Abstract
Pancreatic adenocarcinoma (PDAC) is a lethal and painful disease, which has become one of the most frequent causes of death by malignant diseases around the world. Unfortunately, for the most part, this disease remains incurable. Significant advances in the field of genetics, particularly during the last two decades, has led to the proposal of a progression model, by which this cancer evolves by the accumulation of mutations and deletions in key oncogenes and tumor suppressor genes. This model has been remarkably useful for the development of tumor markers as well as elegant animal models. In spite of these strengths, this model does not take into consideration concepts and methodologies that have been derived from the field of epigenetics nor studies in the field of nuclear structure and function. Since our laboratory has been long been an advocate of these changes as critical for the pathobiology of pancreatic cancer, in this article, we describe an updated, more comprehensive model, which includes these concepts. With the widespread utilization of next generation sequencing for identifying both genetic and epigenetic changes genome-wide, we believe that the framework of this model will help to further identify and validate not only more but better markers for pancreatic cancer. In addition, as opposed to genetic changes, epigenetic alterations are amenable to pharmacological manipulations, consequently the familiarization with this model will help to better understand the potential beneficial effects of this type of therapy for this disease. Thus, we are optimistic that this new integrated paradigm will contribute to advance this field of research not only from a mechanistic point of view, but also from a translational one.
PDAC remains a national health priority and significant therapeutic challenge
This dismal disease ranks 4th as a leading cause of cancer-related deaths in the US, with a median survival of 6 months and a 5-year survival of 3–5% [1]. The bleak prognosis of PDAC is due to an aggressive biology, its rapid dissemination, and late diagnosis, rapidly leading to an incurable stage, for which therapeutic intervention is a challenge. Surgical resection is the only curative modality; however, this only applies to 10% of the patients, with their 5-year survival barely 20% [2]. Notably, these aggressive neoplasms are highly resistant to conventional chemotherapy and radiation [3] with gemcitabine, a nucleoside analog, remaining the standard chemotherapy option for metastatic PDAC [4, 5]. Numerous trials have attempted to improve gemcitabine clinical benefit through alternative schedules or combination with other agents, to no avail [6–8]. Thus, there is an urgent need to develop novel therapies in PDAC, in particular, targeting pathways highly relevant to its pathobiology.
The Genetics Revolution has significantly advanced the field of pancreatic cancer research
Searching for genetic mechanisms, many laboratories discovered oncogenes and tumor suppressor genes for PDAC [9]. These discoveries led to the seminal working model from the John Hopkins’ group [9] that expanded our understanding of the fact that PDAC arises from epithelial cells through accumulation of genetic alterations, driving transitions through increasingly aggressive lesions known as Pancreatic Intraepithelial Neoplasias (PanINs). In particular, mutation of the KRAS oncogene is almost universally found in the majority of PDAC cases [10]. Preneoplastic diseases, like chronic pancreatitis, also harbor initiating KRAS mutations [11], which appear to contribute to its evolution into cancer. This work prompted the development of animal models and tools to study, diagnose, and treat PDAC [12, 13]. Thus, genetic concepts and tools have advanced the field of pancreatic diseases. Moreover, this work has led to characterize oncogenic signals, which, like for instance in the case of kinases, have allowed the development of novel therapeutics tools for this disease. However, in spite of these remarkable achievements, pancreatic cancer remains incurable.
The emergence of a new scientific revolution, Epigenetics, has been further advancing the study of Pancreatic Cancer by generating new tools for their management and treatment
In 1942, C. Waddington coined the term “epigenetics” to refer to inheritance that occurs independently of the coding capacity of DNA [14]. Epigenetic mechanisms confer pluripotent progenitor cells that possess identical genomic DNA, the ability to differentiate into distinct populations. This wide range of differentiation originates from modulating genome expression in manners that are inheritable during cell division. In fact, differently than genetics, epigenetics deals with the inheritance not of the genome but of the mechanisms that regulate the expression of entire gene networks at the right time, right level, and place to define and maintain the integrity of phenotypes. Since Waddington, we have known that a cell has the potential to follow paths of distinct differentiation programs, similar to a ball rolling through different landscapes. Today, we understand that these landscapes are defined by gene expression patterns. Recent Nobel Prize-winning discoveries have revealed that we can induce cells to undergo incredible phenotypic changes by manipulating gene expression in a manner that promotes rapid transit though these landscapes [15]. The generation of iPS cells, which promise to be key for cell-based therapies, involve the manipulation of the epigenetics of the cell, for example, in a manner that leads a fibroblast to convert into an adult pancreatic cell. More importantly, once they divide, these cells will give rise to identical adult pancreatic cells. Thus, we have finally arrived to a fundamental stepping-stone in the field, which will lead to the potential manipulation of the expressed genome for therapeutic purposes in a manner that will revolutionize biology and medicine. However, epigenetics promises much more. For instance, we have learned that similar to genetic aberrations, epigenetic changes are inherited, giving rise to diseases [16]. In addition, environmental insults induce epigenetic modifications to influence health and disease [17]. Thus, through genetics, we inherit the potential to be who we are, but epigenetics transforms this into the reality of who we are in health and in disease. In contrast to genetic alterations, epigenetic mechanisms are amenable to reversal by small molecule drugs, giving rise to the new area of epigenetic therapeutics, with many agents being tested in clinical trials. Thus, epigenetics is promising to provide deeper mechanistic insight into diseases, as well as provide new diagnostic and therapeutic tools for their management.
Crosstalk between Genetics and Epigenetics as a Promising Paradigm in the Field of Pancreatology
Since its inception 25 years ago, our laboratory has helped to promulgate that DNA methylation, histone modification, nucleosome remodeling, and regulatory non-coding RNAs regulate most biological processes that associate to neoplastic transformation in the pancreas. In fact, a significant amount of evidence suggests that epigenetic deregulation is involved in pancreatic cancer development, spreading, and some of their signs and symptoms like thrombosis and cachexia. Epigenetics studies the activation and inactivation of gene networks independent of mutations, therefore, following, we will consider how these mechanisms fit within the genetic-centric paradigm. This exercise has previously led us to propose that a cross talk between genetics and epigenetics is critical for carcinogenesis [18]. In this article, we will review and update this model.
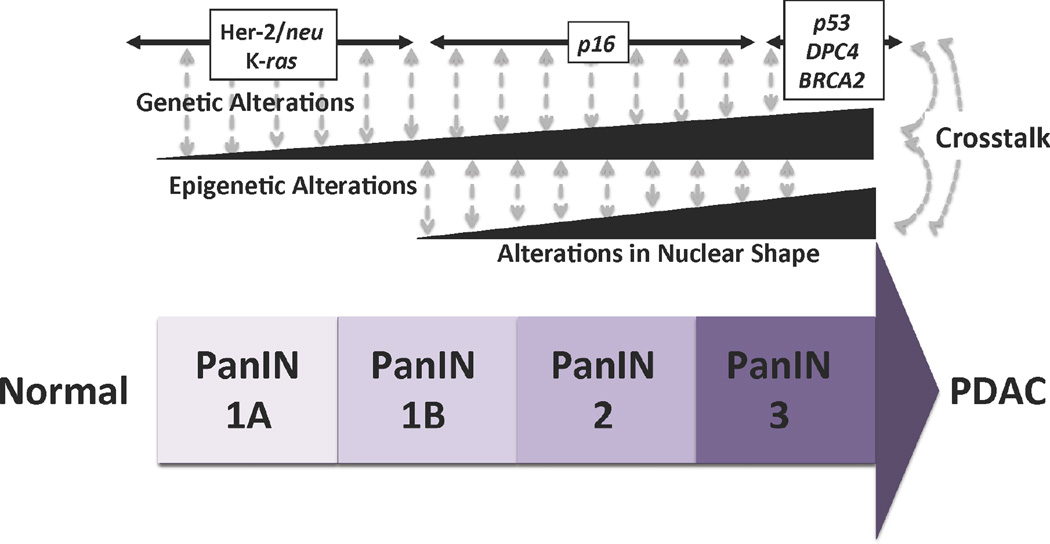
The overarching concept of cancer genetics is that if a gene is over-amplified in cancer, it might have been selected to provide cells with an advantage to grow and survive during neoplastic transformation. On the other hand, similar advantages can be gained by cells via the downregulation of other genes, known as tumor suppressors. Following this premise along with the knowledge available from sequencing, Hruban et al proposed a model in which PDAC arises from epithelial cells through an accumulation of genetic alterations in oncogenes and tumor suppressors [19, 20], which as a result promotes the development of precursor PanIN lesions [21, 22]. Although over time this model increasingly materializes as an incomplete one, it is still highly valuable since it establishes the types of mutations that associate to a particular type of progressive preneoplastic lesion. For instance, the most universal mutation found in pancreatic cancer, oncogenic activation of KRAS is necessary for initiation, but must be complemented later by genetic disruption of tumor suppressor pathways (e.g.: p16, p53, SMAD4, etc.) to give rise to frank invasive cancer [9]. In fact, the validity of this model has been shown in an elegant manner using Genetically Engineered Models (GEM), which have been primarily supported by NIH via the “Mouse Model Consortium” funded by NCI [23]. However, this model does not explain epigenetic changes, which occur between landmark mutations and are responsible of either activating or repressing entire gene expression networks that drive cancer progression. Therefore, in the following paragraphs, we will give examples of these epigenetic mechanisms, namely DNA methylation, histone-based epigenetics, and non-coding RNA epigenetic molecules. Thus, in summary, this updated paradigm for the progression of pancreatic cancer integrates our concept of the “Triple Code Hypothesis” to include two additional types of processes besides genetics, which are epigenetic changes and alterations in nuclear architecture (Figure 1). Our intention in proposing this new paradigm is for investigators in this field to dive into pancreatic cancer with a more in-depth mechanistic approach than using only the tools of molecular pathology.
Figure 1. Crosstalk between Genetics, Epigenetics, and Nuclear Structure in a Revised Comprehensive Progression Model for Pancreatic Cancer.
Our model for the progression of pancreatic cancer incorporates the genetic events described in the previous model as well as epigenetic changes and other alterations caused by changes in nuclear shape. Importantly, this model integrates the concept that these alterations do not occur in isolation, rather genetic alterations crosstalk with epigenetic and nuclear structure changes to give rise to neoplastic transformation, as well as most features of the cancer phenotype and its symptoms.
Mechanistic Basis of Epigenetics: The Nucleosome
Chromatin, which is composed of genomic DNA, histones, non-histone regulatory proteins and both small and long non-coding RNAs are at the mechanistic core of epigenetics [20]. These molecules are packed inside cells in the unit of the nucleosome, which can be viewed as the key nanomachine that senses both environmental and cell-autonomous signals to convert them into a gene regulatory response that ultimately defines distinct phenotypes. In fact, it is at the level of the nucleosome that the processes of “environment-gene interactions” that have been robustly documented by genetic epidemiologists take place [24]. The nucleosome is composed of approximately 150 bp of nuclear DNA wrapped around a histone octamer built from two molecules of each core histone protein: H2A, H2B, H3, and H4 [20]. In addition, a linker histone, known as H1, attaches to the external face of the DNA-histone octamer complex to facilitate further compaction, a process that has vital biological importance. The N-terminal domain of histones, commonly referred to as “histone tails”, extend out from the nucleosome particle, and thereby become easily accessible to epigenetic regulator complexes and serve as the platform on which epigenetic signals are written, read, and erased to codify for the expression of distinct gene expression networks. The body of each core histone, which locates inside of the DNA, is thus less accessible, but under certain circumstances becomes exposed to receive fewer, yet similarly critical, signals. A plethora of studies, originally pioneered independently by C.D. Allis [25] and B.M. Turner [26] revealed that these epigenetic signals, known as “histone marks” are made by covalent chemical modifications. Today, we know that histones receive a large amount of marks, including acetylation of lysines, methylation of lysines and arginines, phosphorylation of serines and threonines, ADP-ribosylation of glutamic acids, and ubiquitination and sumolyation of lysine residues, among others. More importantly, we have learned that it is the type and combination of these marks that serves as the instruction for cells to regulate gene expression in an inheritable manner. This concept is fundamental since it points to the existence of an epigenetic code (histone code) that is used to read the instruction provided by ancestors and cell progenitors to subsequent generations in the form of the genetic code. In fact, aberrant patterns of histone marks are increasingly being associated to clinical phenotype and/or outcome in various cancers [27–29]. In pancreatic cancer, low cellular levels of distinct marks on histone H3, such as dimethyl lysine 4 (H3K4me2), dimethyl lysine 9, or acetyl lysine 18 (H3K18ac), were found to be significant and independent predictors of poor survival, with the most significant predictor of overall survival resulting from combined low levels of H3K4me2 and/or H3K18ac [30]. Histone marks alone, however, are not sufficient for the associated regulatory mechanism. To better understand epigenetics, we have to consider the molecular machinery involved in the deposition, reading, and erasing necessary to give these instructions. Consequently, we subsequently provide insight into these phenomena, which are critical for cells to gain and maintain their normal phenotype.
Sequence Specific Transcription Factors Functions as Adaptor Proteins to link DNA to Epigenetic Regulators
The first types of molecules above the layer of nucleosomes that we have to consider in epigenetics are called sequence specific transcription factors. These proteins are most often modular molecules, which contain specialized domains that mediate their nuclear localization, binding to specific regions of DNA, and coupling to epigenetic regulators [31]. In fact, as reviewed below, most epigenetic regulators are armed with a specialized enzymatic activity that allows them to deposit, read, or erase posttranslational modifications in DNA and associated proteins. These marks function, therefore, as the signaling cascades of epigenetics and are interpreted as instructions for turning on and off extensive networks of genes. For the purpose of this article, we will focus on a selective group of transcription factor proteins that assist the RNA polymerase type II, the enzyme that copies most protein coding genes as well as many, though not all, non-coding RNAs. An example of this type of transcription factor is the tumor suppressor p53, which contains a DNA binding domain to recognize specific DNA sequences in promoters and other important regulatory regions, including enhancers of target genes (e.g. p21) [32]. The most frequent hotspot for mutation of this protein in cancer, including 50–75% of pancreatic cancers [33], is within this DNA binding domain [34]. In addition, this protein also contains a cluster of basic amino acids that function as a nuclear localization signal and a transcriptional regulatory domain that binds to epigenetic regulators, which in turn will mark either the regulatory DNA domains or proteins associated with them. Depending of the chemical nature of these marks, they serve as the initial epigenetic signal to dictate whether the target gene will be either expressed or silenced. Thus, it becomes important to subsequently describe how DNA is stored within the cell nucleus and is eventually sequestered away or accessed by transcription factors, marked, and regulated.
Nucleosome Remodeling Machines and Histone Modifying Enzymes Work in Concert to Regulate Histone Marks
As mentioned above, genomic DNA is highly packed into chromatin within the nucleus of eukaryote cells. It is believed that this sequestration of the genome by a cover of proteins and RNA help to protect it from both chemical and physical insults, as well as maintain many genes in an state of dormancy until necessary for access by the mRNA synthesis machinery to convert them into messages, which are then translated into proteins that help to define phenotypes. However, these associated proteins and RNA have the ability to regulate the genome by directing the expression of genes at the right level, right time, and right place to give rise to particular structures and functions. Among the genome-associated molecular machines that turn on and off genes (epigenetic regulatory complexes), we find two major groups, namely nucleosome remodeling machines and histone modifying enzymes. Nucleosome remodeling machines are multisubunit protein complexes that use energy from ATP to move nucleosomes along the DNA template, thereby exposing or sequestering binding sites that function to specifically recruit other complexes (e.g. histone modifying enzymes) to specific regions of the genome. There are four families of nucleosome remodeling machines, which are classically identified by the type of ATPase subunits [35]. These complexes, which include members of the SWI/SNF (switching defective/sucrose non-fermenting), ISWI (imitation SWI), INO80 (inositol requiring 80), and NuRD (nucleosome remodeling and deacetylation)/Mi-2/CHD (chromodomain, helicase, DNA binding) families, have different subunits of various type and sizes. It has been long known that nucleosome remodeling machines are mutated in many cancers. Due to the nature of these complexes consisting of numerous subunits to perform a single net function, the sum of mutations in individual subunits must be considered to evaluate the prevalence in cancer similar to a single gene, such as TP53. For example, the average incidence of SWI/SNF mutations across all cancer types is nearly 20%, with a frequency of 26% in pancreatic cancer [36].
Histone modifying enzymes are currently considered among the most critical part of the epigenome, besides non-coding RNAs. However, in contrast to non-coding RNAs, today, we know more about the structures, functions, pathobiological roles, and pharmacological manipulation of the histone modifying enzymes. Collectively, they possess enzymatic activities that allow them to deposit epigenomic marks (mark writers) and reverse these reactions when needed (mark erasers), as well as interpret these marks in context (mark readers). Among these proteins, histone methylases, acetylases, and ubiquitin ligases are among the best known and studied histone mark writers. The reactions catalyzed by these enzymes are then reversed by histone code erasers, including deacetylases, demethylases, and deubiquinases. Several histone modifying enzymes are dysregulated in pancreatic cancer. For instance, in recent study by Mazur, et al., SMYD3, MLL5, EZH2, SETD5 and WHSC1L1 were found to be consistently upregulated in pancreatic cancer samples in a screen of 54 known and candidate human lysine methyltransferases, which are histone code writers [37]. Several studies had previously found EZH2 overexpression in pancreatic cancer [38–40]. Interestingly, the oncogenic mutant KRAS signal was found to increase the expression of EZH2 [41]. Furthermore, EZH2 suppresses the p16INK4 tumor suppressor gene, which is critical during injury-induced regeneration during pancreatitis and therefore, contributes to the progression to pancreatic cancer [42]. In addition, EZH2 has been characterized to directly affect the maintenance of the pancreatic cancer stem cell phenotype, which is also associated with its H3K27me3 catalytic activity [43]. Overexpression of some histone code erasers, such as the histone demethylases KDM2B [44] and LSD1 [45], enhance pancreatic cancer growth, while loss of the KDM6B histone demethylase associates with PDAC aggressiveness [46]. Probably the first class of histone modifying enzymes identified to be dysregulated in PDAC was the histone deacetylases (HDACs). For instance, Class I HDACs were strongly expressed in a subset of PDACs from a larger cohort of 82 samples. Strong nuclear immunoreactivity for HDAC1, 2 and 3 was observed for 32%, 63% and 79% of PDAC cases, respectively [47]. In another expression profile of class I HDACs, HDAC1, HDAC2, HDAC3 and HDAC8 were positive in 17 (85%), 18 (90%), 20 (100%) and 18 (90%) of 20 pancreatic cancer cases, respectively, as observed by immunohistochemistry [48]. Further studies in PDAC have linked elevated HDAC1 and HDAC2 levels with poor tumor differentiation and overall survival [49–51]. Ouaïssi, et al. reported that approximately 80% of examined PDAC samples had a significant increase of HDAC7 at the RNA and protein levels [52]. Notably, HDAC7 levels were reduced in chronic pancreatitis, serous cystadenoma, and intraductal papillary mucinous tumor of the pancreas (IMPN) samples, suggesting that HDAC7 overexpression can discriminate pancreatic adenocarcinoma from other pancreatic diseases. Therefore, we urge the reader to carefully study the histone marks and the type of instructions that each provides as well as the associated histone modifying enzymes, which are responsible of their regulation [53–56]. This suggestion is of particular importance since a large amount of drugs, which are used to manipulate these pathways, are being developed and tested at an unprecedented rate [57–59]. Many other compounds such as neuroepileptics and other psychotropic drugs, which have been used for several decades, have potent epigenetic effects, which further support the need for readers to get familiarized with this important family of epigenetic regulators. Similarly, as discussed below, there are writers, readers and erasers of marks on DNA in addition to histones, which contribute to the instructions dictated to entire gene expression networks. Thus, in summary, the final outcome of these cascades are codified by the type and combination of marks controlled in context by writers, readers, and erasers enzymes in response to either cell-autonomous clues or environmental signals.
Marking the Genome by Methylation
The methylation of DNA was the first epigenetic modification to be discovered. Indeed, for decades, the methylation of CpG dinucleotides has been known to play a key role in X-chromosome inactivation, silencing of transposable DNA elements, and imprinting [60]. Methylation occurs in both, promoters and along gene bodies [61], although the functional role of the latter remains poorly understood. Today, there are several types DNA methylation marks known to exist, including the best known that occurs by the covalent addition of a methyl (CH3) group at the 5-carbon of the cytosine ring resulting in 5-methylcytosine (5mC), as well as its oxidized forms, 5-hydroxymethylcytosine (5hmC), 5-formylcytosine and 5-carboxylcytosine [62]. Sterically, when present in promoters and similar regulatory elements, the 5mC mark protrudes into the major groove of DNA, which is the main region recognized by the molecular machinery that regulates gene expression, and thereby inhibits transcription. For instance, methylation of the E-box (CACGTG) prevents n-Myc from binding to the EGFR promoter [63]. In addition, a significant amount of information exists on the type of writers, readers, and erasers of these marks [62, 64, 65]. Methylated CpG islands form a docking site for a family of methyl binding proteins (MBP), which read these marks to interfere with RNA synthesis to result in gene silencing. In the gene body, however, DNA methylation correlates with active transcription, splicing, and elongation [66], though the detailed molecular mechanisms for this phenomenon are undefined. Similarly important, in contrast to the widely assumed notion that DNA methylation is a stable epigenetic mark, active methylation-demethylation cycles also occur. In fact, dynamic regulation of DNA methylation is mainly achieved through a cyclical enzymatic cascade comprised of cytosine methylation by a group of writer enzymes called DNMTs, demethylation by Ten eleven translocation (Tet) dioxygenases (TET1, TET2, and TET3), which act as erasers of these marks, and reconstitution of unmethylated cytosines by replication-dependent dilution and base excision during DNA repair. DNMT1 functions as a maintenance methyltransferase responsible for faithfully reproducing the level and pattern of methylation during somatic cell division [64]. DNMT3a and DNMT3b are involved in adding de novo methyl groups to DNA, in particular during development [64]. The methyltransferase 3-like protein (DNMT3L) does not have enzymatic activity, but works as a necessary partner for the others DNMTs to perform their function. Interestingly, while Dnmt1 has up to a 50-fold preference for hemimethylated CpG sites present at the replication fork, it also appears to promote de novo methylation at non-CpG cytosines [64]. For performing the methylation reaction, all these enzymes utilize a derivative of the amino acid methionine, namely S-Adenosylmethionine (SAM), as a methyl donor. The product of this reaction gives S-adenosylhomocysteine (SAH), which later becomes homocysteine to be catabolized or remethylated to methionine. Notably, therefore, because of the use of these cofactors, epigenetically driven methylation reactions are influenced by metabolism and nutritional intake [67]. Thus, besides signaling cascades, these two processes can influence DNA methylation and impact gene expression.
The effects of DNA methylation on pancreatic cancer have been extensively studied, both in experimental models and in human tissues. The results of these studies have revealed an increase in promoter-associated CpG island methylation, which often results in the silencing of tumor suppressor genes [20, 64, 68]. This phenomenon has led to the development and utilization of several drugs which function as DNMT inhibitors and, which partially due to their effects on reactivating tumor suppressor genes, can slow down the progression of pancreatic cancer in both mice and humans [69, 70]. However, promoter hypermethylation is accompanied by the concomitant hypomethylation of repetitive sequences corresponding to dormant retrotransposons, which can become activated. Hypomethylation of these sequences also becomes more pronounced upon treatment with demethylating drugs [71], though the effect of these processes on the pathobiology of cancer is not completely clear. Interestingly, levels of 5hmC have been found reduced in pancreatic cancer and other cancer types along with a concomitant reduction in the expression of all three TET genes [72]. Besides its mechanistic importance and its relevance for cancer treatment, DNA methylation has been studied as a marker for the early detection of cancer. Cancer-specific DNA methylation patterns can be measured in DNA from detached tumor cells that are released to the blood, in pancreatic juice, or feces [73–76]. Unfortunately, in spite of its promise, there is not yet a clinically applicable assay for this purpose.
Epigenetic Regulation by non-coding RNAs
Recent advances in sequencing technologies have revealed that the genome is extensively transcribed, yielding a large repertoire of noncoding RNAs. These include long RNAs (lncRNAs), and many small noncoding RNAs such as miRNAs (microRNAs), siRNAs (small interfering RNAs), and piRNAs (Piwi-interacting RNAs). These mRNA-like molecules do not code for proteins but rather play key regulatory roles in a variety of cellular processes by modulating the levels and translation of other RNAs, including those coding for proteins [77]. Thus, it becomes important to briefly describe the biochemical constitution and function of these molecules. The most popular among these molecules, miRNAs are small single-stranded molecules (20 – 25 nt) that arise from pre-miRNA, which are characterized by the presence of a hairpin structure [78, 79]. This hairpin is processed to give rise to a mature miRNA, which is used to assemble an RNA induced silencing complex (RISC), containing key regulatory proteins such as Dicer. Once released, the miRNA hybridizes in a complementary manner to target mRNAs via their 3’UTR to subsequently induce cleavage by Argonaute, the catalytic component of RISC, and cause its silencing. Distinct from miRNAs in size, lack of sequence conservation, and increased complexity, small 24 – 30 nt long piRNAs interact in an RNA-protein complex with Piwi proteins, thus imparting their name [79]. These small RNAs are characterized by the presence of a uridine base at the 5’end and a 2’-O-methyl modification at the 3’ end. Piwi proteins are a subclass of Argonaute protein family and are key to the biogenesis of these molecules [80]. The function of these molecules has been ascribed to epigenetic and post-transcriptional silencing of transposable elements during germ line development [81]. Perhaps one of the most rapidly advancing fields within RNA research is the study of lncRNAs. These non-protein coding transcripts are longer than 200 nt in length which, like protein coding RNAs, undergo splicing and polyadenylation [82]. A subgroup of lncRNAs, named large intergenic non-coding RNAs (lincRNAs), have long been known by their role in epigenetic gene silencing, such Xist (X-inactive specific transcript). However, differently than miRNAs, lncRNAs appear to regulate gene expression and translation in various ways without a common mode of action. Thus far, lncRNA function has been categorized into four types of molecular mechanisms: signal, decoy, guide and scaffold [82]. The lncRNAs assigned to the signal type function as molecular signals of transcriptional activity. For the decoy type, the lncRNAs bind to and titrate away other regulatory RNAs or proteins. As a guide, the lncRNAs serve to localize ribonucleoprotein complexes to specific targets, while the scaffold lncRNAs provide a structural platform for the assembly of relevant proteins and/or RNA components. Finally, enhancer RNAs (eRNAs) and Promoter-associated RNAs (PARs) are the most recent types of non-coding RNA to be described [83]. In fact, the role of these molecules in epigenetics is just beginning to be uncovered. Enhancer RNAs are 800 nt in length on average, which up to now have been shown to be only transcriptional activators. Promoter-associated RNAs are non-coding transcripts that range from 16 nt to 200 nt, and they are expressed near the vicinity of promoters. Most of these molecules appear to associate with highly expressed genes and have short half-lives. Though investigations on the functional significance of these molecules have just started, thus far, they are believed to mediate transcriptional activation and repression.
As a result of numerous studies, PDAC cell lines, tissues, and blood samples have been extensively profiled for miRNA expression levels and compared to both normal and chronic pancreatitis samples in order to determine a miRNA expression signature that is associated with PDAC. Recently, however, Ma, et al. performed a comprehensive meta-review of published studies in PDAC to evaluate a total of 538 tumor and 206 noncancerous control samples [84]. This analysis revealed a meta-signature of seven up- and three down-regulated miRNAs, namely miR-155, miR-100, miR-21, miR-221, miR- 31, miR-143, and miR-23a with increased expression and miR-217, miR-148a and miR- 375 with decreased expression. In addition, alterations in miRNA levels are able to modulate chemosensitivity or radiosensitivity of PDAC cells in a variety of settings, with certain miRNAs serving as indicators of chemotherapy efficacy[85]. Therefore, miRNAs continue to be an active area of investigation for diagnostic biomarkers and therapeutic targets in PDAC. During the past 2–3 years, a few studies in pancreatic cancer on lncRNAs have emerged. For instance, expression of HOTAIR, a lincRNA that associates with Polycomb Repressive Complex 2 (PRC2) and its overexpression correlates with poor survival in several cancers, was found increased in pancreatic tumors, in particular more aggressive tumors, compared with control tissue [86]. Furthermore, knockdown of HOTAIR in PDAC cell lines resulted in decreased cell proliferation, altered cell cycle progression, induced apoptosis, reduced cell invasion and inhibited tumor growth in xenografts. Similarly, overexpression of the lncRNAs MALAT1, HULC and PVT1 have been associated with poor outcome in PDAC patients [87–89], while in another study that evaluated lncRNAs by microarray found that patients with high expression levels of the lncRNA BC008363 had significantly improved survival rates than those with lower levels [90]. Another lncRNA frequently downregulated in PDAC, ENST00000480739, was found to suppress tumor invasion and metastasis through regulation of HIF-1α upon re-expression [91], suggesting that lncRNAs may serve as a focus of future therapies.
Shaping Gene Expression through Nuclear Architecture
One of the most universal hallmarks of cancer cells is visible morphological alteration of the nuclei detectable by light microscopy on routine staining, and in fact, is often utilized by pathologists to grade and specify cancer type and stage, such as the transition of PanIN 1B to PanIN 2 [92]. Changes in nuclear structure include increased size, distortions in shape, and alterations in the internal organization of the nucleus [93, 94]. The spatial arrangement of chromosomes and other nuclear components, which defines the nuclear architecture, imparts a scaffold to organize the regulation of distinct functional processes. Any observed alterations in nuclear architecture may result from changes in the nuclear matrix, higher order chromatin folding, and/or the spatial arrangement of nucleic acid metabolism. Therefore, these changes, as seen in cancer, have the potential to impact on the fidelity of genome replication, chromatin organization, as well as gene expression. Studies have indicated that nuclear morphometry may serve as a prognostic indicator in non-resectable pancreatic cancer [95], as well as provide important pre-operative information in assessing pancreas resectability [96]. Another report demonstrated that there is significant deformation of the chromosome 8 territory in a small cohort of PDAC samples compared to histologically normal ductal epithelium [97]. However, the detailed evaluation of common alterations in nuclear architecture in PDAC along with the mechanistic links and its specific impact on nuclear functions remains in its infancy. Nevertheless, it is our opinion that this field is perhaps the most promising for future advances in epigenetics and pancreatic cancer, and therefore, it is important to underscore its importance in the current article in an attempt to stimulate future research.
Epigenetics Opens a New Era for Pancreatic Cancer Markers and Novel Therapeutic Modalities
Working primarily with pancreatic cells, our laboratory has contributed to the better understanding of some key aspects of the current paradigm for epigenetics. Working with transcription factors, for instance, we cloned and characterized several members of the KLF family of epigenetic regulators from the human pancreas. We subsequently have found that these proteins work at the intersection of metabolism and cancer. More importantly, thus far, the work performed on these proteins has provided a comprehensive model of epigenetic pathways in the human pancreas leading to the characterization of several histone acetylases, deacetylases, methylases, and reader proteins, among others. For this purpose, we will underscore the importance of KLF11 and KLF14. Although we identified these proteins in the context of their ability to suppress pancreatic cancer cell growth [98–100], it was later discovered through our work and others that these sequence-specific transcription factors function primarily through the regulation of metabolic gene networks [101–104]. This is important due to the key relationship between metabolism and cancer found uniquely in pancreatic cancer (the Diabetes-PaCa connection). Interestingly, the tumor suppression function of KLF11 is inactivated in PDAC by methylation-mediated gene silencing [105]. On the other hand, mutations in the protein impair certain distinct functions of KLF11 to give rise to Juvenile Diabetes (MODY VII) [106]. More importantly, alterations in a KLF11 binding site within the insulin promoter are responsible of neonatal diabetes [107]. A highly related member of the family, KLF14, is associated with obesity and diabetes, as well as basal cell carcinoma [104, 108]. Studies on these proteins led us to define that they function as a link between the DNA sequence they recognize and histone modifying enzymes. They are modular proteins, which contain small domains that serve as docking platforms for the Sin3-HDAC complex, HATs, WW- and WD40 domain proteins, histone methyltransferases and chromodomain reader molecules [103, 107, 109–112]. In addition, these proteins also heterodimerize with NFκB and PPARγ, which bring additional complexes along for regulatory purposes [102, 113, 114]. For instance, by coupling to the HP1-SUV39H1 as well as to the Sin3-HDAC pathway, these proteins are able to mark the promoters of metabolic gene networks to affect cell survival and growth [101]. In this regard, it is important to take into consideration that oncogenic activation, for instance of KRAS, in the pancreas leads to distinct metabolic changes [115]. KLF proteins are necessary for antagonizing KRAS [98]. Thus, it is likely that these proteins act early during the PDAC initiation process to change metabolic profiles of transforming cells. Noteworthy, however, we have also devoted significant efforts to directly characterize some of the histone code readers, writers, and erasers whether in the context of functioning with or without recruitment by a specific transcription factor. In 2001, we described a widely known domain for the recruitment of histone deacetylases [112]. This mechanism, which we discovered in the exocrine pancreas, is also used by many tumor suppressor genes, including Mad1, an antagonist of the Myc oncogene [116]. Subsequently, we characterized how histone code readers work to mediate growth stimuli downstream of growth factor receptor pathways [117]. Next, we isolated new Polycomb-type writer complexes from the pancreas and defined their growth-promoting role in pancreatic cells [118]. Together, we know that these molecular cascades are essential to define the cancer phenotype, thereby becoming attractive targets for developmental therapeutics.
Conclusion
The current article seeks to promote a change in the conceptual bias, which currently affects the pancreatic cancer field. In this regard, for instance, many researchers and practitioners still see pancreatic cancer exclusively as a disease of epithelial exocrine cells, which become transformed by the accumulation of genetic alterations. We have combined this view with solid observations from our laboratory and others, which reveal that genetic alterations crosstalk with epigenetic and nuclear structure changes to give rise to not only neoplastic transformation, but also to determine most features of the cancer phenotype and its symptoms (Figure 1). This new framework has significant mechanistic value as we seek to comprehend how this disease originates and evolves. This updated paradigm for the progression of pancreatic cancer integrates the concept that the patterns of gene expression networks to define the pancreatic cancer phenotype are dictated by the combination of genetic, epigenetic and nuclear structure instructions according to our “Triple Code Hypothesis”, which considers that all three of these codes contribute to the development and progression of this disease (Figure 2). More importantly, however, we know that many epigenetic alterations are significantly ameliorated by a new type of therapeutics, which target the epigenome. In fact, promising epigenetics-based therapies are currently being evaluated through different types of trials. Thus, the concepts discussed here should fuel a new era of studies, which promise to provide the medical community with new tools to diagnose and treat this dismal disease.
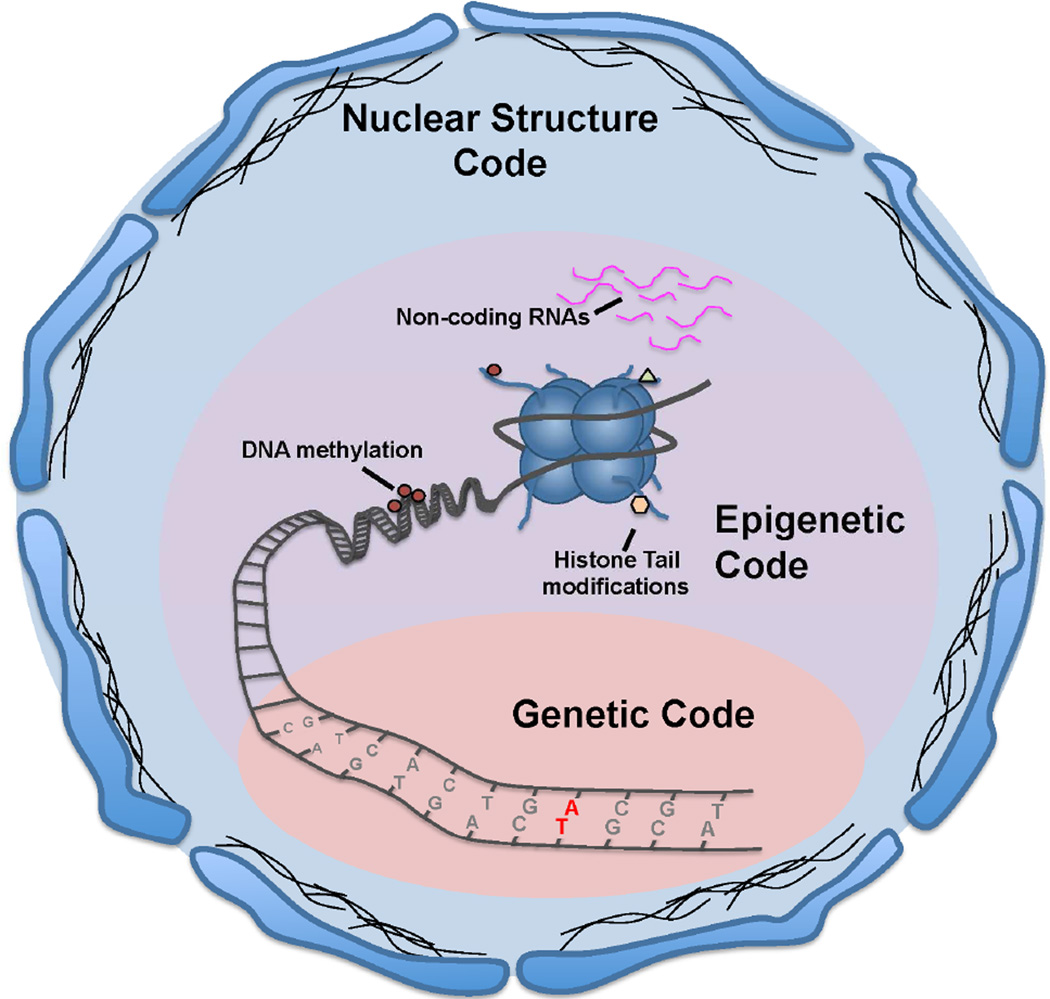
Figure 2. Integration of Instructions from Genetics, Epigenetics and Nuclear Architecture into the “Triple Code Hypothesis”.
Our comprehensive model for the development and progression of PDAC is based upon our understanding that gene expression networks are regulated by the combination of instructions dictated by genetics, epigenetics and nuclear architecture, which we have coined the “Triple Code Hypothesis”. Alterations in the Genetic Code form the foundation of the well-known DNA-centric hypothesis for the establishment and maintenance of the cancer phenotype, which includes mutations and deletions. The Epigenetic Code takes into account changes in DNA methylation, non-coding RNA molecules, and chromatin via histone modifications and the writers, readers, and erasers of the Histone Code. Finally, the Nuclear Structure Code, which includes the nuclear matrix and higher-order chromatin organization, impacts the fidelity of genome replication, chromatin organization, as well as gene expression.
KEY POINTS.
-
-
Many researchers and practitioners still see pancreatic cancer exclusively as a disease of epithelial exocrine cells, which become transformed by the accumulation of genetic alterations.
-
-
Genetic alterations crosstalk with epigenetic and nuclear structure changes to give rise to not only neoplastic transformation, but also to determine most features of the cancer phenotype and its symptoms
-
-
This updated paradigm for the progression of pancreatic cancer integrates the concept that the patterns of gene expression networks to define the pancreatic cancer phenotype are dictated by the combination of genetic, epigenetic and nuclear structure instructions according to the “Triple Code Hypothesis”, which considers that all three of these codes contribute to the development and progression of this disease.
-
-
Many epigenetic alterations are significantly ameliorated by a new type of therapeutics, which target the epigenome; promising epigenetics-based therapies are currently being evaluated through different types of trials.
Acknowledgements
Work in the author’s laboratories is supported by funding from the National Institutes of Health R01 DK52913 (R.U.) and R01 CA178627 (G.L.), as well as the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567) and the Mayo Clinic SPORE in Pancreatic Cancer (P50 CA102701).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest:
The authors declare that they have no competing interests.
References
- 1.Iovanna J, Mallmann M, Goncalves A, Turrini O, Dagorn J. Current knowledge on pancreatic cancer. Front Oncol. 2012;2 doi: 10.3389/fonc.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma C, Eltawil K, Renfrew P, Walsh M, Molinari M. Advances in diagnosis, treatment and palliation of pancreatic carcinoma: 1990–2010. World J Gastroenterol. 2011;17:867–897. doi: 10.3748/wjg.v17.i7.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu X, Zhang Y, Chen C, Yao Q, Li M. Targeted drug delivery in pancreatic cancer. Biochim Biophys Acta. 2010;1805:97–104. doi: 10.1016/j.bbcan.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burris H, Moore M, Andersen J, Green M, Rothenberg M, Modiano M, Cripps M, Portenoy R, Storniolo A, Tarassoff P, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 5.Heinemann V. Gemcitabine: Progress in the Treatment of Pancreatic Cancer. Oncology. 2001;60:8–18. doi: 10.1159/000055290. [DOI] [PubMed] [Google Scholar]
- 6.Rocha Lima C, Green M, Rotche R, Miller W, Jeffrey G, Cisar L, Morganti A, Orlando N, Gruia G, Miller L. Irinotecan Plus Gemcitabine Results in No Survival Advantage Compared With Gemcitabine Monotherapy in Patients With Locally Advanced or Metastatic Pancreatic Cancer Despite Increased Tumor Response Rate. Journal of Clinical Oncology. 2004;22:3776–3783. doi: 10.1200/JCO.2004.12.082. [DOI] [PubMed] [Google Scholar]
- 7.Louvet C, Labianca R, Hammel P, Lledo G, Zampino M, Andre T, Zaniboni A, Ducreux M, Aitini E, Taieb J, et al. Gemcitabine in Combination With Oxaliplatin Compared With Gemcitabine Alone in Locally Advanced or Metastatic Pancreatic Cancer: Results of a GERCOR and GISCAD Phase III Trial. Journal of Clinical Oncology. 2005;23:3509–3516. doi: 10.1200/JCO.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 8.Scheithauer W, Schull B, Ulrich-Pur H, Schmid K, Raderer M, Haider K, Kwasny W, Depisch D, Schneeweiss B, Lang F, Kornek G. Biweekly high-dose gemcitabine alone or in combination with capecitabine in patients with metastatic pancreatic adenocarcinoma: a randomized phase II trial. Annals of Oncology. 2003;14:97–104. doi: 10.1093/annonc/mdg029. [DOI] [PubMed] [Google Scholar]
- 9.Hruban RH, Goggins M, Parsons J, Kern SE. Progression Model for Pancreatic Cancer. Clinical Cancer Research. 2000;6:2969–2972. [PubMed] [Google Scholar]
- 10.Kanda M, Matthaei H, Wu J, Hong SM, Yu J, Borges M, Hruban RH, Maitra A, Kinzler K, Vogelstein B, Goggins M. Presence of Somatic Mutations in Most Early-Stage Pancreatic Intraepithelial Neoplasia. Gastroenterology. 2012;142:730–733. e739. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivera JA, Rall CJN, Graeme-Cook F, Castillo CFn-d, Shu P, Lakey N, Tepper R, Rattner DW, Warshaw AL, Rustgi AK. Analysis of K-ras oncogene mutations in chronic pancreatitis with ductal hyperplasia. Surgery. 1997;121:42–49. doi: 10.1016/s0039-6060(97)90181-1. [DOI] [PubMed] [Google Scholar]
- 12.Herreros-Villanueva M, Hijona E, Cosme A, Bujanda L. Mouse models of pancreatic cancer. World J Gastroenterol. 2012;18:1286–1294. doi: 10.3748/wjg.v18.i12.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez-Mancera PA, Guerra C, Barbacid M, Tuveson DA. What We Have Learned About Pancreatic Cancer From Mouse Models. Gastroenterology. 2012;142:1079–1092. doi: 10.1053/j.gastro.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Waddington CH. The epigenotype. 1942. International journal of epidemiology. 2012;41:10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 15.Colman A. Profile of John Gurdon and Shinya Yamanaka, 2012 Nobel Laureates in Medicine or Physiology. Proceedings of the National Academy of Sciences. 2013;110:5740–5741. doi: 10.1073/pnas.1221823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 17.Ho S-M, Johnson A, Tarapore P, Janakiram V, Zhang X, Leung Y-K. Environmental Epigenetics and Its Implication on Disease Risk and Health Outcomes. ILAR Journal. 2012;53:289–305. doi: 10.1093/ilar.53.3-4.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomberk G, Urrutia R. Pancreatic Cancer. Springer New York: 2010. Epigenetics and its Applications to a Revised Progression Model of Pancreatic Cancer; pp. 143–169. [Google Scholar]
- 19.Lomberk G, Mathison AJ, Grzenda A, Urrutia R. The sunset of somatic genetics and the dawn of epigenetics: a new frontier in pancreatic cancer research. Current Opinion in Gastroenterology. 2008;24:597–602. doi: 10.1097/MOG.0b013e32830b111d. 510.1097/MOG.1090b1013e32830b32111d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCleary-Wheeler AL, Lomberk GA, Weiss FU, Schneider Gn, Fabbri M, Poshusta TL, Dusetti NJ, Baumgart S, Iovanna JL, Ellenrieder V, et al. Insights into the epigenetic mechanisms controlling pancreatic carcinogenesis. Cancer letters. 2013;328:212–221. doi: 10.1016/j.canlet.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maitra A, Fukushima N, Takaori K, Hruban RH. Precursors to invasive pancreatic cancer. Adv Anat Pathol. 2005;12:81–91. doi: 10.1097/01.pap.0000155055.14238.25. [DOI] [PubMed] [Google Scholar]
- 22.Scarlett CJ, Salisbury EL, Biankin AV, Kench J. Precursor lesions in pancreatic cancer: morphological and molecular pathology. Pathology. 2011;43:183–200. doi: 10.1097/PAT.0b013e3283445e3a. [DOI] [PubMed] [Google Scholar]
- 23.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:654–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 24.Mazzio EA, Soliman KFA. Basic concepts of epigenetics: Impact of environmental signals on gene expression. Epigenetics. 2012;7:119–130. doi: 10.4161/epi.7.2.18764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 26.Turner BM. Histone acetylation and an epigenetic code. BioEssays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 27.Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, Wang Q, Chia D, Goodglick L, Kurdistani SK. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174:1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esteller M. The necessity of a human epigenome project. Carcinogenesis. 2006;27:1121–1125. doi: 10.1093/carcin/bgl033. [DOI] [PubMed] [Google Scholar]
- 29.Kanai Y, Arai E. Multilayer-omics analyses of human cancers: Exploration of biomarkers and drug targets based on the activities of the International Human Epigenome Consortium. Frontiers in Genetics. 2014;5 doi: 10.3389/fgene.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manuyakorn A, Paulus R, Farrell J, Dawson NA, Tze S, Cheung-Lau G, Hines OJ, Reber H, Seligson DB, Horvath S, et al. Cellular Histone Modification Patterns Predict Prognosis and Treatment Response in Resectable Pancreatic Adenocarcinoma: Results From RTOG 9704. Journal of Clinical Oncology. 2010;28:1358–1365. doi: 10.1200/JCO.2009.24.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lomberk G, Urrutia R. The family feud: turning off Sp1 by Sp1-like KLF proteins. Biochemical Journal. 2005;392:1–11. doi: 10.1042/BJ20051234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freeman JA, Espinosa JM. The impact of post-transcriptional regulation in the p53 network. Briefings in Functional Genomics. 2013;12:46–57. doi: 10.1093/bfgp/els058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koorstra J-BM, Hustinx SR, Offerhaus GJA, Maitra A. Pancreatic Carcinogenesis. Pancreatology. 2008;8:110–125. doi: 10.1159/000123838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes & Cancer. 2011;2:466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lusser A, Kadonaga JT. Chromatin remodeling by ATP-dependent molecular machines. BioEssays. 2003;25:1192–1200. doi: 10.1002/bies.10359. [DOI] [PubMed] [Google Scholar]
- 36.Shain AH, Pollack JR. The Spectrum of SWI/SNF Mutations, Ubiquitous in Human Cancers. PLoS ONE. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazur PK, Reynoird N, Khatri P, Jansen PWTC, Wilkinson A, Liu S, Barbash O, Van Aller GS, Huddleston M, Dhanak D, et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature. 2014;510:283–287. doi: 10.1038/nature13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ougolkov AV, Bilim VN, Billadeau DD. Regulation of Pancreatic Tumor Cell Proliferation and Chemoresistance by the Histone Methyltransferase Enhancer of Zeste Homologue 2. Clinical Cancer Research. 2008;14:6790–6796. doi: 10.1158/1078-0432.CCR-08-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qazi AM, Aggarwal S, Steffer CS, Bouwman DL, Weaver DW, Gruber SA, Batchu RB. Laser capture microdissection of pancreatic ductal adeno-carcinoma cells to analyze EzH2 by Western Blot analysis. Methods Mol Biol. 2011;755:245–256. doi: 10.1007/978-1-61779-163-5_20. [DOI] [PubMed] [Google Scholar]
- 40.Toll AD, Dasgupta A, Potoczek M, Yeo CJ, Kleer CG, Brody JR, Witkiewicz AK. Implications of enhancer of zeste homologue 2 expression in pancreatic ductal adenocarcinoma. Hum Pathol. 2010;41:1205–1209. doi: 10.1016/j.humpath.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Fujii S, Fukamachi K, Tsuda H, Ito K, Ito Y, Ochiai A. RAS oncogenic signal upregulates EZH2 in pancreatic cancer. Biochemical and Biophysical Research Communications. 2012;417:1074–1079. doi: 10.1016/j.bbrc.2011.12.099. [DOI] [PubMed] [Google Scholar]
- 42.Lasfargues C, Pyronnet S. EZH2 links pancreatitis to tissue regeneration and pancreatic cancer. Clinics and Research in Hepatology and Gastroenterology. 2012;36:323–324. doi: 10.1016/j.clinre.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 43.van Vlerken LE, Kiefer CM, Morehouse C, Li Y, Groves C, Wilson SD, Yao Y, Hollingsworth RE, Hurt EM. EZH2 Is Required for Breast and Pancreatic Cancer Stem Cell Maintenance and Can Be Used as a Functional Cancer Stem Cell Reporter. Stem Cells Translational Medicine. 2013;2:43–52. doi: 10.5966/sctm.2012-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tzatsos A, Paskaleva P, Ferrari F, Deshpande V, Stoykova S, Contino G, Wong K-K, Lan F, Trojer P, Park PJ, Bardeesy N. KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. The Journal of Clinical Investigation. 2013;123:727–739. doi: 10.1172/JCI64535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin Y, Zhu W, Xu W, Zhang B, Shi S, Ji S, Liu J, Long J, Liu C, Liu L, et al. LSD1 sustains pancreatic cancer growth via maintaining HIF1α-dependent glycolytic process. Cancer Letters. 2014;347:225–232. doi: 10.1016/j.canlet.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto K, Tateishi K, Kudo Y, Sato T, Yamamoto S, Miyabayashi K, Matsusaka K, Asaoka Y, Ijichi H, Hirata Y, et al. Loss of histone demethylase KDM6B enhances aggressiveness of pancreatic cancer through downregulation of C/EBPα. Carcinogenesis. 2014;35:2404–2414. doi: 10.1093/carcin/bgu136. [DOI] [PubMed] [Google Scholar]
- 47.Lehmann A, Denkert C, Budczies J, Buckendahl A-C, Darb-Esfahani S, Noske A, Muller B, Bahra M, Neuhaus P, Dietel M, et al. High class I HDAC activity and expression are associated with RelA/p65 activation in pancreatic cancer in vitro and in vivo. BMC Cancer. 2009;9:395. doi: 10.1186/1471-2407-9-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakagawa M, Oda Y, Eguchi T, Aishima S, Yao T, Hosoi F, Basaki Y, Ono M, Kuwano M, Tanaka M, Tsuneyoshi M. Expression profile of class I histone deacetylases in human cancer tissues. Oncol Rep. 2007;18:769–774. [PubMed] [Google Scholar]
- 49.Miyake K, Yoshizumi T, Imura S, Sugimoto K, Batmunkh E, Kanemura H, Morine Y, Shimada M. Expression of hypoxia-inducible factor-1alpha, histone deacetylase 1, and metastasis-associated protein 1 in pancreatic carcinoma: correlation with poor prognosis with possible regulation. Pancreas. 2008;36:e1–e9. doi: 10.1097/MPA.0b013e31815f2c2a. [DOI] [PubMed] [Google Scholar]
- 50.Wang W, Gao J, Man XH, Li ZS, Gong YF. Significance of DNA methyltransferase-1 and histone deacetylase-1 in pancreatic cancer. Oncol Rep. 2009;21:1439–1447. doi: 10.3892/or_00000372. [DOI] [PubMed] [Google Scholar]
- 51.Fritsche P, Seidler B, Schüler S, Schnieke A, Göttlicher M, Schmid RM, Saur D, Schneider G. HDAC2 mediates therapeutic resistance of pancreatic cancer cells via the BH3-only protein NOXA. Gut. 2009;58:1399–1409. doi: 10.1136/gut.2009.180711. [DOI] [PubMed] [Google Scholar]
- 52.Ouaissi M, Sielezneff I, Silvestre R, Sastre B, Bernard JP, Lafontaine JS, Payan MJ, Dahan L, Pirro N, Seitz JF, et al. High histone deacetylase 7 (HDAC7) expression is significantly associated with adenocarcinomas of the pancreas. Ann Surg Oncol. 2008;15:2318–2328. doi: 10.1245/s10434-008-9940-z. [DOI] [PubMed] [Google Scholar]
- 53.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Research. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Izzo A, Schneider R. Chatting histone modifications in mammals. Briefings in Functional Genomics. 2010;9:429–443. doi: 10.1093/bfgp/elq024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim YZ. Altered Histone Modifications in Gliomas. Brain Tumor Research and Treatment. 2014;2:7–21. doi: 10.14791/btrt.2014.2.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ivanov M, Barragan I, Ingelman-Sundberg M. Epigenetic mechanisms of importance for drug treatment. Trends in Pharmacological Sciences. 2014;35:384–396. doi: 10.1016/j.tips.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 58.Rotili D, Mai A. Targeting Histone Demethylases: A New Avenue for the Fight against Cancer. Genes & Cancer. 2011;2:663–679. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 60.Bird AP, Wolffe AP. Methylation-Induced Repression—Belts, Braces, and Chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 61.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 62.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perini G, Diolaiti D, Porro A, Della Valle G. In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12117–12122. doi: 10.1073/pnas.0409097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Subramaniam D, Thombre R, Dhar A, Anant S. DNA Methyltransferases: A Novel Target for Prevention and Therapy. Frontiers in Oncology. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parry L, Clarke AR. The Roles of the Methyl-CpG Binding Proteins in Cancer. Genes Cancer. 2011;2:618–630. doi: 10.1177/1947601911418499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liyanage VRB, Jarmasz JS, Murugeshan N, Del Bigio MR, Rastegar M, Davie JR. DNA Modifications: Function and Applications in Normal and Disease States. Biology. 2014;3:670–723. doi: 10.3390/biology3040670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaelin WG, McKnight SL. Influence of Metabolism on Epigenetics and Disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP, Hruban RH, Goggins M. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835–1839. [PubMed] [Google Scholar]
- 69.Shakya R, Gonda T, Quante M, Salas M, Kim S, Brooks J, Hirsch S, Davies J, Cullo A, Olive K, et al. Hypomethylating therapy in an aggressive stroma-rich model of pancreatic carcinoma. Cancer research. 2013;73:885–896. doi: 10.1158/0008-5472.CAN-12-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Missiaglia E, Donadelli M, Palmieri M, Crnogorac-Jurcevic T, Scarpa A, Lemoine NR. Growth delay of human pancreatic cancer cells by methylase inhibitor 5-aza-2'-deoxycytidine treatment is associated with activation of the interferon signalling pathway. Oncogene. 2005;24:199–211. doi: 10.1038/sj.onc.1208018. [DOI] [PubMed] [Google Scholar]
- 71.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matsubayashi H, Canto M, Sato N, Klein A, Abe T, Yamashita K, Yeo CJ, Kalloo A, Hruban R, Goggins M. DNA methylation alterations in the pancreatic juice of patients with suspected pancreatic disease. Cancer Res. 2006;66:1208–1217. doi: 10.1158/0008-5472.CAN-05-2664. [DOI] [PubMed] [Google Scholar]
- 74.Fukushima N, Walter KM, Uek T, Sato N, Matsubayashi H, Cameron JL, Hruban RH, Canto M, Yeo CJ, Goggins M. Diagnosing pancreatic cancer using methylation specific PCR analysis of pancreatic juice. Cancer Biol Ther. 2003;2:78–83. doi: 10.4161/cbt.183. [DOI] [PubMed] [Google Scholar]
- 75.Dauksa A, Gulbinas A, Barauskas G, Pundzius J, Oldenburg J, El-Maarri O. Whole blood DNA aberrant methylation in pancreatic adenocarcinoma shows association with the course of the disease: a pilot study. PLoS One. 2012;7:e37509. doi: 10.1371/journal.pone.0037509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kisiel JB, Yab TC, Taylor WR, Chari ST, Petersen GM, Mahoney DW, Ahlquist DA. Stool DNA testing for the detection of pancreatic cancer: assessment of methylation marker candidates. Cancer. 2012;118:2623–2631. doi: 10.1002/cncr.26558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Santosh B, Varshney A, Yadava PK. Non-coding RNAs: biological functions and applications. Cell Biochemistry and Function. 2015;33:14–22. doi: 10.1002/cbf.3079. [DOI] [PubMed] [Google Scholar]
- 78.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. The Journal of Pathology. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 79.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seto AG, Kingston RE, Lau NC. The Coming of Age for Piwi Proteins. Molecular Cell. 2007;26:603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 81.Malone CD, Hannon GJ. Small RNAs as Guardians of the Genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li X, Wu Z, Fu X, Han W. lncRNAs: Insights into their function and mechanics in underlying disorders. Mutation Research/Reviews in Mutation Research. 2014;762:1–21. doi: 10.1016/j.mrrev.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 83.Kaikkonen MU, Lam MTY, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovascular Research. 2011;90:430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ma M-Z, Kong X, Weng M-Z, Cheng K, Gong W, Quan Z-W, Peng C-H. Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: meta-analysis, experimental validation and clinical significance. Journal of Experimental & Clinical Cancer Research : CR. 2013;32:71–71. doi: 10.1186/1756-9966-32-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun T, Kong X, Du Y, Li Z. Aberrant MicroRNAs in Pancreatic Cancer: Researches and Clinical Implications. Gastroenterology Research and Practice. 2014;2014:386–561. doi: 10.1155/2014/386561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang CS, Yu W, Cui H, Wang YJ, Zhang L, Han F, Huang T. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015 [PubMed] [Google Scholar]
- 88.Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumour Biol. 2014 doi: 10.1007/s13277-014-2850-8. [DOI] [PubMed] [Google Scholar]
- 89.Peng W, Gao W, Feng J. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Med Oncol. 2014;31:346. doi: 10.1007/s12032-014-0346-4. [DOI] [PubMed] [Google Scholar]
- 90.Li J, Liu D, Hua R, Zhang J, Liu W, Huo Y, Cheng Y, Hong J, Sun Y. Long non-coding RNAs expressed in pancreatic ductal adenocarcinoma and lncRNA BC008363 an independent prognostic factor in PDAC. Pancreatology. 2014;14:385–390. doi: 10.1016/j.pan.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 91.Sun YW, Chen YF, Li J, Huo YM, Liu DJ, Hua R, Zhang JF, Liu W, Yang JY, Fu XL, et al. A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1[alpha] in pancreatic ductal adenocarcinoma. Br J Cancer. 2014;111:2131–2141. doi: 10.1038/bjc.2014.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hruban RH, Adsay NV, Albores–Saavedra J, Compton C, Garrett ES, Goodman SN, Kern SE, Klimstra DS, Klöppel G, Longnecker DS, et al. Pancreatic Intraepithelial Neoplasia: A New Nomenclature and Classification System for Pancreatic Duct Lesions. The American Journal of Surgical Pathology. 2001;25 doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 93.Reddy KL, Feinberg AP. Higher order chromatin organization in cancer. Seminars in Cancer Biology. 2013;23:109–115. doi: 10.1016/j.semcancer.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chow K-H, Factor RE, Ullman KS. The nuclear envelope environment and its cancer connections. Nat Rev Cancer. 2012;12:196–209. doi: 10.1038/nrc3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Linder S, Lindholm J, Falkmer U, Blasjo M, Sundelin P, von Rosen A. Combined use of nuclear morphometry and DNA ploidy as prognostic indicators in nonresectable adenocarcinoma of the pancreas. Int J Pancreatol. 1995;18:241–248. doi: 10.1007/BF02784948. [DOI] [PubMed] [Google Scholar]
- 96.Vasilescu C, Giza DE, Petrisor P, Dobrescu R, Popescu I, Herlea V. Morphometrical differences between resectable and non-resectable pancreatic cancer: a fractal analysis. Hepatogastroenterology. 2012;59:284–288. doi: 10.5754/hge11277. [DOI] [PubMed] [Google Scholar]
- 97.Timme S, Schmitt E, Stein S, Schwarz-Finsterle J, Wagner J, Walch A, Werner M, Hausmann M, Wiech T. Nuclear position and shape deformation of chromosome 8 territories in pancreatic ductal adenocarcinoma. Anal Cell Pathol (Amst) 2011;34:21–33. doi: 10.3233/ACP-2011-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fernandez-Zapico ME, Lomberk GA, Tsuji S, DeMars CJ, Bardsley MR, Lin Y-H, Almada LL, Han J-J, Mukhopadhyay D, Ordog T, et al. A Functional Family-Wide Screening of SP/KLF Proteins Identifies a Subset of Suppressors of KRAS-Mediated Cell Growth. The Biochemical journal. 2011;435:529–537. doi: 10.1042/BJ20100773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fernandez-Zapico ME, Mladek A, Ellenrieder V, Folch-Puy E, Miller L, Urrutia R. An mSin3A interaction domain links the transcriptional activity of KLF11 with its role in growth regulation. Embo j. 2003;22:4748–4758. doi: 10.1093/emboj/cdg470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Truty MJ, Lomberk G, Fernandez-Zapico ME, Urrutia R. Silencing of the transforming growth factor-beta (TGFbeta) receptor II by Kruppel-like factor 14 underscores the importance of a negative feedback mechanism in TGFbeta signaling. J Biol Chem. 2009;284:6291–6300. doi: 10.1074/jbc.M807791200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Calvo E, Grzenda A, Lomberk G, Mathison A, Iovanna J, Urrutia R. Single and combinatorial chromatin coupling events underlies the function of transcript factor krüppel-like factor 11 in the regulation of gene networks. BMC Molecular Biology. 2014;15:10–10. doi: 10.1186/1471-2199-15-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Loft A, Forss I, Siersbaek MS, Schmidt SF, Larsen AS, Madsen JG, Pisani DF, Nielsen R, Aagaard MM, Mathison A, et al. Browning of human adipocytes requires KLF11 and reprogramming of PPARgamma superenhancers. Genes Dev. 2015;29:7–22. doi: 10.1101/gad.250829.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lomberk G, Grzenda A, Mathison A, Escande C, Zhang JS, Calvo E, Miller LJ, Iovanna J, Chini EN, Fernandez-Zapico ME, Urrutia R. Kruppel-like factor 11 regulates the expression of metabolic genes via an evolutionarily conserved protein interaction domain functionally disrupted in maturity onset diabetes of the young. J Biol Chem. 2013;288:17745–17758. doi: 10.1074/jbc.M112.434670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Civelek M, Lusis AJ. Conducting the metabolic syndrome orchestra. Nature genetics. 2011;43:506–508. doi: 10.1038/ng.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fernandez-Zapico M, Molina J, Ahlquist D, Urrutia R. Functional characterization of KLF11, a novel TGFb-regulated tumor suppressor for pancreatic cancer. Pancreatology. 2003;3:436. [Google Scholar]
- 106.Neve B, Fernandez-Zapico ME, Ashkenazi-Katalan V, Dina C, Hamid YH, Joly E, Vaillant E, Benmezroua Y, Durand E, Bakaher N, et al. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4807–4812. doi: 10.1073/pnas.0409177102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bonnefond A, Lomberk G, Buttar N, Busiah K, Vaillant E, Lobbens S, Yengo L, Dechaume A, Mignot B, Simon A, et al. Disruption of a Novel Krüppel-like Transcription Factor p300-regulated Pathway for Insulin Biosynthesis Revealed by Studies of the c.-331 INS Mutation Found in Neonatal Diabetes Mellitus. The Journal of Biological Chemistry. 2011;286:28414–28424. doi: 10.1074/jbc.M110.215822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stacey SN, Sulem P, Masson G, Gudjonsson SA, Thorleifsson G, Jakobsdottir M, Sigurdsson A, Gudbjartsson DF, Sigurgeirsson B, Benediktsdottir KR, et al. New common variants affecting susceptibility to basal cell carcinoma. Nature genetics. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Seo S, Lomberk G, Mathison A, Buttar N, Podratz J, Calvo E, Iovanna J, Brimijoin S, Windebank A, Urrutia R. Kruppel-like factor 11 differentially couples to histone acetyltransferase and histone methyltransferase chromatin remodeling pathways to transcriptionally regulate dopamine D2 receptor in neuronal cells. J Biol Chem. 2012;287:12723–12735. doi: 10.1074/jbc.M112.351395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lomberk G, Mathison AJ, Grzenda A, Seo S, DeMars CJ, Rizvi S, Bonilla-Velez J, Calvo E, Fernandez-Zapico ME, Iovanna J, et al. Sequence-specific recruitment of heterochromatin protein 1 via interaction with Kruppel-like factor 11, a human transcription factor involved in tumor suppression and metabolic diseases. J Biol Chem. 2012;287:13026–13039. doi: 10.1074/jbc.M112.342634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fernandez-Zapico ME, van Velkinburgh JC, Gutierrez-Aguilar R, Neve B, Froguel P, Urrutia R, Stein R. MODY7 gene, KLF11, is a novel p300-dependent regulator of Pdx-1 (MODY4) transcription in pancreatic islet beta cells. J Biol Chem. 2009;284:36482–36490. doi: 10.1074/jbc.M109.028852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang J-S, Moncrieffe MC, Kaczynski J, Ellenrieder V, Prendergast FG, Urrutia R. A Conserved α-Helical Motif Mediates the Interaction of Sp1-Like Transcriptional Repressors with the Corepressor mSin3A. Molecular and Cellular Biology. 2001;21:5041–5049. doi: 10.1128/MCB.21.15.5041-5049.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yin KJ, Fan Y, Hamblin M, Zhang J, Zhu T, Li S, Hawse JR, Subramaniam M, Song CZ, Urrutia R, et al. KLF11 mediates PPARgamma cerebrovascular protection in ischaemic stroke. Brain. 2013;136:1274–1287. doi: 10.1093/brain/awt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fan Y, Guo Y, Zhang J, Subramaniam M, Song CZ, Urrutia R, Chen YE. Kruppel-like factor-11, a transcription factor involved in diabetes mellitus, suppresses endothelial cell activation via the nuclear factor-kappaB signaling pathway. Arterioscler Thromb Vasc Biol. 2012;32:2981–2988. doi: 10.1161/ATVBAHA.112.300349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nature reviews Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 117.Lomberk G, Bensi D, Fernandez-Zapico ME, Urrutia R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat Cell Biol. 2006;8:407–415. doi: 10.1038/ncb1383. [DOI] [PubMed] [Google Scholar]
- 118.Grzenda A, Lomberk G, Svingen P, Mathison A, Calvo E, Iovanna J, Xiong Y, Faubion W, Urrutia R. Functional characterization of EZH2β reveals the increased complexity of EZH2 isoforms involved in the regulation of mammalian gene expression. Epigenetics & Chromatin. 2013;6:3–3. doi: 10.1186/1756-8935-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]