Abstract
The physiological effects of anesthetics have been ascribed to their interaction with hydrophobic sites within functionally relevant CNS proteins. Studies have shown that volatile anesthetics compete for luciferin binding to the hydrophobic substrate binding site within firefly luciferase and inhibit its activity (Franks, N. P., and Lieb, W. R. (1984) Nature 310, 599–601). To assess whether anesthetics also compete for ligand binding to a mammalian signal transduction protein, we investigated the interaction of the volatile anesthetic, halothane, with the Rho GDP dissociation inhibitor (RhoGDIα), which binds the geranylgeranyl moiety of GDP-bound Rho GTPases. Consistent with the existence of a discrete halothane binding site, the intrinsic tryptophan fluorescence of RhoGDIα was quenched by halothane (2-bromo-2-chloro-1,1,1-trifluoroethane) in a saturable, concentration-dependent manner. Bromine quenching of tryptophan fluorescence is short range and W192 and W194 of the RhoGDIα are located within the geranylgeranyl binding pocket, suggesting that halothane binds within this region. Supporting this, N-acetyl-geranylgeranyl cysteine reversed tryptophan quenching by halothane. Short chain n-alcohols (n<6) also reversed tryptophan quenching, suggesting that RhoGDIα may also bind n-alkanols. Consistent with this, E193 was photo-labeled by 3-azibutanol. This residue is located in the vicinity of, but outside, the geranylgeranyl chain binding pocket, suggesting that the alcohol binding site is distinct from that occupied by halothane. Supporting this, N-acetyl-geranylgeranyl cysteine enhanced E193 photo-labeling by 3-azibutanol. Overall, the results suggest that halothane binds to a site within the geranylgeranyl chain binding pocket of RhoGDIα, whereas alcohols bind to a distal site that interacts allosterically with this pocket.
Despite the routine use of volatile anesthetics in surgical procedures for over 150 years, the sites and mechanism of action remain largely unresolved. It has been proposed that these agents interact with hydrophobic sites within functionally relevant CNS proteins (1, 2), and examples of such sites within ion channels and receptors have been noted (3). Two mechanisms have been proposed for anesthetic action on protein structure/function (3). In the first mechanism, anesthetics bind selectively to a subset of the conformations that are available to a protein, and thereby act as classical allosteric agents. Such a mechanism has been shown to apply to the interaction of anesthetics with the nicotinic acetylcholine receptor (4, 5), and also with protein kinase C (PKC), where general anesthetics and alcohols appear to allosterically modulate phorbol ester binding to the C1 domains (6–8). In the second mechanism, anesthetics interact directly with binding sites that are normally utilized by endogenous ligands. Experimental evidence supporting this second mechanism comes exclusively from non-mammalian systems where kinetic studies suggest that alcohols and anesthetics compete for the substrate binding sites for luciferin on the firefly luciferase (9) and for aldehyde on the bacterial luciferase (10, 11).
To test whether this second mechanism might also apply to the interaction of anesthetics with a mammalian protein, we chose Rho GDP dissociation inhibitor α (RhoGDIα),1 which is a pivotal regulator of the biological activities the Rho family of low molecular weight GTPases (Rho GTPases) and shares with the Rho GTPases a central role in the regulation of the cytoskeleton (12–15). The choice of RhoGDIα was also based on the fact that in common with other protein targets for anesthetics, it contains a hydrophobic pocket. This pocket is contained within the C-terminal immunoglobulin-like fold of RhoGDIα and is lined by a number of hydrophobic residues, including W192 and W194 (16, 17). It is formed by two β-sheets that undergo a rigid-body re-orientation upon binding of the C-terminal geranylgeranyl (GG) chain of the Rho GTPases (16), for which it displays a high degree of structural specificity (18). This interaction contributes to the stabilization of the RhoGDIα–Rho GTPase complex, which maintains the Rho GTPase in an inactive, cytosolic GDP-bound state (19–22) It also mediates the extraction of the GDP-bound Rho GTPases from the membrane by sequestering the GG chain, which results in the cycling of the Rho GTPases back into the cytosol as well as to other membrane compartments (23, 24).
In the present study, we investigated whether alcohols and anesthetics interact with the hydrophobic binding pocket within the C-terminal domain of RhoGDIα and assessed ensuing effects on GG chain binding to this pocket. Two strategies were used to locate anesthetic sites. First, we took advantage of the presence of two tryptophans within the GG chain binding pocket (W192 and W194) to monitor the interaction of the halogenated anesthetic, halothane, based on quenching of the emission fluorescence of tryptophan, a method utilized previously in studies of anesthetic binding to serum albumins (25, 26), and α-helical bundle proteins (27, 28). Second, we employed photo-labile alcohols to locate the site of alcohol action (6, 29–32).
EXPERIMENTAL PROCEDURES
Materials
Grace’s insect media, phosphate buffered saline (PBS), fetal bovine serum (FBS), 5-hexadecanoylaminofluorescein (HAF), 3’-O-(N-methylanthraniloyl)-β:γ-imidoguanosine 5’-triphosphate trisodium salt (MANT-GMPPNP), 2'-(or-3')-O-(N-methylanthraniloyl)guanosine 5'-diphosphate, disodium salt (MANT-GDP) and Spodoptera frugiperda (Sf9) cells, and primers were each obtained from Invitrogen Technologies (Carlsbad, CA). The L1 Sensor Chip was purchased for use in a Biacore™ 2000 surace plasmon resonance (SPR) system (Biacore Inc, Piscataway, NJ). Protease Inhibitor Cocktail, guanosine 5’-[β, γ-imido] triphosphate trisodium salt (GMPPNP), halothane (2-bromo-2-chloro-1,1,1-trifluoroethane), n-alkanols, and all other research grade chemicals were from Sigma-Aldrich (St. Louis, MO). The n-alkanol concentration ranges used were normalized on the basis of alcohol hydrophobicity, estimated from octanol/water partition coefficients, as previously described (7). N-acetyl geranylgeranyl cysteine (AGGC) was from Calbiochem (La Jolla, CA); 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS) were from Avanti Polar Lipids Inc (Alabaster, AL). The photo-labile alcohol, 3-azibutanol was synthesized according to the method of Church et al (33), and 3-azioctanol was synthesized as described previously (34). Baculovirus encoding Human Cdc42 N-terminal tagged with 6xHis was a kind gift from Dr. R. A. Cerione (Department of Molecular Medicine, Veterinary Medical Center, Cornell University, NY); baculovirus encoding 6xHis tagged RhoA was prepared as described previously (35). Baculovirus containing RhoGDIα N-terminal tagged with GST was custom manufactured by Orbigen (San Diego, CA). Molecular graphics images were produced using the UCSF Chimera package (36) from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081).
Purification of RhoGDIα
Log phase Sf9 cells in 1 L of Grace’s insect media and 10% v/v FBS were infected with baculovirus encoding RhoGDIα-GST at a multiplicity of infection of 5 and incubated for 4 days. The cells were then washed once with PBS, pelleted, and lysed using a Dounce homogenizer in Buffer A (10 mM HEPES, pH 8.0, 150 mM NaCl, 5 mM MgCl2) containing protease inhibitor cocktail. The cell lysate was homogenized and centrifuged at 3500g for 15 min at 4°C in a Sorvall SS-34 rotor to remove whole cell debris and nuclei. The resultant supernatant was centrifuged at 100,000g for 60 min in a Beckman Ti70 rotor to separate cytosolic from membrane and cytoskeleton fractions. The cytosolic fraction (supernatant) was then incubated at 4°C for 60 min with gentle rocking with 1 mL of glutathione agarose resin that had been pre-equilibrated in Buffer A. The resin was then packed into a 5 mL column attached to an ÄKTApurifier™ FPLC system (GE Healthcare, Piscataway, NJ) and washed with Buffer A until the absorbance of the flow-through at 280 nm was <0.01 absorbance units. RhoGDIα was liberated from the GST tag by thrombin cleavage while the fusion protein was still attached to the glutathione agarose resin. Cleavage was carried out by passing 0.1 units/mL α-thrombin in Buffer A though the column at 25°C for 2 hrs. The fractions containing protein were pooled and applied to a size exclusion column in order to remove pro-thrombin and α-thrombin. The pooled eluted protein was dialyzed against Buffer A for 36 hours with buffer changes every 12 hours. Protein concentrations were determined by Bradford protein assay (BioRad, Faraday, CA) and protein were stored at −80°C in Buffer A containing 20% glycerol. The concentrated RhoGDIα stock was diluted for experiments with either Buffer A or Buffer B (20 mM Tris/HCl, pH 7.5, 5 mM MgCl2, 50 mM NaCl).
Purification of RhoGDIα in complex with Rho GTPases
In order to circumvent problems arising from the low aqueous solubility of the Rho GTPases that results from the hydrophobic nature of the C-terminal GG chain, Cdc42 and RhoA were expressed and purified as a 1:1 stoichiometric complex with RhoGDIα. In this complex the geranylgeranyl chain of each Rho GTPase is shielded from the aqueous environment by insertion into the hydrophobic binding pocket on RhoGDIα (16, 17). The complex was purified as described previously (35), except that the Rho GTPase–RhoGDIα complexes were liberated from the GST tag by thrombin cleavage while the fusion protein was still attached to glutathione–agarose. Cleavage was carried out by passing 0.1 units/mL α-thrombin in Buffer A though the column at 25°C for 2 hrs. The fractions containing protein were pooled and applied to a size exclusion column in order to remove pro-thrombin and α-thrombin.
Halothane binding assay
The interaction of halothane with RhoGDIα was determined from measurements of the corresponding quenching of the intrinsic emission tryptophan fluorescence, as described previously (25, 26). Briefly, RhoGDIα (500 nM) was placed into a screw-top sealed 3 mL cuvette with 2.8 mL of Buffer B. The calculated amount of halothane (0–800 µM) was then added to the cuvette in a volume calculated to minimize headspace, which was then promptly sealed. After incubation for 60 min at room temperature, the steady-state emission spectrum of tryptophan was collected using an ISS upgraded SLM 48000 (ISS, Champaign, IL). The excitation wavelength (295 nm) was chosen so as to minimize the contribution of energy transfer from tyrosine residues to the observed fluorescence signal (37). The effects of AGGC on halothane binding was determined by incubating the required concentrations of AGGC (0–300 µM) with 500 nM RhoGDIα in the presence or absence of 1 mM halothane in 0.5 mL Buffer B for 30 min in gas-tight Hamilton syringes. The mixture was then diluted into 2 mL of Buffer B containing 500 nM RhoGDIα in a 3 mL cuvette, which was promptly sealed prior to recording the tryptophan emission spectra. For determinations of the effects of n-alkanols on halothane binding, the required concentration of halothane was initially added to RhoGDIα (500 nM) in a screw-top sealed 3 mL cuvette with 2.8 mL Buffer B, and the fluorescence intensity at 330 nm upon excitation at 295 nm was monitored as a function of time. After allowing equilibration, n-alkanols were titrated into the cuvette in 10 µL aliquots from concentrated stock solutions in Buffer B. In a separate experiment the operation and timing of each alcohol addition was mimicked by identical additions of Buffer B, in order to control for the effects of halothane evaporation on the fluorescence signal. The increase in baseline fluorescence, due to evaporation of halothane and loss of tryptophan quenching, was found to be <5% of the initial quenched signal. The contribution of inner filter effects to the observed signal was judged to be negligible, based on the observation that halothane had a molar extinction coefficient of <100 M−1 cm−1 at 295 nm (37). The same batch of RhoGDIα was used for each individual halothane quenching experiment, because some batch-to-batch variability was apparent in the extent of tryptophan quenching.
Preparation of Vesicles
Large unilamellar vesicles (LUV) were prepared from lipids as described previously (38). Briefly, chloroform solutions of POPC and POPS (4:1, molar) at a total lipid concentration of 500 µM were mixed in a test tube and the solvent was evaporated under a stream of nitrogen to form a homogenous thin film. The required volume of buffer (10 mM HEPES, pH 7.4 and 150 mM NaCl) was then added and the lipids were allowed to hydrate for 15 min at 25°C. Following this, multilamellar vesicles were formed by vortexing for 1 min. Large unilamellar vesicles of 100 nm diameter (LUV) were prepared from multilamellar vesicles by the extrusion technique, using an Avestin Liposofast Extruder (MM Developments, Ottawa, Canada), also as previously described (39).
Surface Plasmon Resonance determinations
The dissociation of Cdc42 or RhoA from the complex with RhoGDIα was quantified based on the time dependence of the association of the Rho GTPases with membranes immobilized on the hydrophobic surface of an L1 sensor chip in a Biacore™ 2000 (Biacore, Inc., Piscataway, NJ), as described previously (35). Briefly, the surface of the L1 sensor chip was initially cleaned by 2 injections of 10 mM CHAPS and POPC/POPS LUV were then captured on this surface using a flow rate of 5 µL/min and a contact time of 15 min. All measurements were performed at 25°C using a running buffer consisting of 10 mM HEPES, pH 7.4 and 150 mM NaCl. Previous studies have shown that the L1 chip surface is completely covered under the conditions used and that the resultant lipid surface resembles a rough bilayer structure (40, 41). The POPC/POPS surface was then conditioned with sequential injections of 10 mM glycine (pH 1.5) and 10 mM NaOH to remove loosely associated vesicles and to minimize baseline drift. The RhoA–RhoGDIα or RhoGDIα–Cdc42 complex was then diluted to the required final concentration, which was typically 0.5 µM unless otherwise stated, in Buffer A containing 50 µM GMPPNP and the required concentration of n-alkanol. Following this, the free Mg2+ concentration was reduced from 5 mM to <0.5 µM by chelation with 14.3 mM EDTA in order to trigger nucleotide exchange and initiate complex dissociation. The resultant mixture was then immediately injected over the POPC/POPS surface at a flow rate of 10 µL/min, and the increase in the SPR signal (response) due to capture of geranylgeranylated Rho GTPases was monitored as a function of time. The L1 chip surface was regenerated with three 10 µL injections of CHAPS at a flow rate of 5 µL/min.
Analysis of fluorescence quenching data
The extent of tryptophan quenching by halothane (Q) was calculated from the fractional decrease in fluorescence intensity, FQ / F0, where FQ and F0 are the tryptophan fluorescence intensities measured with and without halothane, respectively. These values were determined by averaging >10 data points at the corresponding emission wavelength maximum. The affinity constant (KD) and Hill coefficient (h) for halothane binding were obtained by plotting values of FQ / F0 against halothane concentration and fitting this data to the following modified Hill equation using non-linear least-squares regression analysis:
| (Eq. 1) |
where Fmax is the fluorescence intensity at maximal quenching. The effects of n-alkanols and AGGC on halothane binding to RhoGDIα were determined from plots of the recovery in tryptophan fluorescence as a function of ligand concentration. Fluorescence recovery was equal to (FL – F0) / (F0 – FH) where, F0 is the tryptophan fluorescence intensity without halothane or ligand (n-alkanol or AGGC), FH is the intensity with halothane, and FL is the intensity with halothane and ligand. The concentration of ligand required for a 50% recovery in tryptophan fluorescence (EC50) and the maximal extent of fluorescence recovery (Fmax) was calculated from fits to the following expression using non-linear least-squares regression analysis:
| (Eq. 2) |
The apparent inhibitory constants for each ligand (KI) were determined from the concentration dependent effects of the ligands on the KD for halothane binding, according to the following expression (42):
| (Eq. 3) |
The estimated incremental changes in the apparent free energy of n-alkanol binding upon addition of a methylene unit were calculated from values of KI obtained for each alcohol using Eq. 3, according to:
| (Eq. 4) |
Where R is the universal gas constant, T is the temperature (Kelvin), and n is the n-alkanol chain length. Goodness of fit was assessed from values of average squared residual (χ2).
Analysis of SPR data
Whereas the Biacore™ system allows affinity constants to be determined from the ratio of association and dissociation rate constants, the rate of dissociation of Cdc42 from the POPC/POPS membranes was found to be too slow for accurate determination of a dissociation rate constant. The binding constant for the interaction of Cdc42 with membranes was therefore determined from measurements of the extent of Cdc42 binding at equilibrium (Req,Cdc42) as a function of the concentration of RhoGDIα–Cdc42 complex injected (35). Values of Req,Cdc42 were obtained by fitting the response (Rt) verses time curves for each analyte concentration to an integrated first-order rate equation using non-linear regression analysis:
| (Eq. 5) |
Where R0 is the initial response and kobs is the observed first order rate constant. Values of Req,Cdc42 as a function of n-alkanol concentration were then fitted to a modified Hill equation:
| (Eq. 6) |
Where R0 is the initial response in the absence of analyte, Rmax is the response corresponding to maximal occupation of membrane binding sites, Kapp is the apparent association constant, and h is the Hill coefficient. Goodness of fit was assessed from values of average squared residual (χ2).
Extraction of Cdc42 from membranes by RhoGDIα
The extraction of Cdc42 from membranes by RhoGDIα was determined from the accompanying decrease in fluorescence energy transfer (FRET) between the MANT-fluorophore of MANT-GMPPNP bound to Cdc42 and the membrane probe, HAF, based on a previously described method (23, 43). For these experiments, Cdc42 was expressed in Sf9 cells and purified in the absence of RhoGDIα using procedures that were identical to that described above for the RhoGDIα–Cdc42 complex, except that the purification by glutathione affinity chromatography was omitted. Cdc42 was initially loaded with MANT-GMPPNP as described previously (43). Briefly, 14.3 mM EDTA was added to 60 nM Cdc42 in Buffer B containing 2 µM MANT-GMPPNP, which triggers nucleotide exchange by reducing the free Mg2+ concentration from 5 mM to <0.5 µM. To this were added LUV composed of POPC/POPS (4:1, molar) containing 2 µM HAF (300 µM total lipid concentration) and 50 mM n-butanol as required, and the resultant solution was allowed to equilibrate for 30 min. The emission fluorescence intensity of MANT-GMPPNP was then measured at 25°C as a function of time at 430 nm upon excitation at 355 nm using an ISS modified LSM 48000 multi-frequency phase and modulation fluorimeter. Upon attaining a stable baseline, the extraction of membrane bound Cdc42 was initiated by the addition of 60 nM RhoGDIα in Buffer B, and monitored by measuring the recovery in the emission fluorescence intensity due to the de-quenching of MANT-GMPPNP as a function of time.
Fluorescence-based nucleotide exchange assay
The effects of n-alkanols on the intrinsic nucleotide exchange rates were determined based on a previously described method (23). Briefly, Cdc42 (100 nM) was incubated with 300 nM MANT-GDP in the presence and absence of the required concentration of n-alkanol for 30 min in 1.5 ml of Buffer B, and the MANT fluorescence intensity at 430 nm upon excitation at 360 nm was measured as a function of time. Nucleotide exchange was initiated by adding 2 µM GDP in the presence of 16 mM EDTA and the resultant fluorescence intensity decrease was measured as a function of time.
Photo-affinity labeling
RhoGDIα (2.8 nM) in water (150 µL) was incubated for 30 min without or with AGGC (550 nM) in the presence of 3-azibutanol or 3-azioctanol, which was added from an aqueous solution to yield final alcohol concentrations of 0.01, 0.1 and 1.0 mM in a total reaction volume of 210 µL. This solution was then irradiated on ice at 365 nm using a hand held Black-Ray UV lamp model UVL-56 at a distance of ~ 1 cm for 45 min in a 96-well glass plate. The photoreaction was then quenched by the addition of SDS-PAGE sample buffer.
In-gel tryptic digestion
Approximately 10 µg of photolabeled RhoGDIα was separated by SDS-polyacrylamide gel electrophoresis using a 10% polyacrylamide gel followed by Coomassie blue staining. The band corresponding to the molecular weight of RhoGDIα was cut into ~2 mm3 pieces and incubated overnight at room temperature with 200 µL of 50 mM ammonium bicarbonate solution, and then washed with acetonitrile for 10 min. The solvent was then removed under vacuum and the gel pieces were rehydrated with aqueous 50 mM ammonium bicarbonate solution containing 12.5 ng/µL modified sequencing-grade trypsin (Promega, Madison, WI) at 4°C. After 45 min, the excess trypsin solution was removed and replaced with a volume of 50 mM ammonium bicarbonate solution that just covered the gel pieces, which were then incubated at 37°C overnight. Peptides were then extracted from the gel pieces by first removing the ammonium bicarbonate solution and then performing two consecutive 20 min incubations in a solution composed of 50% v/v acetonitrile and 5% v/v formic acid. The extracts were combined, dried under vacuum for ~1 hr and stored at 4°C until analysis.
Online HPLC and Mass Spectrometry
LC-MSMS experiments were performed on an LTQ-FT mass spectrometer (ThermoFinnigan, San Jose, CA). This is a fully integrated hybrid mass spectrometer consisting of a linear ion trap mass spectrometer combined with a Fourier transform ion cyclotron resonance mass spectrometer. Peptides obtained from tryptic digestion were initially reconstituted in 5 µL of an aqueous solution composed of 2.5% v/v acetonitrile, 0.1% v/v formic acid. A nano-scale reverse-phase HPLC capillary column was created by packing 5 µm C18 spherical silica (pore size 200 Å) into a 12 cm fused silica capillary tube of 75 µm inner diameter with a flame-drawn tip fabricated in-house (44). The column was attached to the HPLC system and peptides were eluted by a linear concentration gradient (acetonitrile with 0.1% formic acid increasing from 10 to 38% over 30 min followed by a step increase to 100%. Each eluted peptide was subjected to electrospray ionization. The m/z of each isolated peptide was determined to high accuracy by FTMS. The remainder of the sample was fragmented and its tandem mass spectrum (MSMS) was analyzed to determine its sequence. The position of photo-incorporation was analyzed using the Bioworks browser 3.1 module of the program Xcalibur (ThermoFinnigan, San Jose, CA).
RESULTS
Direct interaction of halothane with RhoGDIα
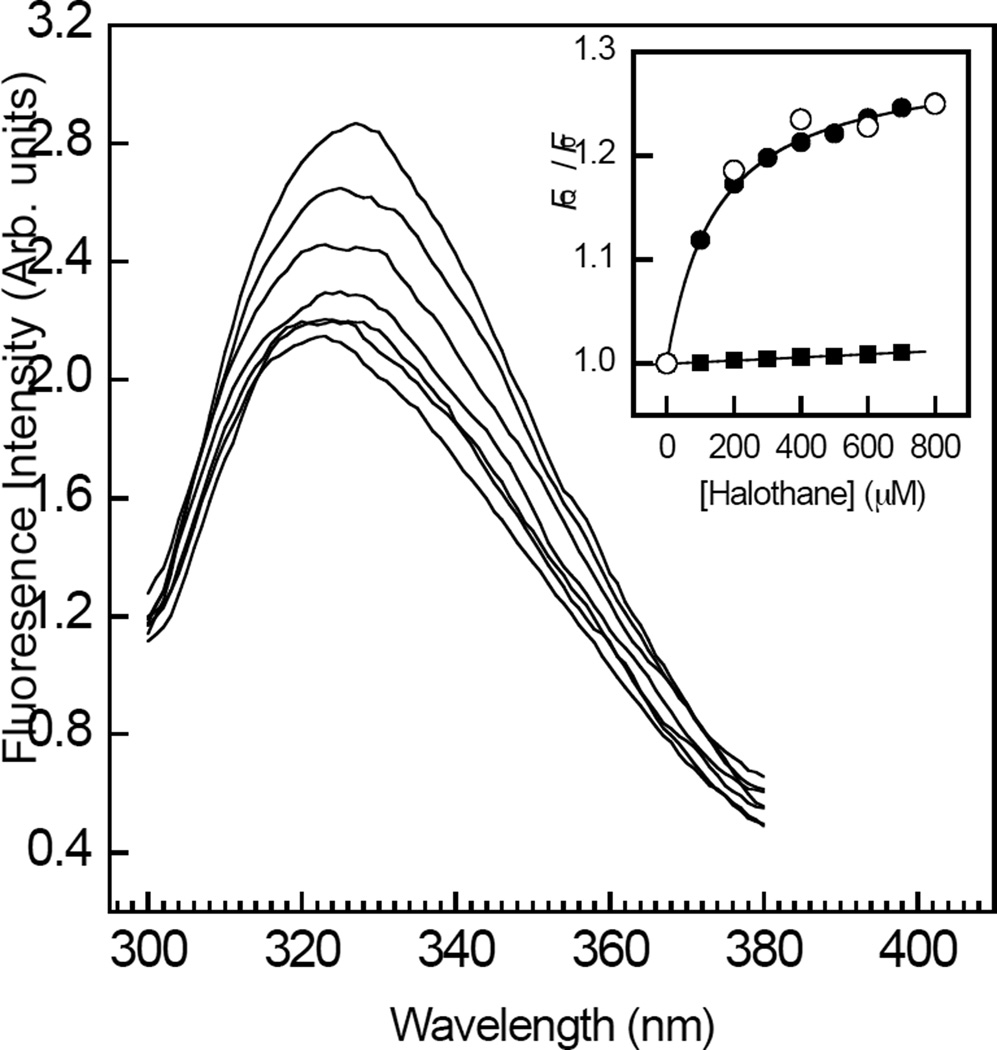
The fluorescence emission spectra of RhoGDIα, obtained upon excitation at 295 nm in the absence of halothane, consisted of an asymmetric peak centered at 327 nm (Figure 1). Consistent with a direct interaction of halothane with RhoGDIα, incubation of the protein with increasing levels of the anesthetic resulted in a quenching of fluorescence emission intensity. The anesthetic also induced a slight dose dependent blue shift in the emission maxima (~2 nm), indicating a small decrease in the dielectric local to the tryptophan residues of RhoGDIα (Figure 1). The tryptophan fluorescence intensity decreased as a function of halothane concentration to a plateau value corresponding to ~30 % of the original signal (Figure 1, inset, ●). Fitting this hyperbolic curve to a modified Hill equation (Eq. 1), yielded values of KD = 147±7 µM, and h = 0.91±0.2, suggesting the presence of one or more non-interacting halothane binding sites on RhoGDIα. The quenching effect of halothane on free L-tryptophan fluorescence intensity was reduced compared to that on RhoGDIα tryptophan fluorescence (Figure 1, inset, ▪), and was a linear function of anesthetic concentration, contrasting with the saturating effect of halothane. Fitting this data to the Stern-Volmer equation: FQ / F0 = 1 + KSV [halothane], using linear regression, yielded a quenching constant, KSV = 15.7±0.2 M−1, which is close to a previous value (26). The possibility that the observed decrease in tryptophan emission might have resulted from a quenching effect of halothane on tyrosine to tryptophan resonance energy transfer (37), was ruled out by the finding that the halothane concentration-response curve was unaffected upon increasing the excitation wavelength to 305 nm (Figure 1, inset, ○).
Figure 1.
Concentration dependent effects of halothane on the steady-state tryptophan emission fluorescence of RhoGDIα. Tryptophan emission fluorescence spectra of RhoGDIα (500 nM) were acquired upon excitation at 295 nm in the presence of increasing levels of halothane (0–800 µM). Inset, the fluorescence intensities at maximal wavelengths obtained using an excitation wavelength of 295 nm (●) or 305 nm (○) were plotted as a function of halothane concentration. The solid curve corresponds to a least squares non-linear regression fit of the data obtained using an excitation wavelength of 295 nm to Eq. 1. Also shown is the concentration dependent effect of halothane on the fluorescence intensities at maximal wavelength obtained for free L-tryptophan upon excitation at 295 nm (▪). The solid line through the data was obtained using linear regression. The data are representative of triplicate experiments. The fits of each individual replicate experiment yielded similar parameter values and errors. Other details are given under “Experimental Procedures.”
AGGC inhibits halothane binding to RhoGDIα
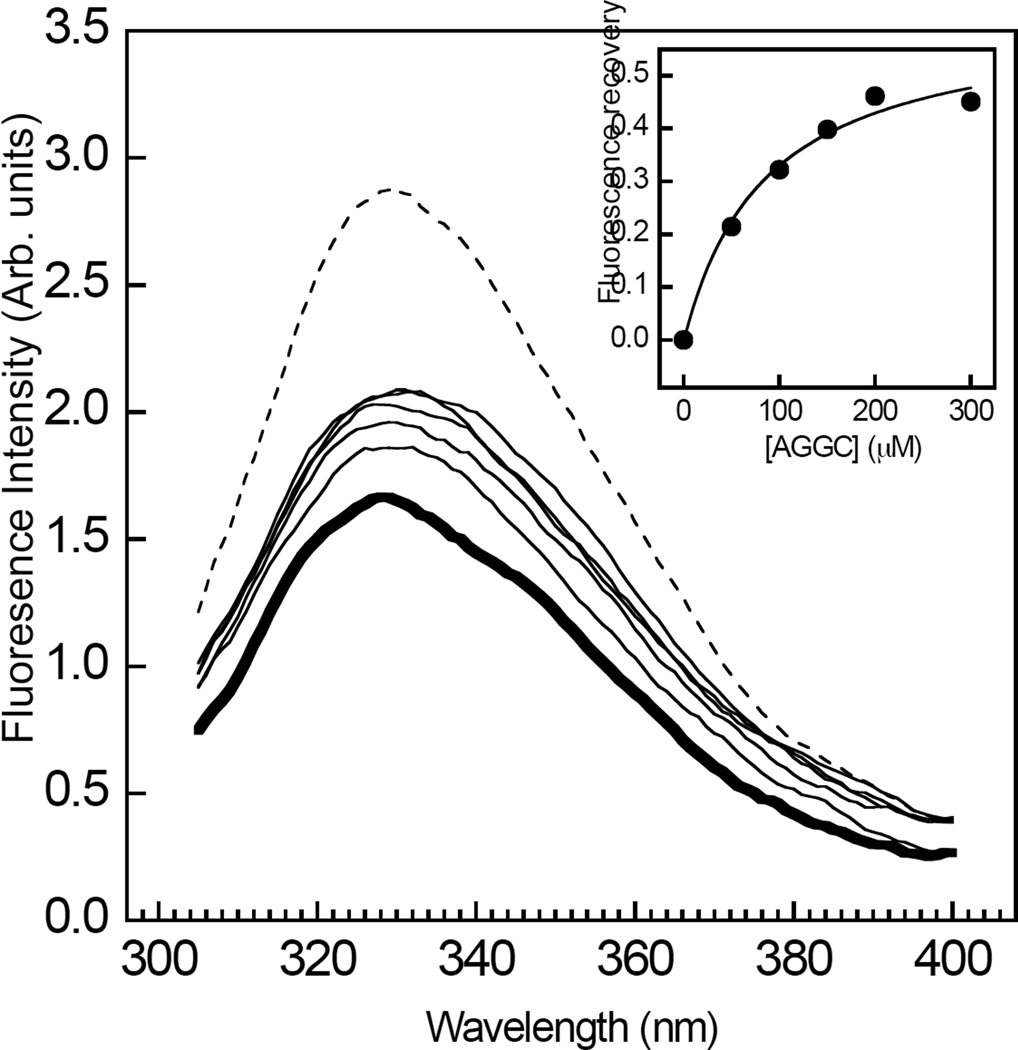
In order to address the question whether the interaction of halothane with RhoGDIα competes with GG chain binding, the quenching of RhoGDIα tryptophan fluorescence by the anesthetic was determined in the presence of increasing levels of the ligand, AGGC. Although lacking the methoxy group that is attached during post-translational modification, this compound mimics the geranylgeranylated C-terminal cysteine of Rho GTPases and has been shown to bind specifically to RhoGDIα (18). Consistent with the results shown in Figure 1, RhoGDIα tryptophan fluorescence was quenched by 1 mM halothane (Figure 2). The titration of AGGC elicited a saturable decrease in the extent of tryptophan quenching (Figure 2). Plotting the fluorescence intensities at each emission maxima against AGGC concentration yielded a hyperbolic concentration-response curve (Figure 2, inset). Fitting this data to Eq. 2 yielded values of EC50 = 65±15 µM, h = 1.2±0.5 and Fmax = 0.5±0.1. The value of KI for the displacement of halothane by AGGC, obtained by substituting EC50 into Eq. 3, was 10 µM, which is close to the apparent dissociation constant for this compound reported in a previous study (18). The possibility that the apparent de-quenching effect of AGGC might have resulted from a change in the local environment of the tryptophans upon binding of the ligand was ruled out in separate control experiments showing that tryptophan fluorescence and emission maxima wavelength were unaffected by AGGC in the absence of halothane (results not shown).
Figure 2.
Concentration dependent effects of AGGC on the halothane induced quenching of RhoGDIα tryptophan emission fluorescence. The steady-state tryptophan emission fluorescence spectra of RhoGDIα (500 nM) were obtained with (bold) or without (dashed) 1 mM halothane, in the presence of increasing concentrations of AGGC (0–300 µM). Inset, the recovery in emission fluorescence from quenching by 1 mM halothane in the presence of AGGC was plotted as a function of AGGC concentration. The solid curve corresponds to a fit of the data obtained to Eq. 2 using least squares non-linear regression analysis. Data are representative of at least three independent experiments. The fits of each individual replicate experiment yielded similar parameter values and errors. Other details are described in “Experimental Procedures.”
Effects of n-alkanols on halothane binding
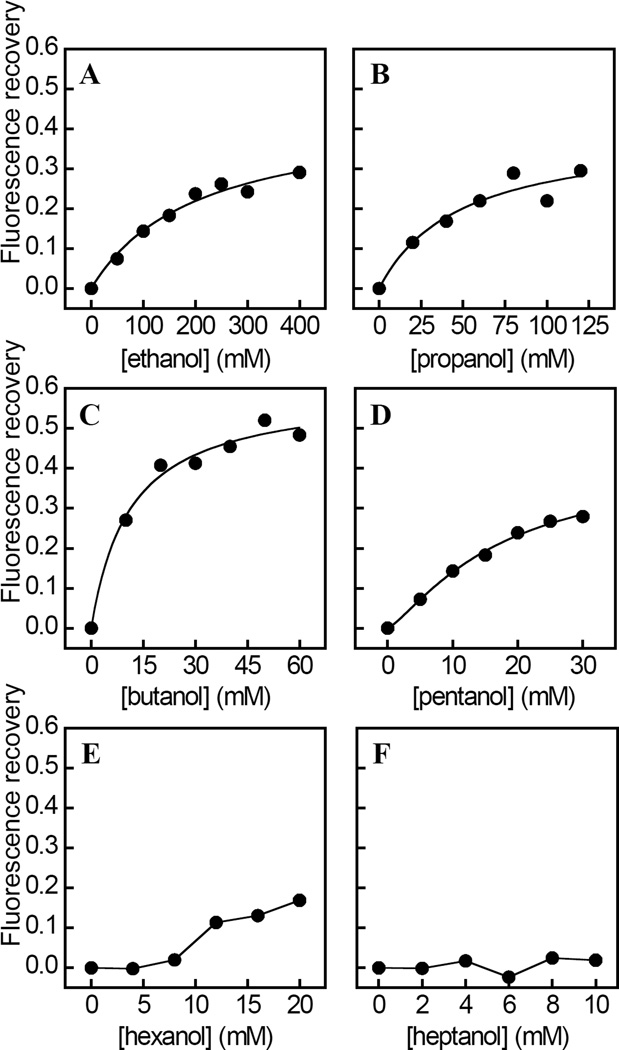
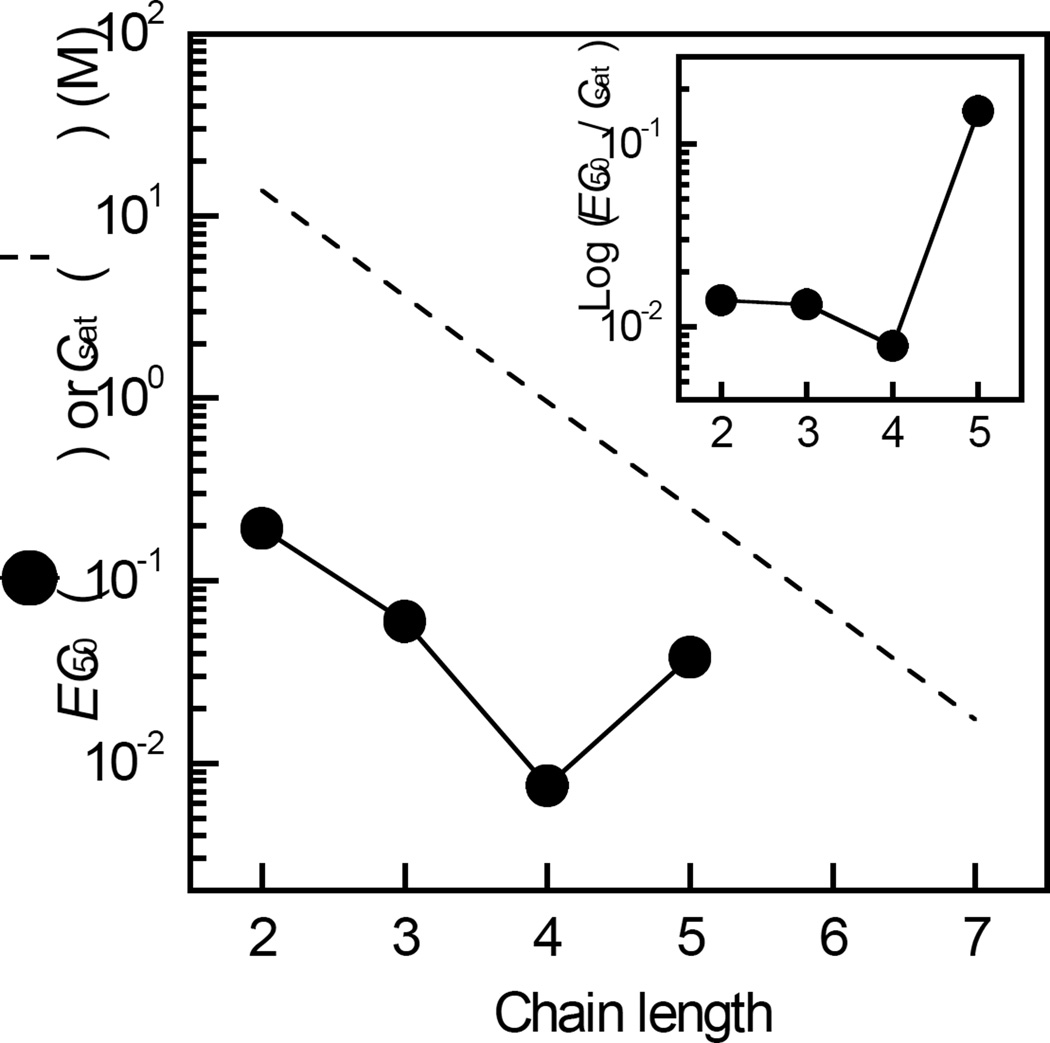
The homologous series of n-alkanols of increasing chain length (n) are anesthetics and have been used to probe the geometry and structure of anesthetic binding sites within a variety of proteins (9, 45–49). The addition of increasing levels of n-alkanols of n=2 to 5 to RhoGDIα in the presence of halothane (0.2 mM) in each case resulted in a saturable and concentration dependent decrease in the extent of tryptophan fluorescence quenching (Figure 3A–3D). By contrast, control experiments indicated that the tryptophan emission fluorescence of RhoGDIα measured without halothane was unaffected by these n-alkanols within the concentration ranges used (results not shown). Strikingly, the effect of n-hexanol did not reach saturation within the concentration range used and n-hepthanol had negligible effects on halothane-induced tryptophan quenching (Figure 3E– 3F). In keeping with this, values of EC50, calculated from fits of the corresponding dose-response curves to Eq. 2 (Table 1), decreased as a non-linear function of n (Figure 4), as revealed by a plot of EC50/Csat against n (Figure 4, inset). By contrast, values of Fmax were independent of n (Table 1). The apparent non-linear relationship between EC50 and n was also evident in the values of KI for the displacement of halothane by each n-alkanol, obtained from Eq. 3 (Table 1), and also by the estimated incremental binding free energy change (ΔΔG) upon addition of a methylene unit, calculated using Eq. 4. Thus, the value of ΔΔG was −3 kJ mol−1 for n=2 to 3, which compares favorably with the value of −2 kJ mol−1 predicted for the partitioning of a methylene group to a hydrophobic site within a protein (50). By contrast, the values of ΔΔG were −5 kJ mol−1 for n=3 to 4, and +4 kJ mol−1 for n=4 to 5, indicating a respective increase and decrease in binding energy relative to those predicted based on hydrophobic interactions alone. The Hill coefficients obtained from fits of the corresponding dose-response data (Figure 3), were found to be ~1 and independent of n, indicating that each of the n-alkanols interact with at least one equivalent binding site.
Figure 3.
Chain length dependent effects of a homologous series of n-alkanols on the halothane induced quenching of RhoGDIα tryptophan emission fluorescence. A–F, the steady-state emission fluorescence intensity of RhoGDIα (500 nM) at 330 nm upon excitation at 295 nm was measured as a function of the concentration of n-alkanols with chain lengths, n=2 to n=7. The effects of the n-alkanols are expressed as the recovery in emission fluorescence from quenching by 0.2 mM halothane in the presence of each n-alkanol concentration. The solid curves represent fits of the data to Eq. 2 using least squares non-linear regression analysis. Calculated values of EC50, h and Fmax are listed in Table 1. Data are representative of three independent experiments. The fits of each individual replicate experiment yielded similar parameter values and errors. Other details are described in “Experimental Procedures.”
Table 1.
Chain Length Dependent Effects of n-Alkanols on Halothane Binding and RhoGDIα– Cdc42 Complex Dissociation.
| Halothane Bindinga |
Cdc42–RhoGDIα Dissociationb |
|||||
|---|---|---|---|---|---|---|
| n | EC50, mM | h | Fmax | KIc, mM | Kapp (mM) | h |
| 2 | 100 ± 58 | 1.4 ± 0.4 | 44 ± 5 | 42 ± 24 | 70 ± 21 | 1.2 ± 0.2 |
| 3 | 48 ± 4.5 | 1.2 ± 0.2 | 40 ± 9 | 20 ± 2 | ndd | ndd |
| 4 | 10 ± 3 | 1.1 ± 0.1 | 51 ± 4 | 4.2 ± 1.2 | 7.5 ± 1.3 | 1.5 ± 0.3 |
| 5 | 38 ± 19 | 1.1 ± 0.2 | 41 ± 8 | 16 ± 8 | ndd | ndd |
Values of EC50, Fmax, h for the displacement of halothane binding were obtained from fits of the dose-response curves shown in Figure 3 to Eq. 2.
Kapp and h for the potentiation of Cdc42– RhoGDIα dissociation were obtained from dose-response curves shown in Figure 6 using Eq. 6.
Values of KI were calculated using Eq. 3.
not determined. See “Experimental Procedures” section for other details.
Figure 4.
Chain length dependent effects of n-alkanols on the quenching of RhoGDIα tryptophan fluorescence by halothane (EC50). The values of EC50 were calculated from least squares non-linear regression fits of the n-alkanol dose response curves shown in Figure 3 to Eq. 2. A plot of values of Csat for each n-alkanol (63) is shown for comparison of EC50 values with aqueous solubilities (dashed line). Inset, the non-linear dependence of EC50 on n as illustrated by a plot of EC50 / Csat against n. Data are representative of three independent experiments. Other details are described in “Experimental Procedures.
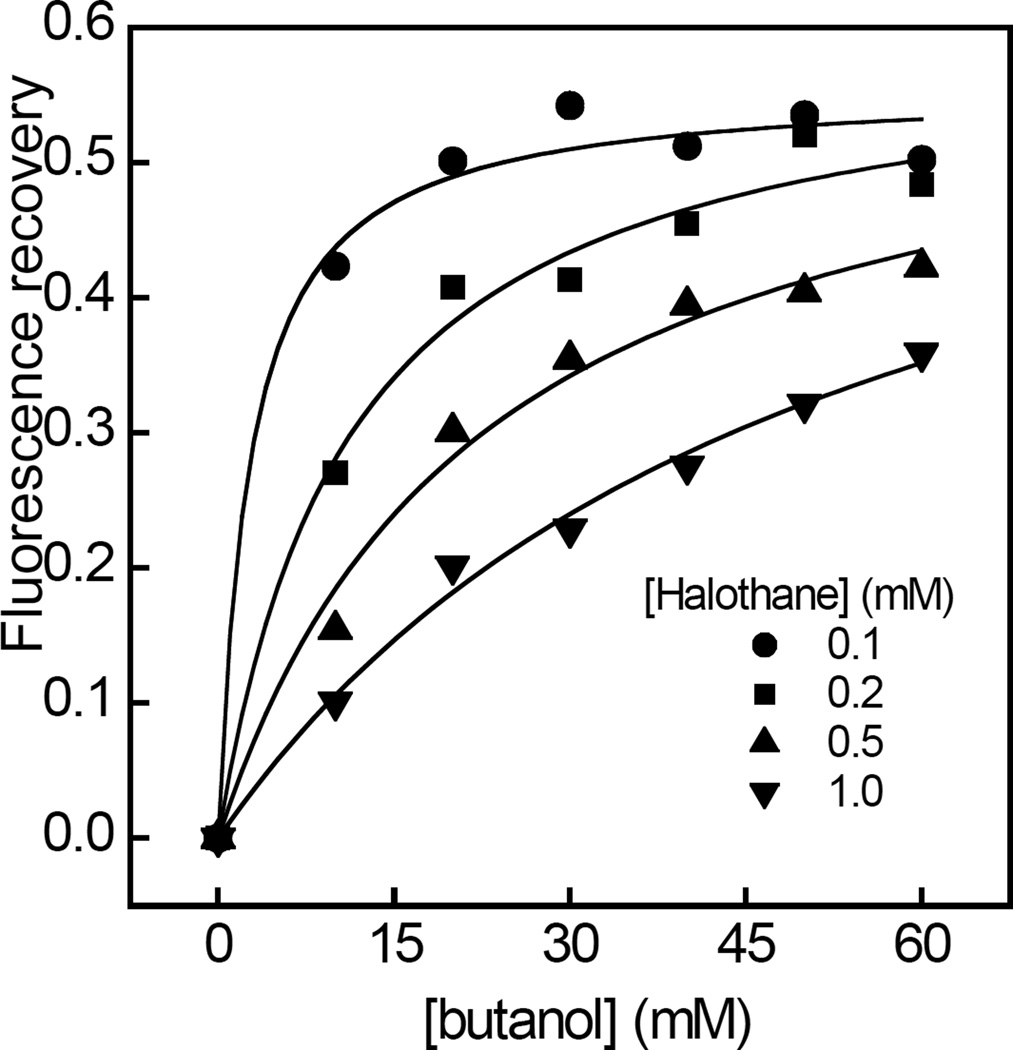
In order to further investigate the mechanism underlying the displacement of halothane binding by n-alkanols, the concentration-response curves for n-butanol induced tryptophan fluorescence recovery were determined in the presence of increasing levels of halothane (Figure 5). It was found that the values of EC50, obtained from fits of each dose-response curve to Eq. 2, increased as a function of halothane concentration, whereas values of Fmax were independent of halothane concentration and corresponded to ~50% recovery in halothane induced quenching (Table 2). The calculated values of h were found to be ~1 and also independent of halothane concentration.
Figure 5.
Concentration dependent effects of n-butanol on the quenching of RhoGDIα tryptophan emission fluorescence by varying levels of halothane. Tryptophan emission fluorescence recovery at 330 nm upon excitation at 290 nm was measured as a function of n-butanol concentration in the presence of 0.1 mM (●), 0.2 mM (▪), 0.5 mM (▲) and 1 mM (▼) halothane. The solid curves represent fits of the data to Eq. 2 using least squares non-linear regression analysis. Values of EC50, h and Fmax are listed in Table 2. Data are representative of three independent experiments. The fits of each individual replicate experiment yielded similar parameter values and errors. Other details are described in “Experimental Procedures.”
Table 2.
Dependence of the EC50, h, and Fmax for the Displacement of Halothane by n-Butanol on Halothane Concentration.
Azialcohols photo-affinity label RhoGDIα
In order to provide further evidence for an alcohol and anesthetic binding site on RhoGDIα and to identify residues that may contribute to its structure, photo-affinity labeling was performed utilizing the diazirinyl n-alkanols, 3-azibutanol and 3-azioctanol (6, 29–32). RhoGDIα was photolabeled with and without AGGC in the presence of 10 and 100 µM azialcohol in two independent experiments, digested with trypsin, and subjected to mass spectrometry. Two photolabeled peptides were generated in the presence but not in the absence of AGGC, at 100 µM 3-azibutanol, which was the lowest concentration at which photolabeling was detected. This indicates that AGGC enhanced the photoincorporation of 3-azibutanol. The characterization of these peptides is shown in Table 3 (Peptides 1 and 2). Photolabeling of RhoGDIα with 3-azioctanol yielded 1 peptide, which was generated in the presence and absence of AGGC, and at 3-azioctanol concentrations as low as 10 µM (Table 3; Peptide 2). The percentage of RhoGDIα that was sequenced was 79.4% (Table 4), so there is a small chance that other photolabeled peptides were missed.
Table 3.
Residues in RhoGDI-α Photolabeled by 3-Azioctanol and 3-Azibutanol Identified in Peptide Fragments Following Trypsinolysis and LTQ–FT Mass Spectrometry.
| Agent | [Alcohol] µMa |
AGGC | Residue Photolabeled |
Peptideb | Charge | Rt, min | Xcorr | MH+c | Accuracyd ppm |
|---|---|---|---|---|---|---|---|---|---|
| 3-azibutanol | 100 | + | E193 | [1] | +3 | 23.45 | 5.29 | 2437.60 | 2.46 |
| 100 | – | Photolabeled peptide not detected | |||||||
| 3-azibutanol | 100 | + | E163 | [2] | +2 | 22.52 | 5.06 | 1824.96 | 3.28 |
| 100 | – | Photolabeled peptide not detected | |||||||
| 3-azioctanol | 10 | + | E163 | [2] | +2 | 18.82 | 2.95 | 1880.90 | 0.00 |
| 10 | – | [2] | +2 | 18.84 | 4.30 | 1880.90 | 0.00 | ||
| Unlabeled peptide f | – | E193 | [1] | +3 | 16.15 | 4.77 | 2365.54 | 0.00 | |
| – | E163 | [2] | +2 | 21.02 | 4.75 | 1752.89 | 0.00 | ||
Lowest 3-azialcohol concentration where photoincorporation was detected.
Peptides photolabeled by 3-azialcohols: [1]=RhoGDIα(181–199)=FTDDDKTDHLSWEWNLTIK; [2]= RhoGDIα(153–167)=AEEYEFLTPVEEAPK. Photolabeled residues are indicated in bold.
Representative observed masses which include increases due to photolabeling by 3-azioctanol (128 Dalton) or 3-azibutanol (72 Dalton).
Accuracy is the difference between the calculated and observed mass expressed in ppm.
Unlabeled peptides are those derived from that fraction of RhoGDIα which was not photolabeled during the experiment with 100 µM azibutanol in the presence of AGGC. See “EXPERIMENTAL PROCEDURES” section for other details
Table 4.
Sequence Coverage of RhoGDIα Following In-Gel Digestion with Trypsin.a
|
001 MAEQEPTAEQ LAQIAAENEE DEHSVNYKPP AQKSIQEIQE LDKDDESLRK YKEALLGRVA VSADPNVPNV 071 VVTGLTLVCS SAPGPLELDL TGDLESFKKQ SFVLKEGVEY RIKISFRVNR EIVSGMKYIQ HTYRKGVKID 141 KTDYMVGSYG PRAEEYEFLT PVEEAPKGML ARGSYSIKSR FTDDDKTDHL SWEWNLTIKK DWKD | |||||
|---|---|---|---|---|---|
| m/z |
Accuracyb (ppm) |
||||
| Position | Sequence | Charge | Observed | Calculated | |
| 34–49 | SIQEIQELDKDDESLR | 2 | 959.4602 | 959.4736 | 14.0 |
| 34–50 | SIQEIQELDKDDESLRK | 2 | 1023.5096 | 1023.5211 | 11.2 |
| 51–58 | YKEALLGR | 1 | 949.5387 | 949.5465 | 8.2 |
| 53–58 | EALLGR | 1 | 658.3844 | 658.3883 | 5.9 |
| 59–99 | VAVSADPNVPNVVVTGLTLVCSSAPGPLELDLTGDLESFKK | 4 | 1039.5316 | 1039.5526 | 20.2 |
| 99–105 | KQSFVLK | 1 | 849.5158 | 849.5193 | 4.1 |
| 100–105 | QSFVLK | 1 | 721.4184 | 721.4243 | 8.2 |
| 106–111 | EGVEYR | 1 | 752.3508 | 752.3573 | 8.6 |
| 114–117 | ISFR | 1 | 522.3012 | 522.3034 | 4.2 |
| 118–127 | VNREIVSGMK | 1 | 1132.617 | 1132.6143 | 2.4 |
| 121–127 | EIVSGMK | 1 | 763.394 | 763.4019 | 10.3 |
| 128–134 | YIQHTYR | 1 | 980.4887 | 980.4948 | 6.2 |
| 128–135 | YIQHTYRK | 1 | 1108.5764 | 1108.5898 | 12.1 |
| 139–152 | IDKTDYMVGSYGPR | 2 | 801.3766 | 801.385 | 10.5 |
| 142–152 | TDYMVGSYGPR | 2 | 623.2784 | 623.2821 | 5.9 |
| 153–167 | AEEYEFLTPVEEAPK | 2 | 876.4141 | 876.4223 | 9.4 |
| 168–172 | GMLAR | 1 | 547.299 | 547.3021 | 5.7 |
| 173–178 | GSYSIK | 1 | 654.3412 | 654.3457 | 6.9 |
| 179–199 | SRFTDDDKTDHLSWEWNLTIK | 3 | 870.0851 | 870.0884 | 3.8 |
| 181–199 | FTDDDKTDHLSWEWNLTIK | 3 | 789.0358 | 789.0441 | 10.5 |
| 181–200 | FTDDDKTDHLSWEWNLTIKK | 3 | 831.7341 | 831.7424 | 10.0 |
| 187–199 | TDHLSWEWNLTIK | 3 | 548.2857 | 548.279 | 12.2 |
The sequence of the RhoGDIα is shown in single letter code. The proteolytic fragments sequenced by MSMS are shown in bold. The sequence coverage by amino acid count is 79.4%;
Accuracy is the difference between the calculated and observed m/z expressed in ppm of the calculated value.
Photolabeling of E193
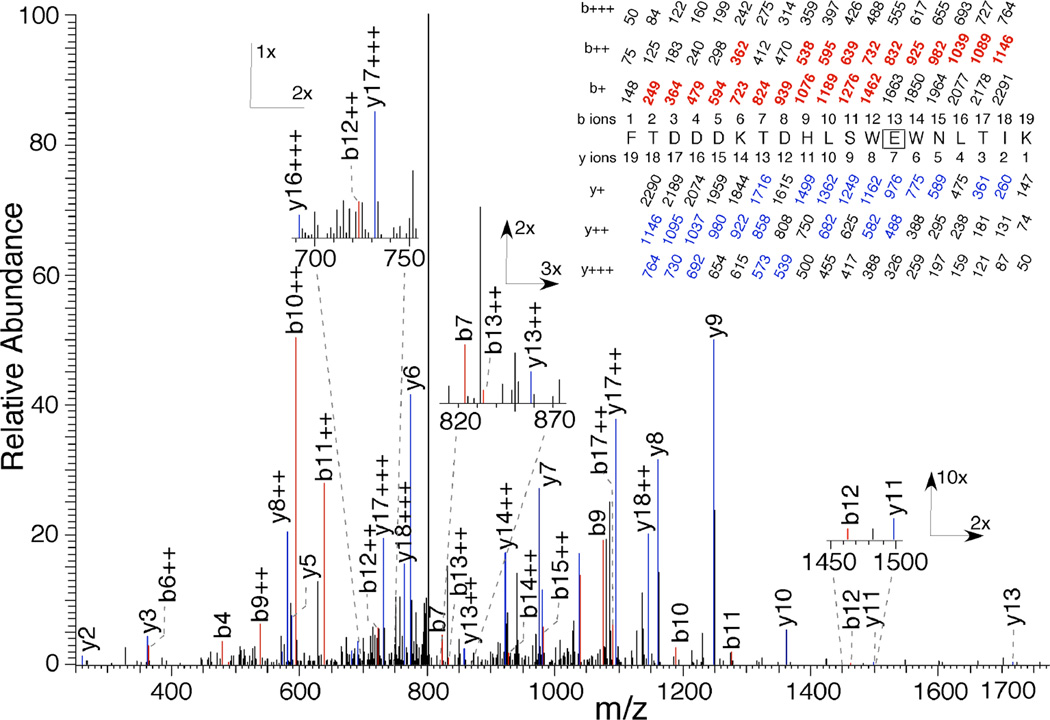
The peptides identified above were fragmented by collision with an inert gas to yield peptide fragments with an intact N-terminus, called b-ions, or an intact C-terminus, called y-ions. The MSMS pattern for Peptide 1 obtained from photolabeling with 100 µM 3-azibutanol in the presence of AGGC is shown in Figure 6. The site of photoincorporation was identified as E193 based on the observed run of strong y-ions from y10 to y5 with loss of modification between y7 and y6. This assignment was confirmed by a run of doubly charged b-ions from b17++ to b9++ with loss of photoincorporation between b13++ and b12++. This residue was not photolabeled by 3-azioctanol at levels up to 1 mM.
Figure 6.
Identification of E193 as a site of photoincorporation of 3-azibutanol (100 µM) on RhoGDIα (2.8 nM) in the presence of AGGC (550 nM) using LC–MSMS. After photolabeling, RhoGDIα was digested with trypsin and subject to HPLC–MSMS as described in the text. Inset, a tryptic peptide bearing a single 72 Da modification and starting at F181 of RhoGDIα. The predicted charge/mass ratios of ions with an intact N-terminus (b-ions) or C-terminus (y-ions) are shown above and below the sequence, respectively, with the indicated charge. The site of photoincorporation for 3-azibutanol was inferred from the MSMS spectrum. The experimentally observed values are colored and in bold and their position indicated on the spectrum. The photolabeled residue is boxed and the statistics are given in Table 3.
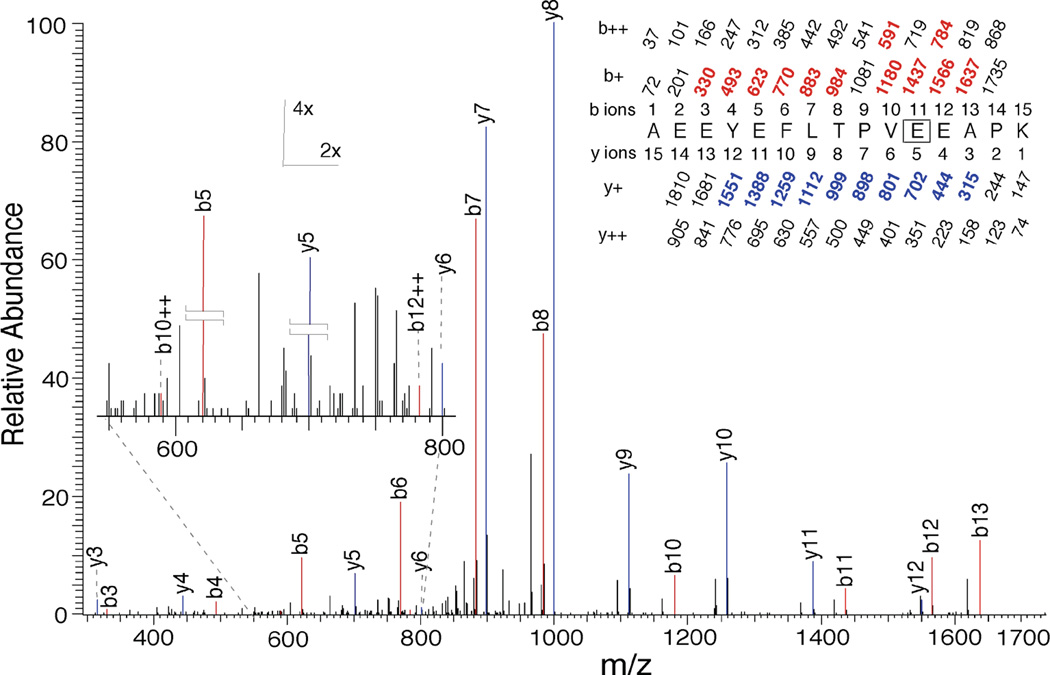
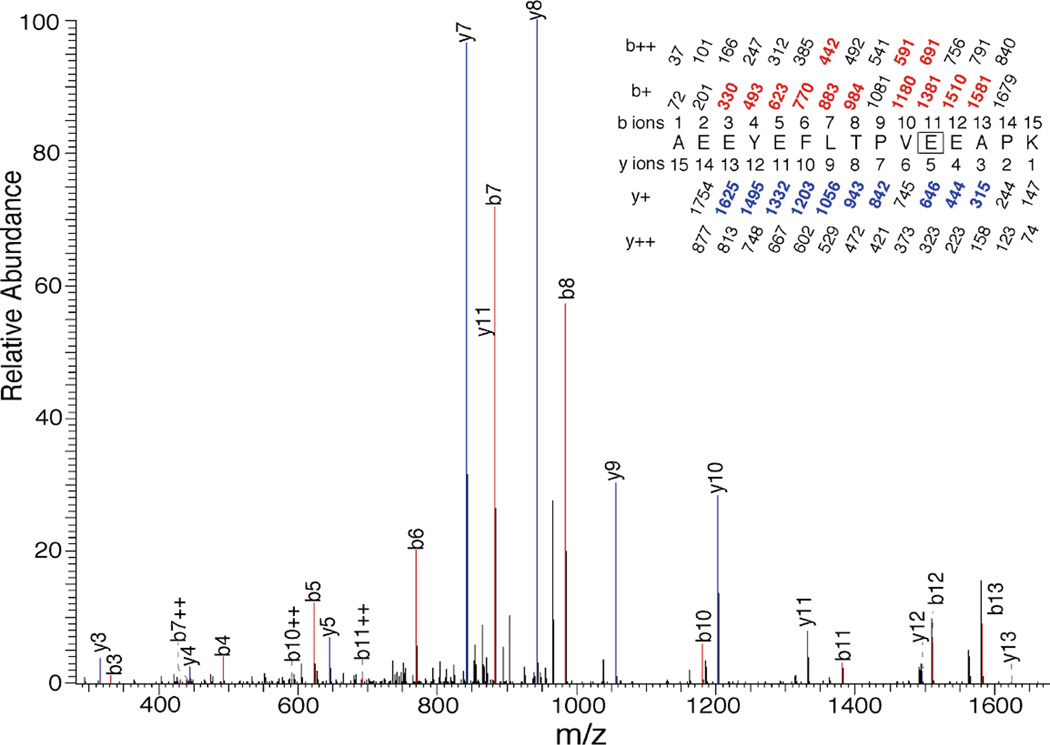
Photolabeling of E163
The MSMS pattern for Peptide 2 obtained from photolabeling of RhoGDIα with 10 µM 3-azioctanol is shown in Figure 7. The doubly charged peptide of m/z = 1733.81 yielded a sequence of y-ions from y12 to y3 with loss of modification between y5 and y4, thus identifying E163 as the site of photoincorporation of 3-azioctanol. This was confirmed by the presence of series of strong b-ions from b13 to b4 (except b9), where modification was lost between b11 and b10. This residue was identified in the presence and absence of AGGC. The corresponding residue photolabeled by 3-azibutanol was only labeled in the presence of AGGC (Figure 8).
Figure 7.
Identification by LC-MSMS of E163 as a photoincorporation site for 3-azioctanol (10 µM) on RhoGDIα (2.8 nM) in the absence of AGGC. After photolabeling, RhoGDIα was digested with trypsin and subject to HPLC–MSMS as described in the text. A tryptic peptide bearing a single 128 Dalton modification and starting at A153 of RhoGDIα had the sequence shown in the inset. Statistics are given in Table 3. The site of photoincorporation of 3-azibutanol was inferred from this MSMS spectrum. In the inset, the predicted charge/mass ratios of ions with an intact N-terminus (b-ions) or C-terminus (y-ions) are shown above and below the sequence, respectively, with the indicated charge. The photolabeled residue is boxed. The experimentally observed values are colored and in bold and their position indicated on the spectrum.
Figure 8.
Identification by LC-MSMS of E163 as a photoincorporation site for 3-azibutanol (100 µM) on RhoGDIα (2.8 nM) in the absence of AGGC. After photolabeling, RhoGDIα was digested with trypsin and subject to HPLC–MSMS as described in the text. A tryptic peptide bearing a single 72 Dalton modification and starting at A153 of RhoGDIα had the sequence shown in the inset. Statistics are given in Table 3. The site of photoincorporation for 3-azibutanol was inferred from this MSMS spectrum. The identified b- and y-ions are indicated above the MS/MS spectrum using the same conventions as in Figure 7.
n-Alkanols enhance Rho GTPase–RhoGDIα complex dissociation
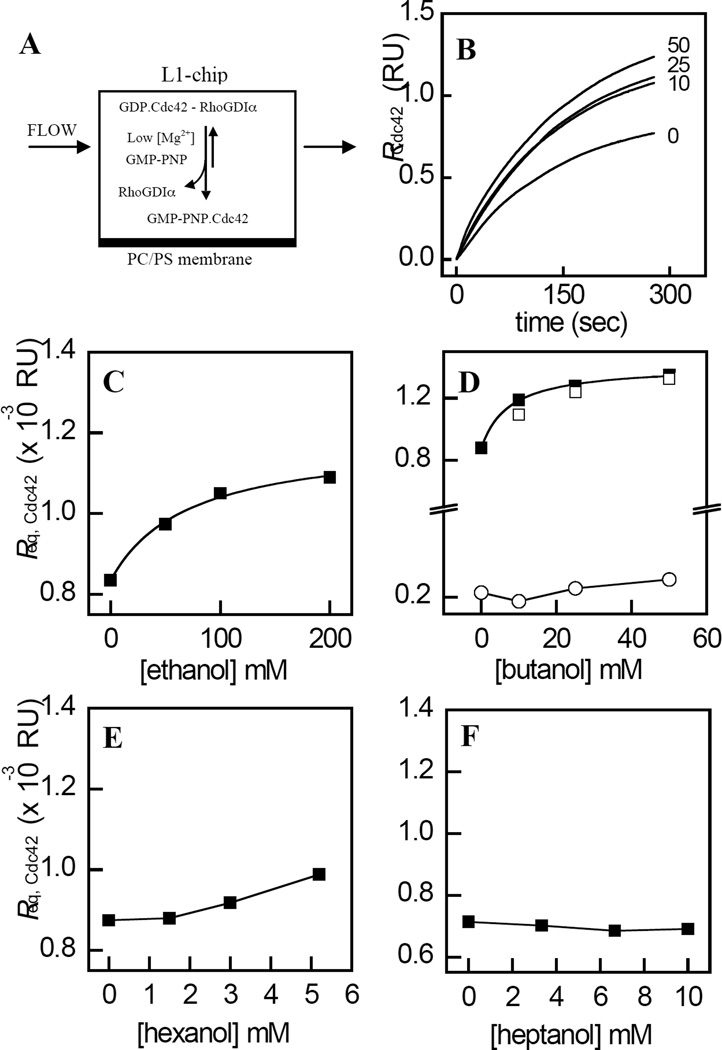
Rho GTPase activation is mediated by the dissociation of the Rho GTPase - RhoGDIα complex, which facilitates the association of the RhoGTPases with membranes (Figure 9A). Since this process requires the transfer of the GG chain from the C-terminal binding pocket within RhoGDIα to the membrane interior (19), we investigated the effects of n-alkanols on the kinetics of the dissociation of Cdc42 from its complex with RhoGDIα. The dissociation of the RhoGDIα–Cdc42 complex was triggered by GDP-GMPPNP exchange on Cdc42 induced by chelating the Mg2+ ion that serves to stabilize GDP binding (23, 51). Consistent with the results of our previous study (35), the injection of Cdc42 in complex with RhoGDIα in the presence of GMPPNP and excess EDTA resulted in a time dependent increase in binding to the captured POPC/POPS membranes (Figure 9B).
Figure 9.
Enhanced membrane-association of Cdc42 by n-alkanols resulting from the destabilization of the RhoGDIα–Cdc42 complex. A, the association of Cdc42 with POPC/POPS (4:1, molar) membranes immobilized on a Biacore L1 chip, was measured from the increase in response (binding) that occurred upon injection of the RhoGDIα– Cdc42 complex in the presence of EDTA and GMPPNP. B, representative time courses for the concentration dependent effects of n-butanol on Cdc42 membrane-association. C– F, values of Req,Cdc42, calculated from fits of the time courses obtained for each concentration of n-alkanols with chain lengths 2, 4, 6, and 7 to Eq. 5, were plotted as function of n-alkanol concentration. Also shown is the effect of n-butanol on Req,Cdc42 values obtained in the absence of EDTA and GMPPNP (Panel D , ○), and the concentration dependent effects of 3-azibutanol on Req,Cdc42 (Panel C, □). The solid curves represent fits of the data to Eq. 6 using least squares non-linear regression analysis. Values of EC50 and h are listed in Table 1. Data are representative of three independent experiments. The fits of each individual replicate experiment yielded similar parameter values and errors. Other details are described in “Experimental Procedures.”
The addition of increasing levels of n-butanol prior to injection of the RhoGDIα–Cdc42 complex over the membrane surface, resulted in a concentration dependent increase in the value of Req for Cdc42 binding (Figure 9B). Plotting values of Req,Cdc42 as a function of ethanol (Figure 9C) or n-butanol concentration (Figure 9D, ▪), in each case yielded a hyperbolic curve. Fitting these data to Eq. 6 yielded values of Kapp= 50±21 mM and 7.5±1.3 mM for ethanol and n-butanol, respectively, which are close to the corresponding values of KI obtained for the displacement of halothane binding by each n-alkanol. Notably, n-butanol had negligible effects on the association of Cdc42 with membranes in the absence of EDTA and GMPPNP (Figure 9D, ○). Consistent with the non-linear chain length dependent effects of the n-alkanols on halothane binding (Figure 3), the association of Cdc42 with membranes was unaffected by n-hexanol (Figure 9E) and n-heptanol (Figure 9F). The concentration dependent effects of 3-azibutanol on Req,Cdc42, which were found to be similar to that obtained for n-butanol (Figure 9D, □), thus validating the use of this alcohol in photo-labeling experiments (see above). The observed non-linear chain length dependent effects of the n-alkanols on the membrane association of Cdc42, argues against the possibility that the observed effects resulted from a non-specific perturbation of the membrane structure, since such effects have been shown to be a linear function of chain length within the range used in the present study (52).
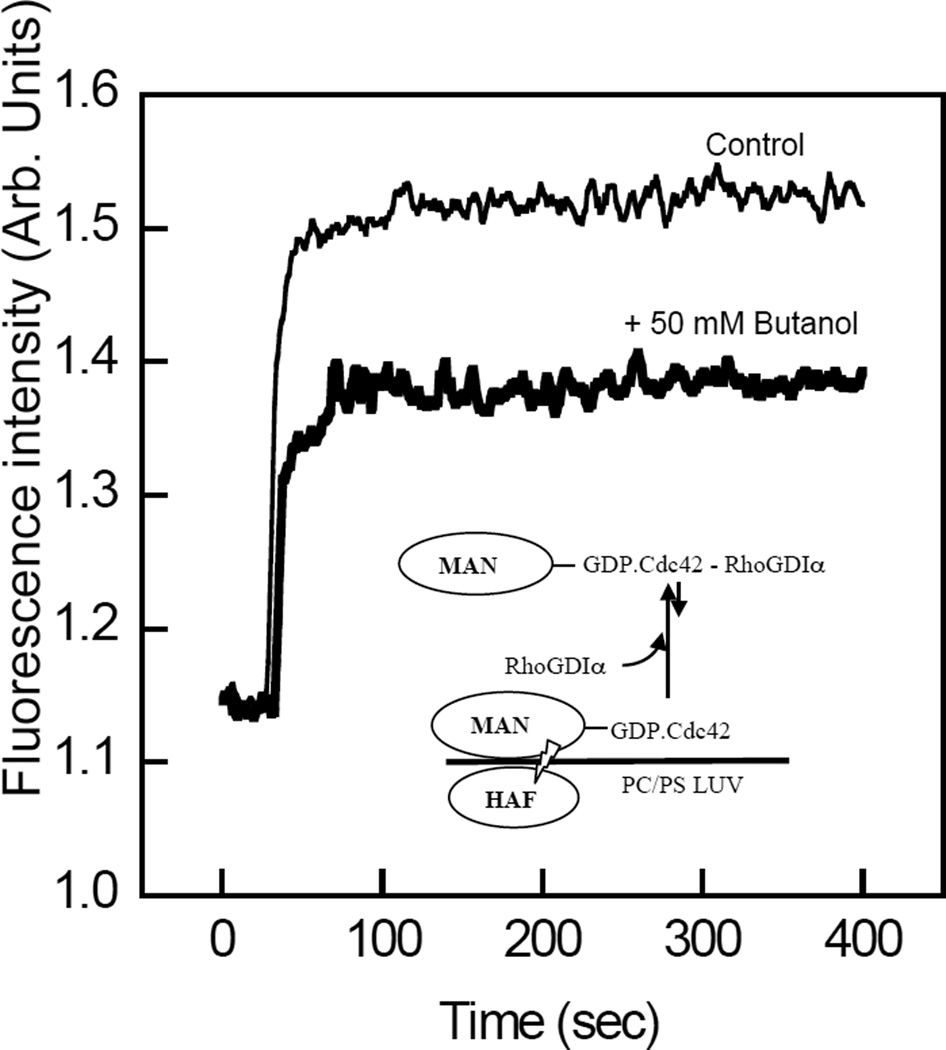
n-Alkanols inhibit the extraction of Cdc42 from membranes by RhoGDIα
It was hypothesized that the extraction of Cdc42 from membranes by RhoGDIα would be inhibited by n-alkanols, since this process involves transfer of the GG chain from the membrane interior to the GG chain binding pocket. In order to address this, the extraction of membrane-bound Cdc42 by RhoGDIα was determined from measurements of FRET between the MANT-donor fluorophore of MANT-GMPPNP loaded onto Cdc42 and the membrane-associated acceptor fluorophore of HAF (35, 43). Consistent with the results of our previous study (35), the addition of POPC/POPS (80:20, molar) labeled with 2 µM HAF quenched the emission fluorescence intensity of MANT-GMPPNP loaded on Cdc42 by ~50% (results not shown). The addition of RhoGDIα (60 nM) resulted in a rapid increase in MANT emission fluorescence due to a decrease in FRET (Figure 10), as reported previously (35, 43). Importantly, the presence of n-butanol (50 mM) attenuated the de-quenching effect of RhoGDIα on MANT fluorescence, suggesting a corresponding reduction in the membrane extraction of Cdc42 in the presence of the n-alkanol (Figure 10). The possibility that the potentiated level of Cdc42 membrane association induced by ethanol and n-butanol may have resulted from modified nucleotide exchange kinetics was ruled out by the finding that neither the rate nor the extent of exchange of MANT-GDP for GDP were affected by 50 mM butanol (results not shown).
Figure 10.
n-Butanol inhibits the extraction of Cdc42 from membranes by RhoGDIα. A, The emission fluorescence intensity of MANT-GMPPNP (2 µM) loaded on Cdc42 (60 nM), at 430 nm upon excitation at 355 nm, was measured in the presence of LUV composed of POPC/POPS (4:1, molar) containing 2 µM HAF. The recovery of fluorescence from quenching due to MANT–HAF FRET upon addition of 60 nM RhoGDIα was monitored as a function of time in the presence (bold) and absence of 50 mM n-butanol. Results are representative of experiments carried out at least three times. Other details are described in “Experimental Procedures.”
DISCUSSION
In this study, we addressed the hypothesis that anesthetics compete with endogenous ligand binding to hydrophobic sites within mammalian proteins. Evidence supporting this was provided by the finding that halothane and n-alkanols bound to sites contained within RhoGDIα and that interact with GG chain binding. However, the results also suggest that halothane and n-alkanols bind to distinct sites within RhoGDIα and that differing mechanisms underlie the effects on GG chain binding. In the case of halothane, the simplest explanation for our data is that halothane bound in the GG binding pocket because its effect was inhibited by the GG chain analog, AGGC. However, this appears not to be the case for the short chain alcohols because AGGC enhanced their association with RhoGDIα. Below, we first critically assess the experimental evidence for these assertions and then discuss the wider implications of the results of this study for the molecular mechanisms of anesthetic action.
Evidence supporting the existence of a discrete halothane binding site within RhoGDIα was provided by the observation that halothane quenched the intrinsic tryptophan fluorescence of RhoGDIα in a concentration dependent and saturable manner. The quenching of tryptophan fluorescence by the heavy atom bromine in halothane (CF3CHClBr) is a short-range effect (~5 Å) (37). Structural studies have shown that W192 and W194 are contained within the C-terminal Rho GTPase GG chain binding pocket, and the indole side-chain of W194 makes contact (~ 3.5 Å) with carbons C15 and C17 of the GG chain (16, 17, 53). Although the indole ring system of W194 is in van der Waals contact, W192 and W204 are more distant from the GG chain (8–10 Å from the terminal methyl C19/20 atoms), and are shielded from the GG chain by L88 and L86 respectively. The partial observation quenching of tryptophan fluorescence by halothane observed in the present study has also been reported previously for the beta-barrel protein porcine odorant binding protein (54). The partial quenching may have resulted from an interaction of halothane with a subset of the 3 tryptophan residues or a rapid and transient interaction with all 3 residues. Fluorescence lifetime measurements would be required to address this, which is beyond the scope of this manuscript. Taken together, these observations suggest that the halothane binding site resides within a region of the molecule that is occupied by the GG chain, perhaps close to W194. Consistent with this hypothesis, AGGC inhibits the ability of halothane to quench the intrinsic tryptophan fluorescence of RhoGDIα. However, it is not possible to rule out a negative heterotrophic interaction of halothane with a site that is outside the GG chain binding pocket but close to one of the tryptophans. Furthermore, the finding that AGGC only partially reversed halothane–induced fluorescence quenching (Figure 2) indicates either that halothane binds at more than one site, or that AGGC exerts an allosteric action on the halothane site.
The finding that n-alkanols inhibited the quenching of intrinsic tryptophan fluorescence by halothane provides evidence that these agents also interact with RhoGDIα. The alkanol chain length dependence of this inhibitory effect indicates that the interaction of n-alkanols with RhoGDIα is highly specific, with a chain length cut-off at n=6, and an optimal chain length for binding at n=4. The finding that the apparent EC50 for this action increased with halothane concentration (Figure 5), suggests that the displacement of halothane is mediated either by a competitive, or a negative allosteric mechanism. Distinguishing between these two possibilities kinetically would require the EC50 for the displacement of halothane by n-alkanols to be determined as a function of halothane concentrations that span several orders of magnitude, which for technical reasons is not experimentally feasible. However, the observation that n-alkanols only reversed a fraction of the halothane induced fluorescence quenching and this fraction varied with chain length (Figure 3), strongly suggests an allosteric interaction between the halothane and alcohol sites. Furthermore, it is unlikely that the n-alkanols bind to a site within the GG–binding pocket, because none of the photolabeled residues resided within this pocket, and AGGC enhanced photolabeling by 3-azibutanol.
The observation that 3-azibutanol photolabeled both E163 and E193 at the lowest concentration used, and that this occurred only in the presence of AGGC, suggests that the short chain n-alkanols bind to allosteric sites in the vicinity of both of these residues. In order to determine which residue contributes to the allosteric action on halothane–induced fluorescence quenching, we utilized 3-azioctanol, which possesses a chain length that is too long to reverse the quenching effect of halothane. The finding that E193 was not photolabeled by 3-azioctanol even at the highest concentration used, indicates that the short chain n-alkanol binding site that allosterically reverses fluorescence quenching is in the vicinity to E193 and not E163. Whereas 3-azioctanol photolabeled E163 at the lowest concentration examined (10 µM), this photoincorporation was unaffected by AGGC, unlike 3-azibutanol. This prompts the intriguing prediction that the extra four carbon atoms stabilize binding in a conformationally–insensitive fashion.
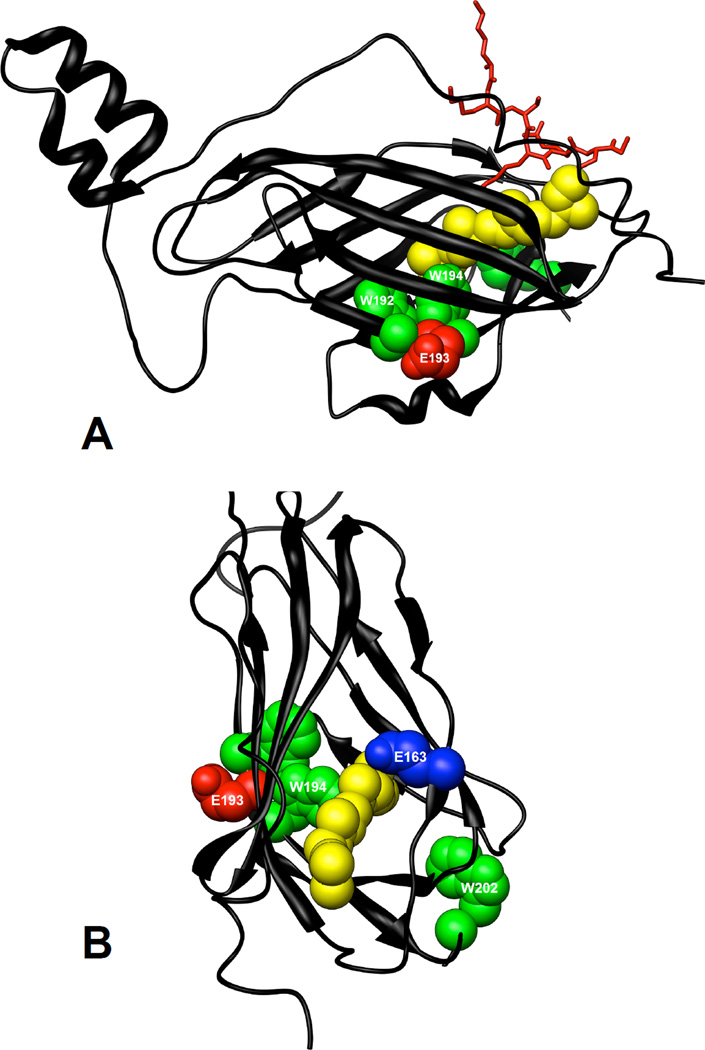
The solution state structure of RhoGDIα has been determined by NMR in both Rho GTPase–bound and –free forms (17, 53), and the structures of the free C-terminal domain of RhoGDIα (55, 56) and the full length protein bound to Rho GTPases have been solved by X-ray crystallography at a resolution of 2.6 Å (16, 57, 58). W192 and W194 are on the β9-strand facing the GG pocket and E193 is on the surface of the protein, facing in the opposite direction (see Figure 11). In a previous study, the effect of the insertion of the GG chain into the GG chain binding pocket was assessed by comparing the structure of the free C-terminal globular fold of RhoGDIα with that of the Cdc42-bound full length protein (16). Insertion of the GG chain induces a structural rearrangement that moves the indole ring nitrogen (Nε) of W194 1.8 Å, and in doing so moves the backbone in this region 1.0 Å outwards. This structural shift also results in a large displacement of E193, the site of photolabeling by 3-azibutanol, and radically changes the surface topology at this residue. Although speculative, this change in the orientation of E193, and in its local topology, might well account for the observed enhancement in the degree of photoincorporation of 3-azibutanol in the presence of AGGC. Structural determinations of RhoGDIα in complex with AGGC are required to resolve this issue fully.
Figure 11.
Location of tryptophans and residues photolabeled by 3-azibutanol within the structure of RhoGDIα. The crystal structure of RhoGDIα in complex with Cdc42 (not shown) was from Hoffman et al (16). Tryptophans, W192, W194 and W202 are shown in green, residues photolabeled by 3-azibutanol are shown in red and the GG chain of Cdc42 is shown in yellow. The C-terminal backbone of Cdc42 (K183-C188) is shown in red. The Protein Data Bank file number of the structure used is 1DOA and the molecular graphics images were produced using the UCSF Chimera package (36)
The initiating event in Rho GTPase mediated signaling corresponds to the release of the Rho GTPases from the inactive cytosolic complex, which is driven by the transfer of the GG chain from the binding pocket within RhoGDIα into the membrane (51). Conversely, the extraction of Rho GTPases from membranes by RhoGDIα terminates Rho GTPase signaling, and results from the transfer of the GG chain from the membrane interior into the binding pocket within RhoGDIα to form the inactive cytosolic complex (12, 16). The observation that ethanol, n-butanol and 3-azibutanol each potentiated the extent of Cdc42 binding to membranes at equilibrium, and that n-butanol inhibited the extraction of Cdc42 from membranes (Figures 9 and 10), suggests that the interaction of these agents with RhoGDIα promotes the dissociation of the RhoGDIα-Cdc42 complex and thus acts on both the initiating and terminating steps in Rho GTPase signaling. Furthermore, the observed lack of an effect of n-hexanol and n-heptanol on the membrane-association of Cdc42 is reminiscent of the non-linear chain length dependent effects of the n-alkanols on halothane binding. Although this suggests the alcohol site that acts on complex stability may be similar to that which mediates the displacement of halothane binding, our photolabeling studies were based on the RhoGDIα and its complex with AGGC, not on the full RhoGDIα–Cdc42 complex. It would be premature to draw conclusions about the nature and location of the alcohol site on the RhoGDIα–Cdc42 complex. However, it is interesting to note the 3-azibutanol site at E163 is in contact with R186 of Cdc42 (16).
Along with the insertion of the GG chain into its binding pocket, the RhoGDIα–Cdc42 complex is also stabilized by an interaction between the N-terminal regulatory arm of RhoGDIα and the switch 1 and 2 regions of the Rho GTPases (16, 53). The conformation of the switch regions and thus their interaction with RhoGDIα are sensitive to GDP-GTP exchange on the Rho GTPases (59). Thus, the finding that nucleotide exchange was an absolute requirement for the potentiation of membrane association of Cdc42 by n-butanol indicates that the corresponding destabilizing effect on the RhoGDIα–Cdc42 complex requires the release of the interaction between the switch regions and the N-terminal regulatory arm of RhoGDIα, and is therefore coupled to GDP–GTP nucleotide exchange activation cycle of the Rho GTPase.
In conclusion, the results provide evidence for the existence of a halothane site within RhoGDIα and support a mechanism in which the anesthetic interacts directly with binding sites that are normally utilized by the GG chain. Moreover, this interaction modified the functional activation of the Rho GTPases, Although caution should be engendered in concluding that an identical interaction between anesthetics and RhoGDIα occurs in the cellular environment, it can be speculated that if anesthetics and alcohols destabilize the RhoGDIα–Cdc42 complex in the cellular environment, this might act to sustain signaling to downstream effectors by enhancing the membrane association of Cdc42. It is also notable that the affinity of halothane binding to RhoGDIα (KD = 147±7 µM) is within pharmacologically relevant ranges (60). Because RhoGDIα belongs to an extended family of proteins that also possess hydrophobic clefts or pockets that bind isoprenyl chains, it is also intriguing to speculate that members of this diverse class of proteins might also contain functional anesthetic binding sites. Some potential candidates might include, RhoGDIγ, Ly/D4GDI, RabGDI isoforms (61), as well as other isoprenyl chain binding proteins, such as geranylgeranyl and farnesyl transferases (62). Finally, RhoGDIα constitutes a novel and powerful model for studies of anesthetic–protein interactions by virtue of its solubility, abundance, and the availability of high resolution structural information (16, 17, 53, 55–58).
ACKNOWLEDGEMENTS
We would like to thank Dr. Christopher D. Stubbs (Oxford Brookes University, Oxford, United Kingdom) for formative contributions to this work, Ms. Kathryn Harrison and Ms. Brigid A. Stagliano for their expert technical assistance, Dr. Richard A. Cerione (Cornell University, New York) for providing the Cdc42 baculovirus, and Dr. S. Shaukat Husain for synthesizing 3-azibutanol and 3-azioctanol. Cojen Ho and Sivananthaperumal Shanmugasundararaj contributed equally to this study.
Footnotes
This work was supported by US Public Health Service grants AA010990 (SJS) and GM069726 (KWM).
Abbreviations: AGGC, N-acetyl-geranylgeranyl cysteine; FC, flow cell; FRET, fluorescence resonance energy transfer; GMPPNP, guanosine 5’-[β, γ-imido] triphosphate trisodium salt; HAF, 5-hexadecanoylaminofluorescein; Halothane, 2-bromo-2-chloro-1,1,1-trifluoroethane; LUV, large unilamellar vesicles; MANT-GDP, 3’-O-(N-methylanthraniloyl)-guanosine 5’-diphosphate trisodium salt; MANT-GMPPNP, 3’-O-(N-methylanthraniloyl)-β:γ-imidoguanosine 5’-triphosphate trisodium salt; POPC, I-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine; POPS, I-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine; RhoGDIα, α-isoform of Rho GDP-dissociation inhibitor; SPR, surface plasmon resonance.
REFERENCES
- 1.Franks NP, Lieb WR. Molecular and cellular mechanisms of general anaesthesia. Nature. 1994;367:607–614. doi: 10.1038/367607a0. [DOI] [PubMed] [Google Scholar]
- 2.Franks NP, Lieb WR. Which molecular targets are most relevant to general anaesthesia? Toxicol Lett. 1998;100–101:1–8. doi: 10.1016/s0378-4274(98)00158-1. [DOI] [PubMed] [Google Scholar]
- 3.Miller KW. The nature of sites of general anaesthetic action. Br J Anaesth. 2002;89:17–31. doi: 10.1093/bja/aef167. [DOI] [PubMed] [Google Scholar]
- 4.Raines DE. Conformational transitions of the nicotinic acetylcholine receptor as a model for anesthetic actions on ligand-gated ion channels: single and sequential mixing stopped-flow fluorescence studies. Toxicol Lett. 1998;100–101:163–168. doi: 10.1016/s0378-4274(98)00181-7. [DOI] [PubMed] [Google Scholar]
- 5.Arias HR, Kem WR, Trudell JR, Blanton MP. Unique general anesthetic binding sites within distinct conformational states of the nicotinic acetylcholine receptor. Int Rev Neurobiol. 2003;54:1–50. doi: 10.1016/s0074-7742(03)54002-8. [DOI] [PubMed] [Google Scholar]
- 6.Das J, Addona GH, Sandberg WS, Husain SS, Stehle T, Miller KW. Identification of a general anesthetic binding site in the diacylglycerol-binding domain of protein kinase Cdelta. J Biol Chem. 2004;279:37964–37972. doi: 10.1074/jbc.M405137200. [DOI] [PubMed] [Google Scholar]
- 7.Slater SJ, Malinowski SA, Stubbs CD. The nature of the hydrophobic n-alkanol binding site within the C1 domains of protein kinase Calpha. Biochemistry. 2004;43:7601–7609. doi: 10.1021/bi049755z. [DOI] [PubMed] [Google Scholar]
- 8.Slater SJ, Kelly MB, Larkin JD, Ho C, Mazurek A, Taddeo FJ, Yeager MD, Stubbs CD. Interaction of alcohols and anesthetics with protein kinase Calpha. J Biol Chem. 1997;272:6167–6173. doi: 10.1074/jbc.272.10.6167. [DOI] [PubMed] [Google Scholar]
- 9.Franks NP, Lieb WR. Do general anaesthetics act by competitive binding to specific receptors? Nature. 1984;310:599–601. doi: 10.1038/310599a0. [DOI] [PubMed] [Google Scholar]
- 10.Middleton AJ, Smith EB. General anaesthetics and bacterial luminescence. II. The effect of diethyl ether on the in vitro light emission of Vibrio fischeri. Proc R Soc Lond B Biol Sci. 1975;193:173–190. doi: 10.1098/rspb.1976.0038. [DOI] [PubMed] [Google Scholar]
- 11.Middleton AJ, Smith EB. General anaesthetics and bacterial luminescence. I. The effect of diethyl ether on the in vivo light emission of Vibrio fischeri. Proc R Soc Lond B Biol Sci. 1975;193:159–171. doi: 10.1098/rspb.1976.0037. [DOI] [PubMed] [Google Scholar]
- 12.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends in Cell Biology. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Dovas A, Couchman JR. RhoGDI: multiple functions in the regulation of Rho family GTPase activities. Biochem J. 2005;390:1–9. doi: 10.1042/BJ20050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- 15.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman GR, Nassar N, Cerione RA. Structure of the Rho Family GTP-Binding Protein Cdc42 in Complex with the Multifunctional Regulator RhoGDI. Cell. 2000;100:345–356. doi: 10.1016/s0092-8674(00)80670-4. [DOI] [PubMed] [Google Scholar]
- 17.Keep NH, Barnes M, Barsukov I, Badii R, Lian L-Y, Segal AW, Moody PC, Roberts GC. A modulator of rho family G proteins, rhoGDI, binds these G proteins via an immunoglobulin-like domain and a flexible N-terminal arm. Structure. 1997;5:623–633. doi: 10.1016/s0969-2126(97)00218-9. [DOI] [PubMed] [Google Scholar]
- 18.Mondal MS, Wang Z, Seeds AM, Rando RR. The specific binding of small molecule isoprenoids to rhoGDP dissociation inhibitor (rhoGDI) Biochemistry. 2000;39:406–412. doi: 10.1021/bi991856n. [DOI] [PubMed] [Google Scholar]
- 19.Olofsson B. Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling. Cell Signal. 1999;11:545–554. doi: 10.1016/s0898-6568(98)00063-1. [DOI] [PubMed] [Google Scholar]
- 20.Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 21.Bokoch GM, Bohl BP, Chuang TH. Guanine nucleotide exchange regulates membrane translocation of Rac/Rho GTP-binding proteins. J Biol Chem. 1994;269:31674–31679. [PubMed] [Google Scholar]
- 22.Dransart E, Olofsson B, Cherfils J. RhoGDIs revisited: novel roles in Rho regulation. Traffic. 2005;6:957–966. doi: 10.1111/j.1600-0854.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- 23.Nomanbhoy TK, Cerione R. Characterization of the interaction between RhoGDI and Cdc42Hs using fluorescence spectroscopy. J Biol Chem. 1996;271:10004–10009. doi: 10.1074/jbc.271.17.10004. [DOI] [PubMed] [Google Scholar]
- 24.Newcombe AR, Stockley RW, Hunter JL, Webb MR. The interaction between rac1 and its guanine nucleotide dissociation inhibitor (GDI), monitored by a single fluorescent coumarin attached to GDI. Biochemistry. 1999;38:6879–6886. doi: 10.1021/bi9829837. [DOI] [PubMed] [Google Scholar]
- 25.Johansson JS. Binding of the volatile anesthetic chloroform to albumin demonstrated using tryptophan fluorescence quenching. J Biol Chem. 1997;272:17961–17965. doi: 10.1074/jbc.272.29.17961. [DOI] [PubMed] [Google Scholar]
- 26.Johansson JS, Eckenhoff RG, Dutton PL. Binding of halothane to serum albumin demonstrated using tryptophan fluorescence. Anesthesiology. 1995;83:316–324. doi: 10.1097/00000542-199508000-00012. [DOI] [PubMed] [Google Scholar]
- 27.Johansson JS, Rabanal F, Dutton PL. Binding of the volatile anesthetic halothane to the hydrophobic core of a tetra-alpha-helix-bundle protein. J Pharmacol Exp Ther. 1996;279:56–61. [PubMed] [Google Scholar]
- 28.Pidikiti R, Zhang T, Mallela KM, Shamim M, Reddy KS, Johansson JS. Sevoflurane-induced structural changes in a four-alpha-helix bundle protein. Biochemistry. 2005;44:12128–12135. doi: 10.1021/bi050896q. [DOI] [PubMed] [Google Scholar]
- 29.Das J, Zhou X, Miller KW. Identification of an alcohol binding site in the first cysteine-rich domain of protein kinase Cdelta. Protein Sci. 2006;15:2107–2119. doi: 10.1110/ps.062237606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Addona GH, Husain SS, Stehle T, Miller KW. Geometric isomers of a photoactivable general anesthetic delineate a binding site on adenylate kinase. J Biol Chem. 2002;277:25685–25691. doi: 10.1074/jbc.M201303200. [DOI] [PubMed] [Google Scholar]
- 31.Arevalo E, Chiara DC, Forman SA, Cohen JB, Miller KW. Gating-enhanced accessibility of hydrophobic sites within the transmembrane region of the nicotinic acetylcholine receptor’s {delta}-subunit. A time-resolved photolabeling study. J Biol Chem. 2005;280:13631–13640. doi: 10.1074/jbc.M413911200. [DOI] [PubMed] [Google Scholar]
- 32.Husain SS, Ziebell MR, Ruesch D, Hong F, Arevalo E, Kosterlitz JA, Olsen RW, Forman SA, Cohen JB, Miller KW. 2-(3-Methyl-3H–diaziren-3-yl)ethyl 1-(1-phenylethyl)-1H–imidazole-5-carboxylate: a derivative of the stereoselective general anesthetic etomidate for photolabeling ligand-gated ion channels. J Med Chem. 2003;46:1257–1265. doi: 10.1021/jm020465v. [DOI] [PubMed] [Google Scholar]
- 33.Church RFR, Weiss MJ. Synthesis and properties of small functionalized diazirine molecules. Some observations on the reaction of a diaziridine with the iodine-iodide ion system. J Org Chem. 1970;35:2465–2471. [Google Scholar]
- 34.Husain SS, Forman SA, Kloczewiak MA, Addona GH, Olsen RW, Pratt MB, Cohen JB, Miller KW. Synthesis and properties of 3-(2-hydroxyethyl)-3-n-pentyldiazirine, a photoactivable general anesthetic. J Med Chem. 1999;42:3300–3307. doi: 10.1021/jm9806300. [DOI] [PubMed] [Google Scholar]
- 35.Cook AC, Ho C, Kershner JL, Malinowski SA, Moldveen H, Stagliano BA, Slater SJ. Competitive binding of protein kinase Calpha to membranes and Rho GTPases. Biochemistry. 2006;45:14452–14465. doi: 10.1021/bi0612420. [DOI] [PubMed] [Google Scholar]
- 36.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 37.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2nd ed. New York: Plenum Press; 1999. [Google Scholar]
- 38.Slater SJ, Kelly MB, Taddeo FJ, Rubin E, Stubbs CD. Evidence for discrete diacylglycerol and phorbol ester activator sites on protein kinase C. Differences in effects of 1-alkanol inhibition, activation by phosphatidylethanolamine and calcium chelation. J Biol Chem. 1994;269:17160–17165. [PubMed] [Google Scholar]
- 39.MacDonald RC, MacDonald RI, Menco BP, Takeshita K, Subbarao NK, Hu LR. Small-volume extrusion apparatus for preparation of large, unilamellar vesicles. Biochim Biophys Acta. 1991;1061:297–303. doi: 10.1016/0005-2736(91)90295-j. [DOI] [PubMed] [Google Scholar]
- 40.Anderluh G, Besenicar M, Kladnik A, Lakey JH, Macek P. Properties of nonfused liposomes immobilized on an L1 Biacore chip and their permeabilization by a eukaryotic pore-forming toxin. Anal Biochem. 2005;344:43–52. doi: 10.1016/j.ab.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 41.Erb E-M, Chen X, Allen S, Roberts CJ, Tendler SJB, Davies MC, Forsen S. Characterization of the Surfaces Generated by Liposome Binding to the Modified Dextran Matrix of a Surface Plasmon Resonance Sensor Chip. Anal Biochem. 2000;280:29–35. doi: 10.1006/abio.1999.4469. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 43.Nomanbhoy TK, Erickson JW, Cerione RA. Kinetics of Cdc42 membrane extraction by Rho-GDI monitored by real-time fluorescence resonance energy transfer. Biochemistry. 1999;38:1744–1750. doi: 10.1021/bi982198u. [DOI] [PubMed] [Google Scholar]
- 44.Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom. 2001;36:1083–1091. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 45.Slater SJ, Cox KJ, Lombardi JV, Ho C, Kelly MB, Rubin E, Stubbs CD. Inhibition of protein kinase C by alcohols and anaesthetics. Nature. 1993;364:82–84. doi: 10.1038/364082a0. [DOI] [PubMed] [Google Scholar]
- 46.Eckenhoff RG, Tanner JW, Johansson JS. Steric hindrance is not required for n-alkanol cutoff in soluble proteins. Mol Pharmacol. 1999;56:414–418. doi: 10.1124/mol.56.2.414. [DOI] [PubMed] [Google Scholar]
- 47.Dildy-Mayfield JE, Mihic SJ, Liu Y, Deitrich RA, Harris RA. Actions of long chain alcohols on GABAA and glutamate receptors: relation to in vivo effects. Br J Pharmacol. 1996;118:378–384. doi: 10.1111/j.1476-5381.1996.tb15413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peoples RW, Ren H. Inhibition of N-methyl-D-aspartate receptors by straight-chain diols: implications for the mechanism of the alcohol cutoff effect. Mol Pharmacol. 2002;61:169–176. doi: 10.1124/mol.61.1.169. [DOI] [PubMed] [Google Scholar]
- 49.Wood SC, Tonner PH, de Armendi AJ, Bugge B, Miller KW. Channel inhibition by alkanols occurs at a binding site on the nicotinic acetylcholine receptor. Mol Pharmacol. 1995;47:121–130. [PubMed] [Google Scholar]
- 50.Nozaki Y, Tanford C. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J Biol Chem. 1971;246:2211–2217. [PubMed] [Google Scholar]
- 51.Robbe K, Otto-Bruc A, Chardin P, Antonny B. Dissociation of GDP dissociation inhibitor and membrane translocation are required for efficient activation of Rac by the Dbl homology-pleckstrin homology region of Tiam. J Biol Chem. 2003;278:4756–4762. doi: 10.1074/jbc.M210412200. [DOI] [PubMed] [Google Scholar]
- 52.Ho C, Stubbs CD. Effect of n-alkanols on lipid bilayer hydration. Biochemistry. 1997;36:10630–10637. doi: 10.1021/bi9703150. [DOI] [PubMed] [Google Scholar]
- 53.Golovanov AP, Hawkins D, Barsukov I, Badii R, Bokoch GM, Lian L-Y, Roberts GCK. Structural consequences of site-directed mutagenesis in flexible protein domains. NMR characterization of the L(55,56)S mutant of RhoGDI. European Journal of Biochemistry. 2001;268:2253–2260. doi: 10.1046/j.1432-1327.2001.02100.x. [DOI] [PubMed] [Google Scholar]
- 54.Johansson JS, Manderson GA, Ramoni R, Grolli S, Eckenhoff RG. Binding of the volatile general anesthetics halothane and isoflurane to a mammalian beta-barrel protein. Febs J. 2005;272:573–581. doi: 10.1111/j.1742-4658.2004.04500.x. [DOI] [PubMed] [Google Scholar]
- 55.Mateja A, Devedjiev Y, Krowarsch D, Longenecker K, Dauter Z, Otlewski J, Derewenda ZS. The impact of Glu-->Ala and Glu-->Asp mutations on the crystallization properties of RhoGDI: the structure of RhoGDI at 1.3 A resolution. Acta Crystallogr D Biol Crystallogr. 2002;58:1983–1991. doi: 10.1107/s090744490201394x. [DOI] [PubMed] [Google Scholar]
- 56.Czepas J, Devedjiev Y, Krowarsch D, Derewenda U, Otlewski J, Derewenda ZS. The impact of Lys-->Arg surface mutations on the crystallization of the globular domain of RhoGDI. Acta Crystallogr D Biol Crystallogr. 2004;60:275–280. doi: 10.1107/S0907444903026271. [DOI] [PubMed] [Google Scholar]
- 57.Scheffzek K, Stephan I, Jensen ON, Illenberger D, Gierschik P. The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI. Nat Struct Biol. 2000;7:122–126. doi: 10.1038/72392. [DOI] [PubMed] [Google Scholar]
- 58.Longenecker K, Read P, Derewenda U, Dauter Z, Liu X, Garrard S, Walker L, Somlyo AV, Nakamoto RK, Somlyo AP, Derewenda ZS. How RhoGDI binds Rho. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 9):1503–1515. doi: 10.1107/s090744499900801x. [DOI] [PubMed] [Google Scholar]
- 59.Takai Y, Sasaki T, Matozaki T. Small GTP-binding proteins. Physiol Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 60.Eger EI, 2nd, Fisher DM, Dilger JP, Sonner JM, Evers A, Franks NP, Harris RA, Kendig JJ, Lieb WR, Yamakura T. Relevant concentrations of inhaled anesthetics for in vitro studies of anesthetic mechanisms. Anesthesiology. 2001;94:915–921. doi: 10.1097/00000542-200105000-00032. [DOI] [PubMed] [Google Scholar]
- 61.Wu SK, Zeng K, Wilson IA, Balch WE. Structural insights into the function of the Rab GDI superfamily. Trends Biochem Sci. 1996;21:472–476. doi: 10.1016/s0968-0004(96)10062-1. [DOI] [PubMed] [Google Scholar]
- 62.Lane KT, Beese LS. Thematic review series: lipid posttranslational modifications. Structural biology of protein farnesyltransferase and geranylgeranyltransferase type I. J Lipid Res. 2006;47:681–699. doi: 10.1194/jlr.R600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 63.Bell GH. Solubilities of normal aliphatic acids, alcohols and alkanes in water. Chem Phys Lipids. 1973;10:1–10. [Google Scholar]