Abstract
CD4+ T cells play a central role in controlling the adaptive immune response by secreting cytokines to activate target cells. Naïve CD4+ T cells differentiate into at least four subsets, Th1, Th2, Th17, and inducible regulatory T cells, each with unique functions for pathogen elimination. The differentiation of these subsets is induced in response to cytokine stimulation, which is translated into Stat activation, followed by induction of master regulator transcription factors. In addition to these factors, multiple other transcription factors, both subset specific and shared, are also involved in promoting subset differentiation. This review will focus on the network of transcription factors that control CD4+ T cell differentiation.
1 Introduction
An effective immune response is vital in the protection against invading foreign pathogens. CD4+ T cells play a pivotal role in host defense by secreting cytokines to drive appropriate immune responses. Classified by cytokine secretion profile, CD4+ T cells are subdivided into four major subsets. Th1 cells secret IFNγ to clear intracellular pathogens while Th2 cells secret IL-4, IL-5, and IL-13 to clear helminthes and extracellular pathogens (Zhou et al. 2009). Th17 cells, originally identified as the causative cell type in the experimental autoimmune encephalitis (EAE, a mouse model of multiple sclerosis), are characterized by the secretion of IL-17 and are involved in the clearance of extracellular bacteria and fungi (Korn et al. 2009). Regulatory T cells (Tregs), including thymus derived Tregs (tTregs) and peripherally induced Tregs (pTregs), secrete anti-inflammatory cytokines including TGFβ and IL-10 and act to suppress immune responses to prevent damage to the host (Josefowicz et al. 2012). At steady state, Tregs are indispensable for maintaining self-tolerance thus preventing autoimmunity through multiple mechanisms. Besides Th1, Th2, Th17, and iTreg cells, some CD4+ T cells reside within the B cell follicle and are thus named T follicular cells (Tfh); these cells express the chemokine receptor CXCR5 and produce large amounts of IL-21 (Crotty 2011). Tfh cells function by providing help to B cells. However, the relationship between Tfh cells and classical Th1, Th2, and Th17 effector cells is not certain since some Tfh cells are capable of producing either IFNγ or IL-4 (Lee et al. 2012a; Yusuf et al. 2010). In addition, regulatory T cells expressing the key transcription factor Foxp3 have been also found in B cell follicles (Chung et al. 2011; Linterman et al. 2011) and Th17 cells have been shown to convert to Tfh cells in Peyer’s patches and provide help to B cells, thus increasing IgA production (Hirota et al. 2013). Thus, it remains unclear whether Tfh cells represent a separate subset or whether they differentiate from other CD4+ T cell subsets. Furthermore, it has been shown that IL-21-expressing Tfh cells may give rise to memory cells that can further differentiate into conventional effector cells during recall responses (Luthje et al. 2012). Finally, Th9 and Th22 cells have also been characterized as separate subsets recently, based on the expression of IL-9 and IL-22, respectively (Jabeen and Kaplan 2012; Duhen et al. 2009), but their relationship to Th2 and Th17 cells, respectively, requires further investigation. Together, these subsets orchestrate the clearance of pathogens while preventing damage to the host.
The induction and maintenance of each CD4+ subset is controlled by the cytokine environment, which activates signal transducers and activators of transcription (Stat) pathways to induce the expression of the master regulator transcription factors. The Stat and master regulator controlling each subset have been defined as follows, Stat4/Tbet (Th1), Stat6/Gata3 (Th2), Stat3/RORγt (Th17), Stat5/FoxP3 (Treg), and Stat3/Bcl6 (Tfh), and have been widely studied (Zhu et al. 2010). Although these factors are essential for the differentiation of a particular subset, the master regulators do not act alone but are instead a component of a larger transcriptional network. Multiple transcription factors can interact directly or indirectly to control gene expression programs. Direct interaction of transcription factors can increase transcriptional activity by increasing recruitment of additional transcription factors or transcriptional machinery to target genes. Conversely, direct interaction may inhibit gene expression by blocking the binding of transcription factors to target genes. Many transcription factors also recruit chromatin and histone modifying enzymes to increase or decrease accessibility of binding sites for other transcription factors. Finally, multiple binding sites within a gene may allow for cooperation between multiple transcription factors or, conversely, allow competitive inhibition between factors, where the binding of one transcription factor may block the binding of another. In this case, the balance of transcription factors will determine the pattern of gene expression. These types of interactions allow cells to alter gene expression in response to changes in the balance of transcription factors, allowing a level of plasticity within CD4+ T cell subsets. In order to fully understand the regulation of CD4+ T cell differentiation and the plasticity potential within each, a thorough understanding of the entire network of transcription factors is required.
2 Signals for Differentiation
The initial steps in CD4+ T cell differentiation, after activation through T cell receptors, relies upon binding of cytokines to their cognate receptors, resulting in the activation of receptor associated Janus kinases (Jaks), which phosphorylate the intracellular domain of cytokine receptors to provide specific docking sites for Stat binding (Leonard and O’Shea 1998; O’Shea and Plenge 2012). Upon recruitment to the receptor, activated Jaks phosphorylate and activate Stats, which in turn induce the expression of genes involved in the initial differentiation of each subset, including the induction of the master regulators. The master regulators activate expression of the polarizing cytokine, which induces a positive feedback loop to strengthen the differentiation toward a given subset.
2.1 Th1: Stat4, Stat1
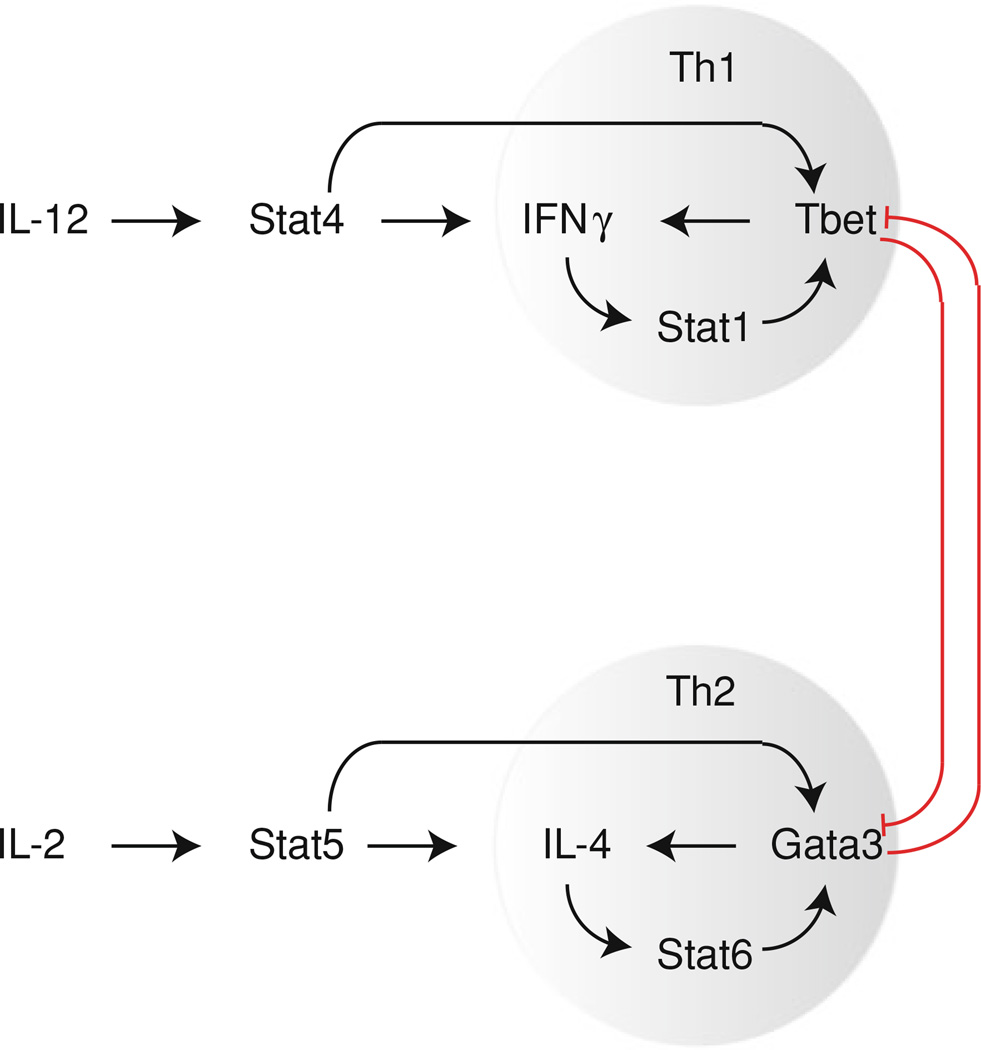
Th1 differentiation is generally induced by IL-12 secreted from antigen presenting cells (Fig. 1). IL-12 signals through Stat4 and the importance of this pathway in Th1 differentiation is demonstrated by Stat4 deficient mice, which show impaired Th1 responses with decreased IFNγ production (Kaplan et al. 1996b; Cai et al. 2000; Thierfelder et al. 1996). Stat4 activates the expression of many Th1-specific genes, including Ifng (Lund et al. 2004; Hoey et al. 2003), Il12rb2, and Tbx21 encoding Tbet (Hoey et al. 2003). In addition to directly inducing Tbet expression, Stat4 also indirectly upregulates Tbet by inducing IFNγ expression, which signals through Stat1 to activate Tbet expression (Lighvani et al. 2001; Afkarian et al. 2002). In turn, Tbet binds to and activates IFNγ expression (Matsuda et al. 2007; Lovett-Racke et al. 2004; Jenner et al. 2009) and the loss of Tbet expression correlates with decreased IFNγ expression (Szabo et al. 2002; Long et al. 2006). As such, during Th1 differentiation, a positive feedback loop is generated between IFNγ and Tbet, reinforcing the Th1 gene expression program. Indeed, while Stat4 deficient mice have defects in Th1 differentiation, treatment with IFNγ resulted in increased Tbet expression (Lighvani et al. 2001) and treatment of wild type cells with either IL-12 or IFNγ-induced Tbet expression (Zhu et al. 2012), indicating Th1 differentiation can occur in the absence of IL-12. However, a study of the contributions of IL-12 and IFNγ to Th1 differentiation demonstrated that while IFNγ stimulation induced Th1 differentiation, recall responses of Th1 cells required IL-12 stimulation, indicating the essential role of IL-12/Stat4 for maintaining Th1 cells (Schulz et al. 2009). Both Stat4 and Tbet are required for maximal IFNγ production and while loss of either one resulted in decreased IFNγ production, loss of both Tbet and Stat4 resulted in a complete block of IFNγ production, indicating cooperation between these factors (Zhu et al. 2012). However, the targets of Stat4 and Tbet are not completely overlapping, pointing to a wider role for Stat4-induced gene expression programs in Th1 differentiation, beyond the induction of IFNγ and Tbet expression (Thieu et al. 2008). Surprisingly, Stat4−/− Stat6−/− double deficient mice cultured under nonpolarizing conditions were capable of secreting IFNγ, indicating that in the absence of Th2 differentiation, cells can activate expression of Th1 specific genes without signaling through Stat4 (Kaplan et al. 1998). However, whether such IFNγ production is Tbet dependent has not been determined.
Fig. 1.
Positive feedback regulation of CD4+ T cell differentiation. Cytokine induced Stat activation initiates CD4+ T cell differentiation by activating expression of master regulators. A positive feedback loop is generated by increased expression of activating cytokines in response to master regulation expression, thus reinforcing expression of the master regulator and the subset specific gene expression program
2.2 Th2: Stat6, Stat5
Differentiation of Th2 cells requires stimulation with IL-4 (Le Gros et al. 1990; Swain et al. 1990; Hsieh et al. 1992; Seder et al. 1992), particularly in vitro, and in the absence of IL-4, cells stimulated through the T cell receptor are driven predominately toward Th1 differentiation (Seki et al. 2004). IL-4 signals through Stat6 and mice deficient in Stat6 show limited Th2 cell differentiation (Kaplan et al. 1996a) and Th2 cytokine production, both in response to Th2 polarizing conditions in vitro (Shimoda et al. 1996) and in response to Th2-inducing pathogens (Takeda et al. 1996). Furthermore, expression of a constitutively active form of Stat6 drives cells toward Th2 differentiation, even under conditions that strongly favor Th1 polarization (Kurata et al. 1999; Zhu et al. 2001). Th1 cells expressing constitutively active Stat6 have altered chromatin structure at both Gata3 regulatory regions (Onodera et al. 2010) and the Th2 cytokine locus (Lee and Rao 2004) resembling that found in Th2 cells, indicating that Stat6 can induce chromatin modifications to increase expression of Th2 specific genes. Similar to Th1 cells, Th2 commitment is also driven by a positive feedback loop, in which IL-4 induces further IL-4 production (Le Gros et al. 1990; Swain et al. 1990; Hsieh et al. 1992; Seder et al. 1992). IL-4 signaling through Stat6 induces Gata3 expression (Kurata et al. 1999; Zhu et al. 2001) and Gata3 in turn, drives the expression of IL-4. Gata3 deficient cells fail to produce IL-4 at early time points after T cell stimulation and show no Th2 differentiation (Yamane et al. 2005). Although Stat6 has a clearly demonstrated role in Th2 differentiation, further study of these mice revealed that while Th2 recall responses were severely impaired (Finkelman et al. 2000), initial Th2 differentiation did occur normally (Finkelman et al. 2000; Jankovic et al. 2000). Indeed, IL-2 in combination with low peptide stimulation was shown to drive early Th2 differentiation in the absence of IL-4 signals (Yamane et al. 2005). IL-2 signals through Stat5 and Stat5 deficient mice were found to have fewer Th2 cells, demonstrating the importance of this pathway in Th2 differentiation (Kagami et al. 2001). Stat5 has been shown to bind to the IL-4 gene (Hural et al. 2000) and to induce IL-4Rα expression (Liao et al. 2008). Furthermore, blocking IL-2 during Th2 polarization blocked Th2 differentiation (Cote-Sierra et al. 2004) while constitutively active Stat5 increased IL-4 production in the absence of IL-2 (Zhu et al. 2003). However, IL-2 and Stat5 do not upregulate Gata3 expression during initial differentiation (Yamane et al. 2005; Cote-Sierra et al. 2004; Zhu et al. 2003). Instead, IL-2 induces initial IL-4 production by naïve T cells, which in turn stimulates Gata3 expression via Stat6, thus activating the positive feedback between Gata3 and IL-4 to strengthen the commitment to the Th2 subset (Fig. 1). In fully committed Th2 cells, however, Stat5 activation is important for maintaining Gata3 expression (Guo et al. 2009).
2.3 Th17: Stat3
Th17 differentiation is induced by stimulation with IL-6 and TGFβ (Yang et al. 2007) and mice with deletions of either cytokine or associated receptors have a block in Th17 differentiation (Nishihara et al. 2007; Li et al. 2007b; Ivanov et al. 2006; Korn et al. 2007), indicating the requirement for both cytokines in Th17 differentiation. Since TGFβ also induces Treg differentiation, IL-6 is required to divert cells from becoming T regulatory cells. IL-6 signals through Stat3 and constitutively active Stat3 is capable of inducing Th17 differentiation and increasing RORγt expression, although this requires costimulation with TGFβ (Zhou et al. 2007). In contrast, Stat3 deficient mice resemble IL-6 deficient mice, showing an inability to generate Th17 cells (Yang et al. 2007; Mathur et al. 2007; Durant et al. 2010) and increased resistance to EAE induction (Harris et al. 2007). Furthermore, human patients with mutations in Stat3 also show diminished Th17 populations and RORγt production (Milner et al. 2008; Ma et al. 2008). The loss of IL-6 (Korn et al. 2007), IL-21 (Nurieva et al. 2007) or Stat3 (Yang et al. 2007) results not only in decreased Th17 populations but also increased Treg populations, demonstrating the reciprocal regulation of Th17 and Treg differentiation. Stat3 binds to and activates multiple Th17 genes, including cytokines IL-17 and IL-21 as well as transcription factors RORγt, Batf, IRF4, Ahr, and Maf (Durant et al. 2010). Although IL-17 is the hallmark cytokine produced by Th17 cells, it is not capable of driving Th17 differentiation (Korn et al. 2009). Instead, IL-21 may play this role as it is capable of replacing IL-6 in the differentiation of Th17 cells and blocking IL-21 under Th17 conditions reduces Th17 differentiation (Wei et al. 2007; Korn et al. 2007). Furthermore, while IL-6 induces initial IL-21 expression, subsequent IL-21 expression is under autoregulation as IL-6 deficient cells were induced to express IL-21 by treatment with IL-21 (Zhou et al. 2007). The importance of IL-21 in Th17 differentiation is demonstrated by IL-21R deficient mice, which had decreased Th17 differentiation (Zhou et al. 2007). Interestingly, although initial induction of IL-21 expression by IL-6 required only Stat3, the maintenance of Th17 differentiation by IL-21 required both Stat3 and RORγt (Nurieva et al. 2007). Furthermore, the induction of Th17 differentiation by constitutively active Stat3 was diminished in the absence of RORγt (Zhou et al. 2007), indicating cooperation between Stat3 and RORγt in Th17 differentiation, either at overlapping targets or by activation of unique targets required for Th17 differentiation. Together, these studies indicate that, similar to Th2 cells, Th17 differentiation also proceeds through an initiation stage of Th17 differentiation followed by the maintenance of the Th17 expression program. Initial IL-6 stimulation activates Stat3, resulting in the increased expression of Th17 cytokines, including IL-21 as well as Th17 transcription factors. In turn, IL-21, signaling through Stat3, is required for the maintenance of Th17 differentiation.
IL-23 is also involved in Th17 differentiation as treatment of cells with IL-23 induces IL-17 production (Aggarwal et al. 2003) while IL-23 receptor deficient mice produce less IL-17 and show decreased susceptibility to EAE induction (McGeachy et al. 2009). Although naïve CD4+ T cells do not express IL-23 receptor, IL-6 induces IL-23R expression, which is further augmented by treatment with both IL-6 and IL-23 (Ghoreschi et al. 2010). Both of these cytokines signal through Stat3 and stimulation with IL-6 and IL-23 resulted in increased binding of Stat3 to the Il23r locus. IL-23 treatment in combination with IL-6 and IL-1β can also drive Th17 differentiation in the absence of TGFβ stimulation (Ghoreschi et al. 2010). Th17 cells induced by IL-23 have a unique phenotype compared to TGFβ-induced Th17 cells as IL-23-induced Th17 cells express both RORγt and Tbet and are more pathogenic when transferred into Rag2−/− mice. Th17 cells generated with TGFβ produce IL-10, which may be the reason for low pathogenicity of such cells (Lee et al. 2012b). Furthermore, addition of TGFβ to IL-23 driven Th17 differentiation resulted in decreased expression of the IL-23 receptor. Thus TGFβ may also limit Th17 pathogenicity by downregulating IL-23 responsiveness. Interestingly, it has been recently reported that Th17 cells produce TGFβ3, which is IL-23 dependent, and TGFβ3 induces pathogenic Th17 cells (Lee et al. 2012b).
2.4 Treg: Stat5
Differentiation of iTregs is induced by stimulation with IL-2 and TGFβ (Josefowicz et al. 2012). Both IL-2Rα deficient mice and Stat5 deficient mice had decreased Treg populations while constitutively active Stat5 rescued Treg populations in IL-2Rα deficient mice (Burchill et al. 2007). Stat5 binds directly to the FoxP3 promoter to induce the Treg differentiation program (Burchill et al. 2007; Yao et al. 2007). Stat5 requires histone deacetylase activity to induce Treg differentiation (Burchill et al. 2008), indicating that in addition to directly activating transcription, Stat5 also acts to remodel chromatin structure to change accessibility of additional factors. IL-2 signaling through Stat5 also blocks Th17 differentiation and the loss of IL-2 signaling resulted in decreased Treg populations while Th17 populations were increased (Laurence et al. 2007). Stat5 binds directly to the Il17 promoter in the same region where Stat3 binds, resulting not only in decreased Stat3 binding but also in loss of permissive histone modifications within the Il17 gene (Yang et al. 2011). Interestingly, Stat3 can also bind to the Foxp3 promoter to inhibit expression (Xu et al. 2010), indicating that the balance between Stat3 and Stat5 levels not only induces the differentiation program of one subset but also actively inhibits the other.
2.5 Tfh: Stat3
Tfh cells are defined by their localization within germinal centers and the surface expression of the chemokine receptor CXCR5 (Crotty 2011). IL-6 signaling through Stat3 has been reported to induce Tfh differentiation (Eddahri et al. 2009; Nurieva et al. 2008). Although IL-6 is involved in both Th17 and Tfh differentiation, Th17 requires both IL-6 and TGFβ while Tfh requires only IL-6 (Nurieva et al. 2008). IL-6-induced Tfh differentiation results in upregulation of IL-21 expression (Dienz et al. 2009; Nurieva et al. 2008), which plays an important role in B cell maturation within germinal centers (Zotos et al. 2010). Indeed, the importance of IL-21 production by Tfh cells has been demonstrated by a decrease in antibody production in mice with T cell-specific deletions of IL-21 (Dienz et al. 2009; Zotos et al. 2010). However, the effect of IL-21 deficiency on the Tfh population size is unclear as some groups have shown decreased Tfh populations in IL-21 and IL-21 receptor deficient mice (Vogelzang et al. 2008; Nurieva et al. 2008) while others have shown normal induction of Tfh differentiation (Zotos et al. 2010), with an accelerated contraction of Tfh cells after stimulation (Linterman et al. 2010). As such, it is possible that similar to Th17 differentiation, IL-6 is required for the initiation of Tfh differentiation while IL-21 is involved in the maintenance of this subset. However, there may be other factors involved in Tfh differentiation, as both IL-6 deficient mice and Stat3 deficient mice showed decreased Tfh populations early in differentiation that equalized at later time points (Choi et al. 2013). Deletion of both Stat3 and Stat1 prevented Tfh differentiation at all time points, indicating the importance of multiple signaling pathways in Tfh differentiation. In addition to upregulation of IL-21 in early Tfh differentiation, IL-6 treatment is also required for downregulation of IL-2Rα expression (Choi et al. 2013). Treatment with either IL-2 or constitutively active Stat5 blocked Tfh differentiation (Ballesteros-Tato et al. 2012; Nurieva et al. 2012) while deletion of either Stat5 or IL-2Rα increased Tfh differentiation (Johnston et al. 2012). Stat5 acts to block Tfh differentiation by binding directly to the Bcl6 promoter to block Stat3 binding and activation of Bcl6 expression (Oestreich et al. 2012).
3 Master Regulators of CD4 T Cell Differentiation
The induction of CD4+ T cell differentiation is generally thought of as a two-step process. As discussed above, cytokine signaling through the Stat transcription factors is an essential first step in the differentiation process, as evidenced by the loss of specific CD4+ subsets in various Stat knockout mice. However, a major role of Stat activity during CD4+ T cell differentiation is to induce the expression of the so-called “master regulators” of CD4+ T cell differentiation. In general, a master regulator is identified when the enforced expression of this factor alone can induce differentiation into a given subset while deletion of the factor prevents differentiation into this subset from naïve CD4+ cells.
3.1 Tbet
The Tbox family member Tbet controls Th1 differentiation (Szabo et al. 2000), as evidenced by increased IFNγ production in response to Tbet overexpression, under both neutral and Th2 priming conditions (Szabo et al. 2000). In addition, Tbet deficient cells express very low levels of IFNγ and are unable to suppress IL-4 and IL-5 expression under neutral priming conditions (Szabo et al. 2002).
3.1.1 Autoactivation of Tbet
Tbet has been shown to bind to its own promoter (Kanhere et al. 2012), indicating a possible requirement for Tbet autoactivation under certain circumstances (Mullen et al. 2001). However, a study overexpressing Tbet in Stat1 deficient cells failed to show upregulation of endogenous Tbet expression in response to retroviral-induced Tbet expression (Afkarian et al. 2002). Tbet binding to its own promoter may cooperate with Stat1 but cannot induce expression in the absence of Stat1, as the loss of Stat1 leads to the loss of Tbet (Lovett-Racke et al. 2004). Likewise, a reporter mouse expressing ZsGreen under the control of the Tbet promoter showed similar levels of ZsGreen expression in both control and Tbet deficient cells in response to IL-12 and IFNγ stimulation, indicating a minimal role for autoactivation in Tbet expression (Zhu et al. 2012). However, the ZsGreen Tbet reporter on a Tbet−/− Stat4−/− double deficient background showed substantial loss of reporter expression during T. gondii infection, indicating that Tbet may autoactivate expression under certain conditions (Zhu et al. 2012). These results indicate that while Tbet may bind its own promoter, autoregulation of Tbet expression occurs only during specific conditions possible at later stage of differentiation or at memory stage when extrinsic stimulating cytokines become limiting.
3.1.2 Mechanism of Action
Large-scale ChIP-sequencing studies have identified binding sites of Tbet throughout the genome, including many immune regulatory genes, as well as cytokine and cytokine receptor genes (Zhu et al. 2012; Kanhere et al. 2012). However, although Tbet is generally considered a transcriptional activator, Tbet has also been reported to target genes for repression, including Socs1, Socs3, and Tcf7, which all show increased expression in response to Tbet deficiency (Oestreich et al. 2011). Tbet overexpression alone did not inhibit these genes, indicating that Tbet does not function to directly repress these genes. Instead, Tbet interacts with Bcl6 (Oestreich et al. 2012) and recruits this repressor to target genes to decrease gene expression.
In addition to directly binding target genes to induce expression, Tbet also induces changes in chromatin structure to increase or decrease gene accessibility. Although expression of a dominant negative mutant of Tbet blocked IFNγ expression when expressed in cells during early stages of Th1 differentiation, expression of this mutant in Th1-committed cells failed to have an effect on IFNγ expression (Mullen et al. 2002). This was shown to correlate with the loss of DNase hypersensitivity site I in the early Th1 cells, whereas the Th1-committed cells maintained this site even in the presence of the mutant Tbet, indicating the importance of gene accessibility for maintenance of gene expression. Subsequent work has demonstrated multiple interactions between Tbet and various chromatin-remodeling enzymes. In naïve T cells, the histone deacetylase Sin3A was associated with the 5’ CNS of Ifng, although as Th1 differentiation proceeded, Sin3A levels at the 5’ CNS decreased and overexpression of Tbet correlated with removal of Sin3A from Ifng (Chang et al. 2008). Tbet has also been shown to interact with the demethylases Jmjd3 and UTX as well as the SWI/SNF remodeling complex (Miller et al. 2010). Tbet recruits the RbBp5 component of the H3K4 methyl-transferase complex to induce permissive H3K4me2 marks at the promoters of Cxcr3 and Ifng, as well as the Jmjd3 H3K27-demethyltransferase to remove the repressive H3K27me3 marks (Miller et al. 2008). Furthermore, loss of Tbet correlated with loss of permissive H3K4me1 marks at the control regions of Ifng, Il12rb2, and Cxcr3 while the Th17 gene Ccr6 had less repressive H3K27me3 marks (Zhu et al. 2012). By altering the chromatin landscape, Tbet increases accessibility of Th1 target genes to allow other factors to bind and strengthen gene expression while also decreasing accessibility of non-Th1 genes.
3.1.3 Inhibition of Other Subsets
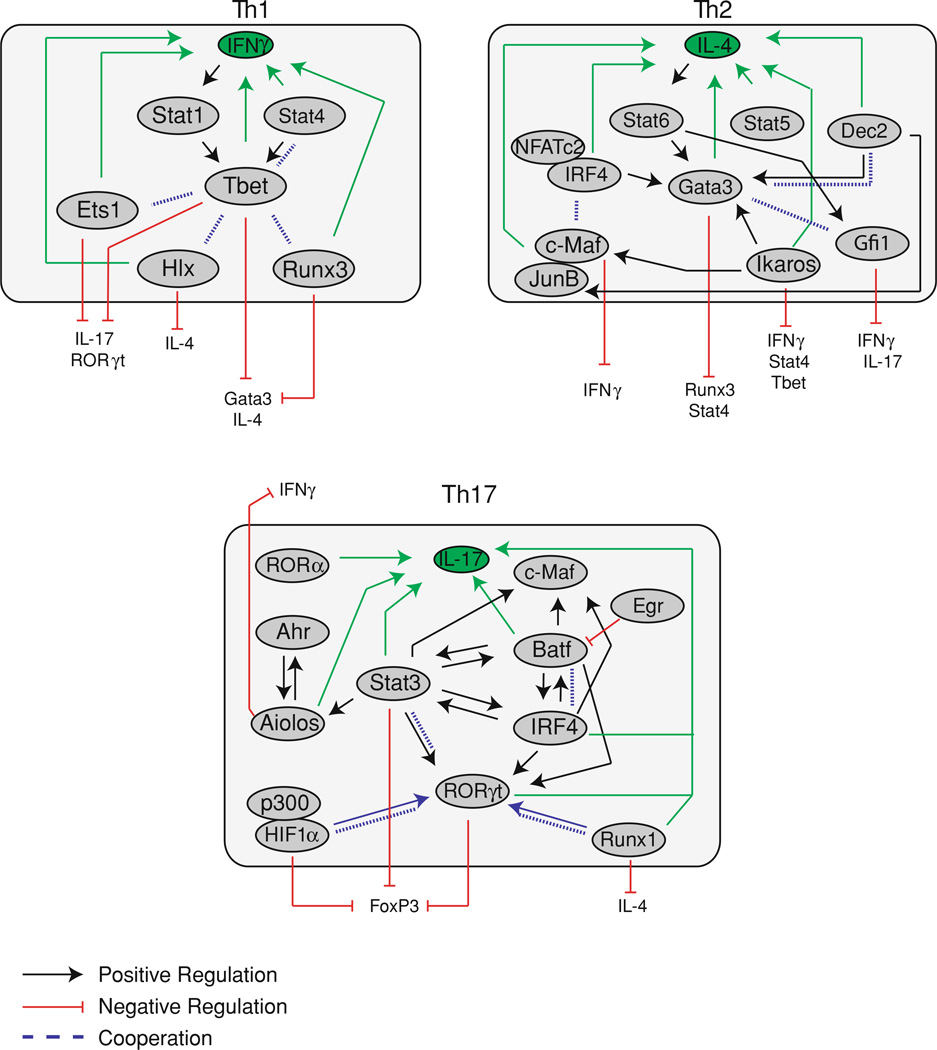
Tbet also play an essential role in the inhibition of other subsets during Th1 differentiation. While Tbet downregulates IL-4 and Gata3 expression (Zhu et al. 2012) in Th1 cells, Gata3 is both expressed and bound to target genes in Th1 cells (Jenner et al. 2009; Kanhere et al. 2012; Wei et al. 2011). Genome-wide analyses indicate that a number of genes are bound by Gata3 in both Th1 and Th2 cells, and include both Th1 and Th2-specific genes. In Th1 cells, many of these sites are also bound by Tbet, which acts to control gene expression as expression of co-occupied genes normally expressed in Th1 cells were increased in response to Tbet over-expression in Ifng−/− Tbet−/− double deficient cells while genes that are normally expressed in Th2 cells showed decreased expression (Jenner et al. 2009). Although some targets are shared in both Th1 and Th2 cells, the overall pattern of Gata3 distribution in Th1 cells is different from Th2 cells, where Gata3 shows decreased binding to Th2-specific genes and increased binding to Th1-specific genes (Kanhere et al. 2012). However, many of the Th1-specific genes bound by Gata3 in Th1 cells do not have a Gata motif at the binding sites but contain binding motifs for either Tbet or Runx family proteins, indicating that Tbet and Runx3 contribute to the localization of Gata3 in Th1 cells. Indeed, Runx3 is able to interact with Gata3 (Yagi et al. 2010), as does phosphorylated Tbet, and this interaction inhibits Th2 differentiation by blocking the binding of Gata3 to target genes (Hwang et al. 2005b). Increased levels of phosphorylated Tbet correlated with increased IFNγ expression while inhibition of Tbet phosphorylation resulted in increased Th2 differentiation (Chen et al. 2011a). Tbet also binds directly to the Gata3 gene and increases restrictive histone modifications to repress Gata3 expression (Zhu et al. 2012). Thus, inhibition of Th2 differentiation by Tbet involves both inhibition of Gata3 expression and sequestering Gata3 from many, but not all, Th2-specific genes. In addition, since Tbet also recruits Gata3 to many genes, including those expressed in Th1 cells, perhaps Tbet commandeers the activating functions of Gata3 to drive part of the Th1 gene program.
Tbet also interacts with many transcription factors to inhibit differentiation of the other CD4+ subsets. Tbet deficient cells express elevated levels of IL-17, indicating a role for Tbet in the inhibition of Th17 differentiation (Mathur et al. 2006). This has been proposed to occur via the binding of Tbet to Runx1 and Runx3, which prevents the binding of these transcription factors to their targets, including the Rorc gene (encoding RORγt) (Lazarevic et al. 2011). Furthermore, Runx1 also forms a complex with RORγt, which is required for full activity of RORγt (Zhang et al. 2008). In this way, binding of Runx1 to Tbet blocks the formation of active RORγt complexes required for Th17 differentiation. Indeed, coexpression of Tbet with RORγt resulted in decreased RORγ activity (Villarino et al. 2010), likely by sequestering Runx1 away from an interaction with RORγt. Finally, as described above, Tbet also binds to Bcl6, which blocks Bcl6-mediated repression of genes required for Tfh differentiation (Oestreich et al. 2011). Taken together, Tbet blocks the differentiation of other subsets by binding to and inhibiting multiple transcription factors.
3.2 Gata3
Th2 differentiation requires the transcription factor Gata3, which, in addition to expression during T cell development, is also expressed at substantial higher levels in Th2 cells (Zhang et al. 1997; Zheng and Flavell 1997). Gata3 deficiency leads to loss of Th2 cytokine expression (Pai et al. 2004; Zhu et al. 2004) and humans heterozygous for GATA3 have decreased frequencies of Th2 cells (Skapenko et al. 2004). In contrast, Gata3 overexpression drives Th2 differentiation (Ouyang et al. 2000; Lee et al. 2000), characterized by increased Th2 cytokine expression and increased endogenous Gata3 expression (Lee et al. 2000). Furthermore, Gata3 overexpression also inhibits Th1 differentiation, as cells cultured under Th1 polarizing conditions continue to express Th2 cytokines (Zheng and Flavell 1997) while Gata3 transgenic mice show decreased responses to delayed type hyper-sensitivity, a Th1 immune response (Nawijn et al. 2001).
3.2.1 Mechanism of Action
Gata3 binds directly to the Il4 enhancer to drive IL-4 production (Agarwal et al. 2000) and also to its own promoter to autoactivate expression (Ouyang et al. 2000). In this way, the positive feedback loop within Th2 differentiation is driven by both Gata3-induced expression of IL-4, which signals through Stat6 to activate Gata3 expression and by autoactivation of Gata3. Although Gata3 autoactivation plays a minimal role when a sufficient amount of IL-4 stimulation is present, as demonstrated by comparable expression levels of Gata3 in both wild type cells and cells expressing mutant Gata3 lacking the exon 4 encoded zinc fingers (Wei et al. 2011), autoactivation may be important for maintaining Gata3 expression when IL-4 signaling ceases.
Gata3 is required continuously for maintenance of Th2 differentiation as loss of Gata3 even after Th2 differentiation leads to decreased Th2 cytokine production and cell proliferation as well as increased IFNγ expression (Zhu et al. 2004). In addition to Th2 cytokines, Gata3 also binds to control regions of many transcription factors and many genes involved in T cell signaling to promote Th2 differentiation (Wei et al. 2011). Surprisingly, ChIP-Seq analysis identified Gata3 binding sites within multiple CD4+ T cell subsets, indicating a role for Gata3 beyond Th2 differentiation. These Gata3 binding sites were shown to be in close proximity to a number of other transcription factor binding sites, including Runx, Ets and AP-1, indicating cooperation of Gata3 with other transcription factors. In Th2 cells, Gata3 binding colocalizes with the binding of Ets family member Fli1 to the DNA and the loss of Gata3 resulted in loss of Fli1 binding at 75 % of the shared binding sites. These results indicate that while Gata3 targets many genes in Th2 cells, it may also play a role in regulation of other subsets by acting within larger transcription factors complexes. Indeed, Tregs lacking Gata3 expression have reduced suppressive activity during inflammation (Wohlfert et al. 2011) and Gata3 has also been shown to be involved in CD8 T cell memory (Wang et al. 2013).
Similar to Tbet, Gata3 also exerts control over transcription programs by modifying chromatin structure. Indeed, overexpression of Gata3 correlated with increased DNase hypersensitivity sites at the Il4 promoter (Ouyang et al. 2000; Lee et al. 2000), indicating increased accessibility for transcriptional activation. Furthermore, deletion of Gata3 resulted in decreased histone acetylation at the Il5 promoter, increased methylation of the Il4 gene and increased histone acetylation at the Ifng promoter (Yamashita et al. 2004). Both Gata3 and Stat6 associate with the Ifng promoter in Th2 cells to alter chromatin structure as the loss of Stat6 correlated with the loss of repressive H2K27me2 histone modifications at Ifng while overexpression of Gata3 increased H3K27me2 marks and decreased permissive H3K9me marks (Chang and Aune 2007). The loss of Gata3 also resulted in decreased permissive H3K4me2 marks and increased restrictive H3K27me3 marks at Th2-specific genes and decreased repressive H3K27me3 marks at both Tbet and Ifng control regions under Th2 conditions (Wei et al. 2011). At the genome level, although only ~10 % of Gata3-bound genes have altered expression upon Gata3 deletion,~50 % of all Gata3-bound genes have epigenetic changes around the Gata3 binding sites indicating a direct role of Gata3 in chromatin remodeling.
The control of the Th2 cytokine cluster requires a three-dimensional conformation of chromatin to bring the promoters for Il-4, Il-5 and Il-13 into close proximity with the locus control region (Spilianakis and Flavell 2004). Gata3 binds to a region within the LCR to induce long-range intrachromosomal interactions of the Th2 locus, as overexpression of Gata3 in nonlymphoid cells-induced permissive chromatin conformation at the Th2 locus, with the LCR in close proximity to the IL-4 cytokine cluster, resembling the structure found in Th2 cells (Spilianakis and Flavell 2004). Thus, by modulating repressive and permissive marks on histones and DNA as well as inducing permissive chromatin interactions, Gata3 controls transcriptional activation and repression of genes in Th2 cells.
3.2.2 Regulation of Other Subsets
Gata3 also plays a role in inhibiting Th1 differentiation, as Gata3 overexpressing cells show decreased Th1 polarization (Zheng and Flavell 1997). Gata3 acts to block Th1 differentiation by decreasing Stat4 expression, as Gata3 expression during Th2 differentiation correlated with decreased expression of Stat4 (Usui et al. 2003). In addition to Tbet and Stat4, Runx3 also plays a role in inducing IFNγ expression (Djuretic et al. 2007; Kohu et al. 2009; Yagi et al. 2010) and Gata3 binds directly to Runx3 (Yagi et al. 2010; Kohu et al. 2009) to inhibit Runx3-mediated IFNγ expression (Yagi et al. 2010). Furthermore, deletion of Gata3 resulted in increased expression of Th17-related genes (Wei et al. 2011). Both the downregulation of gene expression and the direct inhibition of other transcription factors are important functions of Gata3 in inhibiting the differentiation of other subsets while inducing the differentiation of Th2 cells.
3.2.3 Plasticity of Th2 Cells
Although Gata3, when expressed at higher levels, suppress Th1 and Th17-related transcription factors, it has been reported that Gata3 expressed at intermediated levels may tolerate the expression of Tbet. There is evidence that committed Th2 cells are capable of upregulating Tbet and producing IFNγ without losing their capacity to produce IL-4 (Hegazy et al. 2010). After 3 weeks of in vitro Th2 differentiation, LCMV-specific CD4+ T cells were transferred into naïve C57BL/6, followed by LCMV infection 30 days post-transfer. Upon LCMV infection, these in vitro differentiated Th2 cells gained the capacity to produce IFNγ in addition to IL-4 to become IFNγ+IL-4+ double producing cells. These cells also upregulated Tbet expression to similar levels found in Th1 cells and Th2 cells differentiated from Tbet deficient mice failed to produce IFNγ in this model, indicating the importance of activating the Th1 transcriptional program. Indeed, although these Th2 cells initially produced high levels of Gata3, upon transfer and LCMV infection, Gata3 levels decreased while Tbet levels increased and this pattern was maintained up to 60 days post-infection.
A subset of cells has also been identified that produce both Th2 cytokines IL-4, IL-5, and IL-13 as well as IL-17 and express high levels of both Gata3 and RORγt (Wang et al. 2010). Th2 cells stimulated with IL-1β, IL-6, and IL-21 resulted in the induction of IL-17 expression. This plasticity between Th2 and Th17 plays a role in disease settings as patients with asthma had higher expression of IL-17 and IL-22 from sorted Th2 cells upon restimulation. In addition, intranasal exposure to allergens-induced IL-17 producing Th2 cells specifically in the lung and these cells persisted in the lung after exposure.
3.3 RORγt/RORα
Th17 differentiation is controlled by the orphan nuclear receptor RORγt. Mice deficient in RORγt have limited Th17 differentiation (Ivanov et al. 2006; Volpe et al. 2008) while overexpression of RORγt induced IL-17 expression in the absence of Th17 polarizing cytokines (Ivanov et al. 2006). As Th17 cells are the causative agents of EAE, the loss of RORγt and subsequent loss of Th17 cells was also protective against EAE induction. RORγt binds directly to the Il17a/Il17f gene to induce expression (Ichiyama et al. 2008). In addition to RORγt, another family member, RORα, is also expressed in Th17 cells and overexpression of RORα also increased IL-17 production (Yang et al. 2008). RORα binds to the CNS2 of Il17 and overexpression of both RORα and RORγt acted synergistically to increase IL-17 expression, while deletion of both RORγt and RORα resulted in a complete block in Th17 differentiation. Although RORγt is clearly required for Th17 differentiation, it functions within a larger complex of transcription factors (Ciofani et al. 2012). Genome-wide ChIP analysis demonstrated that RORγt functions in cooperation with Batf, IRF4, and Stat3; RORγt appears to regulate gene expression driven by these factors, both by reinforcing Th17-specific genes such as Il17a and Il17f and by attenuating expression of a number of genes induced by Batf, IRF4, and Stat3. However, genes attenuated by RORγt remain expressed in Th17 cells, indicating that RORγt plays a role in fine-tuning the expression of many target genes during Th17 differentiation. RORγt is also involved in the inhibition of both Th1 and Treg differentiation as overexpression of RORγt resulted in decreased IFNγ production (Ivanov et al. 2006) while the loss of RORγt resulted in increased FoxP3 expression (Burgler et al. 2010).
3.3.1 Plasticity of Th17 Cells
Although Th17 cells are considered a unique CD4+ T cell subset, Th17 cells have been shown to produce IFNγ (Mathur et al. 2006; Shi et al. 2008; Lee et al. 2009; Bending et al. 2009). After in vitro differentiation, both Th1 and Th17 cells were transferred into NOD/Scid mice and diabetes development was monitored (Bending et al. 2009). Although Th17 cell transfer showed a slight lag in the induction of diabetes, Th17 transfer resulted in the development of diabetes and analysis of cells isolated from peripheral lymph nodes showed increased IFNγ production and loss of IL-17 production compared to original transplanted Th17 cells. Although these transplanted cells were cultured under Th17 inducing conditions, the possibility remained that a small population of undifferentiated cells could be responsible for diabetes induction. To demonstrate that Th17 cells can convert to IFNγ producers, a subsequent in vitro study used an IL-17f reporter mouse to sort IL-17 expressing Th17 cells and demonstrated that treatment of these cells with IL-12 increased IFNγ production (Lee et al. 2009). These cells also lost RORγt expression while gaining Tbet expression and Th17 cells deficient in either Stat4 or Tbet failed to upregulate IFNγ in response to Th1 polarizing conditions, indicating a requirement for the activation of the Th1 transcriptional program (Lee et al. 2009).
The switch from IL-17 to IFNγ production appears to be important during EAE as suppression of Tbet expression by siRNA after EAE induction resulted in decreased disease severity (Gocke et al. 2007). The use of the IL-17 fate reporter mouse has shown that the majority of the CNS infiltrating IFNγ-producing cells in MOG-induced EAE arises from IL-17 producing cells, and cells that have produced IL-17 produce the highest levels of cytokines upon restimulation (Hirota et al. 2011). Th17 cells have also been shown to become IFNγ+IL17+ double producers (Ivanov et al. 2006; Lee et al. 2009; Lexberg et al. 2010; Villarino et al. 2010), which express both Tbet and RORγt (Lexberg et al. 2010). Interestingly, cells isolated from spinal cords of EAE mice show a larger population of IFNγ+IL-17+ compared to IL-17 single positive. Potentially, this double positive population is composed of those Th17 cells in the process of converting to Th1 cells. This switch of Th17 cells to Th1 cells in the CNS appears to drive the pathogenesis of EAE and demonstrates the importance of CD4+ T cell subset balance in the maintenance of a healthy individual.
3.4 FoxP3
Mutations in the FOXP3/Foxp3 gene were identified in X-linked neonatal diabetes mellitus, entropathy and endocrinopathy syndrome (IPEX) patients (Wildin et al. 2001) and in the scurfy mouse, respectively (Brunkow et al. 2001), representing a human condition and a mouse model characterized by severe autoimmunity. FoxP3 is expressed in CD4+CD25+ T cells and cells overexpressing FoxP3 show poor proliferation and very little cytokine production in response to T cell receptor stimulation (Hori et al. 2003). In addition, FoxP3 overexpressing cells are capable of suppressing the activation of CD4+CD25− T cells and preventing the development of inflammatory bowel disease (IBD) induced by transferring naive CD4+CD25− T cells into lymphopenic hosts (Hori et al. 2003; Fontenot et al. 2003). In contrast, CD4+CD25+ cells deficient in FoxP3 expression failed to suppress CD4+CD25− T cells (Fontenot et al. 2003; Gavin et al. 2007; Williams and Rudensky 2007) and a mixed bone marrow chimera experiment demonstrated that development of suppressive T regulatory cells required FoxP3 expression (Fontenot et al. 2003).
3.4.1 Mechanism of Action
Genome-wide analysis has identified a large number of FoxP3 target genes, including genes involved in regulating transcription as well as genes involved in epigenetic modification (Zheng et al. 2007). Furthermore, analysis of FoxP3 binding partners identified interactions with a large number of transcription factors, as well as candidates involved in chromatin organization and modification (Rudra et al. 2012). Not only does FoxP3 bind to these factors at the protein level, but ChIP-Seq analysis of FoxP3 binding sites showed that FoxP3 also binds to the regulatory regions of these factors, thus controlling the expression of many of its binding partners. In agreement with these genome-wide studies showing interactions of FoxP3 with chromatin modifying enzymes, many FoxP3 target genes were found to have permissive histone modifications. Furthermore, FoxP3 interacts with the histone acetyltransferase TIP60 as well as histone deacetylase 7 (Li et al. 2007a) and disruption of this interaction leads to increased acetylation at the Il2 promoter, which is normally repressed in T regulatory cells (Bettini et al. 2012). However, a study comparing chromatin modifications in naïve T cells with those in FoxP3 expressing cells found no major changes in chromatin modifications before and after FoxP3 expression, indicating that other factors, such as FoxO proteins, are likely required to modify the chromatin landscape in T regulatory cells prior to FoxP3 binding (Samstein et al. 2012).
FoxP3 also exploits direct interaction with NFAT to target specific genes during Treg differentiation (Wu et al. 2006). Expression from an IL-2 reporter construct containing NFAT:AP-1 binding sites was inhibited in the presence of FoxP3. This inhibition required NFAT, as FoxP3 bound to the Il2 promoter sequence only in the presence of the NFAT DNA binding domain. Furthermore, mutation of FoxP3 residues to disrupt NFAT binding resulted in increased IL-2 expression in response to T cell stimulation. The importance of this interaction between NFAT and FoxP3 was demonstrated by a failure of FoxP3 mutant cells to protect against diabetes when transferred into NOD mice.
3.4.2 Regulation of Other Subsets
FoxP3 also plays an active role in inhibiting both Th2 and Th17 differentiation by inhibiting Gata3 and RORγt to block IL-4 and IL-17 expression (Zeng et al. 2009). FoxP3 also binds to phosphorylated Stat3, resulting in the recruitment of FoxP3 to Il6 regulatory regions (Chaudhry et al. 2009). This results in the repression of IL-6 expression, which further blocks Th17 differentiation. In addition, FoxP3 binds to RORγt (Zhou et al. 2008; Lochner et al. 2008; Ichiyama et al. 2008) to inhibit IL-17 expression (Ichiyama et al. 2008; Zhou et al. 2008) by decreasing RORγt binding to the Il17 promoter, which correlates with decreased permissive histone modifications (Ichiyama et al. 2008). FoxP3 also binds to the RORγt: Runx1 complex to inhibit IL-17 expression, as mutation of the RORγt or Runx1 binding site within FoxP3 blocks IL-17 inhibition (Zhang et al. 2008). RORγt in turn has been shown to inhibit Treg differentiation by binding directly to the Foxp3 promoter to inhibit FoxP3 expression (Burgler et al. 2010).
3.4.3 Plasticity of Treg Cells
Stimulation of Treg cells with IL-6 resulted in increased IL-17 expression and the development of both IL-17+FoxP3+ double positive cells and IL-17 single positive cells, indicating that the conversion of Treg cells to Th17 cells includes a double positive population (Xu et al. 2007). Furthermore, two populations of FoxP3 expressing cells have been reported based on the expression level of FoxP3 (Tartar et al. 2010). Those cells expressing high levels of FoxP3 had no detectable RORγt while cells expressing intermediate levels of FoxP3 also expressed high levels of RORγt. These FoxP3 intermediate cells upreguated IL-17 production in response to Th17 polarizing conditions while FoxP3 high expressing cells did not. Furthermore, polarization of the FoxP3 intermediate population under Th17 conditions led to increased RORγt expression while Treg polarization led to increased FoxP3 expression, indicating that FoxP3+RORγt+ have the capacity to become either Th17 or Treg cells, depending on the cytokine environment.
An analysis of proteins interacting with FoxP3 identified Gata3, which both interacts with and is upreguated by FoxP3 (Rudra et al. 2012). Gata3 and FoxP3 are coexpressed in Treg cells in the lamina propria and T cell receptor stimulation of Treg cells resulted in upregulation of Gata3 expression (Wohlfert et al. 2011). Gata3 promotes Treg differentiation by binding directly to the Foxp3 enhancer to increase FoxP3 expression (Wohlfert et al. 2011; Wang et al. 2011). Competitive transfer experiments of wild type and Gata3-deficient bone marrow cells in Rag1−/− mice resulted in much fewer FoxP3 expressing cells arising from Gata3 deficient cells, indicating the importance of Gata3 in Treg differentiation. These Gata3 deficient Treg cells showed decreased suppressive activity and failed to protect against a transfer model of inflammatory bowel disease (Wohlfert et al. 2011; Wang et al. 2011). Gata3 deficient cells also expressed lower levels of both FoxP3 and FoxP3 target genes, including Cd25, Ctla4, and Gitr (Wang et al. 2011) and had decreased FoxP3+CD25+ populations under conditions of inflammation (Wohlfert et al. 2011). Gata3 also binds to the Tbet and Rorc control regions in Treg cells to repress these genes, inhibiting Th1 and Th17 differentiation (Wohlfert et al. 2011). Finally, FoxP3 and Gata3 cooperate to control gene expression as both factors target a number of shared genes in Treg cells and the specific deletion of either factor resulted in altered expression of the shared target genes (Rudra et al. 2012). Thus, Gata3 controls expression of transcription factors within Treg cells to maintain Treg phenotype while also preventing Th1 and Th17 differentiation from Tregs.
3.5 Bcl6
The debate concerning the validity of characterizing follicular T cells as a unique subset was strengthened by the identification of the master regulator Bcl6 (Chtanova et al. 2004). A microarray analysis of Tfh cells, defined as CD57+CXCR5+CD4+ T cells, identified Bcl6 as being preferentially expressed in Tfh cells compared to Th1 and Th2 cells (Chtanova et al. 2004). Subsequent work demonstrated that Bcl6 deficient T cells fail to differentiate into CXC5+PD-1+ Tfh cells while overexpres-sion of Bcl6 increased Tfh differentiation (Yu et al. 2009a; Nurieva et al. 2009; Johnston et al. 2009). Since Bcl6 is a transcriptional repressor, the mechanism of action to induce Tfh differentiation is not likely to rely on activation of transcription. Instead, Bcl6 has been shown to inhibit expression of various microRNAs, which are likely responsible for suppressing Tfh differentiation (Yu et al. 2009a). Indeed, predicted binding sites for microRNAs downregulated by Bcl6 were identified in the 3’ UTR of Cxcr5, Cxcr4, and Pd1, all genes expressed in Tfh cells.
In addition to promoting Tfh differentiation, Bcl6 overexpression also blocks Th1, Th2, and Th17 differentiation by decreasing expression of Tbet, Gata3, and RORγt (Yu et al. 2009a; Nurieva et al. 2009). Bcl6 binds directly to Tbet and Rorc promoters to inhibit expression of these genes. However, Bcl6 is negatively regulated by Blimp1, as Blimp1 overexpression leads to decreased Bcl6 expression and CXCR5+ Tfh differentiation (Johnston et al. 2009). Blimp1 is expressed in non-Tfh subsets (Crotty et al. 2010), where expression is upreguated in response to T cell stimulation, indicating a mechanism by which other subsets inhibit Tfh differentiation.
3.5.1 Plasticity of Tfh Cells
Although Tfh genes are expressed during early Th1 differentiation, IL-2 production by Th1 cells inhibits expression of Tfh genes at later time points (Oestreich et al. 2012). While Bcl6 levels are decreased after Th1 polarization, removal of IL-2 resulted in elevated expression of both Bcl6 and CXCR5 and decreased Blimp1 expression. Furthermore, after culture of Th1 cells under low IL-2 levels to induce Tfh gene expression, the addition of higher IL-2 levels resulted in decreased Bcl6 and CXCR5 expression with increased Blimp1 expression, indicating plasticity in the expression of the Tfh genes. This change in Blimp1 expression in response to IL-2 correlated with increased binding of Blimp1 to Cxcr5 in high IL-2 conditions. In this way, there is a balance between Th1 and Tfh cells, where they appear capable of converting in response to changes in the cytokine environment.
4 Additional Factors Involved in CD4+ T Cell Differentiation
As discussed above, much of the work on the transcriptional regulation of CD4+ T cell differentiation has centered on the idea of a two-step process with the initiation of differentiation induced by Stat activation in response to cytokine stimulation followed by the activation of the master regulator for each subset. However, many studies have identified roles for additional transcription factors within CD4+ T cell differentiation programs and suggest that rather than two factors controlling differentiation, a network of factors are likely involved in fine-tuning the expression and repression of various genes to determine subset gene control.
4.1 Factors Induced by T Cell Stimulation
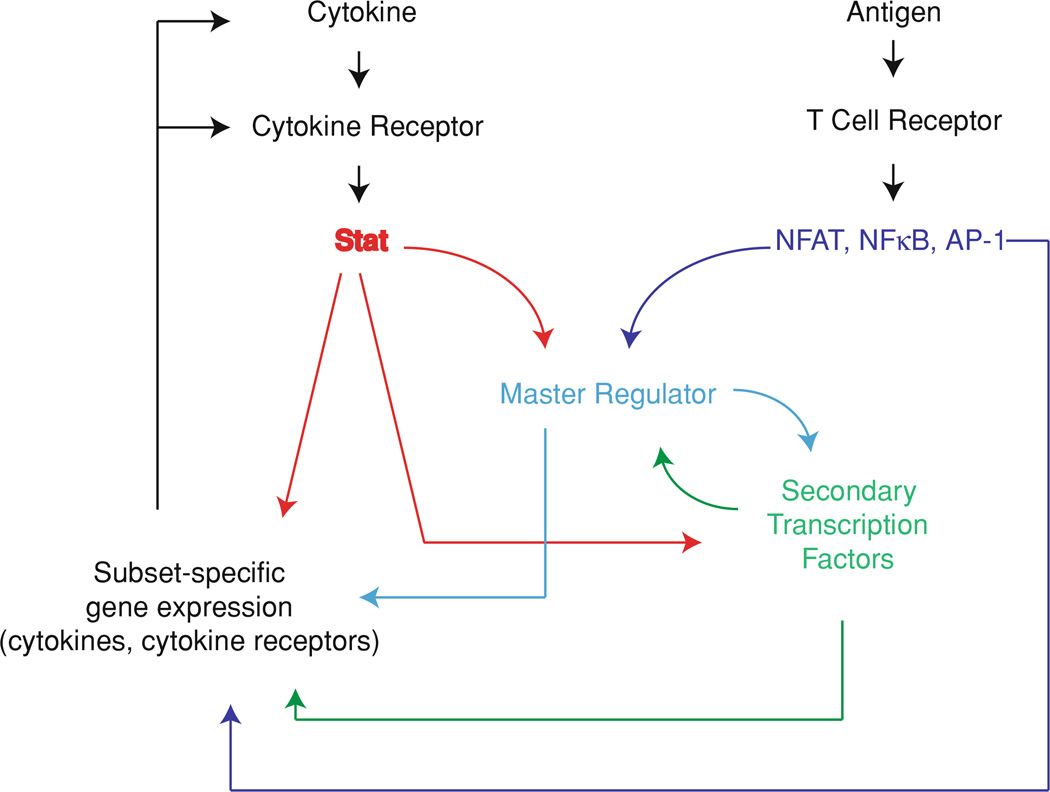
Although cytokine-induced Stat activation plays a critical role in the induction of master regulators, signaling through T cell receptor is also essential for T cell differentiation. Stimulation through the T cell receptor results in the activation of the NFAT, NFκB, and AP-1 families of transcription factors. These factors cooperate with Stat-induced expression of subset-specific cytokines and master regulators. A summary of the interaction of transcription factor activation by T cell signaling with cytokine-mediated Stat activation and the resulting differentiation program is shown in Fig. 2.
Fig. 2.
Cooperation between cytokine signaling and T cell receptor signaling during CD4+ T cell differentiation. NFAT, NFκB, and AP-1 activation in response to T cell stimulation activates expression of master regulators and cytokines to drive CD4+ T cell differentiation. The strength of antigenic stimulation results in altered patterns of transcription factor activation, which cooperates with cytokine-induced signaling to drive subset-specific differentiation
4.1.1 NFAT Proteins
There are five members of the NFAT family of transcription factors, NFAT1 (NFATp or NFATc2), NFAT2 (NFATc or NFATc1), NFAT3 (NFATc4), NFAT4 (NFATc3), and NFAT5 (Macian 2005). In response to stimulation of the T cell receptor, phospholipase C is activated and cleaves phosphatidylinositol 4, 5 bis-phosphate into inositol 1, 4, 5 triphosphate (InsP3) and diacylglycerol. InsP3 in turn triggers the release of intracellular calcium stores, which activates the calcium release activated calcium channels at the cell surface, resulting in elevated cyto-plasmic calcium levels. Intracellular calcium binds to calmodulin, resulting in the activation of calcineurin, which dephosphorylates NFATs, allowing nuclear translocation of these transcription factors. Among the targets of NFAT are the cytokines involved in driving CD4+ T cell differentiation, as the deletion of NFATc1 and NATc2 resulted in decreased production of IL-2, IFNγ, IL-4, and IL-5 in response to T cell stimulation (Peng et al. 2001).
NAFT binding sites have been identified within the Ifng promoter and super-shift analysis demonstrates NFAT binding at these sites (Sica et al. 1997). Furthermore, overexpression of a constitutively active form of NFATc1 resulted in increased IFNγ expression and decreased IL-4 expression, thus polarizing cells to become Th1 (Porter and Clipstone 2002). Similarly, deletion of NFAT1 on an IL-4-deficient background also resulted in decreased IFNγ production accompanied by increased susceptibility to L. major infection resulting in increased lesion size and parasite count (Kiani et al. 2001). NFAT also appears to negatively regulate Th2 responses as mice deficient in NFAT1 (Kiani et al. 1997) or both NFATc2 and NFATc3 (Rengarajan et al. 2002b) produce higher levels of IL-4 and decreased levels of IFNγ in response to nonpolarizing T cell stimulation, and IL-4 expression is maintained for prolonged periods of time after stimulation. Nfat c2−/− Nfat c3−/− double deficient mice also produce higher levels of the Th2 cytokine IL-5 even on an IL-4 deficient background, indicating that NFAT regulates Th2 differentiation independently of IL-4 expression. Similarly, deletion of NFATp and NFAT4 resulted in increased IL-4, IL-5, IL-6, and IL-10 production, while IFNγ and IL-2 production was reduced (Ranger et al. 1998b). These results indicate a role for NFAT family members in the induction of IFNγ production and suppression of Th2 cytokine expression during Th1 differentiation.
In addition to cytokine environment, the strength of T cell stimulation is thought to contribute to Th1/Th2 differentiation pathways, where higher signal strength induces Th1 differentiation while weaker signaling induces Th2 differentiation (Constant and Bottomly 1997). The level of T cell stimulation is translated through intracellular signaling cascades into unique patterns of transcription factor activation. Stimulation with a low affinity peptide was shown to increase the ratio of NFATc to NFATp in differentiating T cells, which corresponded with increased IL-4 production (Brogdon et al. 2002). In agreement with these results, deletion of NFATc resulted in decreased IL-4 production, likely due to the loss of NFAT binding to the Il4 promoter (Yoshida et al. 1998; Ranger et al. 1998a).
NFAT family members have also been shown to bind to the Il17 promoter (Liu et al. 2004; Gomez-Rodriguez et al. 2009; Hermann-Kleiter et al. 2012) and expression of a hyper-activatable NFAT1 resulted in increased IL-17 production in response to Th17 polarizing conditions. Although these mice produced increased levels of IL-17 in vitro, they were less susceptible to EAE induction, which may be due to an increased Treg population, indicating a role for NFAT in Treg differentiation. Indeed, NFAT has been shown to drive FoxP3 expression and Treg differentiation by binding directly to the Foxp3 promoter and enhancer regions (Mantel et al. 2006; Tone et al. 2008). Deletion of NFAT2, NFATc2, and NFAT1 or NFAT1 and NFAT4 resulted in decreased Treg populations, although the Treg cells that were produced remained functional (Vaeth et al. 2012).
Together, these results demonstrate roles for the NFAT family members in driving the expression of multiple subset-specific cytokines as well as FoxP3. Whether NFAT proteins play an important role in regulating the expression of other master transcription factors is understudied. In addition to signal strength inducing specific NFAT family members, subset-specific functions of NFAT family members may arise from cooperation with other factors expressed during differentiation. NFAT1 has been shown to have CD4+ subset-specific target binding, binding to either the Il4 or Ifng promoter in Th2 or Th1 cells, respectively (Agarwal et al. 2000). This specificity in binding is likely dependent on the chromatin structure at these genes and the relative accessibility of binding sites, thus relying on other factors for subset-specific transcriptional activity.
4.1.2 NFκB
The NFκB family of transcription factors is composed of 5 family members, RelA (p65), NFκB1 (p50/p105), NFκB2 (p52/p100), c-Rel, and RelB (Hayden and Ghosh 2012). Stimulation of the T cell receptor leads to the activation of PKCθ, which activates a signaling cascade resulting in NFκB activation [reviewed in (Hayden and Ghosh 2012)], which in turn regulates expression of subset-specific cytokines, cytokine receptors, and transcription factors. To test the role of NFκB during T cell differentiation, transgenic mice were generated to express a mutant form of IκBα, which is resistant to degradation and acts as a dominant inhibitor of NFκB activity (Aronica et al. 1999). These mice showed decreased delayed type hypersensitivity responses, decreased IFNγ production, and increased IL-4 production. These mice also showed decreased levels of phosphorylated Stat4 and decreased Tbet expression (Corn et al. 2003). Similarly, deficiency in RelB also resulted in decreased IFNγ production along with loss of Tbet expression and decreased levels of phosphorylated Stat4 (Corn et al. 2005), indicating the importance of NFκB activation during Th1 differentiation. Indeed, p50 has been shown to bind directly to the Ifng promoter to drive cytokine expression (Lai et al. 2011). However, in contrast to this cooperation between cytokine-induced transcription factors and T cell receptor-induced factors, Tbet has been shown to inhibit members of the NFκB family (Hwang et al. 2005a). Tbet binds to c-Rel to block binding of c-Rel to target genes, resulting in altered gene expression patterns. One target of c-Rel is Il2, which is downregulated in Th1 cells, allowing a switch to IFNγ production. In this way, Tbet acts to block expression of non-Th1 genes by activated NFκB.
Mice deficient in the p50 subunit of NFκB show decreased Th2 differentiation, with decreased IL-4, IL-5 and IL-13 production as well as decreased Gata3 expression with no change in Tbet expression (Das et al. 2001). Similarly, deficiency in the IκB family member Bcl3 also resulted in decreased IL-4 and IL-5 production along with decreased Gata3 expression. Analysis of the Gata3 promoter demonstrated that activated T cell extracts were capable of binding to a putative NFκB binding site and this was blocked by addition of unlabeled NFκB probe and was shifted by an antibody against p50, thus verifying the ability of p50 to bind to Gata3 regulatory regions. However, NFκB appears to be required only for early induction of Gata3 expression, as inhibition of nuclear translocation of NFκB after 4 days of Th2 polarization showed no effect on Gata3 expression (Das et al. 2001).
NFκB family members also play a role in Th17 differentiation as deletion of c-Rel resulted in decreased expression of IL-17 along with decreased susceptibility to EAE induction (Chen et al. 2011b). These mice also had decreased RORγt expression. Two NFκB binding sites were identified upstream of the Rorc transcription start site and a reporter assay using the Rorc promoter demonstrated cooperation between c-Rel and RoRγt in driving expression from the Rorc promoter. Similarly, deletion of the NFκB adapter Carma1 resulted in decreased expression of IL-17a, IL-17f, IL-21, IL-22, IL-23R accompanied by decreased susceptibility to EAE induction (Molinero et al. 2012). The IκB family member IκBζ was shown to be expressed at the highest levels in Th17 cells and deletion of this gene resulted in decreased expression of IL-17f, IL-21, IL-22, and IL-23R while overexpression resulted in increased IL-17f, IL-21, and IL-23R expression (Okamoto et al. 2010). While there was no change in the expression of RORγt or RORα in these mice, RORγt cooperates with IκBζ in driving cytokine expression as there was decreased binding of IκBζ to CNS2 of the Il17 promoter in the absence of RORγt. Together, these results demonstrate a requirement for NFκB activation to drive optimal expression of Th17 cytokines, cytokine receptors, and transcription factors.
In addition to decreased Th17 differentiation in Carma1 deficient mice, there was also an increase in Treg populations, indicating a role for NFκB in Treg differentiation. Indeed, c-Rel deficient mice show decreased Treg populations in the thymus, spleen, and lymph nodes (Isomura et al. 2009; Ruan et al. 2009). NFκB drives FoxP3 expression as both c-Rel and p65 increased expression of a reporter construct containing the Foxp3 promoter, although neither p50 nor RelB increased expression of the reporter (Ruan et al. 2009). Deletion of the putative Rel-NFAT binding sites in the FoxP3 reporter construct inhibited the c-Rel driven expression. Thus, NFκB induces FoxP3 expression to promote Treg differentiation.
Taken together, these results demonstrate roles for NFκB family members in driving expression of subset-specific transcription factors, cytokines, and cytokine receptors. While some genes are controlled by specific family members, such as p65 and c-Rel driving FoxP3 expression while neither p50 nor RelB can, NFκB family members do not necessarily show subset specificity as p50 has been shown to play a role in both Th1 and Th2 differentiation. In this case, it is likely that the NFκB family members activated by the T cell receptor cooperate with those factors induced by the cytokine environment, either through gene accessibility induced by Stats and master regulators or by regulating expression of genes also targeted by Stats and master regulators.
4.1.3 AP-1
The AP-1 transcription factor is a heterodimeric protein composed of members of the Jun (cJun, JunB, JunD), Fos, Maf, and ATF (including Batf) families (Shaulian and Karin 2002). These factors are activated in response to the induction of the MAP kinase cascade upon T cell stimulation. The pattern of AP-1 formation appears to play a role in CD4+ T cell differentiation as strong T cell stimulation resulted in the formation of Fos-Jun dimers while partial ERK inhibition resulted in formation of Jun-Jun dimers and increased IL-4 production (Jorritsma et al. 2003). JunB in particular has been shown to drive IL-4 expression in cooperation with c-Maf (Li et al. 1999).
Batf is a member of the AP-1 family of transcription factors and is expressed in Th1, Th2, and Th17 cells (Schraml et al. 2009). However, generation of Batf deficient mice resulted in a specific defect in Th17 differentiation, while Th1 and Th2 differentiation proceeded normally. These mice showed increased resistance to EAE and microarray analysis demonstrated decreased expression of multiple Th17 associated genes, including Rorc, Rora, Ahr, Il21, and Il17. Surprisingly, overexpression of RORγt in Batf deficient cells failed to rescue IL-17 expression under Th17 polarizing conditions, indicating a specific requirement for Batf during Th17 differentiation, independent of RORγt expression.
Batf has been shown to activate transcription as part of a larger complex, including interferon regulatory factor 4 (IRF4) and JunB (Li et al. 2012; Glasmacher et al. 2012). As IRF4 binds weakly to DNA on its own, IRF4 ChIP-Seq analysis was performed in T cells to identify transcription factor binding sites associated with IRF4 binding. This analysis identified AP-1 binding sites as the motif most highly associated with IRF4 binding. EMSA analysis of DNA sequences containing tandem IRF4 and AP-1 binding sites demonstrated binding by Th17 extracts while supershift analysis identified binding of IRF4, JunB, and Batf. These three factors bind in a cooperative manner to target many genes during Th17 differentiation, including Il17a, Il21, IL23r, and IL12rb1 (Li et al. 2012; Glasmacher et al. 2012) and the loss of any one of these factors inhibits Th17 differentiation. Similar to Batf deficient mice, IRF4 deficient mice also show a block in Th17 differentiation, with decreased IL-17 production and RORγt expression (Brustle et al. 2007). The loss of either Batf or IRF4 resulted in decreased DNA binding of the partner factor, indicating the requirement for both factors for efficient DNA binding and target gene expression (Li et al. 2012; Glasmacher et al. 2012). Furthermore, overexpression of IRF4, Batf, or JunD alone showed limited binding to target DNA while coexpression of all three showed strong binding (Li et al. 2012). Thus, Th17 differentiation requires cooperation among these factors, as the loss of any one prevents the binding of the others, resulting in decreased gene expression.
Batf is negatively regulated by early growth response factor-2 (Egr-2), a member of the Egr zinc finger transcription factor family (Miao et al. 2013). Cells deficient in Egr-2 showed a specific increase in IL-17 production, while IFNγ and IL-2 production remained unchanged in response to nonpolarizing T cell stimulation. This function of Egr-2 was specific to Th17 differentiation, as loss of Egr-2 did not increase IL-17 production under either Th1 or Th2 polarizing conditions. Although nuclear extracts from Egr-2 deficient cells had an elevated level of Batf capable of binding to the Il17 promoter, there was no significant increase in total Batf expression in these cells. Instead, Egr-2 was shown to physically interact with Batf and this interaction is proposed to block Batf binding to the Il17 promoter, thus decreasing IL-17 production. Egr-2 expression is upreguated in response to IL-6 and TGFβ-induced Th17 differentiation and acts to negatively regulate the Th17 differentiation. This plays an important role in vivo, as Egr-2 deficient mice had an increased susceptibility to EAE induction and MS patient CD4+ T cells showed both decreased Egr-2 expression and increased IL-17 and Batf expression, indicating the importance of Egr-2 expression for regulating proper Th17 responses.
4.2 Interferon Regulatory Factor 4
IRF4 is expressed in both B cells and T cells and expression of IRF4 is upreguated in response to T cell stimulation (Biswas et al. 2010). Cells deficient in IRF4 produce lower levels of IL-4 under conditions of nonpolarizing T cell stimulation and when polarized toward Th2 (Rengarajan et al. 2002a; Lohoff et al. 2002). These cells also show limited Gata3 upregulation in response to IL-4 stimulation, although Stat6 was activated, indicating normal IL-4 signaling. However, over-expression of Gata3 rescued IL-4 production in IRF4 deficient cells. In a study of the ability of IRF4 to drive expression from the Il4 promoter, it was shown that overexpression of IRF4 only induced expression of a reporter when coexpressed with NFATc2, while coexpression of IRF4, NFATc2, and c-Maf drove the greatest activation of reporter expression. As such, IRF4 acts as a component of a larger transcription factor complex to support IL-4 production and the importance of IRF4 in this complex demonstrated by the greatly diminished IL-4 production in IRF4-deficient cells (Rengarajan et al. 2002a; Lohoff et al. 2002). IRF4 also interacts with PU.1 and this interaction acts to block binding of IRF4 to target genes, including Il4 and Il10, to decrease IRF4-driven expression of these genes (Ahyi et al. 2009). While PU.1 is expressed in Th2 cells, there is heterogeneity in the expression of PU.1 across the Th2 population, allowing for differential regulation of IL-4 and IL-10 within the subset (Chang et al. 2005).
An additional CD4+ T cell subset has recently been defined as Th9, characterized by the production of IL-9, induced by stimulation with IL-4 and TGFβ (Chen et al. 2011b). Both IRF4 and PU.1 have been shown to play a role in Th9 differentiation. Stimulation under Th9 promoting conditions leads to the upregulation of IRF4 expression, which correlates with increased IL-9 production, likely facilitated by the direct binding of IRF4 to the Il9 promoter (Staudt et al. 2010). IRF4 deficient cells fail to produce IL-9 but produce elevated IFNγ after treatment with IL-4 and TGFβ. Similarly, PU.1 also binds directly to the Il9 promoter and PU.1 deficient cells show greatly decreased IL-9 production in response to IL-4 and TGFβ treatment, with no effect on Th17 or iTreg induction (Chang et al. 2010). Furthermore, overexpression of PU.1 resulted in increased IL-9 production in the absence of polarizing cytokines. Although both PU.1 and IRF4 are involved in Th9 differentiation, these two factors are activated independently, as IRF4 expression requires functional Stat6, indicating a reliance on IL-4 signaling for expression, while PU.1 is expressed independent of IL-4 signaling (Goswami et al. 2012). In the absence of functional Stat6, PU.1 bound the Il9 promoter normally, indicating no requirement for IRF4 in PU.1 binding, however, the decreased IL-9 expression in the absence of IRF4 demonstrates cooperation between these two factors in IL-9 expression and Th9 differentiation.
IRF4 is also involved in both Th17, as discussed above, and Treg differentiation. IRF4 deficient cells show limited differentiation into IL-17 producing cells under Th17 polarizing conditions with decreased RORγt expression. IRF4 deficient mice are resistant to EAE induction (Brustle et al. 2007). Furthermore, IRF4 deficient cells fail to suppress FoxP3 expression under Th17 polarizing conditions, indicating a role for IRF4 in the inhibition of Treg differentiation. However, generation of a Treg-specific deletion of IRF4 resulted in lymphoproliferative disease, with increased CD4+ T cell populations in lymph nodes with lymphocyte infiltration in pancreas, stomach, and lung (Zheng et al. 2009). Although these results point to a loss of Treg function, IRF4 deficient mice did not fail to generate FoxP3 expressing cells and actually had elevated Treg populations in lymph nodes. However, the CD4+ T cells in the lymph nodes were more activated and showed a specific increase in IL-4 production, indicating a loss of Th2 regulation with the loss of IRF4 in Treg cells. FoxP3 and IRF4 interact with each other and may bind together to regulate target genes. In the absence of IRF4, expression of 20 % of Treg-specific genes was decreased while 7 % was increased. In particular, Icos, Maf, Ccr8, and Il1rl1, all involved in Th2 differentiation, were decreased in IRF4 deficient Treg cells. However, it is unclear how the interaction between IRF4 and FoxP3 and the induction of Th2-specific genes in Treg cells acts to suppress Th2 responses.
IRF4 also plays a role in Tfh differentiation, as IRF4 deficient mice fail to form germinal centers and have significantly decreased GC B cell and Tfh cell populations 202. The transfer of wild type CD4+ T cells into IRF4 deficient mice rescued germinal center formation, indicating the requirement of IRF4 in T cells for proper Tfh differentiation and germinal center formation. During Tfh differentiation, IRF4 cooperates with Stat3 to regulate gene expression (Kwon et al. 2009). Examination of the binding sites of IRF4 and Stat3 showed a high degree of overlap between these two factors, where 76 % of Stat3 binding sites in Tfh cells were also bound by IRF4 and in the absence of IRF4, there was decreased or complete loss of Stat3 binding to these sites. Thus, although Stat3 plays an important role in Tfh differentiation, there remains a requirement for IRF4 for proper target binding.
4.3 Runx
The Runx family of transcription factors has also been shown to play a role in CD4+ T cell differentiation. Runx3 binds to the Ifng promoter to induce expression (Djuretic et al. 2007; Yagi et al. 2010) and overexpression of Runx3 increased IFNγ production (Djuretic et al. 2007; Kohu et al. 2009; Yagi et al. 2010) while Runx3 deficiency resulted in decreased IFNγ production (Djuretic et al. 2007). In addition, both Runx1 (Naoe et al. 2007; Kitoh et al. 2009) and Runx3 (Djuretic et al. 2007) bind to the Il4 silencer to inhibit IL-4 expression, as demonstrated by decreased IL-4 expression in response to Runx3 overexpression (Yagi et al. 2010). Runx1 is expressed predominately in naïve T cells while Runx3 is expressed in Th1 cells (Naoe et al. 2007) and these factors have been proposed to play a role in limiting IL-4 production in these subsets. In addition to the interaction with the Il4 silencer, Runx3 also binds to Gata3 (Yagi et al. 2010; Kohu et al. 2009) to inhibit Gata3-mediated gene expression (Kohu et al. 2009). While expression of excess Gata3 can prevent the Runx3-mediated induction of IFNγ expression, it does not prevent IL-4 silencing, indicating the importance of Runx3 in the inhibition of Th2 differentiation (Yagi et al. 2010).
The Il17 promoter contains two binding sites for Runx1 upstream of a RORγt binding site and mutation of either site decreased expression driven by the Il17 promoter (Zhang et al. 2008). Indeed, both RORγt and Runx1 bind directly to the Il17 promoter and Runx1 and RORγt also coprecipitate with each other, indicating a physical interaction in addition to closely spaced binding sites. While overex-pression of Runx1 increased the activity of the Il17 promoter, this was inhibited by mutation of the RORγt site, indicating that RORγt is required for full Runx1 activity. Similarly, while overexpression of both Runx1 and RORγt induced very strong Th17 differentiation under both neutral and Th17 polarizing conditions, overexpression of a dominant negative mutant of Runx1 with RORγt decreased IL-17 differentiation below that induced by RORγt overexpression alone, indicating a requirement for cooperation between these factors during Th17 differentiation. Furthermore, Runx1 expression levels correlate with RORγt expression levels, as overexpression of Runx1 resulted in increased RORγt expression while siRNA knockdown of Runx1 resulted in decreased RORγt. This indicates a possible role for Runx in the regulation of RORγt expression.
The Runx family also plays a role in FoxP3 expression as Runx1, Runx3, and the co-factor Cbfβ bind to the Foxp3 promoter. Deletion of either Runx1 or Cbfβ resulted in decreased FoxP3 expression (Kitoh et al. 2009; Bruno et al. 2009; Rudra et al. 2009; Klunker et al. 2009) while Runx3 overexpression increased FoxP3 expression (Bruno et al. 2009). Furthermore, Runx deficient mice showed signs of lymphoproliferation (Rudra et al. 2009) and these cells failed to prevent colitis when transferred into Scid mice (Kitoh et al. 2009), indicative of a loss of functional Treg cells. The effect of Runx deficiency on Treg populations appears to primarily depend on the loss of FoxP3 expression in the absence of Runx, as cotransfer of FoxP3-expressing cells from Cbfβ-deficient mice or control mice resulted in accelerated loss of FoxP3 expressing cells from Cbfb-deficient cells compared to control cells, while the remaining Cbfβ−/− FoxP3 cells retained suppressive function (Rudra et al. 2009).
Runx1 also associates with the nuclear orphan receptor Nr4a2 in Treg cells and the expression of both factors act synergistically in FoxP3 reporter assays (Sekiya et al. 2011). Nr4a2 binds directly to the Foxp3 promoter and enhancer in Treg cells and is capable of driving Foxp3 promoter and enhancer reporter constructs. Furthermore, loss of Nr4a2 resulted in increased susceptibility to inflammatory bowel disease and failed to be protective in a transfer model of colitis. Both Runx and Nr4a2 are involved in modifying the chromatin landscape of the Foxp3 gene. The deletion of Cbfβ in T regulatory cells resulted in the loss of H3K4me3 and H3K9me3 permissive chromatin modifications (Rudra et al. 2009), while over-expression of Nr4a2 resulted in increased histone 4 acetylation and increased levels of H3K4me3. While FoxP3 has also been shown to bind to its own promoter in a Runx-dependent manner, this binding is dependent on DNA demethylation (Zheng et al. 2010). However, neither Runx nor Nr4a2 removes DNA methylation at the Foxp3 gene, indicating that additional factor(s) are required for the initial demethylation to allow DNA binding at the Foxp3 promoter.
FoxP3 binds to Runx to alter the binding of Runx factors to target genes (Ono et al. 2007). In effector cells, Runx1 binds to and activates expression of Il2, which is suppressed in Treg cells. FoxP3 inhibits Runx1 activity as coexpression of Runx1 and Foxp3 decreased the Runx1-mediated expression of an Il2 promoter-driven reporter construct. Furthermore, the deletion of the region of FoxP3 required for Runx1 binding also resulted in Treg cells that failed to suppress effector cells, indicating the importance of the Runx1-FoxP3 interaction in regulating Treg function. Similarly, the coexpression of FoxP3 with Runx1 decreased the induction of IL-17 expression under Th17 polarizing conditions (Zhang et al. 2008). Thus, by binding to Runx1, FoxP3 inhibits the expression of a number of Runx1 target genes. The mechanism of repression may occur by blocking Runx1 binding to target genes, or conversely, by utilizing Runx1-depending binding to target FoxP3 to genes, resulting in direct repression.
4.4 Ets
Ets1 deficient mice have decreased cytokine production under Th1 polarizing conditions, along with decreased Stat4 levels (Grenningloh et al. 2005). Although the expression of Tbet during Th1 differentiation remained unchanged in Ets1 deficient cells, Tbet was upreguated to a lesser extent during Th1 recalls responses resulting in decreased IFNγ production. To determine if elevated Tbet expression could rescue the defect in recall IFNγ production, these cells were transduced to express Tbet. However, this failed to rescue IFNγ production, indicating a requirement for both Tbet and Ets1 for optimal IFNγ expression. Both Tbet and Ets1 bind to the Ifng promoter and coexpression of both factors acted synergis-tically to upregulate expression of IFNγ. In this way, Ets1 cooperation with Tbet is important to drive IFNγ production for proper Th1 differentiation and recall responses.
Ets1 also plays a role as a negative regulator of Th17 differentiation, as Ets1 deficient cells produce more IL-17, IL-22, IL-23R, and RORγt in response to Th17 polarizing conditions (Moisan et al. 2007). This effect was reversed by introduction of Ets1 after Th17 differentiation. The proposed mechanism of Th17 inhibition by Ets1 involves the IL-2-dependent downregulation of Th17 differentiation. While addition of IL-2 to wild type cells resulted in approximately 5-fold reduction in Il7 producing cells, Ets1-deficient cells treated with IL-2 resulted in only a 2-fold reduction in IL-17 producing cells. As these cells do not show any deficiency in Stat5 signaling, Ets1 was proposed to act downstream of Stat5 to inhibit Th17 differentiation in response to IL-2 treatment.
Ets-1 also binds to the Foxp3 promoter to drive FoxP3 expression (Polansky et al. 2010). In addition, Ets-1 has also shown a broader role in Treg differentiation by binding to many FoxP3 targets (Samstein et al. 2012). To identify accessible promoters in T regulatory cells, FoxP3− and FoxP3+ cells were subjected to DNase digest followed by high throughput sequencing to identify areas of accessible DNA. More than 99 % of DNase hypersensitivity sites identified were accessible in both subsets. Likewise, 98 % of FoxP3 binding sites were accessible in FoxP3− cells, indicating FoxP3-independent opening of chromatin. To look for other factors that might play a role in FoxP3 mediated transcription, FoxP3 binding sites were examined for other transcription factor binding sites and an enrichment of Ets and Runx binding sites was found, with an enrichment score higher than the FoxP3 binding site, indicating a possible role in the recruitment of FoxP3 to target genes. Indeed, binding of these factors to target genes preceded FoxP3 binding as Ets and Runx were bound to target regions in both FoxP3+ and FoxP3− cells. Since the DNase hypersensitivity of these sites was similar before and after FoxP3 expression, it was hypothesized that another factor may bind in place of FoxP3, thus protecting DNA from digest. In FoxP3− cells, Foxo1 was shown to bind to FoxP3 binding sites, with a subsequent loss of Foxo1 binding in FoxP3+ cells. The displacement of Foxo1 by FoxP3 corresponded to genes that were downregulated in FoxP3 cells. Those few regions of FoxP3 binding that were not accessible in FoxP3− cells were found to have increased accessibility after T cell stimulation and expression from these regions was decreased in calcineurin deficient cells, indicating that FoxP3 relies on other transcription factors to regulate chromatin accessibility and binds to regions already accessible. Interestingly, in addition to binding to FoxP3 target genes, Foxo proteins also play a role in the regulation of FoxP3 expression as both Foxo1 and Foxo3 were shown to bind directly to the Foxp3 promoter to regulate FoxP3 expression, with loss of either resulting in decreased Treg populations (Harada et al. 2010; Ouyang et al. 2010).
4.5 Ikaros
The Ikaros family of zinc finger DNA binding proteins is involved in Th2, Th17, and Treg differentiation. Ikaros appears to play a role in the inhibition of Th1 responses during Th2 differentiation as Ikaros deficient cells cultured under Th2 conditions produced increased levels of IFNγ (Thomas et al. 2010; Umetsu and Winandy 2009). Ikaros binds directly to the Tbet and Ifng promoters (Thomas et al. 2010; Quirion et al. 2009) to repress the expression of these genes, as expression of a dominant negative mutant of Ikaros resulted in increased expression of both Tbet and IFNγ (Thomas et al. 2010). Furthermore, Ikaros also binds directly to the Il4 promoter, potentially activating IL-4 expression, as Ikaros deficient cells produced less IL-4 under both nonpolarizing T cell stimulation (Thomas et al. 2010; Umetsu and Winandy 2009) and during Th2 differentiation (Quirion et al. 2009), although with time IL-4 expression returned to normal levels (Thomas et al. 2010). Ikaros deficient cells show decreased histone acetylation at the Th2 locus both in naïve cells and under Th2 polarizing conditions, indicating that Ikaros may function by altering chromatin accessibility of the Th2 locus during Th2 differentiation (Quirion et al. 2009). Finally, Ikaros deficient cells primed under Th2 inducing conditions also show altered expression patterns of multiple transcription factors involved in Th differentiation, including decreased Gata3 and c-Maf expression, both involved in Th2 differentiation, while Tbet and Stat1, both involved in Th1 differentiation, were upreguated. This suggests a broad role for Ikaros in controlling the expression of multiple genes to strengthen Th2 commitment while inhibiting Th1 differentiation, likely by interacting with unique factors to either activate or repress expression of target genes.
A second member of the Ikaros family, Aiolos, is specifically expressed in Th17 cells (Quintana et al. 2012). In Aiolos deficient cells, there was decreased expression of the Th17 cytokines IL-17a, IL-17f, and IL-21 along with decreased expression of the transcription factors Maf and Ahr. However, Aiolos overex-pression was not sufficient to induce Th17 differentiation under non-Th17 polarizing conditions. Ahr and Stat3, both of which bind to the Aiolos promoter, induce Aiolos expression in Th17 cells as overexpression of Ahr or a constitutively active Stat3 increased expression of an Aiolos expression construct. Similar to Ikaros deficient cells, Aiolos deficient cells also show an altered pattern of chromatin modifications, with fewer restrictive histone modifications and more permissive modifications at the IL-2 promoter, indicating a role for Aiolos in modifying the chromatin landscape of target genes. The importance of Aiolos in the regulation of CD4+ T cell differentiation is demonstrated in a transfer model of colitis where mice receiving Aiolos deficient cells developed a more severe wasting disease compared to control mice, due to increased IFNγ+, IL-2+ and IFNγ+IL17+ cells.
The Ikaros family members, Helios and Eos, are expressed selectively in T regulatory cells. Helios is highly expressed by tTregs and thus may be a marker for these cells (Thornton et al. 2010). However, since Helios expression is detected in inducible Tregs under certain conditions (Gottschalk et al. 2012), Helios expression may be associated with the maturity of Tregs. Eos interacts with FoxP3 and knockdown of Eos in cells overexpressing FoxP3 resulted in loss of FoxP3-mediated IL-2 suppression, indicating a requirement of Eos in FoxP3-mediated gene repression (Pan et al. 2009). Eos carries out this suppressive function by interacting with the transcriptional corepressor C-terminal binding protein-1 (CtBP1), which is found in a complex with FoxP3 and Eos, where depletion of Eos prevented association of CtBP1 with FoxP3. The loss of Eos also resulted in changes in histone acetylation and methylation, as well as DNA methylation at the Il2 promoter, resulting in a pattern that more closely resembled that found in effector cells. A similar result was found with the knockdown of CtBP1, indicating the importance of the recruitment of CtBP1 by Eos to the FoxP3 containing complex. The role of Eos was not limited to the Il2 promoter, as analysis of genome-wide expression changes in Eos deficient cells showed that many genes normally suppressed by FoxP3 were not suppressed in Eos deficient cells while genes normally upreguated by FoxP3 were unchanged in the absence of Eos. Thus, this interaction of Eos and FoxP3 is specifically required for repression of gene expression.
5 Transcription Factor Networks
Given the large number of transcription factors involved in the differentiation of CD4+ T cell subsets, studies have started to look at transcription factor networks in the regulation of differentiation. To examine the interaction of transcription factors during Th17 differentiation, ChIP-Seq was performed with antibodies against Stat3, IRF4, Batf, c-Maf, RORγt, and p300 in cells undergoing Th17 differentiation (Ciofani et al. 2012). There was a high degree of co-localization of these factors at genes expressed in Th17 cells, indicating cooperation during Th17 differentiation. By grouping transcription factor binding sites that clustered in close proximity with each other, it was found that regions where all five of these factors bound showed the highest fold change in expression during Th17 differentiation. Of these factors, IRF4 and Batf were also bound to these regions in nonpolarized cells, leading to speculation that they may be required for the formation of the larger five factor containing complex. Indeed, IRF4 and Batf are codependent, where loss of one prevents binding of the other and this correlates with decreased chromatin accessibility at regions where all five factors bind.
To understand the unique function of each transcription factor during Th17 differentiation, RNA-Seq was performed on control and transcription factor knockout mice to look at genome-wide expression changes. By comparing these results, a network was created to map the interactions of these factors with each other and with target genes during Th17 differentiation. This analysis demonstrated that IRF4, Batf, and Stat3 form a positive feedback loop to strengthen their expression during Th17 differentiation while c-Maf forms a negative feedback loop, being activated by Batf, Stat3, and IRF4, but acting to repress Batf. By comparing the targets identified by ChIP-Seq analysis with the changes in expression profile in knockout mice, it is also possible to identify novel factors involved in subset differentiation. In this study, Etv6, Ncoa2, Skil, Trib3, and the AP-1 family member Fosl2, are each predicted to be involved in Th17 differentiation, although further study is needed to understand how they fit into the defined network of transcription factors.
A second genome-wide analysis of Th17 cells made use of microarray analysis performed at three time points during Th17 differentiation, which allowed for the grouping of genes based on the timing of upregulation after Th17 differentiation as well as the length of upregulation (Yosef et al. 2013). Genes upreguated at the early time point included the transcription factors Batf, IRF4, Stat3, Stat1, and IRF1, which showed sustained expression during Th17 differentiation, while RORγt and Ahr were induced at the intermediate time point. These results correspond well with the positive feedback loop between Batf, IRF4, and Stat3 and the requirement for Batf and IRF4 for the binding of the other transcription factors. Together, these results demonstrate the major transcriptional regulators during Th17 differentiation and allow identification of additional regulators.
Many transcription factors besides the ones described above have been identified for the differentiation of each CD4+ T cell subset including Hlx (Mullen et al. 2002; Martins et al. 2005; Mikhalkevich et al. 2006), Gfi-1 (Zhu et al. 2002, 2006, 2009), Dec2 (Yang et al. 2009), Hif1a (Dang et al. 2011), Ahr (Veldhoen et al. 2008; Quintana et al. 2008; Kimura et al. 2008), Tcf7 (Yu et al. 2009b, 2011; van Loosdregt et al. 2013) and Notch (Shin et al. 2006; Bailis et al. 2013; Minter et al. 2005; Fang et al. 2007; Amsen et al. 2007; Mukherjee et al. 2009; Samon et al. 2008; Auderset et al. 2013; Elyaman et al. 2012). However, their contributions to T cell differentiation in the context of the transcriptional network are poorly understood. Figure 3 shows a schematic outline of the transcription factor networks as currently known for Th1, Th2, and Th17 cells. In order to gain a more thorough understanding of the transcriptional networks controlling the differentiation of all CD4+ T subsets, similar large-scale ChIP-Seq analyses of known transcription factors coupled with knockout studies should be performed for each subset. Based on the studies of the Th17 differentiation pathway, it is likely that these results will also show widespread cooperation between a number of transcription factors to induce subset-specific transcription programs.
Fig. 3.
Schematic summary of transcription factor networks involved in CD4+ T cell differentiation. The differentiation of each CD4+ T cell subset is controlled not only by Stats and master regulators but also by secondary transcription factors that activate subset-specific gene expression, including both cytokines and master regulators. There are many cooperative interactions between the transcription factors expressed within a given subset to strengthen gene expression and inhibition between factors to either regulate gene expression within a subset or to inhibit the differentiation of other subsets
6 Long Intergenic Noncoding RNA
The role of long intergenic noncoding RNA (lincRNA) in T cell differentiation has become a subject of interest recently as lincRNA have been shown to play a role in many cellular functions, including chromatin remodeling, as well as both tran-scriptional repression and activation (Pagani et al. 2013). Mice overexpressing the NeST lincRNA show increased IFNγ expression and while they show increased resistance to Salmonella, they have increased susceptibility to Theiler virus (Gomez et al. 2013). NeST was found predominately in the nucleus of these overexpressing cells and was shown to interact with WDR5, a core subunit of the MLL1–4 and SET1A/1B complexes, which act to methylate H3K4 residues. Indeed, NeST overexpressing mice show increased H3K4me3 marks at the Ifng gene. Thus, by increasing permissive histone methylation, NeST increases IFNγ expression. The expression of lincRNA may be subset specific as NeST showed higher expression in Th1 cells compared to Th2 cells (Collier et al. 2012). This subset-specific expression was driven by the Th1 transcription factors Stat4 and Tbet, as mice deficient in either factor showed decreased NeST expression. A large-scale study of lincRNA expression during T cell development and differentiation recently identified over 1500 lincRNA clusters and found that almost 50 % of those expressed in CD4+ T cells showed subset-specific expression (Hu et al. 2013). A number of these lin-cRNA were bound by subset-specific transcription factors Stat4, Tbet, Stat6, and Gata3, indicating that these factors control the expression not only of protein coding genes but also noncoding regions to regulate T cell differentiation. Stat4 showed stronger binding at lincRNA genes preferentially expressed in Th1 cells and these targets were downregulated in Stat4 deficient cells. Similarly, Stat6 also showed higher binding at lincRNA genes preferentially expressed in Th2 cells and these targets were downregulated in Stat6 deficient cells. Both Tbet and Gata3 bind to subset-specific lincRNA clusters, but the loss of either transcription factor resulted in both upregulation and downregulation of target lincRNA genes, indicating that Tbet and Gata3 both activate and repress lincRNA expression during Th1 or Th2 differentiation, respectively. These results demonstrate that subset-specific transcription factors drive the expression of noncoding genes. More work is required to understand the function lincRNA in the regulation of T cell differentiation.
7 Conclusion
Early work on transcriptional regulation of T cells has focused on the two-factor model of differentiation, in which cytokine stimulation activates the Stat family of transcription factors, which in turn activate the master regulators of CD4+ T cell differentiation. However, it has become clear that these factors do not act alone to induce differentiation but instead interact with a number of other transcription factors for optimal CD4+ T cell differentiation and function. The use of large-scale, genome-wide studies is beginning to shed light on when and how transcription factors are up- or downregulated during development as well as providing information on the interactions between transcription factors at multiple target genes during differentiation. In this way, a picture of the network of transcription factors is beginning to emerge, although more work is needed to generate and validate transcriptional networks for all CD4+ T cell subsets during differentiation. The essential role of transcriptional regulation in maintaining proper CD4+ T cell function is demonstrated by mutations of various transcription factors and the resulting diseases. Mutations of Foxp3 results in the development IPEX autoimmunity, while mutation of STAT3 leads to hyper IgE syndrome, characterized by recurrent staphylococcal skin abscesses, eczema, and pulmonary infections (Yong et al. 2012) and MS patients have been reported to express decreased levels of Egr2, resulting in increased Th17 responses (Miao et al. 2013). These disorders illustrate the importance of transcriptional regulation during CD4+ T cell differentiation and highlight the need for a more thorough understanding of these transcriptional networks to provide insight into the balance of protective immunity versus autoimmunity in humans.
Acknowledgment
The work is supported by the Division of Intramural Research, NIAID, National Institutes of Health, USA.
Footnotes
The authors declare no competing financial interests.
Contributor Information
Darah Christie, Email: darah.christie@nih.gov.
Jinfang Zhu, Email: jfzhu@niaid.nih.gov.
References
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3(6):549–557. doi: 10.1038/ni794. [DOI] [PubMed] [Google Scholar]
- Agarwal S, Avni O, Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity. 2000;12(6):643–652. doi: 10.1016/s1074-7613(00)80215-0. [DOI] [PubMed] [Google Scholar]
- Aggarwal S, Ghilardi N, Xie MH, de Sauvage FJ, Gurney AL. Interleukin-23 promotes a distinct CD4 T cell activation state characterized by the production of interleukin-17. J Biol Chem. 2003;278(3):1910–1914. doi: 10.1074/jbc.M207577200. [DOI] [PubMed] [Google Scholar]
- Ahyi AN, Chang HC, Dent AL, Nutt SL, Kaplan MH. IFN regulatory factor 4 regulates the expression of a subset of Th2 cytokines. J Immun. 2009;183(3):1598–1606. doi: 10.4049/jimmunol.0803302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsen D, Antov A, Jankovic D, Sher A, Radtke F, Souabni A, Busslinger M, McCright B, Gridley T, Flavell RA. Direct regulation of Gata3 expression determines the T helper differentiation potential of Notch. Immunity. 2007;27(1):89–99. doi: 10.1016/j.immuni.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronica MA, Mora AL, Mitchell DB, Finn PW, Johnson JE, Sheller JR, Boothby MR. Preferential role for NF-kappa B/Rel signaling in the type 1 but not type 2 T cell-dependent immune response in vivo. J Immun. 1999;163(9):5116–5124. [PubMed] [Google Scholar]
- Auderset F, Schuster S, Fasnacht N, Coutaz M, Charmoy M, Koch U, Favre S, Wilson A, Trottein F, Alexander J, Luther SA, MacDonald HR, Radtke F, Tacchini-Cottier F. Notch signaling regulates follicular helper T cell differentiation. J Immun. 2013;191(5):2344–2350. doi: 10.4049/jimmunol.1300643. [DOI] [PubMed] [Google Scholar]
- Bailis W, Yashiro-Ohtani Y, Fang TC, Hatton RD, Weaver CT, Artis D, Pear WS. Notch simultaneously orchestrates multiple helper T cell programs independently of cytokine signals. Immunity. 2013;39(1):148–159. doi: 10.1016/j.immuni.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36(5):847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bending D, De la Pena H, Veldhoen M, Phillips JM, Uyttenhove C, Stockinger B, Cooke A. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest. 2009;119(3):565–572. doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettini ML, Pan F, Bettini M, Finkelstein D, Rehg JE, Floess S, Bell BD, Ziegler SF, Huehn J, Pardoll DM, Vignali DA. Loss of epigenetic modification driven by the Foxp3 transcription factor leads to regulatory T cell insufficiency. Immunity. 2012;36(5):717–730. doi: 10.1016/j.immuni.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas PS, Bhagat G, Pernis AB. IRF4 and its regulators: evolving insights into the pathogenesis of inflammatory arthritis? Immunol Rev. 2010;233(1):79–96. doi: 10.1111/j.0105-2896.2009.00864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogdon JL, Leitenberg D, Bottomly K. The potency of TCR signaling differentially regulates NFATc/p activity and early IL-4 transcription in naive CD4+ T cells. J Immunol. 2002;168(8):3825–3832. doi: 10.4049/jimmunol.168.8.3825. [DOI] [PubMed] [Google Scholar]
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- Bruno L, Mazzarella L, Hoogenkamp M, Hertweck A, Cobb BS, Sauer S, Hadjur S, Leleu M, Naoe Y, Telfer JC, Bonifer C, Taniuchi I, Fisher AG, Merkenschlager M. Runx proteins regulate Foxp3 expression. J Exp Med. 2009;206(11):2329–2337. doi: 10.1084/jem.20090226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8(9):958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- Burchill MA, Yang J, Vang KB, Moon JJ, Chu HH, Lio CW, Vegoe AL, Hsieh CS, Jenkins MK, Farrar MA. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28(1):112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178(1):280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- Burgler S, Mantel PY, Bassin C, Ouaked N, Akdis CA, Schmidt-Weber CB. RORC2 is involved in T cell polarization through interaction with the FOXP3 promoter. J Immunol. 2010;184(11):6161–6169. doi: 10.4049/jimmunol.0903243. [DOI] [PubMed] [Google Scholar]
- Cai G, Radzanowski T, Villegas EN, Kastelein R, Hunter CA. Identification of STAT4-dependent and independent mechanisms of resistance to Toxoplasma gondii. J Immunol. 2000;165(5):2619–2627. doi: 10.4049/jimmunol.165.5.2619. [DOI] [PubMed] [Google Scholar]
- Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L, Nguyen ET, Robertson MJ, Perumal NB, Tepper RS, Nutt SL, Kaplan MH. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010;11(6):527–534. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, Klemsz MJ, Kaplan MH. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22(6):693–703. doi: 10.1016/j.immuni.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Chang S, Aune TM. Dynamic changes in histone-methylation ‘marks’ across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat Immunol. 2007;8(7):723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- Chang S, Collins PL, Aune TM. T-bet dependent removal of Sin3A–histone deacetylase complexes at the Ifng locus drives Th1 differentiation. J Immun. 2008;181(12):8372–8381. doi: 10.4049/jimmunol.181.12.8372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326(5955):986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Lee SM, Gao B, Shannon S, Zhu Z, Fang D. c-Abl-mediated tyrosine phosphorylation of the T-bet DNA-binding domain regulates CD4+ T-cell differentiation and allergic lung inflammation. Mol Cell Biol. 2011a;31(16):3445–3456. doi: 10.1128/MCB.05383-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Hardy K, Pagler E, Ma L, Lee S, Gerondakis S, Daley S, Shannon MF. The NF-kappaB transcription factor c-Rel is required for Th17 effector cell development in experimental autoimmune encephalomyelitis. J Immunol. 2011b;187(9):4483–4491. doi: 10.4049/jimmunol.1101757. [DOI] [PubMed] [Google Scholar]
- Choi YS, Eto D, Yang JA, Lao C, Crotty S. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J Immunol. 2013;190(7):3049–3053. doi: 10.4049/jimmunol.1203032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol. 2004;173(1):68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, Fan HM, Liu ZM, Neelapu SS, Dong C. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17(8):983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, Newberry KM, Meadows S, Greenfield A, Yang Y, Jain P, Kirigin FK, Birchmeier C, Wagner EF, Murphy KM, Myers RM, Bonneau R, Littman DR. A validated regulatory network for Th17 cell specification. Cell. 2012;151(2):289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol. 2012;189(5):2084–2088. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constant SL, Bottomly K. Induction of Th1 and Th2 CD4+ T cell responses: the alternative approaches. Annu Rev Immunol. 1997;15:297–322. doi: 10.1146/annurev.immunol.15.1.297. [DOI] [PubMed] [Google Scholar]
- Corn RA, Aronica MA, Zhang F, Tong Y, Stanley SA, Kim SR, Stephenson L, Enerson B, McCarthy S, Mora A, Boothby M. T cell-intrinsic requirement for NF-kappa B induction in postdifferentiation IFN-gamma production and clonal expansion in a Th1 response. J Immunol. 2003;171(4):1816–1824. doi: 10.4049/jimmunol.171.4.1816. [DOI] [PubMed] [Google Scholar]
- Corn RA, Hunter C, Liou HC, Siebenlist U, Boothby MR. Opposing roles for RelB and Bcl-3 in regulation of T-box expressed in T cells, GATA-3, and Th effector differentiation. J Immunol. 2005;175(4):2102–2110. doi: 10.4049/jimmunol.175.4.2102. [DOI] [PubMed] [Google Scholar]
- Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, Hu-Li J, Zhu J, Paul WE. Interleukin 2 plays a central role in Th2 differentiation. Proc Natl Acad Sci U.S.A. 2004;101(11):3880–3885. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11(2):114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang EV, Barbi J, Yang HY, Jinasena D, Yu H, Zheng Y, Bordman Z, Fu J, Kim Y, Yen HR, Luo W, Zeller K, Shimoda L, Topalian SL, Semenza GL, Dang CV, Pardoll DM, Pan F. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146(5):772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A. A critical role for NF-kappa B in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat Immunol. 2001;2(1):45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- Dienz O, Eaton SM, Bond JP, Neveu W, Moquin D, Noubade R, Briso EM, Charland C, Leonard WJ, Ciliberto G, Teuscher C, Haynes L, Rincon M. The induction of antibody production by IL-6 is indirectly mediated by IL-21 produced by CD4+ T cells. J Exp Med. 2009;206(1):69–78. doi: 10.1084/jem.20081571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8(2):145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat Immunol. 2009;10(8):857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun HW, Kanno Y, Powrie F, O’Shea JJ. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32(5):605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddahri F, Denanglaire S, Bureau F, Spolski R, Leonard WJ, Leo O, Andris F. Interleukin-6/STAT3 signaling regulates the ability of naive T cells to acquire B-cell help capacities. Blood. 2009;113(11):2426–2433. doi: 10.1182/blood-2008-04-154682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elyaman W, Bassil R, Bradshaw EM, Orent W, Lahoud Y, Zhu B, Radtke F, Yagita H, Khoury SJ. Notch receptors and Smad3 signaling cooperate in the induction of interleukin-9-producing T cells. Immunity. 2012;36(4):623–634. doi: 10.1016/j.immuni.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27(1):100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelman FD, Morris SC, Orekhova T, Mori M, Donaldson D, Reiner SL, Reilly NL, Schopf L, Urban JF., Jr Stat6 regulation of in vivo IL-4 responses. J Immunol. 2000;164(5):2303–2310. doi: 10.4049/jimmunol.164.5.2303. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+ CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445(7129):771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, Grainger JR, Chen Q, Kanno Y, Watford WT, Sun HW, Eberl G, Shevach EM, Belkaid Y, Cua DJ, Chen W, O’Shea JJ. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467(7318):967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner CJ, Rutz S, Hackney J, Nurieva R, Escalante CR, Ouyang W, Littman DR, Murphy KM, Singh H. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338(6109):975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocke AR, Cravens PD, Ben LH, Hussain RZ, Northrop SC, Racke MK, Lovett-Racke AE. T-bet regulates the fate of Th1 and Th17 lymphocytes in autoimmunity. J Immunol. 2007;178(3):1341–1348. doi: 10.4049/jimmunol.178.3.1341. [DOI] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152(4):743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Rodriguez J, Sahu N, Handon R, Davidson TS, Anderson SM, Kirby MR, August A, Schwartzberg PL. Differential expression of interleukin-17A and −17F is coupled to T cell receptor signaling via inducible T cell kinase. Immunity. 2009;31(4):587–597. doi: 10.1016/j.immuni.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami R, Jabeen R, Yagi R, Pham D, Zhu J, Goenka S, Kaplan MH. STAT6-dependent regulation of Th9 development. J Immunol. 2012;188(3):968–975. doi: 10.4049/jimmunol.1102840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk RA, Corse E, Allison JP. Expression of Helios in peripherally induced Foxp3+ regulatory T cells. J Immunol. 2012;188(3):976–980. doi: 10.4049/jimmunol.1102964. [DOI] [PubMed] [Google Scholar]
- Grenningloh R, Kang BY, Ho IC. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J Exp Med. 2005;201(4):615–626. doi: 10.1084/jem.20041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Wei G, Zhu J, Liao W, Leonard WJ, Zhao K, Paul W. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proc Natl Acad Sci U.S.A. 2009;106(32):13463–13468. doi: 10.1073/pnas.0906988106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Harada Y, Elly C, Ying G, Paik JH, DePinho RA, Liu YC. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J Exp Med. 2010;207(7):1381–1391. doi: 10.1084/jem.20100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TJ, Grosso JF, Yen HR, Xin H, Kortylewski M, Albesiano E, Hipkiss EL, Getnet D, Goldberg MV, Maris CH, Housseau F, Yu H, Pardoll DM, Drake CG. Cutting edge: an in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol. 2007;179(7):4313–4317. doi: 10.4049/jimmunol.179.7.4313. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26(3):203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegazy AN, Peine M, Helmstetter C, Panse I, Frohlich A, Bergthaler A, Flatz L, Pinschewer DD, Radbruch A, Lohning M. Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity. 2010;32(1):116–128. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Hermann-Kleiter N, Meisel M, Fresser F, Thuille N, Muller M, Roth L, Katopodis A, Baier G. Nuclear orphan receptor NR2F6 directly antagonizes NFAT and RORgammat binding to the Il17a promoter. J Autoimmun. 2012;39(4):428–440. doi: 10.1016/j.jaut.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, Garefalaki A, Potocnik AJ, Stockinger B. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol. 2011;12(3):255–263. doi: 10.1038/ni.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Turner JE, Villa M, Duarte JH, Demengeot J, Steinmetz OM, Stockinger B. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. 2013;14(4):372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T, Zhang S, Schmidt N, Yu Q, Ramchandani S, Xu X, Naeger LK, Sun YL, Kaplan MH. Distinct requirements for the naturally occurring splice forms Stat4alpha and Stat4beta in IL-12 responses. EMBO J. 2003;22(16):4237–4248. doi: 10.1093/emboj/cdg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299(5609):1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- Hsieh CS, Heimberger AB, Gold JS, O’Garra A, Murphy KM. Differential regulation of T helper phenotype development by interleukins 4 and 10 in an alpha beta T-cell-receptor transgenic system. Proc Natl Acad Sci U.S.A. 1992;89(13):6065–6069. doi: 10.1073/pnas.89.13.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013;14(11):1190–1198. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hural JA, Kwan M, Henkel G, Hock MB, Brown MA. An intron transcriptional enhancer element regulates IL-4 gene locus accessibility in mast cells. J Immunol. 2000;165(6):3239–3249. doi: 10.4049/jimmunol.165.6.3239. [DOI] [PubMed] [Google Scholar]
- Hwang ES, Hong JH, Glimcher LH. IL-2 production in developing Th1 cells is regulated by heterodimerization of RelA and T-bet and requires T-bet serine residue 508. J Exp Med. 2005a;202(9):1289–1300. doi: 10.1084/jem.20051044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005b;307(5708):430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- Ichiyama K, Yoshida H, Wakabayashi Y, Chinen T, Saeki K, Nakaya M, Takaesu G, Hori S, Yoshimura A, Kobayashi T. Foxp3 inhibits RORgammat-mediated IL-17A mRNA transcription through direct interaction with RORgammat. J Biol Chem. 2008;283(25):17003–17008. doi: 10.1074/jbc.M801286200. [DOI] [PubMed] [Google Scholar]
- Isomura I, Palmer S, Grumont RJ, Bunting K, Hoyne G, Wilkinson N, Banerjee A, Proietto A, Gugasyan R, Wu L, McNally A, Steptoe RJ, Thomas R, Shannon MF, Gerondakis S. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J Exp Med. 2009;206(13):3001–3014. doi: 10.1084/jem.20091411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126(6):1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Jabeen R, Kaplan MH. The symphony of the ninth: the development and function of Th9 cells. Curr Opin Immunol. 2012;24(3):303–307. doi: 10.1016/j.coi.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D, Kullberg MC, Noben-Trauth N, Caspar P, Paul WE, Sher A. Single cell analysis reveals that IL-4 receptor/Stat6 signaling is not required for the in vivo or in vitro development of CD4+ lymphocytes with a Th2 cytokine profile. J Immunol. 2000;164(6):3047–3055. doi: 10.4049/jimmunol.164.6.3047. [DOI] [PubMed] [Google Scholar]
- Jenner RG, Townsend MJ, Jackson I, Sun K, Bouwman RD, Young RA, Glimcher LH, Lord GM. The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proc Natl Acad Sci U.S.A. 2009;106(42):17876–17881. doi: 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209(2):243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325(5943):1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorritsma PJ, Brogdon JL, Bottomly K. Role of TCR-induced extracellular signal-regulated kinase activation in the regulation of early IL-4 expression in naive CD4+ T cells. J Immunol. 2003;170(5):2427–2434. doi: 10.4049/jimmunol.170.5.2427. [DOI] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami S, Nakajima H, Suto A, Hirose K, Suzuki K, Morita S, Kato I, Saito Y, Kitamura T, Iwamoto I. Stat5a regulates T helper cell differentiation by several distinct mechanisms. Blood. 2001;97(8):2358–2365. doi: 10.1182/blood.v97.8.2358. [DOI] [PubMed] [Google Scholar]
- Kanhere A, Hertweck A, Bhatia U, Gokmen MR, Perucha E, Jackson I, Lord GM, Jenner RG. T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nat Commun. 2012;3:1268. doi: 10.1038/ncomms2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996a;4(3):313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996b;382(6587):174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- Kaplan MH, Wurster AL, Grusby MJ. A signal transducer and activator of transcription (Stat)4-independent pathway for the development of T helper type 1 cells. J Exp Med. 1998;188(6):1191–1196. doi: 10.1084/jem.188.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani A, Garcia-Cozar FJ, Habermann I, Laforsch S, Aebischer T, Ehninger G, Rao A. Regulation of interferon-gamma gene expression by nuclear factor of activated T cells. Blood. 2001;98(5):1480–1488. doi: 10.1182/blood.v98.5.1480. [DOI] [PubMed] [Google Scholar]
- Kiani A, Viola JP, Lichtman AH, Rao A. Down-regulation of IL-4 gene transcription and control of Th2 cell differentiation by a mechanism involving NFAT1. Immunity. 1997;7(6):849–860. doi: 10.1016/s1074-7613(00)80403-3. [DOI] [PubMed] [Google Scholar]
- Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci U.S.A. 2008;105(28):9721–9726. doi: 10.1073/pnas.0804231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, Yaguchi H, Kitabayashi I, Tsukada T, Nomura T, Miyachi Y, Taniuchi I, Sakaguchi S. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31(4):609–620. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Klunker S, Chong MM, Mantel PY, Palomares O, Bassin C, Ziegler M, Ruckert B, Meiler F, Akdis M, Littman DR, Akdis CA. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J Exp Med. 2009;206(12):2701–2715. doi: 10.1084/jem.20090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohu K, Ohmori H, Wong WF, Onda D, Wakoh T, Kon S, Yamashita M, Nakayama T, Kubo M, Satake M. The Runx3 transcription factor augments Th1 and down-modulates Th2 phenotypes by interacting with and attenuating GATA3. J Immunol. 2009;183(12):7817–7824. doi: 10.4049/jimmunol.0802527. [DOI] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, Oukka M, Kuchroo VK. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448(7152):484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- Kurata H, Lee HJ, O’Garra A, Arai N. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity. 1999;11(6):677–688. doi: 10.1016/s1074-7613(00)80142-9. [DOI] [PubMed] [Google Scholar]
- Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, Carotta S, Donovan CE, Goldman ML, Tailor P, Ozato K, Levy DE, Nutt SL, Calame K, Leonard WJ. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 2009;31(6):941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W, Yu M, Huang MN, Okoye F, Keegan AD, Farber DL. Transcriptional control of rapid recall by memory CD4 T cells. J Immunol. 2011;187(1):133–140. doi: 10.4049/jimmunol.1002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, Shevach EM, O’Shea JJ. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26(3):371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2011;12(1):96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gros G, Ben-Sasson SZ, Seder R, Finkelman FD, Paul WE. Generation of interleukin 4 (IL-4)-producing cells in vivo and in vitro: IL-2 and IL-4 are required for in vitro generation of IL-4-producing cells. J Exp Med. 1990;172(3):921–929. doi: 10.1084/jem.172.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DU, Rao A. Molecular analysis of a locus control region in the T helper 2 cytokine gene cluster: a target for STAT6 but not GATA3. Proc Natl Acad Sci U.S.A. 2004;101(45):16010–16015. doi: 10.1073/pnas.0407031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O’Garra A, Arai N. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med. 2000;192(1):105–115. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Silva DG, Martin JL, Pratama A, Hu X, Chang PP, Walters G, Vinuesa CG. Interferon-gamma excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity. 2012a;37(5):880–892. doi: 10.1016/j.immuni.2012.10.010. [DOI] [PubMed] [Google Scholar]
- Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, Sobel RA, Regev A, Kuchroo VK. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012b;13(10):991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30(1):92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- Lexberg MH, Taubner A, Albrecht I, Lepenies I, Richter A, Kamradt T, Radbruch A, Chang HD. IFN-gamma and IL-12 synergize to convert in vivo generated Th17 into Th1/Th17 cells. Eur J Immunol. 2010;40(11):3017–3027. doi: 10.1002/eji.201040539. [DOI] [PubMed] [Google Scholar]
- Li B, Samanta A, Song X, Iacono KT, Bembas K, Tao R, Basu S, Riley JL, Hancock WW, Shen Y, Saouaf SJ, Greene MI. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci U.S.A. 2007a;104(11):4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Tournier C, Davis RJ, Flavell RA. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18(2):420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-beta1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007b;26(5):579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, Leonard WJ. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490(7421):543–546. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, Zhao K, Leonard WJ. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat Immunol. 2008;9(11):1288–1296. doi: 10.1038/ni.1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighvani AA, Frucht DM, Jankovic D, Yamane H, Aliberti J, Hissong BD, Nguyen BV, Gadina M, Sher A, Paul WE, O’Shea JJ. T-bet is rapidly induced by interferon-gamma in lymphoid and myeloid cells. Proc Natl Acad Sci U.S.A. 2001;98(26):15137–15142. doi: 10.1073/pnas.261570598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman MA, Beaton L, Yu D, Ramiscal RR, Srivastava M, Hogan JJ, Verma NK, Smyth MJ, Rigby RJ, Vinuesa CG. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J Exp Med. 2010;207(2):353–363. doi: 10.1084/jem.20091738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, Fagarasan S, Liston A, Smith KG, Vinuesa CG. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17(8):975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XK, Lin X, Gaffen SL. Crucial role for nuclear factor of activated T cells in T cell receptor-mediated regulation of human interleukin-17. J Biol Chem. 2004;279(50):52762–52771. doi: 10.1074/jbc.M405764200. [DOI] [PubMed] [Google Scholar]
- Lochner M, Peduto L, Cherrier M, Sawa S, Langa F, Varona R, Riethmacher D, Si-Tahar M, Di Santo JP, Eberl G. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J Exp Med. 2008;205(6):1381–1393. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohoff M, Mittrucker HW, Prechtl S, Bischof S, Sommer F, Kock S, Ferrick DA, Duncan GS, Gessner A, Mak TW. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc Natl Acad Sci U.S.A. 2002;99(18):11808–11812. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Slaiby AM, Hagymasi AT, Mihalyo MA, Lichtler AC, Reiner SL, Adler AJ. T-bet down-modulation in tolerized Th1 effector CD4 cells confers a TCR-distal signaling defect that selectively impairs IFN-gamma expression. J Immunology. 2006;176(2):1036–1045. doi: 10.4049/jimmunol.176.2.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Racke AE, Rocchini AE, Choy J, Northrop SC, Hussain RZ, Ratts RB, Sikder D, Racke MK. Silencing T-bet defines a critical role in the differentiation of autoreactive T lymphocytes. Immunity. 2004;21(5):719–731. doi: 10.1016/j.immuni.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Lund RJ, Chen Z, Scheinin J, Lahesmaa R. Early target genes of IL-12 and STAT4 signaling in th cells. J Immunol. 2004;172(11):6775–6782. doi: 10.4049/jimmunol.172.11.6775. [DOI] [PubMed] [Google Scholar]
- Luthje K, Kallies A, Shimohakamada Y, Belz GT, Light A, Tarlinton DM, Nutt SL. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat Immunol. 2012;13(5):491–498. doi: 10.1038/ni.2261. [DOI] [PubMed] [Google Scholar]
- Ma CS, Chew GY, Simpson N, Priyadarshi A, Wong M, Grimbacher B, Fulcher DA, Tangye SG, Cook MC. Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J Exp Med. 2008;205(7):1551–1557. doi: 10.1084/jem.20080218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5(6):472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- Mantel PY, Ouaked N, Ruckert B, Karagiannidis C, Welz R, Blaser K, Schmidt-Weber CB. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol. 2006;176(6):3593–3602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- Martins GA, Hutchins AS, Reiner SL. Transcriptional activators of helper T cell fate are required for establishment but not maintenance of signature cytokine expression. J Immunol. 2005;175(9):5981–5985. doi: 10.4049/jimmunol.175.9.5981. [DOI] [PubMed] [Google Scholar]
- Mathur AN, Chang HC, Zisoulis DG, Kapur R, Belladonna ML, Kansas GS, Kaplan MH. T-bet is a critical determinant in the instability of the IL-17-secreting T-helper phenotype. Blood. 2006;108(5):1595–1601. doi: 10.1182/blood-2006-04-015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur AN, Chang HC, Zisoulis DG, Stritesky GL, Yu Q, O’Malley JT, Kapur R, Levy DE, Kansas GS, Kaplan MH. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J Immunol. 2007;178(8):4901–4907. doi: 10.4049/jimmunol.178.8.4901. [DOI] [PubMed] [Google Scholar]
- Matsuda JL, George TC, Hagman J, Gapin L. Temporal dissection of T-bet functions. J Immunol. 2007;178(6):3457–3465. doi: 10.4049/jimmunol.178.6.3457. [DOI] [PubMed] [Google Scholar]
- McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O’Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10(3):314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao T, Raymond M, Bhullar P, Ghaffari E, Symonds AL, Meier UC, Giovannoni G, Li S, Wang P. Early growth response gene-2 controls IL-17 expression and Th17 differentiation by negatively regulating Batf. J Immunol. 2013;190(1):58–65. doi: 10.4049/jimmunol.1200868. [DOI] [PubMed] [Google Scholar]
- Mikhalkevich N, Becknell B, Caligiuri MA, Bates MD, Harvey R, Zheng WP. Responsiveness of naive CD4 T cells to polarizing cytokine determines the ratio of Th1 and Th2 cell differentiation. J Immunol. 2006;176(3):1553–1560. doi: 10.4049/jimmunol.176.3.1553. [DOI] [PubMed] [Google Scholar]
- Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22(21):2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol cell. 2010;40(4):594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, Kanno Y, Spalding C, Elloumi HZ, Paulson ML, Davis J, Hsu A, Asher AI, O’Shea J, Holland SM, Paul WE, Douek DC. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452(7188):773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minter LM, Turley DM, Das P, Shin HM, Joshi I, Lawlor RG, Cho OH, Palaga T, Gottipati S, Telfer JC, Kostura L, Fauq AH, Simpson K, Such KA, Miele L, Golde TE, Miller SD, Osborne BA. Inhibitors of gamma-secretase block in vivo and in vitro T helper type 1 polarization by preventing Notch upregulation of Tbx21. Nat Immunol. 2005;6(7):680–688. [PubMed] [Google Scholar]
- Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho IC. Ets-1 is a negative regulator of Th17 differentiation. J Exp Med. 2007;204(12):2825–2835. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero LL, Cubre A, Mora-Solano C, Wang Y, Alegre ML. T cell receptor/CARMA1/ NF-kappaB signaling controls T-helper (Th) 17 differentiation. Proc Natl Acad Sci U.S.A. 2012;109(45):18529–18534. doi: 10.1073/pnas.1204557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Schaller MA, Neupane R, Kunkel SL, Lukacs NW. Regulation of T cell activation by Notch ligand, DLL4, promotes IL-17 production and Rorc activation. J Immunol. 2009;182(12):7381–7388. doi: 10.4049/jimmunol.0804322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, High FA, Hutchins AS, Lee HW, Villarino AV, Livingston DM, Kung AL, Cereb N, Yao TP, Yang SY, Reiner SL. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292(5523):1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- Mullen AC, Hutchins AS, High FA, Lee HW, Sykes KJ, Chodosh LA, Reiner SL. Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nat Immunol. 2002;3(7):652–658. doi: 10.1038/ni807. [DOI] [PubMed] [Google Scholar]
- Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, Taniuchi I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J Exp Med. 2007;204(8):1749–1755. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawijn MC, Dingjan GM, Ferreira R, Lambrecht BN, Karis A, Grosveld F, Savelkoul H, Hendriks RW. Enforced expression of GATA-3 in transgenic mice inhibits Th1 differentiation and induces the formation of a T1/ST2-expressing Th2-committed T cell compartment in vivo. J Immunol. 2001;167(2):724–732. doi: 10.4049/jimmunol.167.2.724. [DOI] [PubMed] [Google Scholar]
- Nishihara M, Ogura H, Ueda N, Tsuruoka M, Kitabayashi C, Tsuji F, Aono H, Ishihara K, Huseby E, Betz UA, Murakami M, Hirano T. IL-6-gp130-STAT3 in T cells directs the development of IL-17+ Th with a minimum effect on that of Treg in the steady state. Int Immunol. 2007;19(6):695–702. doi: 10.1093/intimm/dxm045. [DOI] [PubMed] [Google Scholar]
- Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448(7152):480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, Dong C. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29(1):138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325(5943):1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva RI, Podd A, Chen Y, Alekseev AM, Yu M, Qi X, Huang H, Wen R, Wang J, Li HS, Watowich SS, Qi H, Dong C, Wang D. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J Biol Chem. 2012;287(14):11234–11239. doi: 10.1074/jbc.M111.324046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea JJ, Plenge R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity. 2012;36(4):542–550. doi: 10.1016/j.immuni.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich KJ, Huang AC, Weinmann AS. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Medicine. 2011;208(5):1001–1013. doi: 10.1084/jem.20102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13(4):405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Iwai Y, Oh-Hora M, Yamamoto M, Morio T, Aoki K, Ohya K, Jetten AM, Akira S, Muta T, Takayanagi H. IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature. 2010;464(7293):1381–1385. doi: 10.1038/nature08922. [DOI] [PubMed] [Google Scholar]
- Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, Miyachi Y, Tsukada T, Sakaguchi S. Foxp3 controls regulatory T-cell function by interacting with AML1/ Runx1. Nature. 2007;446(7136):685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- Onodera A, Yamashita M, Endo Y, Kuwahara M, Tofukuji S, Hosokawa H, Kanai A, Suzuki Y, Nakayama T. STAT6-mediated displacement of polycomb by trithorax complex establishes long-term maintenance of GATA3 expression in T helper type 2 cells. J Exp Med. 2010;207(11):2493–2506. doi: 10.1084/jem.20100760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Beckett O, Ma Q, Paik JH, DePinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11(7):618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, Murphy KM. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12(1):27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- Pagani M, Rossetti G, Panzeri I, de Candia P, Bonnal RJ, Rossi RL, Geginat J, Abrignani S. Role of microRNAs and long-non-coding RNAs in CD4(+) T-cell differentiation. Immunol Rev. 2013;253(1):82–96. doi: 10.1111/imr.12055. [DOI] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ho IC. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci U.S.A. 2004;101(7):1993–1998. doi: 10.1073/pnas.0308697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Yu H, Dang EV, Barbi J, Pan X, Grosso JF, Jinasena D, Sharma SM, McCadden EM, Getnet D, Drake CG, Liu JO, Ostrowski MC, Pardoll DM. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325(5944):1142–1146. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SL, Gerth AJ, Ranger AM, Glimcher LH. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity. 2001;14(1):13–20. doi: 10.1016/s1074-7613(01)00085-1. [DOI] [PubMed] [Google Scholar]
- Polansky JK, Schreiber L, Thelemann C, Ludwig L, Kruger M, Baumgrass R, Cording S, Floess S, Hamann A, Huehn J. Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med. 2010;88(10):1029–1040. doi: 10.1007/s00109-010-0642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter CM, Clipstone NA. Sustained NFAT signaling promotes a Th1-like pattern of gene expression in primary murine CD4+ T cells. J Immunol. 2002;168(10):4936–4945. doi: 10.4049/jimmunol.168.10.4936. [DOI] [PubMed] [Google Scholar]
- Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453(7191):65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- Quintana FJ, Jin H, Burns EJ, Nadeau M, Yeste A, Kumar D, Rangachari M, Zhu C, Xiao S, Seavitt J, Georgopoulos K, Kuchroo VK. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat Immunol. 2012;13(8):770–777. doi: 10.1038/ni.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirion MR, Gregory GD, Umetsu SE, Winandy S, Brown MA. Cutting edge: Ikaros is a regulator of Th2 cell differentiation. J Immunol. 2009;182(2):741–745. doi: 10.4049/jimmunol.182.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranger AM, Hodge MR, Gravallese EM, Oukka M, Davidson L, Alt FW, de la Brousse FC, Hoey T, Grusby M, Glimcher LH. Delayed lymphoid repopulation with defects in IL-4-driven responses produced by inactivation of NF-ATc. Immunity. 1998a;8(1):125–134. doi: 10.1016/s1074-7613(00)80465-3. [DOI] [PubMed] [Google Scholar]
- Ranger AM, Oukka M, Rengarajan J, Glimcher LH. Inhibitory function of two NFAT family members in lymphoid homeostasis and Th2 development. Immunity. 1998b;9(5):627–635. doi: 10.1016/s1074-7613(00)80660-3. [DOI] [PubMed] [Google Scholar]
- Rengarajan J, Mowen KA, McBride KD, Smith ED, Singh H, Glimcher LH. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J Exp Med. 2002a;195(8):1003–1012. doi: 10.1084/jem.20011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J, Tang B, Glimcher LH. NFATc2 and NFATc3 regulate T(H)2 differentiation and modulate TCR-responsiveness of naive T(H)cells. Nat Immunol. 2002b;3(1):48–54. doi: 10.1038/ni744. [DOI] [PubMed] [Google Scholar]
- Ruan Q, Kameswaran V, Tone Y, Li L, Liou HC, Greene MI, Tone M, Chen YH. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31(6):932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, deRoos P, Chaudhry A, Niec RE, Arvey A, Samstein RM, Leslie C, Shaffer SA, Goodlett DR, Rudensky AY. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol. 2012;13(10):1010–1019. doi: 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Egawa T, Chong MM, Treuting P, Littman DR, Rudensky AY. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2009;10(11):1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samon JB, Champhekar A, Minter LM, Telfer JC, Miele L, Fauq A, Das P, Golde TE, Osborne BA. Notch1 and TGFbeta1 cooperatively regulate Foxp3 expression and the maintenance of peripheral regulatory T cells. Blood. 2008;112(5):1813–1821. doi: 10.1182/blood-2008-03-144980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, Neph S, Sabo P, Kim JM, Liao W, Li MO, Leslie C, Stamatoyannopoulos JA, Rudensky AY. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151(1):153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, Hatton RD, Stormo GD, Weaver CT, Russell JH, Murphy TL, Murphy KM. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460(7253):405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz EG, Mariani L, Radbruch A, Hofer T. Sequential polarization and imprinting of type 1 T helper lymphocytes by interferon-gamma and interleukin-12. Immunity. 2009;30(5):673–683. doi: 10.1016/j.immuni.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Seder RA, Paul WE, Davis MM, de St Groth BF. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J Exp Med. 1992;176(4):1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki N, Miyazaki M, Suzuki W, Hayashi K, Arima K, Myburgh E, Izuhara K, Brombacher F, Kubo M. IL-4-induced GATA-3 expression is a time-restricted instruction switch for Th2 cell differentiation. J Immunol. 2004;172(10):6158–6166. doi: 10.4049/jimmunol.172.10.6158. [DOI] [PubMed] [Google Scholar]
- Sekiya T, Kashiwagi I, Inoue N, Morita R, Hori S, Waldmann H, Rudensky AY, Ichinose H, Metzger D, Chambon P, Yoshimura A. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat Commun. 2011;2:269. doi: 10.1038/ncomms1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4(5):E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- Shi G, Cox CA, Vistica BP, Tan C, Wawrousek EF, Gery I. Phenotype switching by inflammation-inducing polarized Th17 cells, but not by Th1 cells. J Immunol. 2008;181(10):7205–7213. doi: 10.4049/jimmunol.181.10.7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, Chu C, Quelle FW, Nosaka T, Vignali DA, Doherty PC, Grosveld G, Paul WE, Ihle JN. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380(6575):630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- Shin HM, Minter LM, Cho OH, Gottipati S, Fauq AH, Golde TE, Sonenshein GE, Osborne BA. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. EMBO J. 2006;25(1):129–138. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, Dorman L, Viggiano V, Cippitelli M, Ghosh P, Rice N, Young HA. Interaction of NF-kappaB and NFAT with the interferon-gamma promoter. J Biol Chem. 1997;272(48):30412–30420. doi: 10.1074/jbc.272.48.30412. [DOI] [PubMed] [Google Scholar]
- Skapenko A, Leipe J, Niesner U, Devriendt K, Beetz R, Radbruch A, Kalden JR, Lipsky PE, Schulze-Koops H. GATA-3 in human T cell helper type 2 development. J Exp Med. 2004;199(3):423–428. doi: 10.1084/jem.20031323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5(10):1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Staudt V, Bothur E, Klein M, Lingnau K, Reuter S, Grebe N, Gerlitzki B, Hoffmann M, Ulges A, Taube C, Dehzad N, Becker M, Stassen M, Steinborn A, Lohoff M, Schild H, Schmitt E, Bopp T. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33(2):192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Swain SL, Weinberg AD, English M, Huston G. IL-4 directs the development of Th2-like helper effectors. J Immunol. 1990;145(11):3796–3806. [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100(6):655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Szabo SJ, Sullivan BM, Stemmann C, Satoskar AR, Sleckman BP, Glimcher LH. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295(5553):338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380(6575):627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- Tartar DM, VanMorlan AM, Wan X, Guloglu FB, Jain R, Haymaker CL, Ellis JS, Hoeman CM, Cascio JA, Dhakal M, Oukka M, Zaghouani H. FoxP3+RORgammat+T helper intermediates display suppressive function against autoimmune diabetes. J Immunol. 2010;184(7):3377–3385. doi: 10.4049/jimmunol.0903324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DA, Doherty PC, Grosveld GC, Ihle JN. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382(6587):171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, Kaplan MH. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity. 2008;29(5):679–690. doi: 10.1016/j.immuni.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RM, Chen C, Chunder N, Ma L, Taylor J, Pearce EJ, Wells AD. Ikaros silences T-bet expression and interferon-gamma production during T helper 2 differentiation. J Biol Chem. 2010;285(4):2545–2553. doi: 10.1074/jbc.M109.038794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184(7):3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9(2):194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- Umetsu SE, Winandy S. Ikaros is a regulator of Il10 expression in CD4+ T cells. J Immunol. 2009;183(9):5518–5525. doi: 10.4049/jimmunol.0901284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity. 2003;18(3):415–428. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- Vaeth M, Schliesser U, Muller G, Reissig S, Satoh K, Tuettenberg A, Jonuleit H, Waisman A, Muller MR, Serfling E, Sawitzki BS, Berberich-Siebelt F. Dependence on nuclear factor of activated T-cells (NFAT) levels discriminates conventional T cells from Foxp3+ regulatory T cells. Proc Natl Acad Sci U.S.A. 2012;109(40):16258–16263. doi: 10.1073/pnas.1203870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loosdregt J, Fleskens V, Tiemessen MM, Mokry M, van Boxtel R, Meerding J, Pals CE, Kurek D, Baert MR, Delemarre EM, Grone A, Koerkamp MJ, Sijts AJ, Nieuwenhuis EE, Maurice MM, van Es JH, Ten Berge D, Holstege FC, Staal FJ, Zaiss DM, Prakken BJ, Coffer PJ. Canonical wnt signaling negatively modulates regulatory T cell function. Immunity. 2013;39(2):298–310. doi: 10.1016/j.immuni.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld JC, Stockinger B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453(7191):106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- Villarino AV, Gallo E, Abbas AK. STAT1-activating cytokines limit Th17 responses through both T-bet-dependent and -independent mechanisms. J Immunology. 2010;185(11):6461–6471. doi: 10.4049/jimmunol.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelzang A, McGuire HM, Yu D, Sprent J, Mackay CR, King C. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity. 2008;29(1):127–137. doi: 10.1016/j.immuni.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Volpe E, Servant N, Zollinger R, Bogiatzi SI, Hupe P, Barillot E, Soumelis V. A critical function for transforming growth factor-beta, interleukin 23 and proinflammatory cytokines in driving and modulating human T(H)-17 responses. Nat Immunol. 2008;9(6):650–657. doi: 10.1038/ni.1613. [DOI] [PubMed] [Google Scholar]
- Wang Y, Misumi I, Gu AD, Curtis TA, Su L, Whitmire JK, Wan YY. GATA-3 controls the maintenance and proliferation of T cells downstream of TCR and cytokine signaling. Nat Immunol. 2013;14(7):714–722. doi: 10.1038/ni.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Su MA, Wan YY. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity. 2011;35(3):337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Voo KS, Liu B, Chen CY, Uygungil B, Spoede W, Bernstein JA, Huston DP, Liu YJ. A novel subset of CD4(+) T(H)2 memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma. J Exp Med. 2010;207(11):2479–2491. doi: 10.1084/jem.20101376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, Zhu J, Zhao K. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35(2):299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Laurence A, Elias KM, O’Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem. 2007;282(48):34605–34610. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L, Bricarelli FD, Byrne G, McEuen M, Proll S, Appleby M, Brunkow ME. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27(1):18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat Immunol. 2007;8(3):277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- Wohlfert EA, Grainger JR, Bouladoux N, Konkel JE, Oldenhove G, Ribeiro CH, Hall JA, Yagi R, Naik S, Bhairavabhotla R, Paul WE, Bosselut R, Wei G, Zhao K, Oukka M, Zhu J, Belkaid Y. GATA3 controls Foxp3(+) regulatory T cell fate during inflammation in mice. J Clin Invest. 2011;121(11):4503–4515. doi: 10.1172/JCI57456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, Mathis D, Benoist C, Chen L, Rao A. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126(2):375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- Xu L, Kitani A, Fuss I, Strober W. Cutting edge: regulatory T cells induce CD4+CD25-Foxp3- T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J Immunol. 2007;178(11):6725–6729. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]
- Xu L, Kitani A, Stuelten C, McGrady G, Fuss I, Strober W. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity. 2010;33(3):313–325. doi: 10.1016/j.immuni.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Junttila IS, Wei G, Urban JF, Jr, Zhao K, Paul WE, Zhu J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity. 2010;32(4):507–517. doi: 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H, Zhu J, Paul WE. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J Exp Med. 2005;202(6):793–804. doi: 10.1084/jem.20051304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Ukai-Tadenuma M, Miyamoto T, Sugaya K, Hosokawa H, Hasegawa A, Kimura M, Taniguchi M, DeGregori J, Nakayama T. Essential role of GATA3 for the maintenance of type 2 helper T (Th2) cytokine production and chromatin remodeling at the Th2 cytokine gene loci. J Biol Chem. 2004;279(26):26983–26990. doi: 10.1074/jbc.M403688200. [DOI] [PubMed] [Google Scholar]
- Yang XO, Angkasekwinai P, Zhu J, Peng J, Liu Z, Nurieva R, Liu X, Chung Y, Chang SH, Sun B, Dong C. Requirement for the basic helix-loop-helix transcription factor Dec2 in initial TH2 lineage commitment. Nat Immunol. 2009;10(12):1260–1266. doi: 10.1038/ni.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282(13):9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM, Dong C. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28(1):29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, Kanno Y, O’Shea JJ, Laurence A. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12(3):247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl R, Hennighausen L, Wu C, O’Shea JJ. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109(10):4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong PF, Freeman AF, Engelhardt KR, Holland S, Puck JM, Grimbacher B. An update on the hyper-IgE syndromes. Arthritis Res Ther. 2012;14(6):228. doi: 10.1186/ar4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, Gennert D, Satija R, Shakya A, Lu DY, Trombetta JJ, Pillai MR, Ratcliffe PJ, Coleman ML, Bix M, Tantin D, Park H, Kuchroo VK, Regev A. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496(7446):461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Nishina H, Takimoto H, Marengere LE, Wakeham AC, Bouchard D, Kong YY, Ohteki T, Shahinian A, Bachmann M, Ohashi PS, Penninger JM, Crabtree GR, Mak TW. The transcription factor NF-ATc1 regulates lymphocyte proliferation and Th2 cytokine production. Immunity. 1998;8(1):115–124. doi: 10.1016/s1074-7613(00)80464-1. [DOI] [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009a;31(3):457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Yu Q, Sharma A, Ghosh A, Sen JM. T cell factor-1 negatively regulates expression of IL-17 family of cytokines and protects mice from experimental autoimmune encephalomy-elitis. J Immunol. 2011;186(7):3946–3952. doi: 10.4049/jimmunol.1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Sharma A, Oh SY, Moon HG, Hossain MZ, Salay TM, Leeds KE, Du H, Wu B, Waterman ML, Zhu Z, Sen JM. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nat Immunol. 2009b;10(9):992–999. doi: 10.1038/ni.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuf I, Kageyama R, Monticelli L, Johnston RJ, Ditoro D, Hansen K, Barnett B, Crotty S. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2010;185(1):190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng WP, Chang C, Lai JJ. Immune suppressive activity and lack of T helper differentiation are differentially regulated in natural regulatory T cells. J Immunol. 2009;183(6):3583–3590. doi: 10.4049/jimmunol.0900146. [DOI] [PubMed] [Google Scholar]
- Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272(34):21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9(11):1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89(4):587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458(7236):351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463(7282):808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445(7130):936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30(5):646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8(9):967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, Ziegler SF, Littman DR. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453(7192):236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19(5):739–748. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- Zhu J, Davidson TS, Wei G, Jankovic D, Cui K, Schones DE, Guo L, Zhao K, Shevach EM, Paul WE. Down-regulation of Gfi-1 expression by TGF-beta is important for differentiation of Th17 and CD103+ inducible regulatory T cells. J Exp Med. 2009;206(2):329–341. doi: 10.1084/jem.20081666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Guo L, Min B, Watson CJ, Hu-Li J, Young HA, Tsichlis PN, Paul WE. Growth factor independent-1 induced by IL-4 regulates Th2 cell proliferation. Immunity. 2002;16(5):733–744. doi: 10.1016/s1074-7613(02)00317-5. [DOI] [PubMed] [Google Scholar]
- Zhu J, Guo L, Watson CJ, Hu-Li J, Paul WE. Stat6 is necessary and sufficient for IL-4’s role in Th2 differentiation and cell expansion. J Immunology. 2001;166(12):7276–7281. doi: 10.4049/jimmunol.166.12.7276. [DOI] [PubMed] [Google Scholar]
- Zhu J, Jankovic D, Grinberg A, Guo L, Paul WE. Gfi-1 plays an important role in IL-2-mediated Th2 cell expansion. Proc Natl Acad Sci U.S.A. 2006;103(48):18214–18219. doi: 10.1073/pnas.0608981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Jankovic D, Oler AJ, Wei G, Sharma S, Hu G, Guo L, Yagi R, Yamane H, Punkosdy G, Feigenbaum L, Zhao K, Paul WE. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 2012;37(4):660–673. doi: 10.1016/j.immuni.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr, Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5(11):1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotos D, Coquet JM, Zhang Y, Light A, D’Costa K, Kallies A, Corcoran LM, Godfrey DI, Toellner KM, Smyth MJ, Nutt SL, Tarlinton DM. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J Exp Med. 2010;207(2):365–378. doi: 10.1084/jem.20091777. [DOI] [PMC free article] [PubMed] [Google Scholar]