Abstract
The protein ontology (PRO) is designed as a formal and well-principled open biomedical ontologies (OBO) foundry ontology for proteins. The components of PRO extend from the classification of proteins, on the basis of evolutionary relationships at the full-length level, to the representation of the multiple protein forms of a gene, such as those resulting from alternative splicing, cleavage and/or post-translational modifications, and protein complexes. As an ontology, PRO differs from a database in that it provides description about the protein types and their relationships. In addition, the representation of specific protein types, such as a phosphorylated protein form, allows precise definition of objects in pathways, complexes, or in disease modeling. This is useful for proteomics studies where isoforms and modified forms must be diffierentiated, and for biological pathway/network representation where the cascade of events often depends on a specific protein modification. PRO is manually curated starting with content derived from scientific literature. Only annotation with experimental evidence is included, and is in the form of relationship to other ontologies. In this tutorial, you will learn how to use the PRO resources to gain information about proteins of interest, such as finding conserved isoforms (ortho-isoforms), and different modified forms and their attributes. In addition, it will provide some details on how you can contribute to the ontology via the rapid annotation interface RACE-PRO.
Keywords: Biomedical ontology, Protein ontology, Community annotation, Protein
1. Introduction
Biomedical ontologies have emerged as critical tools in genomic and proteomic research where complex data in disparate resources need to be integrated. In this context, gene ontology (GO) (1) has become the common language to describe biological processes, protein function and localization. Protein or peptides detected in proteomic experiments are usually mapped to database entries, followed by data mining for GO terms and other data with the aim of characterizing the proteomic products (2).
However, there are some issues in capturing scientific knowledge based on the current infrastructure in that most sequence and organism databases provide gene-centric organization: one entry for one gene or canonical gene product. But in reality, many protein forms may derive from a single gene as a result of alternative splicing and/or subsequent posttranslational modifications. These various protein forms may have different properties. Therefore, the functional annotation of a protein may represent composite annotation of several protein forms, which may lead to noisy data mining results, and eventually to misinterpretation of data mining results. This missing infrastructure may also affect interoperability since some of the databases need to represent this level of granularity and create these objects independently, adding complexity to data integration.
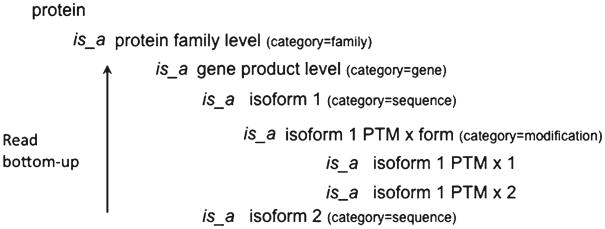
The protein ontology (PRO) (3, 4) is an OBO Foundry ontology that describes the different protein forms and their relationships in order to provide the appropriate framework for tackling the above-mentioned problems. PRO provides a means to refer to a specific protein object and append the corresponding annotations. This means that, for example, posttranslationally modified and unmodified forms of a given protein are two distinct objects in the ontology. Figure 1 shows a schematic representation of the ontology, which is organized in different levels (see Note 1) that can be grouped into four main categories (in decreasing hierarchical order):
Family: a PRO term at this level refers to proteins that can trace back to a common ancestor over the entire length of the protein. The leaf-most nodes at this level are usually families comprising paralogous sets of gene products (of a single or multiple organisms). In Fig. 2, PRO:000000676 is an example of this level. Note that the hierarchy in the ontology (Fig. 2a) reflects the evolutionary relationship of this group (Fig. 2b), HCN1-4 are paralogs that belong to the same homeomorphic family (full-length sequence similarity and have common domain architecture); therefore, in the ontology they are all under the same parent node (PRO:000000676).
Gene: a PRO term at this level refers to the protein products of a distinct gene. A single term at the gene-level distinction collects the protein products of a subset of orthologs for that gene (the subset that is so closely related that its members are considered the same gene). From the example depicted in Fig. 2 the HCN1 gene product (PRO:000000705) would include the proteins of the rat, mouse, rabbit and human HCN1 genes.
Sequence: a PRO term at this level refers to the protein products with a distinct sequence upon initial translation. The sequence differences can arise from different alleles of a given gene, from splice variants of a given RNA, or from alternative initiation and ribosomal frame shifting during translation. One can think of this as a mature mRNA-level distinction. Similarly to the gene product level, this level collects the protein products of a subset of orthologous splice variants for that gene, and we call them ortho-isoforms. Figure 3a shows an example of two nodes at the sequence level, PRO:000003420 and PRO:000003423, corresponding to isoform 1 (p75) and isoform 2 (p52) derived from gene LEDG. In this case literature is the data source for these protein forms. Figure 3b depicts the experimentally determined LEDG gene products (protein known as PC4 and SFRS1-interacting protein) based on the PMID:18708362 (5). Note that, although the experimental data displayed is from human, the article also describes the existence of these isoforms in mouse, so the human and mouse p75 isoforms will be both described by the PRO:000003420 term.
Modification: a PRO term at this level refers to the protein products derived from a single mRNA species that differ because of some change (or lack thereof) that occurs after the initiation of translation (co- and posttranslational). This includes sequence differences due to cleavage and chemical changes to one or more amino acid residues. Figure 3a shows an example of the cleaved version (p38) of isoform 2 (p52) of the LEDG gene. This level represents ortho-modified forms, the presence of posttranslational modifications on equivalent residues in ortho-isoforms.
Fig. 1.
PRO hierarchical organization. The ontology is read from bottom-up. PTM post-translational modification, x type of modification (such as acetylation, phosphorylation).
Fig. 2.
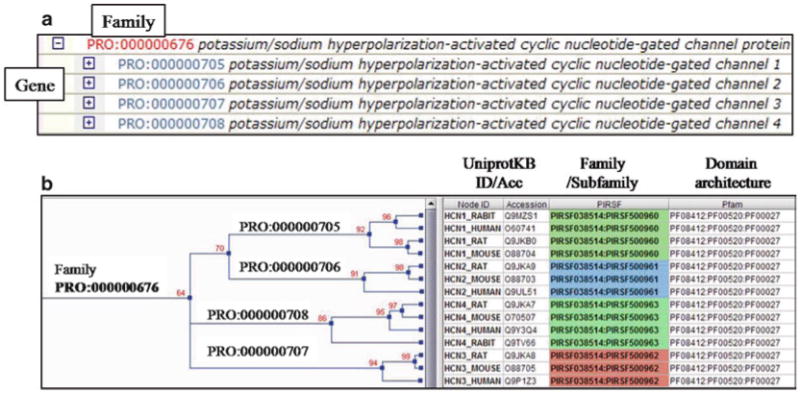
Family category refects the evolution of full-length proteins. (a) PRO ontology terms for the potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel protein. The family and gene product levels are shown. (b) Left panel: neighbor-joining tree showing the evolutionary relation of some representative proteins of the HCN1-HCN4 genes. The PRO IDs of each class is shown. Right panel: display of the corresponding database identifiers for: protein (UniProtKB), family (PIRSF (17)), and domain (Pfam).
Fig. 3.

Protein ontology to describe protein forms. (a) PRO ontology terms for the PC4 and SFRS1-interacting protein (derived from LEDG gene) depicting the isoforms, and modified forms. (b) Literature is the source for PRO forms; the scheme shows the different protein forms derived from the LEDG gene as described in a given article.
1.1. Relevance
We have previously described the various states of proteins involved in the TGF-beta signaling pathway (4), and also in the intrinsic apoptotic pathway (6). In the latter case, one key regulator of apoptosis is Bcl2 antagonist of cell death (Bad, PRO:000002184), whose phosphorylation state determines whether the cell fate is apoptosis or survival. It is generally stated that the BAD unphosphorylated form activates apoptosis and that the phosphorylated form of BAD leads to cell survival. However, the ontology shows that there are at least six distinct phosphorylated forms, which can be phosphorylated via activation of various kinases, such as AKT1, MAPK8 (JNK1), PKA, and CDC2. Although phosphorylation by the first three leads to interaction with the 14-3-3 proteins and cell survival, the outcome of the phosphorylation by CDC2 is the opposite, leading to translocation to the mitochondria an activation of apoptosis. This knowledge is key for the correct interpretation of proteomic results.
Therefore in this tutorial, you will learn how to use the PRO resources to gather this type of information about your protein(s) of interest.
2. Materials
The PRO website is accessible at http://pir.georgetown.edu/pro/pro.shtml.
2.1. Download
The ontology (pro.obo), the annotation (PAF.txt), and mappings to external databases can be downloaded from the ftp site at ftp.pir.georgetown.edu/databases/ontology/pro_obo/. This chapter is based on Release 8.0 v1. The ontology is also available in OBO and OWL formats through the OBO Foundry (7) and Bioportal (8). For general documentation please see http://pir.georgetown.edu/pro/pro_dcmtt.shtml.
2.2. PRO Files
The pro.obo file is in OBO 1.2 format and can be opened with OBO Edit 2.0 (9). This file displays a version information block, followed by a stanza of information about each term. Each stanza in the obo file is preceded by [Term] and it is composed of an ID, a name, synonyms (optional), a definition, comment (optional), cross-reference (optional) and relationship to other terms (see example below).
format-version: 1.2
date: 15:12:2009 13:48
saved-by: cecilia
auto-generated-by: OBO-Edit 2.0
default-namespace: pro
remark: release: 8.0, version 1
[Term]
id: PRO:000000003
name: HLH DNA-binding protein inhibitor
def: “A protein with a core domain composition consisting of a Helix-loop-helix DNA-binding domain (PF00010) (HLH), common to the basic HLH family of transcription factors, but lacking the DNA binding domain to the consensus E box response element (CANNTG). By binding to basic HLH transcription factors, proteins in this class regulate gene expression.” [PRO:CNA
]comment: Category=family.
synonym: “DNA-binding protein inhibitor ID” EXACT []
synonym: “ID protein” RELATED []
xref: PIRSF:PIRSF005808
is_a: PRO:000000001 ! protein
The annotations to PRO terms are distributed in the PAF.txt file. To facilitate interoperability to the best extent, this tab delimited file follows the structure of the gene ontology association (GAF) file. Please read the README file and the PAF guidelines. pdf in the ftp site to learn about the structure of this file. PRO terms are annotated with relation to other ontologies or databases. Currently in use: Gene ontology (GO) to describe processes, function and localization; Sequence ontology (SO) (10) to describe protein features; PSI-MOD (11) to describe protein modifications; MIM (12) to describe disease states; and Pfam (13) to describe domain composition.
2.3. Link to PRO
Use the persistent URL: http://purl.obolibrary.org/obo/PRO_xxxxxxxxx, where PRO_xxxxxxxxx is the corresponding PRO ID with an underscore (_) instead of semicolon (:). Example: link to PRO:000000447 would be http://purl.obolibrary.org/obo/PRO_000000447
3. Methods
3.1. PRO Homepage
The PRO homepage (http://pir.georgetown.edu/pro/pro.shtml) (Fig. 4) is the starting point to navigate through the protein ontology resources. The menu on the left side links to several documents and information pages, as well as to the ftp download page. The functionalities in the homepage include the subheadings: “PRO Browser,” “PRO Entry Retrieval,” “Text Search,” and “Annotation.”
Fig. 4.
PRO homepage (partial snapshot). The left menu links to documentation and downloads, whereas the right part displays the current functionalities.
3.1.1. PRO Browser
The browser is used to explore the hierarchical structure of the ontology (Fig. 5). The icons with a plus and minus signs allow expanding and collapsing nodes, respectively. Next to these icons is a PRO ID, which links to the corresponding entry report, followed by the term name. Unless otherwise stated the implicit relation between nodes is is_a.
Fig. 5.
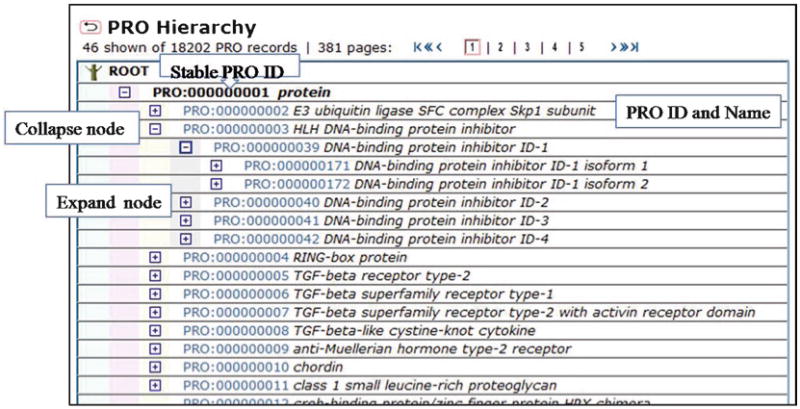
The PRO browser shows the ontology hierarchy. Use icons to expand/collapse nodes, or select an ID to go to the PRO entry view.
3.1.2. PRO Entry
The PRO entry provides an integrated report about the ontology and annotation available for a given PRO term. If you know the PRO ID you can use the “retrieve PRO entry” box in the homepage. Alternatively, you can open an entry by clicking on the PRO ID in any other page (search, browser, etc.). The entry report contains four sections (Fig. 6):
Ontology information: this section displays the information from the ontology about a term (source: the pro.obo file). You can link to the parent node, to the hierarchy, and find the definition and synonyms of the term, among other things.
Information about the entities that were use to create the PRO entry: this section lists the sequences, in the case where category corresponds to gene, sequence or modification, for which some experimental information exists. Taxon information as well as PSI-MOD ID and modification sites are indicated when applicable. In many cases, the modifications sites are unknown and therefore only the PSI-MOD ID is listed. For cleaved products, the protein region is indicated and is underlined in the displayed sequence (Fig. 6b). In the case of category corresponding to family, this section provides a cross-reference to the database that is the source of the class.
Synonymous mappings: this section contains mappings to external databases that link to protein forms as described in the given class (information source: mapping files). This is the case for Reactome (14) entry REACT_13251 which represents the human constitutive active form of ROCK-1 (Fig. 6c).
Annotation: This section shows the annotation of the term with the different ontologies (source: PAF file). These annotations were contributed by the PRO consortium group and by community annotators through submission of RACE-PRO annotations (see Subheading 3.1.4).
Fig. 6.
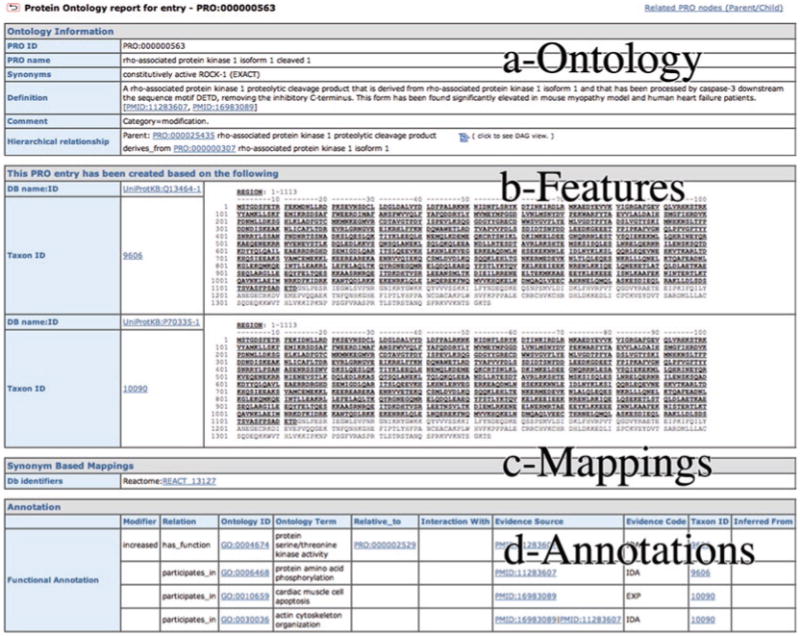
Sample PRO entry report. The different sections are indicated and explained in detail in the text.
3.1.3. Searching PRO
The search can be performed by entering a keyword or ID in the text box provided in the homepage. For example, you could just type the name of the protein for which you want to find related terms. Alternatively, advanced text search is available by clicking on the Search PRO title above the search box on the home page. Advanced text search supports Boolean (AND, OR, NOT) searches, as well as null (not present)/null (present) searches with several field options (see Note 2). Figure 7 shows an example of advanced search, which should retrieve all PRO terms that are in the modification category and contain annotation for protein– protein interaction.
Fig. 7.
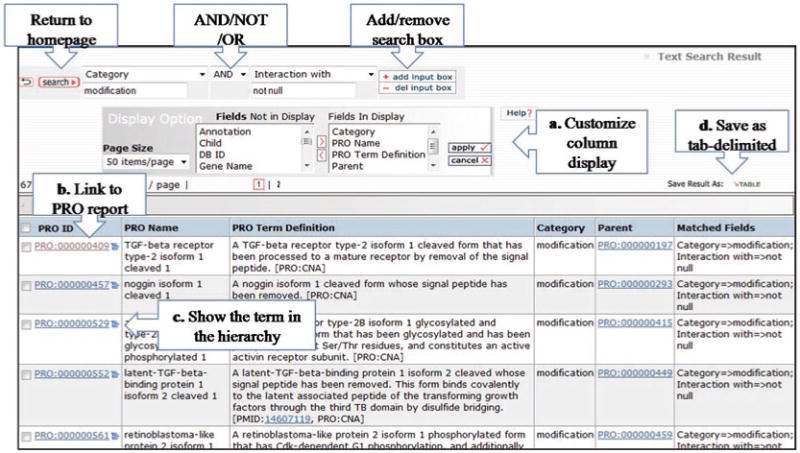
Advance search and result table.
Results are shown in a table format with the following default columns (Fig. 7): the PRO ID, PRO name, PRO term definition, the category, the parent term ID, and the matched field. Some of the functionality in this page includes:
Display Option: to customize result table by adding or removing columns. Use > to add or < to remove items from the list, but always select apply for the changes to take effect.
Link to PRO entry report: the link is available by selecting the PRO ID
Link to hierarchical view: the icon shows the term in the hierarchy, i.e., opens the browser.
Save: the result table as a tab-delimited file.
3.1.4. Annotation
The annotation section is for community interaction. The PRO tracker should be used to request new terms or to change/comment on existing ones. The link is directed to an external page (sourceforge) where you will need to provide the details about the terms of interest. On the other hand, if you have the data and domain knowledge you can directly submit annotation via the rapid annotation interface RACE-PRO as described below.
3.1.4.1. Rapid Annotation Interface RACE-PRO
Follow a few simple steps and become an author of annotations in PRO. As an example of the procedure, the annotation pertinent for the cleaved product p38 from Fig. 3 is shown in Fig. 8. First fill your personal information. This information will not be distributed to any third party, but will only be used for saving your data and for communication purposes.
Fig. 8.
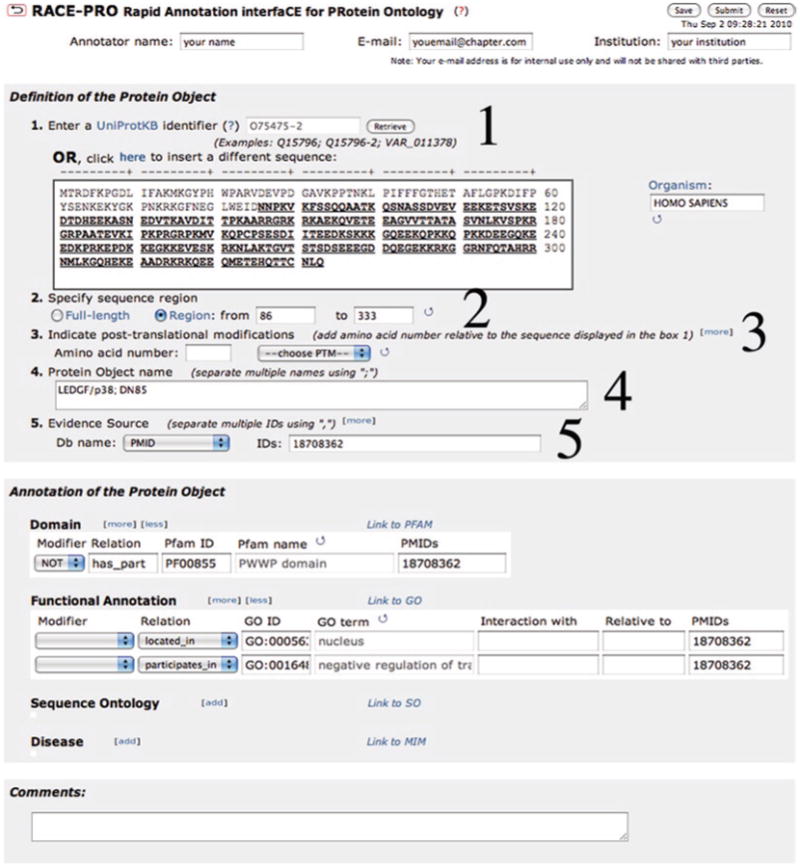
RACE-PRO entry to describe the cleaved product (p38) shown in Fig. 3b.
3.1.4.1.1. Definition of the Protein Object
This block allows you to enter all the information about a protein form along with the source of evidence. It is mandatory to add all the information relevant to this section whenever applicable.
Retrieve the sequence: if you use a UniProtKB identifier (15) and click “Retrieve,” the sequence retrieved is formatted to show the residue numbers, and the organism box is automatically filled. You can use identifiers for isoforms (a UniProtKB accession followed by a dash and a number) as in the example shown here. If you happen to have an identifier from a different database, you can use the ID mapping or batch retrieval services either from the PIR (16) or UniProt (17) websites to obtain the corresponding UniProtKB accession and retrieve the sequence – just be aware of which isoform or variant that you want to describe. Alternatively, you can paste a sequence, but in this case you will need to add the organism name (the link to NCBI taxonomy browser by clicking on the Organism title is provided as help).
Protein region: once the sequence is retrieved, you can select a subsequence in the cases where the protein form you are describing is not the full length, but a cleaved product or a fragment (as is the case of this example). After you do this, click on the circle arrow and the selected region will be underlined.
Selecting the Modification: If you need to describe a modification (or modifications), enter the residue number and the type of modification. If the modification is not in the list, use the “Other” option to add it. These terms will be later mapped to the corresponding PSI-MOD terms. If the modification site is unknown, please enter “?” in the residue number box. Use the [more] or the [less] to add or remove a modification line.
Be aware that the amino acid number should always refer to the sequence displayed in the sequence box. When clicking on the circle arrow, you will see the residues highlighted. Check that these are the ones expected. If there is no information about any posttranslational modification, then do not complete this line (as in the current example).
Protein object name: add names by which this object is referred to in the paper or source of data (separated by;). In the current example, both LEDG/p38 and DN85 are used to refer to the shorter cleaved form of LEDG isoform 2.
Evidence source: add the database (DB) that is the source of the annotation, in this case it is PubMed so we select as PMID. If the DB is not listed use the “Other” option and provide it. In the ID box you can add many IDs for a given DB separated by comma. Use the [more] or [less] to add or remove DB lines.
3.1.4.1.2. Annotation of the Protein Object
Only annotation from experimental data that is pertinent for the protein form (and species) described in the previous section should be added. There are three types of annotation that are based on different databases/ontologies: domain (Pfam), GO, and disease (MIM). If the paper describes the existence of a protein form with no associated properties, then do not fill this section.
All the information about the different columns in the table is described in the PAF guidelines. But below are some clarifications:
Modifiers: used to modify a relation between a PRO term and another term. It includes the GO qualifiers NOT, contributes to plus increased, decreased, and altered (to be used with the relative to column) e.g., “NOT has part PF00085 PWWP domain” is used because LEDG/p85 lacks this domain as determined in the paper, although it is present in the full length form.
Relation to the specific annotation. For some databases/ontologies there is a single relation possible and therefore it is already displayed, for GO we use three depending on the ontology used. Example: located in is used for GO component for subcellular locations, whereas participates in is used for GO biological processes.
Add ID for the specific database/ontology. If you need to search use the “link to..” link. If you enter the ID, the name autofills. Example: The paper shows in Fig. 5 that the p38 interferes with the transactivation potential of the full-length protein. Also the same figure shows the nuclear subcellular localization of this protein form. Then we can search for both GO terms in AMIGO and add the IDs to the annotation table.
The “Interaction with” column is used with the GO term “protein binding” to indicate to the binding partner. Please add the corresponding UniProtKB Acc and/or PRO ID. Examples with “Interaction with” column are found in any of the entry annotations from the PRO terms listed in Fig. 6.7.
The “Relative to” column is used only in conjunction with modifiers of the type increased, decreased and altered. In this column add the reference protein to which the protein form is being compared to. Either provide its UniProtKB Acc, its PRO ID, or its name. The annotation for rho-associated protein kinase 1 isoform 1 cleaved 1 in Fig. 6.6 has one such example: increased has_function GO:0004674 Relative to PRO:000002529 (rho-associated protein kinase 1 isoform 1 unmodified form).
3.1.4.1.3. Comment Section
Just add any comment that clarifies any of the content.
3.1.4.1.4. Saving/Submitting the Annotation
These options are found in the right upper corner of the RACE- PRO form. The save option allows saving the data in case you have not finished and need to complete the annotation later. When you save you are given a REF number; you can insert this number in the UniProtKB identifier box to retrieve your entry. Submit is used when you are done with the entry. You will still have the same reference number. Please keep it for tracking purposes.
3.1.4.1.5. What Happens Next?
An editor from the PRO team will review the entry and send you back comments/suggestions. Then the corresponding PRO term is generated along with the annotations. These will have the corresponding source attribution.
3.2. Conclusion
The PRO website can be used to retrieve information about the various protein forms derived from a given gene and to learn about their relationships. The integrated information for each form can be viewed in the entry report that collects information about the ontology and annotation (whenever available), and also provides mappings to external databases. This website constitutes a highly valuable resource providing a landscape of protein diversity and associated properties that is relevant for proteomics analysis.
4. Notes
Recently PRO has been funded to include protein complexes, so be aware that the structure of the framework may look slightly different in the future but the definition of each of the existing levels should not change. In addition, the PRO ID will soon change from PRO: to PR: to avoid confusion with other existing database identifiers.
- Some search tips:
- If you want to retrieve all the entries from a given category, for example, all the nodes for gene product level, then search selecting the category field and type gene. Search for category has the following options: family, gene, sequence, and modification.
- Some of the search fields are of the type null/not null. This is the case for the ortho-isoform and ortho-modified form. So if you are interested in retrieving the ortho-isoform entries, please select as a search field ortho-isoform and type not null.
- The specifics about what are the options for the DB ID, Modifiers and relations fields are listed in the PAF guidelines (see Subheading 2).
Acknowledgments
PRO Consortium participants: Protein Information Resource, The Jackson Laboratory, Reactome, and the New York State Center of Excellence in Bioinformatics and Life Sciences. PRO is funded by NIH grant #R01 GM080646-01.
References
- 1.The Gene Ontology Consortium. Gene ontology: tool for the unifcation of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li D, Li JQ, Ouyang SG, Wang J, Xu X, Zhu YP, He FC. An integrated strategy for functional analysis in large-scale proteomic research by gene ontology. Prog Biochem Biophys. 2005;32:1026–1029. [Google Scholar]
- 3.Natale D, Arighi C, Barker WC, Blake J, Chang T, et al. Framework for a Protein Ontology. BMC Bioinformatics. 2007;8(Suppl 9):S1. doi: 10.1186/1471-2105-8-S9-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arighi CN, Liu H, Natale DA, Barker WC, Drabkin H, Blake JA, Smith B, Wu CH. TGF-beta signaling proteins and the Protein Ontology. BMC Bioinformatics. 2009;10(Suppl 5):S3. doi: 10.1186/1471-2105-10-S5-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown-Bryan TA, Leoh LS, Ganapathy V, Pacheco FJ, Mediavilla-Varela M, Filippova M, Linkhart TA, Gijsbers R, Debyser Z, Casiano CA. Alternative splicing and caspase-mediated cleavage generate antagonistic variants of the stress oncoprotein LEDGF/p75. Mol Cancer Res. 2008;6:1293–1307. doi: 10.1158/1541-7786.MCR-08-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nchoutmboube J, Arighi CN, Wu CH. BIBM09, IEEE International Conference on Bioinformatics & Biomedicine. Washington, DC: 2009. Data integration and literature mining for the curation of protein forms in the protein ontology (PRO) [Google Scholar]
- 7.URL: http://www.obofoundry.org/.
- 8.URL: http://bioportal.bioontology.org/.
- 9.Day-Richter J, Harris MA, Haendel M, Lewis S Gene Ontology OBO-Edit Working Group. OBO-Edit – an ontology editor for biologists. Bioinformatics. 2007;23:2198–2200. doi: 10.1093/bioinformatics/btm112. [DOI] [PubMed] [Google Scholar]
- 10.Eilbeck K, Lewis SE, Mungall CJ, Yandell M, Stein L, Durbin R, Ashburner M. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 2005;6:R44. doi: 10.1186/gb-2005-6-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.URL: http://psidev.sourceforge.net/mod/.
- 12.URL: http://www.ncbi.nlm.nih.gov/sites/entrez?db=omim.
- 13.Finn RD, Mistry J, Schuster-Bockler B, Griffths-Jones S, Hollich V, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vastrik I, D'Eustachio P, Schmidt E, Joshi-Tope G, Gopinath G, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.URL: http://proteininformationresource.org/pirwww/search/idmapping.shtml.
- 17.Wu CH, Nikolskaya A, Huang H, Yeh LS, Natale DA, Vinayaka CR, Hu Z, Mazumder R, Kumar S, Kourtesis P, Ledley RS, Suzek BE, Arminski L, Chen Y, Zhang J, Cardenas JL, Chung S, Castro-Alvear J, Dinkov G, Barker WC. PIRSF family classification system at the Protein Information Resource. Nucleic Acids Res. 2004;32:D112–D114. doi: 10.1093/nar/gkh097. [DOI] [PMC free article] [PubMed] [Google Scholar]