GFP-Nrf2, r, by the equation: r=(ĪNucleus×ANucleus)/(ĪCell×ACell), where ANucleus and ACell are areas of the nucleus and whole cell, respectively. Ratio r varies from 0 to 1 and ĪNucleus/ĪCell from 0 to >1. (D) Cumulative frequency distribution of GFP-Nrf2 cytoplasm-nuclear translocation oscillation period (histogram distribution shown inset). Median (lower–upper quartile): control - - -, 129 (81–175) min (n=44); +2 μM SFN —, 80 (64–128) min (n=53), p<0.001; Mann–Whitney U test. (E) Cumulative frequency distribution of GFP-Nrf2 cytoplasm-nuclear translocation amplitude (histogram distribution shown inset). Median (lower–upper quartile): control ---, 0.65 (0.35–0.87) arbitrary units (n=44); +2 μM SFN —, 0.40 (0.34–0.59) arbitrary units (n=53), p<0.05; Mann–Whitney U test. (F) Time course of NQO1-ARE transcriptional activity in control (□–□) and SFN stimulated (■—■) HMEC-1 cells in vitro. Data are mean±SEM, n=11; significance—***p<0.001; t-test. Dose response of NQO1-ARE transcriptional response on activator concentration in HMEC-1 cells studied by luciferase reporter assay. Luminescence output is given in RLU. (G, H) Six-hour-treatment-response data were fitted to a logistic regression equation, NQO1-ARE transcriptional response=[Activator]n/([Activator]n+EC50n)+c, where EC50 is the median effective concentration, n the logistic regression coefficient or Hill coefficient, and c the constitutive or background response. (G) SFN, EC50=1.3±0.3 μM, n=1.64±0.58 and c=29.3±3.3 (n=13); and (H) QTN, EC50=8.3±0.5 μM, n=3.81±0.79, c=32.3±3.3 (n=11). Correlation of oscillation amplitude on period. (I) Control cells—correlation coefficient r=0.63 with regression equation: amplitude=0.0028×Period (min)+0.27; p<0.001. (J) Cells activated with 2 μM SFN−correlation coefficient r=0.62 with regression equation: amplitude=0.0035×Period (min)+0.17; p<0.001. GFP, green fluorescent protein; HMEC-1, human microvascular endothelial cell line-1; RLU, relative light unit. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
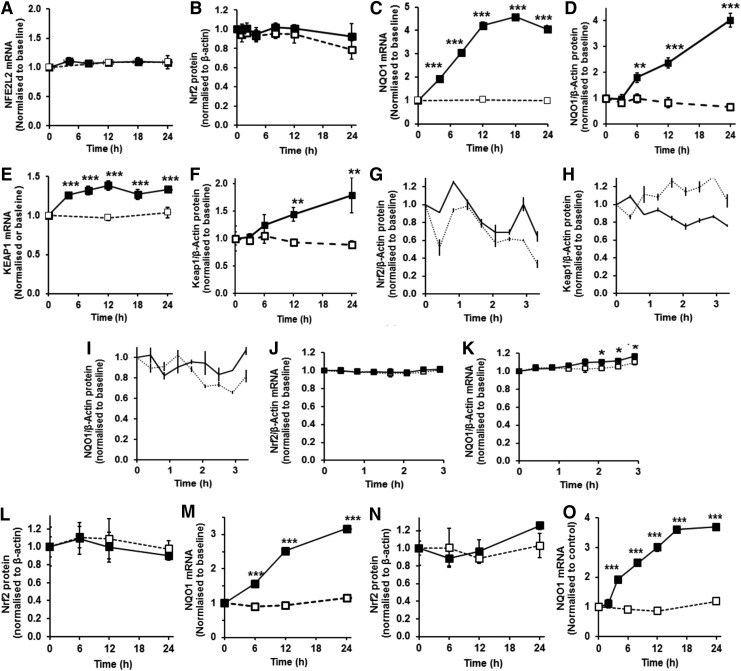
FIG. 4.
Evidence against the accumulation of Nrf2 protein during the activation of ARE-linked gene expression. Cellular Nrf2 and Keap1 during the induction of NQO1 expression by 2 μM SFN: (A) Nrf2 (NFE2L2) mRNA, (B) Nrf2 protein, (C) NQO1 mRNA, (D) NQO1 protein (E) Keap1 mRNA and (F) Keap1 protein, and. Key: □---□, control; ■—■, +2 μM SFN. Data are mean±SD (n=3), normalized to baseline level. Examination of short-period variability of Nrf2, Keap1 and NQO1 in the initial 3 h exposure period: (G) Nrf2 protein, (H) Keap1 protein, (I) NQO1 protein, (J) Nrf2 mRNA and (K) NQO1 mRNA. Key: ····, control; —, +2 μM SFN. Cellular Nrf2 during the induction of NQO1 expression by SFN in human aortic endothelial cells in primary culture: (L) Nrf2 protein, (M) NQO1 mRNA. Key: □---□, control; ■—■, +2 μM SFN. Cellular Nrf2 during the induction of NQO1 expression by SFN in human BJ fibroblasts in primary culture: (N) Nrf2 protein, (O) NQO1 mRNA. Key: □---□, control; ■—■, +1 μM SFN. For (G–O), data are mean with bars showing the range of two estimates; and for (J, K), data are mean±SD (n=3). Significance: (G–I), autocorrelation function analysis for white noise—p>0.05 for all panels; other panels— *p<0.05, **p<0.01 and ***p<0.001 with respect to unstimulated control (t-test).
We next studied the effect of Nrf2 activators, SFN and QTN, on cytoplasm-nucleus translocational oscillations. To establish a concentration that achieves functional Nrf2 activation without cytotoxicity we studied the concentration dependence of Nrf2 transactivational activity using a transfected quinone reductase-ARE (NQO1-ARE) luciferase reporter vector. The NQO1-ARE transcriptional response increased slowly over 6 h in unstimulated cells and with ca. threefold increased rate when cells were stimulated with 2 μM SFN (Fig. 2F). We used the 6 h treatment response as a fixed time point measure of NQO1-ARE transcriptional activity. Stimulant concentration-transcriptional response relationships were explored for SFN (0.5–8 μM) and QTN (5–40 μM). We found median effective concentration EC50 values producing half-maximal increase in transcriptional response were 1.3±0.3 μM SFN and 8.3±0.5 μM QTN (Fig. 2G, H). SFN (2 μM) and QTN (8 μM) were used in subsequent studies. The concentration-response profile for SFN was similar to that previously found using a Nrf2 Neh2 domain-lucerifase reporter (50). Similar activation response was found when cells were cultured short-term without epidermal growth factor (EGF)-indicating that the response was not dependent on EGF signaling. These concentrations do not induce growth arrest, change in distribution of cells in the cell growth cycle or toxicity (56, 66), which is rather typical of much higher concentrations; for example, disturbance of the cell cycle and induction of apoptosis by 12.5–50 μM SFN (3). The low concentrations of SFN and QTN used herein are similar to peak plasma concentrations achieved with a diet containing source materials rich in these bioactive compounds or their precursors associated with health benefit (6, 16). When cells were treated with SFN there was an increase in frequency and decrease in amplitude of the oscillations (Fig. 2C, E and Supplementary Video S2). The period of oscillation was decreased to 80 (64–128) min (−38%, p<0.001) with concomitant decrease in oscillation amplitude to 0.40 (0.34–0.59) arbitrary units (−38%, p<0.05); n=53, Mann–Whitney U. Oscillation period correlated positively with amplitude (Fig. 2I, J). We used a statistical analysis of the autocorrelation function (4) to reject the hypothesis that what we observed are not faster oscillations but a loss of oscillations resulting in stochastic fluctuations. This hypothesis was strongly rejected (Ljung-Box test, p<3×10−16 for all SFN-treated cell time series). Similar oscillations of Nrf2 were found with QTN where the oscillation period was intermediate to that of control and SFN-treated cells (Supplementary Video S3). Removal of SFN from the culture by washing cells and continued culture with fresh medium returned the translocational oscillation period to that of quiescent cells in the subsequent 2–4 h, consistent with the half-life of cell protein bound isothiocyanate adducts (64).
We next sought to validate the detection of translocational oscillations of Nrf2 in HMEC-1 cells in vitro. To test whether the Nrf2 oscillations were associated with cell stress induced by illumination with excitation wavelength light to record the GFP image, we varied the frequency of image frame capture with fixed exposure time of 250 ms from 2 to 10 min in the time-lapse microscopy. The presence of oscillations and oscillation frequency did not change significantly: oscillation frequency—5 min frame interval, 110 (99–118) min (n=6); and 10 min frame interval, 120 (110–130) min (n=7). Hence microscope illumination was not a driver of Nrf2 oscillation. HMEC-1 cells have a dermal microvascular endothelial phenotype and translocational oscillations of Nrf2 may be a response to abnormal high oxygen concentration. To test this we incubated transfected cells under a depleted oxygen atmosphere of 3% oxygen, typical of skin in vivo (61). Translocational oscillations of Nrf2 were found of similar frequency to those characterized above (Supplementary Video S4); oscillation frequency 104 (87–115) min (n=6). To test whether Nrf2 oscillations were an artifact of the characteristics of the HMEC-1 cell line, we transfected human aortic endothelial cells and human BJ fibroblasts (human foreskin fibroblasts originated in the laboratory of J.R. Smith, Baylor College of Medicine, Houston, Texas, USA) fibroblasts in primary culture and recorded similar time lapse images. Therein we found similar cytoplasm-nucleus translocational oscillations of Nrf2 (Supplementary Videos S5 and S6).
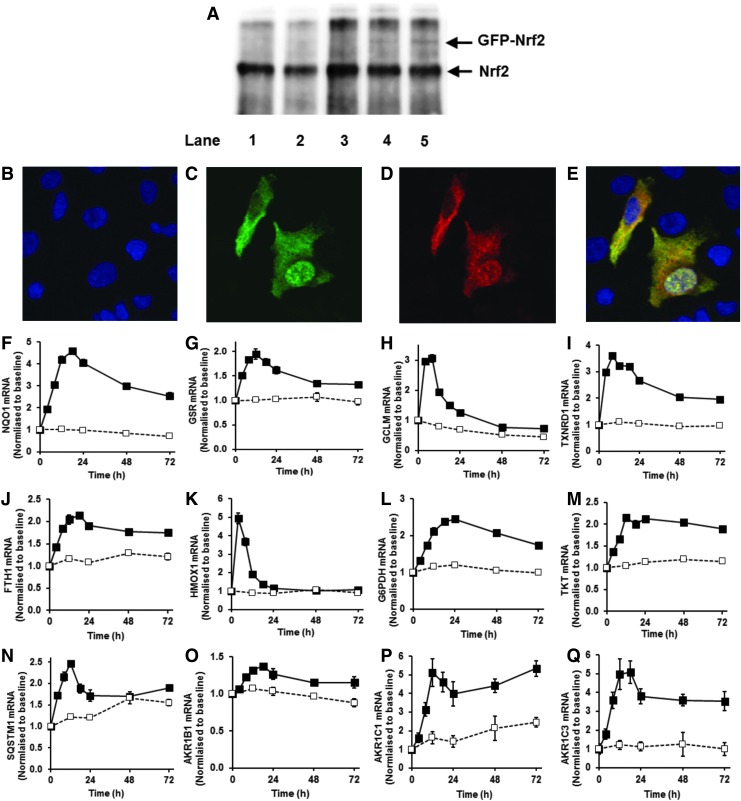
We also made test transfections to determine the extent to which Nrf2-GFP increases the total cellular content of Nrf2 and to check that GFP-Nrf2 faithfully reports the cellular location of Nrf2. Transfection with the vector pcDNA3-EGFP-C4-Nrf2 produces expression of Nrf2-GFP comprising the human Nrf2 domain (sequence mass 68 kDa, with N-terminal tag of GFP, 27 kDa sequence mass). Antibodies raised to the C-terminal domain of Nrf2, EP1808Y, detected both Nrf2 and Nrf2-GFP with equal affinity. Western blotting with EP1808Y antibody detected a band at 98 kDa from control, untransfected HMEC-1 cells (consistent with electrophoretic mobility of Nrf2) (32, 65), and bands at 98 and 125 kDa from transfected HMEC-1 (consistent with electrophoretic mobility of Nrf2 and Nrf2-GFP) (Fig. 3A). Densitometry of these bands produced an intensity ratio of 100:4. Immunoblotting of Nrf2 with H300 antibodies raised to an Nrf2 N-terminal epitope gave similar results—intensity ratio of 100:7, indicating that expression of GFP-Nrf2 produced only a minor increase in the total Nrf2 pool of 4%–7%. Immunofluorescence micrographs of transfected HMEC-1 cells with anti-Nrf2 antibody produced staining for some cells mostly in the cytosol, others mostly nucleus or both cytosol and nucleus. Irrespective of where anti-Nrf2 immunostaining was located (green), immunofluorescence of GFP (red) showed co-localization with Nrf2 (Fig. 3B–E). Addition of stimulants did not change total GFP-Nrf2 fluorescence. Moreover, studies with double transfection of cells to express GFP-Nrf2 and destabilized Venus-ARE-linked transcription reporter with monitoring of Nrf2 translocation and ARE-linked transcription before and after SFN stimulation indicated that cells with increased Nrf2 translocation frequency were those with increased ARE transcription activity.
FIG. 3.
Validation of Nrf2 subcellular location by GFP-Nrf2 reporter and ARE-linked transcriptional response in human vascular endothelial HMEC-1 cells in vitro. (A) Immunoprecipitation with anti-Nrf2 antibody (H300) and Western blot for Nrf2 with EP1808Y antibody. Lanes 1–3, untransfected cells; lanes 4 and 5, cells transfected with pcDNA3-EGFP-C4-Nrf2 expressing GFP-Nrf2. Nrf2 and GFP-Nrf2 bands were at 98 and 125 kDa, respectively. (B–E) Fluorescence microscopy of HMEC-1 cells for detection of Nrf2 and GFP by immunofluorescence. Two cells in the field of view have Nrf2 mostly in the cytoplasm (left) and in the cytoplasm and nucleus (right). (B) Nuclear staining with DAPI, (C) Immunofluorescence for Nrf2 with EP1808Y primary antibody and Alexa Fluor®488 conjugated goat anti-rabbit IgG secondary antibody, (D) Immunofluorescence for GFP with GFP-1020 primary antibody and Alexa Fluor®555 conjugated goat anti-chicken IgG secondary antibody, and (E) overlay image. (F–Q). Changes in gene expression, mRNA levels normalized to housekeeping genes in control (open square, dashed line) and SFN stimulated cells (filled square, solid line). (F) quinone reductase, NQO1, (G) glutathione reductase GSR, (H) γ-glutamylcysteine ligase (regulatory subunit) GCLM, (I) thioredoxin reductase TXNRD1, (J) ferritin FTH1, (K) heme oxygenase HMOX1, (L) glucose-6-phosphate dehydrogenase G6PDH, (M) transketolase TKT, (N) sequestosome-1/p62 SQSTM1 and (O, P, and Q), aldoketo reductase isoforms AK1B1, AK1C1 and AK1C3. Key: dashed line with open squares, control; solid line with filled squares, +2 μM SFN. Data are mean±SD (n=3), normalized to baseline level. Significance: all control and SFN time courses were significantly different in repeated measures analysis and at individual time points (p<0.001, t-test) except for HMOX1 from 24 to 72 h and SQSTM1 at 48 h. Indicators of significance are omitted for clarity. Stimulation of HMEC-1 cells with SFN produced increased expression of ARE-linked genes. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Increased frequency and decreased amplitude of Nrf2 translocational oscillations were associated with increased expression of ARE-linked genes: quinone reductase, glutathione reductase, γ-glutamylcysteine ligase-regulatory subunit, thioredoxin reductase, ferritin and heme oxygenase linked to antioxidant cytoprotective responses (47) (Fig. 3F–K); glucose-6-phosphate dehydrogenase and transketolase linked to metabolic re-programming (41) (Fig. 3L, M); sequestosome-1 (p62) linked to autophagy in healthspan (11) (Fig. 3N), and aldoketo reductase isoforms 1B1, 1C1 and 1C3, linked to carbonyl stress cytoprotective responses (Fig. 3O–Q). The time course profile of mRNA of these ARE-linked genes increased over the initial 6–8 h, matching the period of pharmacological activity of SFN. The pharmacological response of SFN and related isothiocyanates is associated with reversible binding to target protein thiols (protein thiocarbamoylation) which increases over the initial 4 h, remaining above the half-maximal value for 6–8 h and declining thereafter; cf. reversible cell protein binding by phenethyl isothiocyanate (64). Thereafter, mRNA levels return to baseline over periods reflecting half-lives of mRNA and residual increased expression. Response duration may be increased by induction of expression of SQSTM1/p62 which is itself an activator of Nrf2 (28). Increased ARE-linked gene mRNA level was associated with increased expression product, for example, increased NQO1 protein (see Fig. 4C, D).
Activation of the ARE transcriptional response occurs without increase in Nrf2 or decrease in Keap1 protein
It is currently considered that increased Nrf2 transactivational activity is mediated by accumulation of Nrf2 protein in the cell cytosol with CK2-driven entry of Nrf2 into the nucleus and increased ARE-linked gene transcription. We examined mRNA and protein of Nrf2 in HMEC-1 cells with and without treatment with 2 μM SFN and found no change in the initial 24 h (Fig. 4A, B), although Nrf2 transactivational activity was increased in this period by SFN—as evidenced by increased NQO1 expression at the mRNA and protein levels in the initial 24 h period (Fig. 4C, D). In contrast to current mechanistic interpretation, Nrf2 transactivational activity appears not linked to change in cellular levels of Nrf2 protein for SFN concentration-limited responses but rather to change in subcellular localization in translocational oscillations (see Fig. 2B, C, F). Keap1 mRNA was increased at 4 h poststimulation and remained increased thereafter to 24 h poststimulation, and Keap1 protein was not decreased and rather increased at 12 h poststimulation (Fig. 4E, F). Change in cytoplasmic/nuclear translocational oscillation frequency and amplitude of Nrf2 induced by SFN, however, occurred within the initial 6 h poststimulation period (see Fig. 2B, C). Taken together, these observations suggest high frequency, low amplitude oscillations of Nrf2 associated with transactivational activity are not linked to change in Nrf2 or Keap1 protein. Induction of Keap1 expression by SFN was expected as Keap1 is an ARE-linked gene (33).
We also examined levels of Nrf2, Keap1 and NQO1 protein in the initial 24 h of control and stimulant treatments to detect possible oscillatory change in regulatory components and target gene product. Time intervals of 25 min were small relative to the period of Nrf2 translocational oscillations. Inspection of time course profiles in cellular content of Nrf2, Keap1 and NQO1 protein (Fig. 4G–I) and statistical testing (α=0.95, autocorrelation function analysis) show that variation in response is that expected for white noise, implying that the time series are not oscillatory. There was also no evidence of oscillation in levels of Nrf2 mRNA (Fig. 4J), nor in target gene NQO1 mRNA which was increased in SFN-treated cells from 125 min poststimulation, with respect to unstimulated control (Fig. 4K).
The key features of the Nrf2 regulatory mechanism, induction of ARE-regulated gene expression with Nrf2 oscillation without change in total Nrf2 protein concentration, were also found in other cells types—human aortic endothelial cells and human BJ fibroblasts in primary culture (Supplementary Videos S5 and S6 and Fig. 4L–O).
Mathematical models successfully reproduce the oscillations
Mathematical models were developed alongside the experiments in order to guide hypothesis generation and to understand the origin and nature of the oscillations. Initial model development was constrained by the autonomous nature of the oscillations, asymmetric profile of slow nuclear import and rapid export of Nrf2, the positive correlation of oscillation period and amplitude and unchanged Nrf2 mRNA and protein—the rate of synthesis and degradation of Nrf2 protein being assumed equal whether in the cytoplasm or nucleus by including terms for cytoplasmic and nuclear proteolysis of Nrf2. We also propose that the protein threonine phosphatase, PGAM5, associated with Nrf2 and Keap1 in a ternary complex in the cytoplasm (8, 35, 55), dephosphorylates Fyn for its inactivation—Table 1.
Table 1.
Processes and Constraints of the Systems and Mathematical Models of Nrf2 Regulation
| Process | Reaction | Rate | Catalyst/stimulant/mediator |
|---|---|---|---|
| Cytoplasm | |||
| Dissociation of Nrf2:Keap1: PGAM5 complex (I) | Nrf2:Keap1:PGAM5→Nrf2+Keap1+PGAM5 | R1 | |
| Nrf2:Keap1:PGAM5 complex formation | pNrf2+Keap1+PGAM5→Nrf2:Keap1:PGAM5 | R2 | PPase, deacetylase |
| Activation of Fyn | Fyn→aFyn | R3 | GSK3β, SFN, QTN |
| Inactivation of Fyn | aFyn→Fyn | R4 | PGAM5 |
| Dissociation of PGAM5:Nrf2:Keap1 complex (II) | Nrf2:Keap1:PGAM5→Nrf2:Keap1+PGAM5 | R8 | |
| Degradation of Nrf2:Keap1 complex | Nrf2:Keap1→Proteolysis products | R9 | Cytoplasmic proteasome |
| Synthesis of Keap1 | Protein synthesis→Keap1 | R11 | Translation |
| Synthesis of Nrf2 | Protein synthesis→Nrf2 | R12 | Translation |
| Degradation of Keap1 | Keap1→Proteolysis products | R14 | Cytoplasmic proteasome |
| Degradation of Nrf2 exported from the nucleus | pNrf2→Proteolysis products | R17 | Cytoplasmic proteasome |
| Degradation of Nrf2 | Nrf2→Proteolysis products | R19 | Cytoplasmic proteasome |
| Nucleus (NB Nrf2 imported into the nucleus is phosphorylation by CK2) | |||
| Phosphorylation of inactive Nrf2 | iNrf2→pNrf2 | R5 | aFyn |
| Dephosphorylation of inactive Nrf2 | pNrf2→iNrf2 | R6 | PTPase |
| Inactivation of Nrf2 | Nrf2→iNrf2 | R7 | Acetyl transferase |
| Degradation of iNrf2 | iNrf2→Proteolysis products | R10 | Nuclear proteasome |
| Degradation of Nrf2 | Nrf2→Proteolysis products | R13 | Nuclear proteasome |
| Degradation of pNrf2 | pNrf2→Proteolysis products | R15 | Nuclear proteasome |
| Cytoplasm–nuclear translocation | |||
| Nuclear export of pNrf2 | pNrf2 (nucleus)→pNrf2 (cytoplasm) | R16 | crm1, aFyn |
| Nuclear import of Nrf2 | Nrf2 (cytoplasm)→Nrf2 (nucleus) | R18 | Importins α5 and β1, CK2 |
Constraints: 1. Nrf2 oscillations are translocational with no oscillation in total cellular Nrf2 protein. 2. Oscillations are autonomous (occur without stimulation). 3. Oscillations have an asymmetric waveform with slow nuclear import and rapid export. 4. Stimulation, by SFN or QTN increases frequency and decreases amplitude of oscillations (this work). 5. SFN and QTN increase Fyn activity (34, 37, 70). 6. Nrf2 is exported from the nucleus following phosphorylation by Fyn (20). 7. Cytoplasmic Nrf2 is imported into the nucleus by CK2 (1). 8. PGAM5 forms a ternary complex with Keap1 and Nrf2 in the cytoplasm (36). 9. Nrf2 is inactivated in the nucleus by acetylation (40).
Assumptions: ternary complex-bound PGAM5 catalyses the inactivation of Fyn. Inferred biochemical catalysts are: PPase for cytoplasmic dephosphorylation of Nrf2 [removing Tyr-568 phosphorylation of Fyn (20) and Ser phosphorylation of CK2 (1)], PTPase for reversal of phosphorylation of Nrf2 by Fyn in the nucleus, nuclear acetyl transferase and cytoplasmic deacetylase for reversible acetylation of Nrf2 (40, 46), and cytoplasmic and nuclear proteasome for Nrf2 and Keap1 degradation.
aFyn, phosphothreonine-activated Fyn; CK2, casein kinase-2; GSK3β, glycogen synthase kinase 3β; Keap1, Kelch-like erythroid cell-derived protein with CNC homology-associated protein 1; Nrf2, nuclear factor erythroid 2-related factor 2; PPase, protein phosphatase; PTPase, protein tyrosine phosphatase; QTN, quercetin; SFN, sulforaphane.
A key feature of the experimentally observed oscillations is the rapid coordinated exit of the Nrf2 from the nucleus. This is driven by phosphorylation of nuclear Nrf2 by activated Fyn and represents one of the main mechanisms driving the oscillations. After the expulsion from the nucleus, Nrf2 rebinds Keap1 and PGAM5 in the ternary complex but at a level greater than at equilibrium. As a consequence the complex is disrupted by this overshoot from the equilibrium level with consequent release of PGAM5 from the complex. We assume that PGAM5 dephosphorylates and inactivates Fyn and less efficiently when free; thereby release of PGAM5 is associated with activation of Fyn. The combination of phosphorylation and de-phosphorylation of Fyn enables this subsystem to display ultrasensitivity, manifested by the gradual build-up of free PGAM5 and by the delayed and gradual build-up and then a rapid influx of activated Fyn into the nucleus. This phosphorylates nuclear Nrf2 and drives its rapid expulsion from the nucleus ready for the cycle to repeat [Fig. 5A (schematic) and B (regulatory model)]. We modeled ternary complex disruption mathematically in a generic form without discrimination for destabilization mechanism. The model reproduces the key features of experimental observations: asymmetric oscillations and the faster oscillations of decreased amplitude when stimulated (Fig. 5C). A stochastic version of the model simulated using the Gillespie algorithm (14) gives simulations that qualitatively match the experimental time course profiles and transcriptional response well (Supplementary Data S1 section and Fig. 5D).
FIG. 5.
Cell signaling sustaining Nrf2 translocational oscillations for control of ARE-linked gene expression. (A) Schematic diagram. (B) Regulatory model—(i) model of the signaling network and (ii) extended model with inactivation of nuclear Nrf2 introduced. Negative feedback is provided by successive de-phosphorylation and phosphorylation steps, which induce ultra-sensitivity in Fyn activation and the Fyn phosphorylation of nuclear Nrf2 and which produce a delay between Nrf2 nuclear entry and the Fyn phosphorylation which drives nuclear export. Variables y1−y9 are indicated. (C) Simulations of Nrf2 oscillation in the extended model. Two levels of Nrf2 in the nucleus, total (Nrf2+iNrf2+pNrf2, normalized) and (Nrf2, non-normalized), are calculated in the control and SFN-stimulated conditions, which are modeled respectively by small and large values of the parameter (h1) that represents the maximum rate of Fyn activation. Key: (—) active nuclear Nrf2, (- - -) total nuclear Nrf2, h1=0.168; (solid red lines) active nuclear Nrf2, and (dashed red lines) total nuclear Nrf2, h1=0.264. See Supplementary Data for other parameter values and equations. Active nuclear Nrf2 is non-normalized and total nuclear Nrf2 is normalized. (D) Stochastic model in the control condition. The result is smoothed by wavelets in the same way as used in the processing of the video frames. The time scale is given in arbitrary units (a.u.). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Experimental validation of the fyn-glycogen synthase kinase 3β axis in Nrf2/ARE transcriptional response regulation
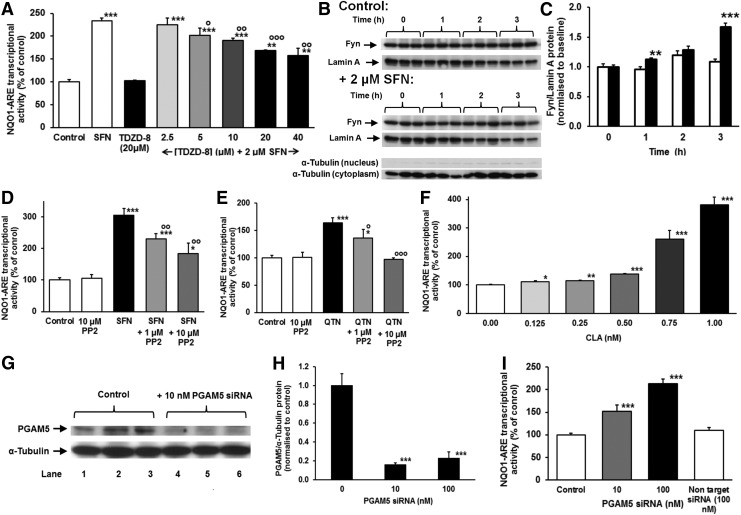
Fyn mediated nuclear export of Nrf2 has a critical role in its refresh rate and thereby transcriptional activity. Upstream activation of Fyn occurs by glycogen synthase kinase 3β (GSK3β) catalyzed phosphorylation on threonine residues (22). We propose that Fyn returns to quiescence by dephosphorylation by threonine phosphatase PGAM5. Inhibition of GSK3β with the specific inhibitor TDZD-8 decreased SFN-activated NQO1-ARE transcriptional response, confirming the role of GSK3β in the SFN activated response (Fig. 6A). Inhibition was, however, partial at 40 μM TDZD-8, which suggests other effects are likely involved in the SFN-activated response. Activation of Nrf2 by SFN increased nuclear localization of Fyn (Fig. 6B, C). Addition of Fyn inhibitor PP2 to HMEC-1 cells stimulated by SFN decreased the proportion of cells exhibiting oscillations, 16/20 versus 3/10 (p=0.012, contingency tables) and decreased NQO1-ARE transcriptional response induced by SFN and QTN (Fig. 6D, E), confirming that Nrf2 oscillations and ARE-regulated transcription are linked to and sustained by active Fyn. Addition of the threonine phosphatase inhibitor calyculin A (CLA) and knockdown of PGAM5 activated the NQO1-ARE transcriptional response (Fig. 6F–I). Consistent with this, knockdown of PGAM5 confers resistance to oxidative cytotoxicity induced by peroxides (63). Protein threonine phosphatase inhibition by CLA provided potent activation of Nrf2 being the first sub-nanomolar activator of the Nrf2 system. The stimulation of NQO1-ARE transcriptional activity by CLA was greater than ca. 80% knockdown of PGAM5. This may be due to effects of CLA other than inhibition of PGAM5 although the sub-nanomolar response is typical of CLA at its specific targets (52).
FIG. 6.
Fyn kinase-PGAM5-GSK3β axis of regulation of Nrf2 transcriptional activity. (A) Effect of TDZD on NQO1-ARE transcriptional activity in the absence and presence of SFN. Data are mean±SD, n=3. Activation of Fyn kinase: (B) Western blot of Fyn and β-actin (loading control) in nuclear extracts of HMEC-1 cells incubated for 0–3 h with and without 2 μM SFN. Western blots of α-tubulin are shown below, indicating minimal contamination of nuclear and cytoplasmic extracts. (C) Nucleus Fyn protein in control and SFN stimulated HMEC-1 cells in vitro. (D, E) Effect of inhibition of Fyn kinase by PP2 on NQO1-ARE transcriptional activity in the absence and presence of SFN and QTN, respectively. (F) Activation of NQO1-ARE transcriptional activity by CLA. (G–I) Activation of NQO1-ARE transcriptional activity by PGAM5 siRNA, respectively. (G, H) Validation of PGAM5 knockdown, (I) effect on NQO1-ARE transcriptional activity. CLA, calyculin A; GSK3β, glycogen synthase kinase 3β. Significance: (A, C–F, H, I), *p<0.05, **p< 0.01 and ***p<0.001 with respect to unstimulated control; and ○p<0.05, ○○p<0.01 and ○○○p<0.001 with respect to SFN (A, C) or QTN (E) stimulated controls (t-test).
Smaller faster oscillations can activate genes
At first sight it appears difficult to explain how a small amount of Nrf2 resident for a short time in the nucleus produces greater gene transcription than a greater amount of Nrf2 in the nucleus resident for longer. Using our model we show that this counterintuitive observation can be explained if we assume that after entering the nucleus in the active state, Nrf2 is deactivated before it is phosphorylated by Fyn and leaves the nucleus to be reactivated in the cytoplasm. Acetylation of Nrf2 in the nucleus was found to decrease ARE-regulated transcription (40). When this mechanism is included into the model we find that for a reasonable set of parameter values the levels of active nuclear Nrf2 are substantially higher with the smaller faster oscillations (Fig. 5C). The way we did this is described (Supplementary Data S1 section). These results suggest the following predictions.
Trapping Nrf2 in the nucleus turns off transcription after the initial quantum is produced
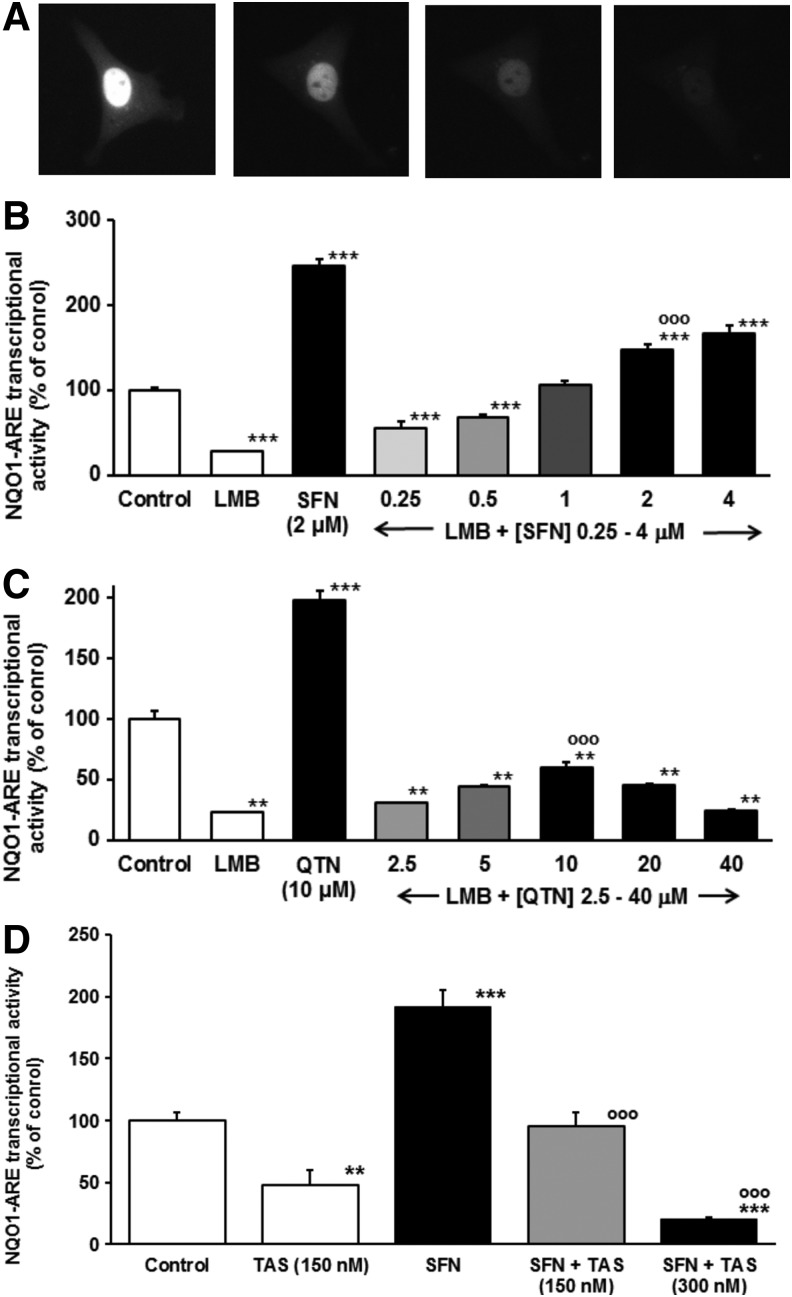
To test the transactivational activity of Nrf2 with prolonged dwell time in the nucleus we trapped Nrf2 in the nucleus by addition of leptomycin B (LMB)—a highly specific and potent inhibitor of crm1 (29) used previously in mechanistic studies of Nrf2 nuclear export (25). Addition of LMB to HMEC-1 cells transfected to express GFP-Nrf2 showed GFP-Nrf2 fluorescence localized in the cell nucleus. Furthermore, with concurrent block of de novo Nrf2 synthesis by addition of cycloheximide, Nrf2 protein was degraded in the nucleus with a half-life of 66 (48–195) min (n=8) (Fig. 7A). A recent estimate of total cellular half-life of Nrf2 was 62 min (48). Nuclear proteolysis of Nrf2, reported previously (18), suggests that Nrf2 is not being spared from degradation even when free of Keap1 and located in the nucleus. This also confirmed that Nrf2 is exported from the nucleus via crm1 and may be degraded within the nucleus. Addition of LMB produced inhibition of constitutive NQO1-ARE transcriptional response and SFN and QTN-induced NQO1-ARE transcriptional response (Fig. 7B, C). Our regulatory model invokes nuclear inactivation of Nrf2, mediated by acetylation as found previously (40). Nrf2 is an established target for reversible acetylation where acetylation is removed by histone deacetylases (46). The deacetylase inhibitor Trichostatin A (TAS) markedly increases acetylation of endothelial cell proteins in the concentration range 100 nM–1 μM (49). Addition of TAS (150 and 300 nM) to HMEC-1 cells inhibited constitutive and SFN-induced ARE-linked transcriptional response (Fig. 7D), consistent with acetylation as a likely cause of inactivation of nucleus trapped Nrf2. These studies indicate that Nrf2 loses its transactivational activity with prolonged dwell time in the nucleus where Nrf2 acetylation and nuclear proteasomal degradation are involved.
FIG. 7.
Trapping of Nrf2 in the nucleus and Nrf2 acetylation block Nrf2 transcriptional activity. (A) Fluorescence microscopy image frames of HMEC-1 cells transfected to express GFP-Nrf2 incubated with LMB for 2 h and cycloheximide (10 μg/ml) added. Frame time (left to right): 0, 40, 80 and 120 min. (B, C) Effect of LMB (2 ng/ml) on NQO1-ARE transcriptional activity in the absence and presence of SFN and QTN, respectively. Cells were preincubated for 2 h with LMB and then 6 h with and without Nrf2 activator. (D) Effect of TAS (150 and 300 nM) on NQO1-ARE transcriptional activity in the absence and presence of SFN (mean±SD, n=3). LMB, leptomycin B; TAS, Trichostatin A. Significance: (A, C–F, H, I), **p<0.01 and ***p<0.001 with respect to unstimulated control; and ○○○p<0.001 with respect to SFN (B, D) or QTN (C) stimulated controls (t-test).
Discussion
In this study we have shown that an important much-studied signaling system uses oscillations to control expression of its target genes and, in addition, we propose a significant revision of the signaling mechanism. The current view of regulation of Nrf2 transactivational activity envisages that stimulants stabilize Nrf2 from proteolysis and increased total cellular Nrf2 protein produces equilibrium shift to increased Nrf2 in the nucleus with resulting increased ARE-linked transcription (2). Nuclear expulsion of Nrf2 after double phosphorylation is also viewed as a route for Nrf2 degradation (22) (Fig. 1). Key to the need for revision was the observation that stimulation of HMEC-1 cells with 2 μM SFN achieved 67% maximal transcriptional response without increase in total cellular Nrf2 protein. This suggests that transcriptional activity of Nrf2 is encoded in the time dependence of its subcellular localization rather than total cellular Nrf2 concentration in basal and stimulated states. Also, trapping of Nrf2 in the nucleus by LMB and Fyn kinase inhibition decreased rather than increased transcriptional activity of Nrf2, suggesting that return of Nrf2 to the cytoplasm has a reactivation function. We found that the Nrf2 signaling system is an autonomous oscillator with translocational oscillations occurring whether stimulants such as SFN are present or absent. When stimulated the frequency of the oscillation increases, the amplitude decreases and the target genes are up-regulated. To gain greater understanding of this we developed a mathematical model of regulation of the Nrf2 system and tested its predictions. As a result we propose a revised mechanism for both the signaling dynamics and the transcriptional response of the Nrf2 system (Fig. 5A). The nuclear translocation period of Nrf2 was 129 min in quiescent cells and decreased to 80 min during activation of the ARE transcriptional response. Recent estimate of Nrf2 half-life in human cells by stable isotopic labelling studies is ca. 5 h (31). This suggests that Nrf2 makes two to three cytoplasmic nucleus translocations before degradation in unstimulated cells and more in stimulated cells. Nuclear translocation of Keap1 occurs in some systems (51) and its translocational oscillation would be worthy of future investigation.
We validated the detection of Nrf2 translocational oscillations. We conclude that GFP-Nrf2 faithfully reported the subcellular localization of Nrf2, GFP-Nrf2 transfection produced only a small marginal increase in the cellular Nrf2 pool unlikely to perturb total Nrf2 localization, Nrf2 translocational oscillations are not dependent on incident illumination nor oxygen concentration in HMEC-1 cells and they occur in multiple cell types—including human cells in primary culture. Oscillations of Nrf2 may have been overlooked in previous studies because higher concentrations of SFN and QTN were used. Previous observations of increased total cellular Nrf2 protein (23, 57) have often used cytotoxic concentrations of SFN and QTN, 25 and 40 μM respectively (23, 39, 66). Cytotoxic concentrations of SFN and QTN may impact differently on cytosol-nucleus translocational oscillations of Nrf2 and, indeed, activate the Nrf2 system indirectly as a defence to toxicity.
For regulatory and signaling systems the most common form of feedback that produces oscillation is where the production of a molecular species represses its own production but does it with a delay and in a manner yielding high sensitivity: the switch-off signal lags the switch-on signal, causing repeated overshoots. Our system is different. It belongs to a class of negative feedback systems where translocation of a molecular species M between two compartments from N to C is enhanced in a sensitive fashion by increased levels of M in N at some previous times: the enhanced leave-now signal for M in N lags the arrival of M, causing M to overshoot repeatedly analogous to the mechanism suggested in (19). In our case, increased free Nrf2 in the cytoplasm causes, after a delay, increased Fyn in the nucleus and hence an increased rate of translocation from the nucleus. We show in the Supplementary Data that, if y represents the total amount of Nrf2 in the nucleus, then one can approximate this system by the differential delay equation
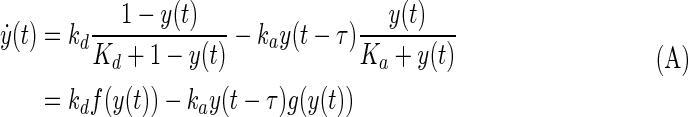 |
The first term describes influx into the nucleus and the second outflux. We describe in the SI how inclusion of the term y(t - τ) induces oscillations for certain values of τ because then the system continually overshoots the state y* where the terms balance. By applying linear stability analysis to this equation, the oscillation period is approximately 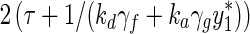 , where γf and γg are the absolute derivative of f(y) and g(y) in Equation (A) at y*. This illustrates the difference between the current nuclear-export driven oscillation model and the conventional synthesis-degradation driven oscillation model. In the latter, the period is controlled only by the degradation rate of the synthesized product, whereas for [Eq. (A)] period is influenced also by the kd γf term and when translated back to the full model this corresponds to the rate of Keap1 liberation from the PGAM5:Nrf2:Keap1 complex.
, where γf and γg are the absolute derivative of f(y) and g(y) in Equation (A) at y*. This illustrates the difference between the current nuclear-export driven oscillation model and the conventional synthesis-degradation driven oscillation model. In the latter, the period is controlled only by the degradation rate of the synthesized product, whereas for [Eq. (A)] period is influenced also by the kd γf term and when translated back to the full model this corresponds to the rate of Keap1 liberation from the PGAM5:Nrf2:Keap1 complex.
The mechanism destabilizing the ternary complex in the basal state likely relates to change in post-translational modifications (PTMs) of Nrf2. Indeed, this is why ternary complex formation is not a simple equilibrium process: the putative form of Nrf2 involved in complex formation (with tyr and ser phosphorylation and lys acetylation) is different from that involved in complex dissociation (with no PTMs or ser phosphorylation as CK2-dependent phosphorylation may destabilize the complex and is constitutively active). It is also likely that modification of Keap1 by Nrf2 activators also influences ternary complex stability (60)—a feature where conventional and oscillatory mechanisms of Nrf2 regulation may concur. Hitherto regulatory control of the Nrf2 has focused on binding and chemical modification of Keap1 by activators but in an Nrf2:Keap1 binary complex. Mutation of C273S, C288S and C155S affecting transcriptional response (69, 72) may do so through effects on ternary complex stability and translocational oscillations. SFN may also affect ternary complex stability through binding and modification of cysteinyl thiol groups of Keap1 (60) but it remains unclear if QTN affects ternary complex stability directly (57). Stimulants such as SFN and QTN may rather act independently of this by up-regulating Fyn activation in the cytoplasm: for SFN through activation of GSK3β (34) and for QTN by suppressing inhibition of Fyn (37, 70). Both these interventions increase Fyn activity, Nrf2 oscillation frequency and refresh-rate. Nuclear export of Nrf2 is nevertheless important to activate the optimum transcriptional response—as evidenced by the effect of crm1 and Fyn inhibition on the Nrf2 transcriptional response.
Nuclear localization of Nrf2 in response to cell stress and nuclear export following phosphorylation by Fyn has been previously observed (21) but these together cannot account for oscillatory Nrf2 shuttling. The crucial new feature that sustains oscillations is linkage of Nrf2 nuclear import and export by the PGAM5-mediated feedback loop which prevents attainment of the equilibrium steady-state with different phosphorylation steps driving Nrf2 relocation.
For oscillatory systems such as the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling system, Stat/Smad/Hes1 and p53, synthesis and degradation of protein components play a key role in the mechanisms generating oscillations. Strong oscillations in the cellular levels of key proteins such as inhibiting factors are observed (30, 42, 71). We found no evidence for oscillation in Nrf2, Keap1 nor NQO1 protein. We conclude Nrf2 translocational oscillations mediate the ARE-linked transcriptional response without oscillation in the cellular content of Nrf2, Keap1 or target gene protein. The oscillatory model, however, predicts increased oscillatory bursts of transcription or suppression of transcription of ARE-linked genes over the period that stimulants remain at pharmacologically competent concentrations in the cell. Translocational oscillations of Nrf2 likely do not lead to detectable oscillations of mRNA and protein of ARE-regulates genes through damping effects of the downstream kinetics of transcription, translation, and mRNA and protein degradation acting as a low-pass filter.
Limitations of this study are lack of in vivo evidence of Nrf2 oscillation although in the primary cell culture studies ex vivo evidence of oscillation is provided. We have not provided a mathematical description of transition between basal and activated states but this may be achieved in future refinements where Nrf2 oscillation frequency as a function of time-dependent receptor-bound stimulant concentration is included. The basis by which subsets of ARE-linked genes are selectively regulated is not explored but Nrf2 translocational oscillation frequency provides a new factor to consider in the mechanism of target gene selection together with Mafs and other members of the Nrf2 interactome (26). The new insight into regulation of Nrf2 explains multiple features unexplained by the conventional mechanism: activation occurs without change in Nrf2 protein concentration, Nrf2 transactivational activity is inhibited by prolonged nuclear residence time (73) and potent activators of Nrf2 may function without directly binding to Nrf2 or Keap1 (54).
Recent research has shown that several key mammalian signaling systems such as ERK, NF-κB, N-FAT/Crz1, Stat/Smad/Hes1 and p53 have dynamic oscillatory or cyclic behavior (5a, 30, 42, 49a, 71) leading to the hypothesis that the temporal profile of the signal is either encoding important information or functioning to enhance the robustness of the response. We show that Nrf2 differs from these in three key ways that throw light on general signaling design principles: (i) for the systems mentioned cycling is induced by stimulation whereas we show that the Nrf2 system is an autonomous oscillator, indicating that oscillatory systems have a role in constitutive and stimulus response control; (ii) except for extracellular-signal-regulated kinase (ERK), regulatory degradation and synthesis of protein components is crucial whereas we show that oscillations of Nrf2 are driven by cyclic PTMs (ERK also does not employ a direct negative feedback loops); and (iii) the Nrf2 system is unique in the way it regulates transcription of target genes. Our mathematical model uses a feedback mechanism with similarities to that used in a minimal mechanism explaining activation of Saccharomyces cerevisiae transcriptional activators Msn2 and Msn4 (19) where a shuttling process receives delayed activation from an upstream signal to induce oscillations.
Materials and Methods
Materials
Antibodies used and other chemicals are given in Supplementary Data S1 section.
Cell culture conditions
Human microvascular HMEC-1 endothelial cells were cultured in MCDB-131 medium with supplements (10% fetal bovine serum, 5 mM l-glutamine, 10 ng/ml EGF, 1 ng/ml of hydrocortisone, 100 U/ml penicillin and 100 μg/ml streptomycin) at 37°C under 5% CO2/air atmosphere. The medium was replaced with fresh medium every 3 days and sub-cultured when near 90% of confluence (65). Cell treatments were: SFN (2 μM), QTN (8 μM), PP2 (5 μM) and LMB (2 ng/ml). For image analysis, the treatment of cells was performed 18 h post-transfection on the microscope stage. HMEC-1 cells (2–3×105) were seeded on glass bottomed 35 mm diameter dishes (MatTek Corporation, Ashland, MA) and cultured at 37°C and 5% CO2/air overnight—reaching 70%–80% confluence. The cells were transfected with 1.0 μg pcDNA3-EGFP-C4-Nrf2 using Lipofectamine 2000 (2 μl) in the ratio vector/Lipofectamine 2000, 1:2. Briefly, the plasmid and Lipofectamine 2000 were diluted with reduced serum medium (Opti-MEM®I; Invitrogen, Paisley, United Kingdom) separately and incubated at room temperature for 5 min, combined together and incubated at room temperature for 20 min. During this period, cells were washed twice with reduced serum medium (1 ml). After incubation, the cells washed once with reduced serum medium and cultured with MCDB-131 without phenol red medium overnight.
For fluorescence imaging, chemical treatments were added to the dish with the fresh MCDB-131 without phenol red and the images were recorded immediately on a time-lapse fluorescence inverted microscope (Cell^R live cell microscopy system; Olympus, Southend-on-Sea, United Kingdom) equipped with a 37°C, 5% CO2 environmental enclosure (Solent, Segensworth, United Kingdom) and a digital camera iXon2 BV888 EMCCD (Andor, Belfast, United Kingdom). Time-lapse videos were created with CellM & CellR imaging software for life science (Olympus). Cells were imaged through a 40× objective using Olympus HQGFP Filter cube 0.9NA. The exposure time was from 50 to 200 ms with autofocus and recording one frame per 2 min. Methods for assessment of cellular content of Nrf2 during transfection for expression of GFP-Nrf2 and subcellular co-localization of Nrf2 and GFP-Nrf2 and construction and use of the dVenus-NQO1-ARE reporter are described in Supplementary Data S1 section.
The dual luciferase reporter assay
HMEC-1 cells were co-transfected with vectors pGL3-NQO1 and pRL-TK (66). HMEC-1 cells (2×105 cells/well) in 24-well plates were incubated overnight and then transfected with 0.5 μg reporter vector and 10 ng pRL-TK plasmid using Lipofectamine 2000 according to manufacturer's instructions. The empty pGL3-basic vector was used as control. After 24 h, SFN or diluted DMSO vehicle was added and incubated for 0–24 h. The cells were then washed once with phosphate-buffered saline (PBS) and assayed for luciferase activity immediately or kept at −80°C until analysis. Cell culture lysis reagent (Promega, Southampton, United Kingdom), 100 μl, was added to each well and the plate placed on the shaker and gently shaking for 30 min. After shaking, the cell lysate was centrifuged (12,000 g, 5 min, 4°C) and 20 μl supernatant used in the analysis. Luciferase activity was determined using a Dual Luciferase Reporter Assay System (Promega) with luminescence measurements with an Orion Microplate Luminometer (Berthold Detection Systems GmbH, Bleichstrasse, Germany). The relative firefly luciferase activities were normalized to Renilla luciferase activities of the co-transfected pRL-TK vector. Studies also employed a stable transfectant NQO1-ARE luciferase reporter cell line (Supplementary Data).
Video image analysis
Time-course cell images were segmented using in-house software, CellTracker (Systems Biology Centre, University of Warwick, United Kingdom, http://go.warwick.ac.uk/celltracker) (10). The determined cellular and nuclear boundaries were checked by the investigator and corrected where necessary. CellTracker then calculated the cellular and nuclear intensities averaged over the respective area. The time-course intensity ratio between the nucleus and the cell were interpolated when there are unprocessed frames (encountered when cytoplasmic and nuclear intensities were similar), then de-noised by using a wavelet (Matlab functions wavedec and wrcoef are used with sym4 and level 2 options). Peaks and troughs were determined by thresholding time course profiles. A peak (trough) occurred when it was flanked by points on the both sides that were lower (higher) at least by the threshold value of 0.2. Each cycle was characterized by its temporal span and height (depth). For each peak (trough), span was defined as the temporal distance between the two flanking troughs (peaks) on each side. Peak height (trough depth) was measured from the point at the peak (trough) time on the baseline connecting the two flanking troughs (peaks) on each side. Within a cell population ca. 30% of cells were found to undergo oscillations for three to eight cycles.
Western blotting assessment of Fyn partitioning between the nucleus and cytoplasm
HMEC-1 cells (5×106) were seeded in 10 cm diameter Petri dishes, cultured overnight and SFN (2 μM) or vehicle (0.002% DMSO) added. The cells were incubated a further 0–6 h, then washed twice with ice cold PBS, dislodged with a scrapper in 1 ml PBS and sedimented by centrifugation. Nuclear and cytosolic protein fractions were prepared using CelLytic™ and NuCLEAR™ extraction kits (Sigma, Poole, United Kingdom) and analyzed by Western blotting (see Supplementary Data S1 section).
Nrf2 system and ARE-linked gene expression by digital mRNA profiling
HMEC-1 cells (5×105 cells/well) were seeded on six-well plates in MCDB-131 medium with supplements (see Cell culture conditions section) and cultured overnight at 37°C under 5% CO2/air. Cells were treated with 2 μM SFN or vehicle (0.002% DMSO) and cultured further for up to 72 h. At desired time point, cells were washed twice with ice-cold PBS and total RNA was extracted using RNeasy Mini Kit (Qiagen, Crawley, United Kingdom). Total RNA (600–800 ng) was analyzed for mRNA copy number of target genes by the NanoString nCounter Gene Expression method (12) (outsourced to Nanostring, Seattle, WA). A custom codeset of genes including three reference genes (β-actin, clathrin heavy chain and β-glucuronidase) was designed.
Statistical analysis
Data are mean±SD for parametric data and median (upper–lower quartile) for nonparametric data. Significance testing of mean and median changes was assessed by Student's t-test and Mann–Whitney U test, respectively. Spearman correlation analysis was performed of nonparametric data. Other tests are described where implemented.
Supplementary Material
Abbreviations Used
- aFyn
phosphothreonine-activated Fyn
- ARE
antioxidant response element
- CK2
casein kinase-2
- CLA
calyculin A
- Cul3
Cullin-3
- EGF
epidermal growth factor
- ERK
extracellular-signal-regulated kinase
- GFP
green fluorescent protein
- GSK3β
glycogen synthase kinase 3β
- Hes1
hairy and enhancer of split-1
- HMEC-1
human microvascular endothelial cell-1 cell line
- iNrf2
nuclear Nrf2 inactivated by acetylation
- Keap1
Kelch-like erythroid cell-derived protein with CNC homology-associated protein 1
- LMB
leptomycin B
- N-FAT
nuclear factor of activated T-cells
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NQO1
quinone reductase
- Nrf2
nuclear factor erythroid 2-related factor 2
- p62
sequestosome-1
- PBS
phosphate-buffered saline
- PGAM5
phosphoglycerate mutase family member 5 protein threonine phosphatase
- pNrf2
nuclear Nrf2 phosphorylated by Fyn on Tyr-568
- PP2
4-amino-5-(4-chlorophenyl)-7-(dimethylethyl) pyrazolo[3,4-d]pyrimidine
- PPase
protein phosphatase
- PTM
post-translational modification
- PTPase
protein tyrosine phosphatase
- QTN
quercetin
- RLU
relative light unit
- SD
standard deviation
- SFN
sulforaphane
- STAT
signal transducer and activator of transcription
- Rbx-1
ring-box 1
- TDZD-8
4-benzyl-2-methyl-1,2,4-thiadiazolidine-3,5-dione
- TAS
Trichostatin A
- Ub
ubiquitin
Acknowledgments
The authors thank the Biosciences and Biotechnology Research Council (BBSRC), United Kingdom for research funding—Diet and Health Research Industry Club (DRINC) project grant No. 07/84, and the European Union Framework Programme-7 for support for the BIOCLAIMS research programme, grant agreement No. 244995 and grant K/ZDS/002442. D.A.R. held an EPSRC Senior Fellowship (GR/S29256/01). We thank Drs. Andrew Blanks (Warwick Medical School, University of Warwick) and Chengjin Du (Systems Biology Centre, University of Warwick) for expert assistance with time-lapse video microscopy and semi-automated image analysis.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Apopa PL, He XQ, and Ma Q. Phosphorylation of nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J Biochem Mol Toxicol 22: 63–76, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Baird L. and Dinkova-Kostova A. The cytoprotective role of the Keap1/Nrf2 pathway. Arch Toxicol 85: 241–272, 2011 [DOI] [PubMed] [Google Scholar]
- 3.Bertl E, Bartsch H, and Gerhauser C. Inhibition of angiogenesis and endothelial cell functions are novel sulforaphane-mediated mechanisms in chemoprevention. Mol Cancer Ther 5: 575–585, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Brockwell PJ. and Davis RA. Introduction to Time Series and Forecasting. New York: Springer, 2002, p. 437 [Google Scholar]
- 5.Burton NC, Kensler TW, and Guilarte TR. In vivo modulation of the Parkinsonian phenotype by Nrf2. Neurotoxicology 27: 1094–1100, 2006 [DOI] [PubMed] [Google Scholar]
- 5a.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 455: 485–490, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke JD, Hsu A, Riedl K, Bella D, Schwartz SJ, Stevens JF, and Ho E. Bioavailability and inter-conversion of sulforaphane and erucin in human subjects consuming broccoli sprouts or broccoli supplement in a cross-over study design. Pharmacol Res 64: 456–463, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins AR, Lyon CJ, Xia XF, Liu JZ, Tangirala RK, Yin F, Boyadjian R, Bikineyeva A, Pratico D, Harrison DG, and Hsueh WA. Age-accelerated atherosclerosis correlates with failure to upregulate antioxidant genes. Circ Res 104: e42–e54, 2009 [DOI] [PubMed] [Google Scholar]
- 8.Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, Layfield R, Foster BJ, Goldring CE, and Park BK. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1-Nrf2 cell defense pathway. J Biol Chem 285: 16782–16788, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullinan SB, Gordan JD, Jin J, Harper JW, and Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol 24: 8477–8486, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du CJ, Marcello M, Spiller DG, White MRH, and Bretschneider T. Interactive segmentation of clustered cells via geodesic commute distance and constrained density weighted Nystrom method. Cytometry A 77A: 1137–1147, 2010 [DOI] [PubMed] [Google Scholar]
- 11.Ebato C, Uchida T, Arakawa M, Komatsu M, Ueno T, Komiya K, Azuma K, Hirose T, Tanaka K, Kominami E, Kawamori R, Fujitani Y, and Watada H. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab 8: 325–332, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Fortina P. and Surrey S. Digital mRNA profiling. Nat Biotechnol 26: 293–294, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Gao L, Wang JK, Sekhar KR, Yin HY, Yared NF, Schneider SN, Sasi S, Dalton TP, Anderson ME, Chan JY, Morrow JD, and Freeman ML. Novel n-3 fatty acid oxidation products activate Nrf2 by destabilizing the association between Keap1 and Cullin3. J Biol Chem 282: 2529–2537, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Gillespie DT. Exact stochastic simulation of coupled chemical-reactions. J Phys Chem 81: 2340–2361, 1977 [Google Scholar]
- 15.Hayashi R, Himori N, Taguchi K, Ishikawa Y, Uesugi K, Ito M, Duncan T, Tsujikawa M, Nakazawa T, Yamamoto M, and Nishida K. The role of the Nrf2-mediated defense system in corneal epithelial wound healing. Free Radic Biol Med 61: 333–342, 2013 [DOI] [PubMed] [Google Scholar]
- 16.Hubbard GP, Wolffram S, de Vos R, Bovy A, Gibbins JM, and Lovegrove JA. Ingestion of onion soup high in quercetin inhibits platelet aggregation and essential components of the collagen-stimulated platelet activation pathway in man: a pilot study. Br J Nutr 96: 482–488, 2006 [PubMed] [Google Scholar]
- 17.Ishii T, Itoh K, Ruiz E, Leake DS, Unoki H, Yamamoto M, and Mann GE. Role of Nrf2 in the regulation of CD36 and stress protein expression in murine macrophages: activation by oxidatively modified LDL and 4-hydroxynonenal. Circ Res 94: 609–616, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, and Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 8: 379–391, 2003 [DOI] [PubMed] [Google Scholar]
- 19.Jacquet M, Renault G, Lallet S, De Mey J, and Goldbeter A. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J Cell Biol 161: 497–505, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain AK, Bloom DA, and Jaiswal AK. Nuclear import and export signals in control of Nrf2. J Biol Chem 280: 29158–29168, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Jain AK. and Jaiswal AK. Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. J Biol Chem 281: 12132–12142, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Jain AK. and Jaiswal AK. GSK-3beta acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. J Biol Chem 282: 16502–16510, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Jeong WS, Keum YS, Chen C, Jain MR, Shen GX, Kim JH, Li WG, and Kong ANT. Differential expression and stability of endogenous nuclear factor E2-related factor 2 (Nrf2) by natural chemopreventive compounds in HepG2 human hepatoma cells. J Biochem Mol Biol 38: 167–176, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Kapeta S, Chondrogianni N, and Gonos ES. Nuclear erythroid factor 2-mediated proteasome activation delays senescence in human fibroblasts. J Biol Chem 285: 8171–8184, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaspar JW. and Jaiswal AK. Tyrosine phosphorylation controls nuclear export of Fyn, allowing Nrf2 activation of cytoprotective gene expression. FASEB J 25: 1076–1087, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katsuoka F, Motohashi H, Ishii T, Aburatani H, Engel JD, and Yamamoto M. Genetic evidence that small Maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol Cell Biol 25: 8044–8051, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, and Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol 26: 221–229, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, Kim M, Nishito Y, Iemura Si, Natsume T, Ueno T, Kominami E, Motohashi H, Tanaka K, and Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213–223, 2010 [DOI] [PubMed] [Google Scholar]
- 29.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, and Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci U S A 96: 9112–9117, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, and Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet 36: 147–150, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Larance M, Ahmad Y, Kirkwood KJ, Ly T, and Lamond AI. Global subcellular characterization of protein degradation using quantitative proteomics. Mol Cell Proteomics 12: 638–650, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau A, Tian W, Whitman SA, and Zhang DND. The predicted molecular weight of Nrf2: it is what it is not. Antioxid Redox Signal 18: 91–93, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee OH, Jain AK, Papusha V, and Jaiswal AK. An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J Biol Chem 282: 36412–36420, 2007 [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Zhang T, Korkaya H, Liu S, Lee HF, Newman B, Yu Y, Clouthier SG, Schwartz SJ, Wicha MS, and Sun D. Sulforaphane, a dietary component of broccoli/broccoli sprouts, inhibits breast cancer stem cells. Clin Cancer Res 16: 2580–2590, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo SC. and Hannink M. PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated keap1-dependent ubiquitin ligase complex. J Biol Chem 281: 37893–37903, 2006 [DOI] [PubMed] [Google Scholar]
- 36.Lo SC. and Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp Cell Res 314: 1789–1803, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lolli G, Cozza G, Mazzorana M, Tibaldi E, Cesaro L, Donella-Deana A, Meggio F, Venerando A, Franchin C, Sarno S, Battistutta R, and Pinna LA. Inhibition of protein kinase CK2 by flavonoids and tyrphostins. A structural insight. Biochemistry 51: 6097–6107, 2012 [DOI] [PubMed] [Google Scholar]
- 38.Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, and Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res 38: 5718–5734, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuo M, Sasaki N, Saga K, and Kaneko T. Cytotoxicity of flavonoids toward cultured normal human cells. Biol Pharmaceut Bull 28: 253–259, 2005 [DOI] [PubMed] [Google Scholar]
- 40.Mercado N, Thimmulappa R, Thomas CMR, Fenwick PS, Chana KK, Donnelly LE, Biswal S, Ito K, and Barnes PJ. Decreased histone deacetylase 2 impairs Nrf2 activation by oxidative stress. Biochem Biophys Res Commun 406: 292–298, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, and Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22: 66–79, 2012 [DOI] [PubMed] [Google Scholar]
- 42.Nelson DE, Ihekwaba AEC, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, and White MRH. Oscillations in NF-kB signaling control the dynamics of gene expression. Science 306: 704–708, 2004 [DOI] [PubMed] [Google Scholar]
- 43.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, and Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem Biophys Res Commun 339: 79–88, 2006 [DOI] [PubMed] [Google Scholar]
- 44.Pearson KJ, Lewis KN, Price NL, Chang JW, Perez E, Cascajo MV, Tamashiro KL, Poosala S, Csiszar A, Ungvari Z, Kensler TW, Yamamoto M, Egan JM, Longo DL, Ingram DK, Navas P, and de Cabo R. Nrf2 mediates cancer protection but not prolongevity induced by caloric restriction. Proc Natl Acad Sci U S A 105: 2325–2330, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pi J, Bai Y, Reece JM, Williams J, Liu D, Freeman ML, Fahl WE, Shugar D, Liu J, Qu W, Collins S, and Waalkes MP. Molecular mechanism of human Nrf2 activation and degradation: role of sequential phosphorylation by protein kinase CK2. Free Radic Biol Med 42: 1797–1806, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rauh D, Fischer F, Gertz M, Lakshminarasimhan M, Bergbrede T, Aladini F, Kambach C, Becker CFW, Zerweck J, Schutkowski M, and Steegborn C. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat Commun 4: 3327, 2013 [DOI] [PubMed] [Google Scholar]
- 47.Reddy NM, Kleeberger SR, Yamamoto M, Kensler TW, Scollick C, Biswal S, and Reddy SP. Genetic dissection of the Nrf2-dependent redox signaling-regulated transcriptional programs of cell proliferation and cytoprotection. Physiol Genom 32: 74–81, 2007 [DOI] [PubMed] [Google Scholar]
- 48.Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, and Zhang DD. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci U S A 108: 1433–1438, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rose JL, Huang H, Wray SF, and Hoyt DG. Integrin engagement increases histone H3 acetylation and reduces histone H1 association with DNA in murine lung endothelial cells. Mol Pharmacol 68: 439–446, 2005 [DOI] [PubMed] [Google Scholar]
- 49a.Shankaran H, Ippolito DL, Chrisler WB, Resat H, Bollinger N, Opresko LK, Wiley HS. Rapid and sustained nuclear-cytoplasmic ERK oscillations induced by epidermal growth factor. Mol Syst Biol 5: 332, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smirnova Natalya A, Haskew-Layton Renee E, Basso M, Hushpulian Dmitry M, Payappilly Jimmy B, Speer Rachel E, Ahn Y-H, Rakhman I, Cole Philip A, Pinto John T, Ratan Rajiv R, and Gazaryan Irina G. Development of Neh2-luciferase reporter and its application for high throughput screening and real-time monitoring of Nrf2 activators. Chem Biol 18: 752–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Z, Zhang S, Chan JY, and Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol 27: 6334–6349, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swingle M, Ni L, and Honkanen R. Small-molecule inhibitors of Ser/Thr protein phosphatases. In: Protein Phosphatase Protocols, edited by Moorhead G. New York: Springer, 2007. pp. 23–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taguchi K, Motohashi H, and Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells 16: 123–140, 2011 [DOI] [PubMed] [Google Scholar]
- 54.Takaya K, Suzuki T, Motohashi H, Onodera K, Satomi S, Kensler TW, and Yamamoto M. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radic Biol Med 53: 817–827, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takeda K, Komuro Y, Hayakawa T, Oguchi H, Ishida Y, Murakami S, Noguchi T, Kinoshita H, Sekine Y, Iemura Si, Natsume T, and Ichijo H. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc Natl Acad Sci U S A 106: 12301–12305, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan W-F, Lin L-P, Li M-H, Zhang Y-X, Tong Y-G, Xiao D, and Ding J. Quercetin, a dietary-derived flavonoid, possesses antiangiogenic potential. Eur J Pharmacol 459: 255–262, 2003 [DOI] [PubMed] [Google Scholar]
- 57.Tanigawa S, Fujii M, and Hou DX. Action of Nrf2 and Keap1 in ARE-mediated NQO1 expression by quercetin. Free Radic Biol Med 42: 1690–1703, 2007 [DOI] [PubMed] [Google Scholar]
- 58.Theodore M, Kawai Y, Yang JQ, Kleshchenko Y, Reddy SP, Villalta F, and Arinze IJ. Multiple nuclear localization signals function in the nuclear import of the transcription factor nrf2. J Biol Chem 283: 8984–8994, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, and Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide array. Cancer Res 62: 5196–5203, 2002 [PubMed] [Google Scholar]
- 60.Wakabayashi N, Dinkova-Kostova AT, Holtzclaw WD, Kang M, Kobayashi A, Yamamoto M, Kensler TW, and Talalay P. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci U S A 101: 2040–2045, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W, Winlove CP, and Michel CC. Oxygen partial pressure in outer layers of skin of human finger nail folds. J Physiol 549: 855–863, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X, Tomso DJ, Chorley BN, Cho HY, Cheung VG, Kleeberger SR, and Bell DA. Identification of polymorphic antioxidant response elements in the human genome. Hum Mol Genet 16: 1188–1200, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Z, Jiang H, Chen S, Du F, and Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148: 228–243, 2012 [DOI] [PubMed] [Google Scholar]
- 64.Xu K. and Thornalley PJ. Involvement of GSH metabolism in the cytotoxicity of the phenethyl isothiocyanate and its cysteine conjugate to human leukaemia cells in vitro. Biochem Pharmacol 61: 165–177, 2001 [DOI] [PubMed] [Google Scholar]
- 65.Xue M, Qian Q, Adaikalakoteswari A, Rabbani N, Babaei-Jadidi R, and Thornalley PJ. Activation of NF-E2-related factor-2 reverses biochemical dysfunction of endothelial cells induced by hyperglycemia linked to vascular disease. Diabetes 57: 2809–2817, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xue M, Rabbani N, Momiji H, Imbasi P, Anwar MM, Kitteringham NR, Park BK, Souma T, Moriguchi T, Yamamoto M, and Thornalley PJ. Transcriptional control of glyoxalase 1 by Nrf2 provides a stress responsive defence against dicarbonyl glycation. Biochem J 443: 213–222, 2012 [DOI] [PubMed] [Google Scholar]
- 67.Xue P, Hou Y, Chen Y, Yang B, Fu J, Zheng H, Yarborough K, Woods CG, Liu D, Yamamoto M, Zhang Q, Andersen ME, and Pi J. Adipose deficiency of Nrf2 in ob/ob mice results in severe metabolic syndrome. Diabetes 62: 845–854, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yagishita Y, Fukutomi T, Sugawara A, Kawamura H, Takahashi T, Pi JB, Uruno A, and Yamamoto M. Nrf2 protects pancreatic beta-cells from oxidative and nitrosative stress in diabetic model mice. Diabetes 63: 605–618, 2014 [DOI] [PubMed] [Google Scholar]
- 69.Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, and Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol Cell Biol 28: 2758–2770, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yokoyama T, Kamata Y, and Ohtsuki K. Casein kinase 2 (CK2)-mediated reduction of the activities of Src family tyrosine kinases in vitro. Biol Pharmaceut Bull 27: 1895–1899, 2004 [DOI] [PubMed] [Google Scholar]
- 71.Yoshiura S, Ohtsuka T, Takenaka Y, Nagahara H, Yoshikawa K, and Kageyama R. Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc Natl Acad Sci U S A 104: 11292–11297, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang DD. and Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol 23: 8137–8151, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou W, Lo SC, Liu JH, Hannink M, and Lubahn DB. ERR-beta: a potent inhibitor of Nrf2 transcriptional activity. Mol Cell Endocrinol 278: 52–62, 2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.